Abstract
Next-generation sequencing is excellently suited to evaluate the abundance of mRNAs to study gene expression. Here we compare two alternative technologies, cap analysis of gene expression (CAGE) and serial analysis of gene expression (SAGE), for the same RNA samples. Along with quantifying gene expression levels, CAGE can be used to identify tissue-specific transcription start sites, while SAGE monitors 3′-end usage. We used both methods to get more insight into the transcriptional control of myogenesis, studying differential gene expression in differentiated and proliferating C2C12 myoblast cells with statistical evaluation of reproducibility and differential gene expression. Both CAGE and SAGE provided highly reproducible data (Pearson's correlations >0.92 among biological triplicates). With both methods we found around 10 000 genes expressed at levels  2 transcripts per million (
2 transcripts per million ( 0.3 copies per cell), with an overlap of 86%. We identified 4304 and 3846 genes differentially expressed between proliferating and differentiated C2C12 cells by CAGE and SAGE, respectively, with an overlap of 2144. We identified 196 novel regulatory regions with preferential use in proliferating or differentiated cells. Next-generation sequencing of CAGE and SAGE libraries provides consistent expression levels and can enrich current genome annotations with tissue-specific promoters and alternative 3′-UTR usage.
0.3 copies per cell), with an overlap of 86%. We identified 4304 and 3846 genes differentially expressed between proliferating and differentiated C2C12 cells by CAGE and SAGE, respectively, with an overlap of 2144. We identified 196 novel regulatory regions with preferential use in proliferating or differentiated cells. Next-generation sequencing of CAGE and SAGE libraries provides consistent expression levels and can enrich current genome annotations with tissue-specific promoters and alternative 3′-UTR usage.
INTRODUCTION
Next-generation sequencing (NGS) platforms have provided us with the technology needed to expand genomic methods to a new scale. Depending on the technology, these machines can produce gigabases of sequences per day. Due to its superior resolution and sensitivity, NGS is increasingly used to replace array technologies, in particular the genome-wide evaluation of chromatin immunoprecipitation (ChIP-seq) and gene expression profiling experiments. Sequence-based expression analysis can be performed using several approaches. The traditional serial analysis of gene expression (SAGE) method (1) starts with capturing RNA poly-A tails with oligo(dT) beads. Double-stranded cDNA synthesis is performed followed by digestion with a restriction enzyme, commonly NlaIII (2). With the fragments resulting from the digestion only the most 3′ fragment is retained. An additional restriction digest is then performed with MmeI (cuts  20 bp downstream) to create a fragment of acceptable length for sequencing. In the original method, short cDNA fragments, each representing the most 3′ NlaIII digestion site of a specific transcript, were concatenated and cloned, followed by traditional sequencing. However, now the concatenation and cloning steps can be omitted. Instead SAGE library sequences are directly equipped with appropriate sequencing linkers and analyzed in next-generation sequencers (3).
20 bp downstream) to create a fragment of acceptable length for sequencing. In the original method, short cDNA fragments, each representing the most 3′ NlaIII digestion site of a specific transcript, were concatenated and cloned, followed by traditional sequencing. However, now the concatenation and cloning steps can be omitted. Instead SAGE library sequences are directly equipped with appropriate sequencing linkers and analyzed in next-generation sequencers (3).
Cap analysis of gene expression (CAGE) (4) is a method specifically designed for the study of gene expression at transcription initiation sites, as it captures 5′-ends of mRNAs. After trapping the 5′-cap-structures of mRNAs, sequences are converted to double-stranded cDNA and equipped with a linker containing a restriction site for the enzyme MmeI (or EcoP15I) that cuts  20 (or 25–27) bp downstream to create a fragment of appropriate length for sequencing and mapping. Thus, where SAGE captures the 3′ most NlaIII digestion site of mRNA and is thus 3′-end biased, CAGE tags represent the ultimate 5′-end of the transcript and indicate the genomic transcription start site (TSS). In both SAGE and CAGE, one transcript is only represented by a single read, and (next-generation) sequencing of SAGE and CAGE libraries is therefore referred to as Digital Gene Expression profiling (3,5). While DeepSAGE and DeepCAGE are alternative names for NGS-based analysis of SAGE and CAGE libraries, we refer to these in this manuscript as SAGE and CAGE. In RNA-Seq (6–8), which starts with random fragmentation of the RNA or cDNA, the entire transcript is sequenced. Consequently, a transcript is commonly represented by multiple reads and the amount of reads is dependent on the transcript length. RNA-Seq gives more detailed information about the structure of the transcripts and alternative splicing, in particular when combined with paired-end sequencing, while CAGE is more suitable for analysis of alternative TSSs and SAGE for analysis of alternative polyadenylation sites.
20 (or 25–27) bp downstream to create a fragment of appropriate length for sequencing and mapping. Thus, where SAGE captures the 3′ most NlaIII digestion site of mRNA and is thus 3′-end biased, CAGE tags represent the ultimate 5′-end of the transcript and indicate the genomic transcription start site (TSS). In both SAGE and CAGE, one transcript is only represented by a single read, and (next-generation) sequencing of SAGE and CAGE libraries is therefore referred to as Digital Gene Expression profiling (3,5). While DeepSAGE and DeepCAGE are alternative names for NGS-based analysis of SAGE and CAGE libraries, we refer to these in this manuscript as SAGE and CAGE. In RNA-Seq (6–8), which starts with random fragmentation of the RNA or cDNA, the entire transcript is sequenced. Consequently, a transcript is commonly represented by multiple reads and the amount of reads is dependent on the transcript length. RNA-Seq gives more detailed information about the structure of the transcripts and alternative splicing, in particular when combined with paired-end sequencing, while CAGE is more suitable for analysis of alternative TSSs and SAGE for analysis of alternative polyadenylation sites.
Myogenesis is an essential process for muscle development and regeneration, with defects resulting in diseases such as muscular dystrophies. To support our studies towards treatment of muscle-related diseases, we have performed extensive analysis of muscle-derived gene expression profiles (9–11). This included the analysis of muscle differentiation using a well-established model, the mouse myoblast cell line (C2C12) (12). Two primary transcription factors regulating this process are MyoD and Myogenin, but many other regulatory elements have been identified [reviewed in (13) and (14)]. For a better understanding of how expression profiles change during adaptation to different biological situations, it is important to consider promoter activities and their regulation. Several bioinformatic approaches have been designed for this, including CORE_TF (15) and oPOSSUM (16), searching for shared transcription factor binding sites in the promoter region. However, these approaches critically depend on correct genome annotations regarding TSSs, which can vary by tissue type. Unfortunately, most studies performed thus far use methods directed at the 3′-end of RNA transcripts [including the well-known oligo(dT)-primed cDNA synthesis]. Consequently gene annotation is weakest at the 5′-end. CAGE is therefore excellently suitable for the identification of alternative TSSs and putative regulatory regions upstream of those TSSs. We applied both CAGE and SAGE to study muscle differentiation to assess their concordance in estimation of gene expression levels and complementarity in gene annotation.
MATERIALS AND METHODS
Cells, RNA isolation and differentiation markers
Proliferating C2C12 mouse myoblasts were grown on collagen-coated plates in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum (FBS). To induce fusion into myotubes, cells were serum deprived by changing to a medium of DMEM supplemented with 2% FBS for 9 days (referred to as differentiated cells).
For CAGE and SAGE, RNA was isolated from proliferating and differentiated cells. RNA was isolated from three independent cultures (biological triplicates). Cells grown in flasks (175 cm ) were harvested by trypsinization and centrifugation before RNA extraction with a Nucleospin RNA L kit from Macherey-Nagel. RNA quality was high, as determined with Agilent's Lab-on-chip total RNA nano assay (RNA integrity number
) were harvested by trypsinization and centrifugation before RNA extraction with a Nucleospin RNA L kit from Macherey-Nagel. RNA quality was high, as determined with Agilent's Lab-on-chip total RNA nano assay (RNA integrity number  9). Myogenic properties of the cells were confirmed in RT–PCR/qPCR experiments–using primer sets (Supplementary Table S1) specific for MyoD1, Myogenin, while the housekeeping genes Gapdh and Hprt were used to control for differences in the amount of input cDNA. RT–PCR experiments were performed using oligo(dT) priming for cDNA synthesis and qPCR was carried out using a Roche Lightcycler 480.
9). Myogenic properties of the cells were confirmed in RT–PCR/qPCR experiments–using primer sets (Supplementary Table S1) specific for MyoD1, Myogenin, while the housekeeping genes Gapdh and Hprt were used to control for differences in the amount of input cDNA. RT–PCR experiments were performed using oligo(dT) priming for cDNA synthesis and qPCR was carried out using a Roche Lightcycler 480.
Library preparation and next-generation sequencing
The CAGE protocol published in Valen et al. (5) was modified to enable direct sequencing on an Illumina platform. Briefly, cDNA was synthesized from total RNA by random priming, and 5′-ends of mRNA within RNA/DNA hybrids were selected by the Cap Trapper method. Then linkers having sequences needed for Illumina sequencing and a recognition site for EcoP15I (proliferating: 5′-CCACCGACAGGTTCAGAGTTCTACAGAGACAGCAG and differentiated: 5′-CCACCGACAGGTTCAGAGTTCTACAGCTTCAGCAG) were ligated to the 3′-end of single-stranded cDNAs. After synthesis of the second cDNA strand, double-stranded cDNA was digested with EcoP15I. A second linker having sequences needed for Illumina sequencing (5′-TCGTATGCCGTCTTCTGCTTGAGCATACGGCAGAAGACGAAC) was ligated to the open 3′-end of the DNA fragments, and ligation products were PCR amplified prior to sequencing.
SAGE libraries were prepared for each individual RNA sample with a FC-102-1005 DGE-Tag Profiling NlaIII SamplePrepKit from Illumina. This involves isolating RNA poly-A tails with oligo(dT) beads, converting into single and then a double-stranded cDNA, performing a first restriction digest with NlaIII (at CATG's) and retaining the 3′ most fragments, adding a 5′-linker (containing a restriction site for MmeI), performing an MmeI digestion and adding a 3′-linker.
Each CAGE and SAGE library was then sequenced on an individual lane on an Illumina Genome Analyzer II for 36 cycles. One CAGE sample from each timepoint was also sequenced a second time with 32 cycles.
Initial sequence analysis
All sequenced lanes were run through the initial Illumina Genome Analyzer Pipeline (Firecrest  Bustard
Bustard  Gerald) for image analysis and quality control, yielding one scarf file per sample (lane). For reads from SAGE samples, the NlaIII recognition sequence ‘CATG' was introduced at the 5′-end with Linux commands. Scarf files were then run through the open-source GAPSS_R pipeline developed in house (www.lgtc.nl/GAPSS). In general, this pipeline takes sequences and has the options to: remove first bases [often of lower quality than other 5′-nucleotides (17)]; edit for linkers (present in the sequence reads when sequencing more cycles than the fragment length); align to a reference genome with Rmap (18); report data as region files (reporting tags in a region, a region defined as a stretch of adjacent nucleotides with aligned reads); and create UCSC Genome Browser (19) (http://genome.ucsc.edu/) viewable wiggle tracks.
Gerald) for image analysis and quality control, yielding one scarf file per sample (lane). For reads from SAGE samples, the NlaIII recognition sequence ‘CATG' was introduced at the 5′-end with Linux commands. Scarf files were then run through the open-source GAPSS_R pipeline developed in house (www.lgtc.nl/GAPSS). In general, this pipeline takes sequences and has the options to: remove first bases [often of lower quality than other 5′-nucleotides (17)]; edit for linkers (present in the sequence reads when sequencing more cycles than the fragment length); align to a reference genome with Rmap (18); report data as region files (reporting tags in a region, a region defined as a stretch of adjacent nucleotides with aligned reads); and create UCSC Genome Browser (19) (http://genome.ucsc.edu/) viewable wiggle tracks.
We ran GAPSS_R with the following parameters: the first base (lower quality) was removed in CAGE samples; CAGE and SAGE samples were edited for 3′ linker sequences (5′-TCGTATGCCGTCTTCTGCTTG for CAGE and 5′-TCGTATGCCGTCTTCTGCTTGAAAAAAAAAAAAAAA for SAGE), permitting one mismatch in the linker [to account for sequencing errors that occur more towards the 3′-end (17) where linkers were edited from]. After linker editing, the majority of CAGE reads were 26 bases in length, whereas SAGE reads were 21 or 22 bases in length (including the ‘CATG'). Alignment was performed against the mouse repeat-masked reference genome build 37 with Rmap v0.41, an alignment tool that reports only unique alignments. This was done to maximize the reliability in the alignment process, although we realize that we remove potentially important expressed regions (20), such as retrotransposons (21). Default settings were used during alignment, except to use FASTA input and permitting two mismatches with CAGE reads and one mismatch with SAGE reads. The choice of mismatches permitted is because longer sequences (CAGE) are more likely to contain a sequencing error as the number of errors increases at later sequencing cycles. Region files were created and for CAGE regions we combined adjacent regions, permitting gaps of maximal 100 bases to cluster TSSs and make sure that newly identified TSSs were well separated from annotated TSSs. We kept all data separated by strand, since both methods preserve information on the transcribed strand. Wiggle files for visualization in the UCSC Genome Browser were also separated by strand.
Custom Perl scripts were run on all CAGE and SAGE region files to create reference region files (strand separated) composed of the overlapping regions from all samples. For CAGE region files, we again permitted gaps of a maximum by 100 bases. Another custom Perl script was used to link all individual region files to their reference region file, reporting the estimated number of tags in each individual region of the reference region file.
Statistical and biological processes analysis
The statistical language R was used for analysis of differential expression for CAGE and SAGE data. A threshold of two tags per million aligned reads (average across all samples) was applied. This filter represents the lower limit for consistent detection given our read depth, and will remove noise and background transcription. In addition, for CAGE data, we excluded regions where all reads started at exactly the same position, resulting in regions of ≤33 nt. These are likely sample preparation artifacts, as even sharply defined TSSs demonstrate some variability in start position, resulting in regions that cover  33 nt. Each region was tested separately with a Bayesian algorithm that takes into account library size (22,23). A Bayesian error rate <0.05 was considered significant. For gene-level tests, all tags overlapping a gene (including 1000 bases upstream and downstream of the gene) were summarized before statistical testing. For the calculation of expression ratios between differentiated and proliferating cells, data were first scaled to the average total number of aligned reads. For analysis of reproducibility, data were square root transformed to stabilize variance between samples, after which the Pearson's correlation coefficient was calculated.
33 nt. Each region was tested separately with a Bayesian algorithm that takes into account library size (22,23). A Bayesian error rate <0.05 was considered significant. For gene-level tests, all tags overlapping a gene (including 1000 bases upstream and downstream of the gene) were summarized before statistical testing. For the calculation of expression ratios between differentiated and proliferating cells, data were first scaled to the average total number of aligned reads. For analysis of reproducibility, data were square root transformed to stabilize variance between samples, after which the Pearson's correlation coefficient was calculated.
To compare differentially expressed genes to previously published microarray data, we took results from Tomczak et al. (12), performed VSN normalization (24) and analyzed data from differentiated versus proliferating cells with limma (25,26) in R. Multiple testing was done according to Benjamini and Hochberg (27). Probes were annotated with NetAffx from the Affymetrix website (www.affymetrix.com) and linked to the CAGE and SAGE top 30 genes based on gene symbols.
To annotate the biological processes, we took the top 30 differentially regulated genes from CAGE and SAGE (with a Bayesian Error rate  1×10−50 and sorted for differentiated cells on a ratio of differentiated to proliferating cells), as well as the microarray data (sorted on adjusted P-value) and ran these against 7689 Gene Ontology (GO) (28,29) Biological Processes in Anni 2.1 (30).
1×10−50 and sorted for differentiated cells on a ratio of differentiated to proliferating cells), as well as the microarray data (sorted on adjusted P-value) and ran these against 7689 Gene Ontology (GO) (28,29) Biological Processes in Anni 2.1 (30).
Sequence annotation
All CAGE and SAGE regions were annotated based on the ElDorado genome annotation (Genomatix, Version 07-2008) for being located in exons, introns or intergenic regions. Regions that covered an exon and neighboring intron or intergenic region were categorized as partial. In addition, a region was categorized as a promoter if it was located in the ElDorado-defined promoter region of a transcript. The distance to the nearest TSS (upstream or downstream) was also calculated. CAGE regions were correlated with CAGE data available in ElDorado [originating from the FANTOM3 project (31)].
CAGE region confirmation
To confirm that our CAGE regions represented newly discovered 5′-ends of transcripts, we designed primers within CAGE regions upstream of eight genes (Bpag, Cpeb1, Junb, Myl1, Pik3ca, Ppt2, Sertad4x and Usp34, primers in Supplementary Table S1). RT–PCR experiments were performed using random hexamer priming for cDNA synthesis and qPCR performed on a Roche Lightcycler 480. To provide additional validity to these CAGE regions, we inspected multiple UCSC tracks [UCSC genes, Ensembl (32) genes, Vega genes, Other RefSeq, AceView Genes, N-SCAN and Transcriptome].
To validate that our novel CAGE regions were indicative of myogenic promoters, we took all differentially expressed CAGE regions (see ‘Results'), expanded or contracted them to a length of 2000 bp, retrieved sequences with Ensembl Perl API scripts and ran them through CORE_TF (15), a program that identifies section overrepresented transcription factor binding sites. For a background sequence, we used 2000 mouse promoters defined as 1000 bp before and 1000 bp after the annotated TSS. A Match (33,34) setting to minimize the sum of false positives and false negatives was used.
We looked into more detail at the upstream CAGE regions of Myl1, a myogenic gene that was confirmed to have differential expression in the differentiation analysis. To this, we performed standard PCR for a primer set that spans the novel CAGE region into the first UCSC exon (Forward-TCAGCCAAAATTCCAAGTTGA, Reverse-CCTCCAGAAGAACCTGTCAGA). We also checked this CAGE region, plus 500 bases upstream sequence, for functional evidence. This was done by taking the mouse sequence, searching for orthologous sequences and identifying conserved patterns of transcription factor binding sites, as has been previously described (35,36).
RESULTS
The biological model and experimental setup
To study gene expression during myogenic differentiation, we used C2C12 mouse myoblasts, a common cell model for myogenesis, combined with next-generation sequencing technology. RNA was isolated from three independent cultures, both of proliferating and differentiated cells. At the latter condition, cells had differentiated into fused and multinucleated myotubes. To confirm successful differentiation, qPCR was performed to determine the expression levels of the genes encoding the late myogenic transcription factor Myogenin and the master myogenic regulator MyoD. Both of these should be expressed at higher levels in differentiated than proliferating cells. qPCR confirmed that cells had started to express Myogenin in differentiated cells and had higher expression of MyoD in differentiated cells (Supplementary Figure S1). CAGE and SAGE libraries were then prepared from all six RNA samples (three independent cell cultures for both proliferating and differentiated cells) and used to determine expression levels based on measurements in the 5′- and 3′-regions of the transcripts, respectively.
Sequencing and alignment characteristics
Each CAGE and SAGE library was sequenced on a single lane of the Illumina Genome Analyzer II. To investigate technical reproducibility, two CAGE samples (one from proliferating and one from differentiated cells) were sequenced in duplicate. After running the Illumina Genome Analyzer Pipeline for image and sequence quality analysis, we obtained on average 4.5 and 6.9 million reads from the CAGE and SAGE libraries, respectively (Table 1). The scarf files, converted to FASTQ format, containing the reads are available at GEO (37) under the accession number GSE21580. We aligned these reads to the repeat-masked mouse reference genome and were able to uniquely map (reporting alignments that are unique to one position in the genome), on average, 1.9 million (42%) and 4.1 million (59%) tags for CAGE and SAGE, respectively (Table 1).
Table 1.
Sequencing results
| No. of reads sequenced | No. of reads aligned | Percent aligned (%) | |
|---|---|---|---|
| CAGE sample | |||
| Prolif-1 | 4 886 341 | 2 086 233 | 42.7 |
| Prolif-1 duplo | 3 933 233 | 1 770 247 | 45.0 |
| Prolif-2 | 5 003 964 | 2 421 443 | 48.4 |
| Prolif-3 | 4 734 605 | 2 062 081 | 43.6 |
| Diff-1 | 4 525 321 | 1 679 081 | 37.1 |
| Diff-1 duplo | 3 101 153 | 1 252 451 | 40.4 |
| Diff-2 | 5 060 041 | 2 195 263 | 43.4 |
| Diff-3 | 4 830 194 | 1 578 087 | 32.7 |
| SAGE sample | |||
| Prolif-1 | 5 941 753 | 3 351 426 | 56.4 |
| Prolif-2 | 7 768 787 | 4 464 057 | 57.5 |
| Prolif-3 | 6 723 476 | 3 878 953 | 57.7 |
| Diff-1 | 9 467 926 | 5 811 947 | 61.4 |
| Diff-2 | 7 269 002 | 4 618 715 | 63.5 |
| Diff-3 | 4 392 416 | 2 494 618 | 56.8 |
Indicators for CAGE and SAGE samples: Prolif for proliferating cells and Diff for differentiating cells, followed by a number representing the biological triplicates. For CAGE there are sequencing duplicates indicated by ‘duplo'. The table contains the number of reads, the number of reads that align uniquely to the repeat-masked genome and the percent aligned.
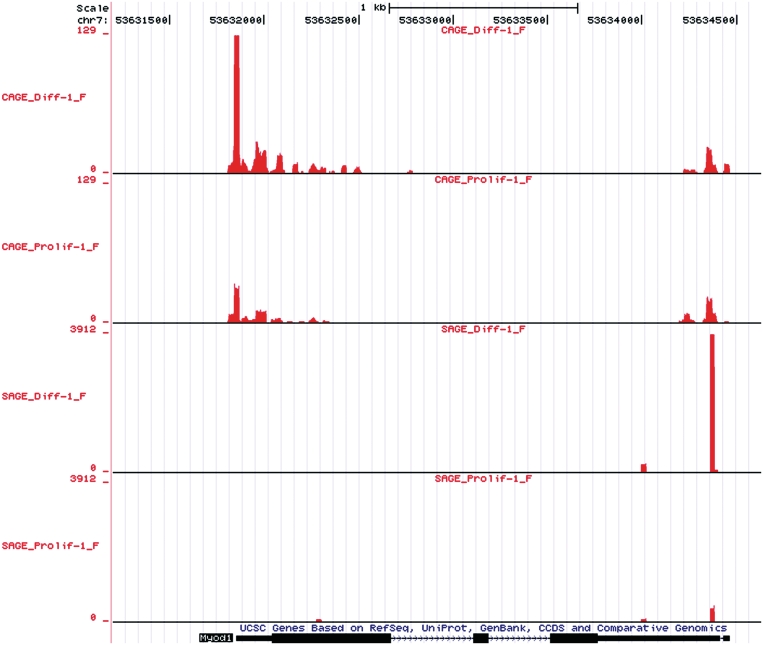
For visual analysis, we constructed UCSC Genome Browser wiggle files. The wiggle files are available at GEO under accession number GSE21580 and at http://www.lgtc.nl/publications/Hestand_2010_CAGE_SAGE_wig/. To retain information on the direction of transcription, there is one file for each strand. In Figure 1, we show an example wiggle track for the Myod1 gene. We clearly see the sharp SAGE peak starting at the most 3′-CATG site followed by 18 additional nucleotides. The CAGE peak at the 5′-end of the transcript is wider, reflecting the variability in the transcription start position. In line with the qPCR experiments, both peaks are larger in differentiated than in proliferating cells. As observed before (31), and observed for many other genes in the current study, CAGE also detects transcription starts in the 3′-region of the gene. Interestingly, this peak is not induced during differentiation, suggesting the formation of an independent transcript in this region.
Figure 1.
CAGE and SAGE wiggle tracks for proliferating (Prolif) and differentiated (Diff) cells in the UCSC Genome Browser for the myogenic marker MyoD. We only display reads aligning to the forward strand, the coding direction for MyoD. Chromosomal positions are indicated at the top. For each track the Y-axis scale corresponds to the number of tags aligned at that genomic position. Scales use a maximum from each relevant technique in this viewing window (129 for CAGE and 3912 for SAGE). There is 5 and 3
and 3 concordance for CAGE and SAGE samples, respectively. CAGE provides broader peaks, reflecting TSSs plus
concordance for CAGE and SAGE samples, respectively. CAGE provides broader peaks, reflecting TSSs plus  26 nt of downstream sequence, wheres SAGE provides discrete peaks. A higher number of tags are in differentiated compared to proliferating samples.
26 nt of downstream sequence, wheres SAGE provides discrete peaks. A higher number of tags are in differentiated compared to proliferating samples.
To account for the variability in TSS positions, we summarized adjacent CAGE reads into regions, while permitting gaps of maximally 100 nt to resolve gaps in alignments due to non-unique genomic sequences. Doing so, the CAGE regions obtained provide us with clearly distinct clusters of TSSs (with median lengths of 314 nt). We identified 742 355 different CAGE regions and 361 655 different SAGE regions. To remove noise and events regarded as background transcription, we applied an expression threshold of two tags-per-million [ 0.3 copies per cell (23)]. Increasing our (low) threshold would have eliminated many of the novel differentially regulated TSSs (defined below; Supplementary Figure S3). We found 41 862 CAGE and 43 512 SAGE regions with expression above the threshold of 2 tags-per-million.
0.3 copies per cell (23)]. Increasing our (low) threshold would have eliminated many of the novel differentially regulated TSSs (defined below; Supplementary Figure S3). We found 41 862 CAGE and 43 512 SAGE regions with expression above the threshold of 2 tags-per-million.
Technical reproducibility and biological overlap
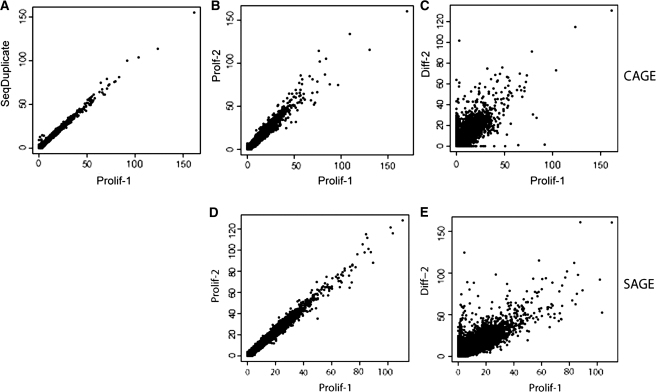
To analyze the technical reproducibility and the similarity between biological replicates, we calculated the correlation between the expression levels for all CAGE regions or SAGE tags. A high correlation was observed between the technical CAGE replicates (median Pearson's and correlation of 0.981) as well as the biological triplicates [median Pearson's and correlation of 0.963 (Figure 2A and B, Supplementary Table S2]. As expected, correlation between proliferating and differentiated cells was lower (median Pearson's correlation of 0.771), (Figure 2C, Supplementary Table S2). Similarly, we observed a high reproducibility for the SAGE experiments (median Pearson's correlation of 0.930) between biological triplicates (Figure 2D and Supplementary Table S2). Again, the correlation between proliferating and differentiated cells (median Pearson's correlation of 0.839) was lower than that between cells from the same condition (Figure 2E and Supplementary Table S2).
Figure 2.
High reproducibility was found in CAGE regions between sequencing duplicates (A) and biological replicates (B). (C) Shows correlation between CAGE samples from proliferating and differentiated cells. High reproducibility can also be found between SAGE biological replicates (D). (E) Shows the correlation between SAGE samples from proliferating and differentiated cells. The plotted values represent the square root of the number of tags per region.
Annotation of regions
We annotated the 41 862 CAGE regions using ElDorado's mouse genome annotation: 9957 regions map to annotated exons, 27 190 partially overlap exons and intronic/intergenic regions, 2368 map to introns and 2347 to intergenic regions. The median number of tags in the exonic and partial regions (63 tags and 90 tags respectively) were higher than in the intronic and intergenic regions (45 tags, and 54 tags, respectively). These data clearly show that our CAGE experiments generally confirm previously annotated transcripts but also detect many (lower abundant) TSSs/transcribed regions that have not yet been identified and/or annotated as such in current genome databases.
Based on ElDorado annotation of our 41 862 CAGE regions, 13 541 of the CAGE regions (32%) contained an annotated TSS, 6331 CAGE regions (15%) were annotated as promoters (i.e. a genomic region surrounding a TSS containing functional elements like transcription factor binding sites that are responsible for the regulation of the expression of the transcript) and 8028 (19%) CAGE regions contained an annotated transcript 3′-end. The 3′-end alignments are consistent with the previously observed (31) significant amount of (shorter) transcripts originating from the 3′-ends of genes. We compared our CAGE results to previous CAGE studies (FANTOM3) contained in ElDorado and identified 31 680 regions (76%) overlapping with at least one on the FANTOM3 CAGE tags. Only 6119 (15%) and 5635 (13%) of these regions were observed in FANTOM3 muscle and heart CAGE libraries, respectively. This is explained by the small size of these muscle and heart libraries (31), together representing only 1% of all available CAGE tags in FANTOM3.
Comparison of CAGE, SAGE and microarray expression data
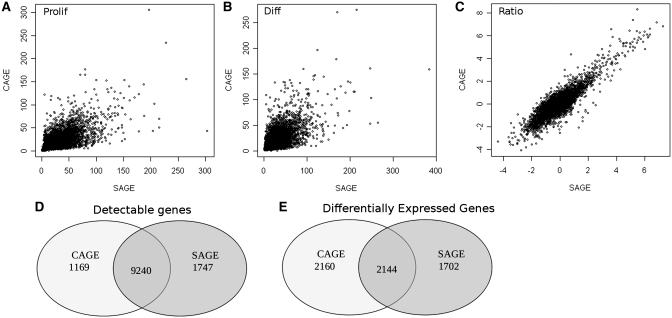
To compare gene expression levels and analyze differential gene expression, we assigned CAGE and SAGE regions to genes (including 1000 bases upstream and downstream of the gene). Expression above a threshold of two transcripts per million ( 0.3 copies per cell) (38) was observed for 10 409 and 10 987 genes, respectively. Expression profiles for both methods showed a high correlation (Figure 3A–C), with 9240 genes being expressed in both methods above two transcripts per million (Figure 3D). Supplementary Figure S2 shows that the relative overlap is even bigger when higher detection thresholds are applied, obviously at the expense of many more genes not reaching the detection threshold. The 4304 genes were differentially expressed between proliferating and differentiated cells (Bayesian error rate
0.3 copies per cell) (38) was observed for 10 409 and 10 987 genes, respectively. Expression profiles for both methods showed a high correlation (Figure 3A–C), with 9240 genes being expressed in both methods above two transcripts per million (Figure 3D). Supplementary Figure S2 shows that the relative overlap is even bigger when higher detection thresholds are applied, obviously at the expense of many more genes not reaching the detection threshold. The 4304 genes were differentially expressed between proliferating and differentiated cells (Bayesian error rate  0.05) according to the CAGE data and 3 46 according to the SAGE data with 2144 genes present in the lists of significant genes (Figure 3E). Most others were just borderline significant according to one of the methods.
0.05) according to the CAGE data and 3 46 according to the SAGE data with 2144 genes present in the lists of significant genes (Figure 3E). Most others were just borderline significant according to one of the methods.
Figure 3.
Correlation of CAGE versus SAGE for proliferating samples (A), differentiated samples (B), and the ratio of proliferating and differentiated cells (C). Values are the square root of the number of tags per gene for A and B. For C, the values are the log ratio of the normalized number of tags per gene in differentiated over proliferating cells. The overlap of detectable genes (D) and differentially expressed genes (E) between CAGE and SAGE is indicated.
We compared the top 30 most differentially expressed genes for both methods (Table 2) to results from a similar microarray data set on myogenic differentiation in the same cell line (12). In general, the genes identified by CAGE and SAGE also demonstrated very significant changes on the microarrays. However, in the top 30, 13 genes identified by CAGE and 10 identified by SAGE were not represented on the array, demonstrating the comprehensive nature of the CAGE- and SAGE-based gene expression profiling techniques. The biological processes controlled by the top 30 CAGE, SAGE and microarray genes were annotated with the Anni2.1 text-mining tool (Table 3). All CAGE- and SAGE-derived GO terms can readily be related to muscle development, whereas 3/10 GO terms associated with the microarray-derived gene list cannot (‘cyclin-dependent protein kinase inhibitor activity’, ‘6-phosphofructokinase’ and ‘tumor suppressor activity').
Table 2.
Differential gene expression
| CAGE gene | Ratio | Microarray P-value | SAGE gene | Ratio | Microarray P-value |
|---|---|---|---|---|---|
| Hfe2 | 4073 | NA | RP23-36P22.5 | 576 | NA |
| Myom3 | 1624 | NA | Neb | 525 | NA |
| Lmod2 | 1305 | NA | Mylpf | 504 | 1.70 × 10−15 |
| Myh7 | 1124 | 5.98 × 10−3 | Ttn | 380 | NA |
| Mb | 908 | 1.07 × 10−14 | Myh3 | 368 | 2.40 × 10−14 |
| RP23-36P22.5 | 735 | NA | Xirp1 | 306 | 2.24 × 10−13 |
| Pygm | 717 | 4.82 × 10−17 | 1110002H13Rik | 263 | NA |
| Myl4 | 614 | 8.86 × 10−20 | Tnnc1 | 232 | 1.24 × 10−11 |
| Synpo2l | 595 | NA | Cav3 | 150 | 3.58 × 10−22 |
| Myh1 | 561 | 3.64 × 10−15 | Cbfa2t3 | 133 | 2.89 × 10−10 |
| Tnni1 | 529 | 2.24 × 10−9 | Chrng | 115 | 4.63 × 10−9 |
| Tnni2 | 442 | 3.20 × 10−11 | Myom2 | 105 | 6.66 × 10−16 |
| Mpa2l | 410 | NA | Tnnt1 | 100 | 1.15 × 10−10 |
| Ctrb1 | 406 | 7.55 × 10−7 | Ryr1 | 92 | 7.03 × 10−14 |
| Ttn | 402 | NA | Apobec2 | 84 | 2.95 × 10−15 |
| Neb | 374 | NA | Cox6a2 | 72 | 2.45 × 10−16 |
| Kcnq4 | 365 | NA | Dio2 | 64 | 2.14 × 10−10 |
| Mylpf | 341 | 1.70 × 10−15 | C1qtnf3 | 52 | 4.36 × 10−5 |
| 1110002H13Rik | 341 | NA | Htr2b | 43 | 3.76 × 10−6 |
| Inpp4b | 328 | NA | Sgcg | 42 | 1.15 × 10−12 |
| Xirp1 | 307 | 2.24 × 10−13 | Fndc5 | 39 | NA |
| Atp2a1 | 304 | 2.06 × 10−14 | Jsrp1 | 36 | NA |
| Casq2 | 297 | 4.74 × 10−6 | Ankrd23 | 36 | NA |
| Cacna1s | 296 | 5.20 × 10−19 | AK031267 | 29 | NA |
| Ces2 | 245 | NA | Sema6a | 26 | 3.08 × 10−3 |
| Cox6a2 | 241 | 2.45 × 10−16 | Lgr5 | 23 | 9.33 × 10−1 |
| Myog | 238 | 2.36 × 10−6 | Pdlim3 | 22 | 3.18 × 10−6 |
| Myh3 | 234 | 2.40 × 10−14 | Klhl31 | 22 | NA |
| Tmem182 | 216 | NA | ORF63 | 21 | NA |
| Tnnc1 | 215 | 1.24 × 10−11 | Gfra2 | 19 | 2.98 × 10−2 |
Top 30 genes from SAGE and CAGE expression data. All genes with a Bayesian error rate  1 × 10−50 were sorted on the ratio (normalized tags from differentiated/proliferating cells) and the highest ratios for differentiated cells displayed. The microarray P-values are adjusted P-values for differential gene expression from a similar experiment [proliferating and differentiated C2C12 cells (12)]. NA, no probe annotation for the gene.
1 × 10−50 were sorted on the ratio (normalized tags from differentiated/proliferating cells) and the highest ratios for differentiated cells displayed. The microarray P-values are adjusted P-values for differential gene expression from a similar experiment [proliferating and differentiated C2C12 cells (12)]. NA, no probe annotation for the gene.
Table 3.
| CAGE GO | SAGE GO | Microarray GO |
|---|---|---|
| (1) Regulation of striated muscle contraction | (1) Regulation of muscle contraction | (1) Cyclin-dependent protein kinase inhibitor activity |
| (2) Cardiac muscle contraction | (2) Cardiac muscle contraction | (2) Myogenesis |
| (3) Myogenesis | (3) Myogenesis | (3) Skeletal muscle development |
| (4) Regulation of muscle contraction | (4) Regulation of striated muscle contraction | (4) Myoblast differentiation |
| (5) Skeletal muscle development | (5) Skeletal muscle development | (5) 6-Phosphofructokinase activity |
| (6) Muscle Development | (6) Myofibril assembly | (6) Muscle Development |
| (7) Striated muscle contraction | (7) Muscle Development | (7) Muscle cell differentiation |
| (8) Myoblast differentiation | (8) Myoblast fusion | (8) Tumor suppressor activity |
| (9) Muscle cell differentiation | (9) Striated muscle contraction | (9) Myofibril assembly |
| (10) Sarcomere organization | (10) Muscle cell differentiation | (10) Heart development |
The top 10 GO biological processes associated with the top 30 genes for CAGE, SAGE and microarray experiments indicate clear muscle relations, with the exception of three (in italics) processes in the microarray data.
Differential TSS use and validation
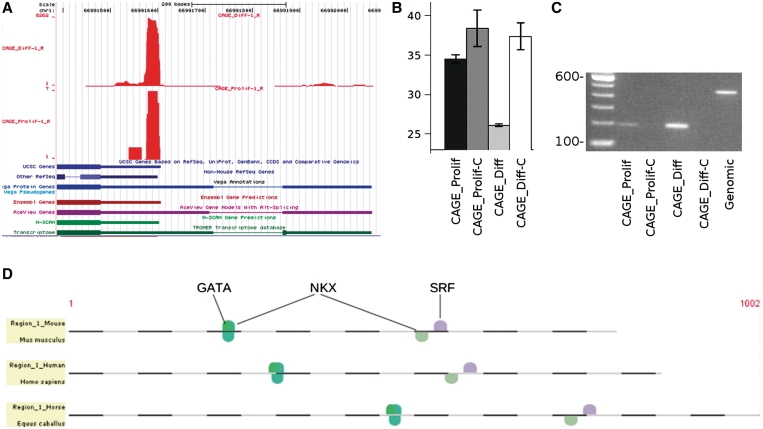
In our CAGE data, we identified 196 regions, 111 regions upstream of the start of a known gene and 85 CAGE regions downstream of an annotated gene, with significantly different numbers of tags in proliferating and differentiated cells (Supplementary Table S3). The differential expression of transcripts originating from seven out of eight of these regions (upstream from genes Bpag, Cpeb1, Junb, Myl1, Pik3ca, Ppt2, Sertad4x and Usp34) were confirmed by RT–PCR/qPCR (Figure 4B and Supplementary Figure S4). To evaluate if these novel exons were contained in a transcript of the gene of interest, we inspected the following tracks in the UCSC Genome Browser: UCSC genes, Ensembl genes, Vega genes, Other RefSeq, AceView Genes, N-SCAN and Transcriptome (Figure 4A and Supplementary Figure S5). In all but Junb we found the CAGE regions overlapping at least one exon from an additional track connected to the gene of interest (Figure 4A and Supplementary Figure S5). This indicates that these CAGE regions usually represented alternative transcripts that are not yet properly annotated in all resources, including the mainstream UCSC and Ensembl annotations. This suggests that the mainstream genome annotation are far from complete and that additional evidence, including our CAGE data, is required to more precisely define transcript structure.
Figure 4.
The UCSC display of (A) UCSC/Ensembl-defined first exon and an upstream Myl1 CAGE region (reverse strand reads only, on which the gene lies) for samples Prolif-1 and Diff-1. The Y-axis indicates the number of tags aligned at each position in the genome. We also display additional track information (UCSC genes, Ensembl genes, Vega genes, Other RefSeq, AceView Genes, N-SCAN and Transcriptome), several of which confirm the presence of the upstream CAGE region. (B) qPCR with primers within the CAGE region for Prolif, Prolif-C (reverse transcriptase control), Diff and Diff-C (reverse transcriptase control). The qPCR results are plotted as threshold cycle (Cp) values (lower = higher expression), with bars indicating a range of one SD between technical duplicates. (C) Standard PCR on agarose gel with forward primer in the novel CAGE region and reverse primer in the conventional exon 1. Comparison with the genomic control verifies the presence of an intron of 200 bases. A 100-bp ladder is included. (A–C) are consistent with higher expression in differentiated than proliferating cells. (D) Cross-species conserved muscle-specific transcription factor binding sites around and upstream of the Myl1 CAGE region support its role as a promoter for this region.
To support that differential transcription in the 196 CAGE regions is regulated by myogenic transcription factors, we searched for overrepresented transcription factor binding sites and found the binding sites for the master regulators MyoD (P-value 6.49 × 10−03 from CORE_TF's binomial test), Myogenin (P-value: 3.87×10−02) and the Ebox motif (P-value 6.02×10−03) [frequently found in muscle promoters (39,40)] to be significantly overrepresented in 2000 bp of sequence composed of the CAGE and surrounding regions (Supplementary Table S4).
For one of these novel CAGE regions, Myl1, we confirmed by standard RT–PCR that there is a transcript extending from the novel CAGE region into the UCSC-defined exon 1 (Figure 4C). The CAGE sequencing, RT–PCR/qPCR within the region, and the standard PCR into exon 1 all confirmed that this transcript is only present in differentiated cells, explaining why it is missing in standard genome annotations. For functional evidence that this region is used as a promoter, we also looked for conserved transcription factor binding sites in and upstream of this region. Within the Genomatix Suite, we identified orthologous sequence regions from human and horse corresponding to the CAGE region and 5′-upstream (promoter) sequence. In this area, we identified conserved binding sites for NKX, GATA and SRF (Figure 4D), all of which are known to be involved in the regulation of muscle genes (41). This makes it likely that the region directly upstream of the novel exon 1 is used as an alternative promoter.
DISCUSSION
Using CAGE and SAGE methods with next-generation sequencing, we have measured gene expression levels during myogenic differentiation and identified muscle-specific TSSs. By elucidating promoter regions and regulation in these myogenic cells, we hope to better understand the process of muscle development and regeneration, providing clues to cure muscle-related illnesses. Since biologists and clinicians often study (first) exons and 5′-promoter regions, it is crucial to know the positions of TSSs in the genome. Our data will help them identify potentially pathogenic mutations in transcripts and promoters used during myogenic differentiation, which might have been overlooked with current genome annotations. On a technical level, this is the first time CAGE and SAGE have been evaluated using the same RNA samples.
We found both the technically demanding CAGE method and the slightly less laborious SAGE method to be extremely robust. Biological triplicates with independent sample preparations and sequencing runs were found to have high correlations (Figure 2, Supplementary Table S2). This is in line with previous findings in the FANTOM4 CAGE study (42) and our previous (23) finding with SAGE. Higher technical reproducibility also enhances the ability to verify low expressed genes, which was an obstacle in microarray analysis. The high quality of the data implies that more investments should be made in biological than technical replicates.
This study also highlights other advantages over microarrays. For a third of the top 30 genes, (13/31 CAGE genes and 10/30 SAGE genes, Table 2), there was no probe on the microarray. Finding many more significant genes not interrogated by the microarrays stresses the more comprehensive transcript profiling by next-generation sequencing-based methods. We also found more muscle-related biological processes associated with the top 30 CAGE and SAGE genes compared to the microarray top 30 genes (Table 3), indicating the higher relevance of the top hits for the process under study.
The data provided by these methods have expanded our knowledge of muscle-specific transcription. Only 32% of the analyzed CAGE regions contained an annotated TSS, indicating that we discovered many novel TSSs. Seventy-six percent of CAGE regions matched known FANTOM3 CAGE tags. The high overlap with previous FANTOM CAGE regions indicates that our CAGE regions reflect true TSSs. However, <20% of our regions matched known muscle-related CAGE tags. This is likely due to the lower sequencing depth in the previous FANTOM3 muscle studies. This shows that we have greatly enriched the annotation of muscle-related TSSs and that there is currently a lack of information on tissue-specific TSS usage. To exemplify this point, we identified 196 intergenic regions significantly different between proliferating and differentiated cells, indicating muscle-specific alternative promoter and first exon usage. We found overrepresentation of muscle-specific transcription factor binding sites for MyoD and Myogenin and E-boxes in these regions, indicating that the identified regions potentially serve as promoters. Several of these were verified by PCR and additional UCSC track evidence. This, also taking into account the overall lower level of expression compared to known TSSs (Supplementary Figure S3), shows that our methods are reliable to detect rare transcriptional events.
This is the first study to compare NGS of CAGE and SAGE libraries from the same RNA samples. Gene expression measurements by CAGE and SAGE are generally consistent. The high correlation between methods (Figure 3A–C), large overlap between genes detected (Figure 3D) and differential gene lists (Figure 3E), and gene involvement in similar biological pathways (Table 3) indicate that these methods are both useful for expression profiling. However, in studies into promoter regulation, one should preferably use CAGE, whereas in studies regarding micro-RNA regulation, RNA transcript stability and alternative polyadenylation one should preferably use SAGE.
Of the 4304 and 3846 genes differentially expressed between proliferating and differentiated cells with CAGE and SAGE, respectively, over half (2144) of the genes are identical. More changes in CAGE than SAGE levels could indicate that alternative promoter usage is more common than that of alternative 3′-ends. The detection of genes by one technique, but not the other, is mostly inherent to the use of thresholds, as application of higher thresholds than the one applied (2 transcripts per million) resulted in a higher percentage-wise overlap. Alternatively, a minority of transcripts may be missed entirely by one of the methods due to the absence of a CATG site in the transcript (SAGE) or because of our filtering for non-unique sequences, which was done to increase the reliability of the mapping. For both techniques, we frequently detected multiple regions in the same gene. Seventy-five percent of the genes had multiple SAGE tags with abundance above the threshold of two transcripts per million. In our previous paper (23), we discussed that this is probably not a technical artifact but most likely due to different 3′-ends and usage of multiple polyadenylation sites.
Similar to previous studies (31), we found a large number of CAGE tags aligning to the 3′-end of known transcripts. This phenomenon has been previously validated by the rapid amplification of cDNA ends (RACE) method and explained as potential 3′-derived regulatory non-coding RNAs (31). This type of non-coding RNAs, frequently derived from regions in or around the 3′-UTR, have been reviewed before (43). Gustincich et al. (43) also report that they tend to have an additional gene downstream on the opposite strand, indicating a sense–antisense mechanism or protection. With additional analysis steps, CAGE could serve as a method for identification of non-coding RNAs. In addition, these should be recognized as a potential source of erroneous expression levels measured in SAGE and 3′-based microarrays.
Likewise, 67% of the genes contained multiple CAGE regions. These observations are consistent with the finding of many (short) transcripts from exonic regions in a tiling array study (44). Apart from alternative TSSs, resulting in alternative RNA isoforms with different first exons, coding for different protein isoforms, these CAGE regions may represent degradation products of the mRNA. This phenomenon was previously referred to as ‘exon painting' (44). Examples of genes, where nearly all exons are covered by CAGE tags are Col1a1 and Col1a2 (Supplementary Figure S6A and B, respectively). It is not likely that these are random degradation products, given the high RNA integrity in all samples, the observation of genes with a highly abundant peak at the 5′-end without any exon painting (Supplementary Figure S6C and D) and the high reproducibility of the exon painting patterns in independent CAGE sample preparations. This suggests that there is a biological explanation for the exon painting phenomenon. From our study, it is highly likely that many of these shorter transcripts contain a cap structure. The process of recapping of transcript fragments has been documented before (44). Fejes-Toth et al. (44) propose that long RNAs are spliced into mature and translatable RNAs, but that these mature RNAs can also be further processed. This further processing involves cleavage into smaller RNA fragments and possible modification by additional 5′-capping (44). The presence of exon painting complicates the identification of novel TSSs and is the reason why we focused on the discovery of novel TSSs in intergenic regions and did not report alternative TSSs within annotated genes. A positive consequence of the exon painting phenomenon is that the CAGE technique gives additional information on the exon structure of many genes.
The large data yield and reproducibility should serve as an example of the advantages of applying next-generation sequencing to CAGE and SAGE techniques. This work has provided a substantial increase in our knowledge of myogenic TSSs and expression. These methodologies should be expanded to other tissues and processes in the future to enrich our knowledge of the transcriptional regulation and to enrich current genome annotations. As demonstrated in this manuscript, with the use of biological replicates, appropriate techniques and sequencing depth, and proper analysis (e.g. thresholds) it is possible to reliably monitor gene expression and rare transcriptional events.
SUPPLEMENTARY DATA
Supplementary Data are availble at NAR online.
FUNDING
Centre for Medical Systems Biology within the framework of the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO); Center for Biomedical Genetics (in The Netherlands). Funding for open access charge: Center for Biomedical Genetics.
Conflict of interest statament. A. K., M. S., W. v W., M. S., T. W., and M. H. declare that they have competing financial interests. All other authors have declared no conflict of interest.
ACKNOWLEDGEMENTS
We wish to thank Michel Villerius, Michiel van Galen, and Ivo Fokkema for their computational assistance. We also wish to thank Rolf Vossen for advice with the Lightcycler 480. We would also like to thank Henk P. J. Buermans and Emile J. de Meijer for wet-lab assistance and providing primers for expression analysis.
REFERENCES
- 1.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 2.Harbers M, Carninci P. Tag-based approaches for transcriptome research and genome annotation. Nat. Methods. 2005;2:495–502. doi: 10.1038/nmeth768. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen KL, Hogh AL, Emmersen J. DeepSAGE–digital transcriptomics with high sensitivity, simple experimental protocol and multiplexing of samples. Nucleic Acids Res. 2006;34:e133. doi: 10.1093/nar/gkl714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shiraki T, Kondo S, Katayama S, Waki K, Kasukawa T, Kawaji H, Kodzius R, Watahiki A, Nakamura M, Arakawa T. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl Acad. Sci. USA. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valen E, Pascarella G, Chalk A, Maeda N, Kojima M, Kawazu C, Murata M, Nishiyori H, Lazarevic D. Genome-wide detection and analysis of hippocampus core promoters using DeepCAGE. Genome Res. 2009;19:255–265. doi: 10.1101/gr.084541.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morin R, Bainbridge M, Fejes A, Hirst M, Krzywinski M, Pugh T, McDonald H, Varhol R, Jones S, Marra M. Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. Biotechniques. 2008;45:81–94. doi: 10.2144/000112900. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, vanBaren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sterrenburg E, Turk R, Hoen PAC't, van Deutekom JCT, Boer JM, van Ommen G-JB, den Dunnen JT. Large-scale gene expression analysis of human skeletal myoblast differentiation. Neuromuscul. Disord. 2004;14:507–518. doi: 10.1016/j.nmd.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 10.Turk R, Sterrenburg E, van derWees CGC, deMeijer EJ, deMenezes RX, Groh S, Campbell KP, Noguchi S, vanOmmen GJB, den Dunnen JT, et al. Common pathological mechanisms in mouse models for muscular dystrophies. FASEB J. 2006;20:127–129. doi: 10.1096/fj.05-4678fje. [DOI] [PubMed] [Google Scholar]
- 11.Jelier R, Hoen PAC't, Sterrenburg E, den Dunnen JT, van Ommen G-JB, Kors JA, Mons B. Literature-aided meta-analysis of microarray data: a compendium study on muscle development and disease. BMC Bioinformatics. 2008;9:291. doi: 10.1186/1471-2105-9-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomczak KK, Marinescu VD, Ramoni MF, Sanoudou D, Montanaro F, Han M, Kunkel LM, Kohane IS, Beggs AH. Expression profiling and identification of novel genes involved in myogenic differentiation. FASEB J. 2004;18:403–405. doi: 10.1096/fj.03-0568fje. [DOI] [PubMed] [Google Scholar]
- 13.Pownall ME, Gustafsson MK, Emerson CPJ. Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu. Rev. Cell Dev. Biol. 2002;18:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- 14.Sartorelli V, Caretti G. Mechanisms underlying the transcriptional regulation of skeletal myogenesis. Curr. Opin. Genet. Dev. 2005;15:528–535. doi: 10.1016/j.gde.2005.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hestand MS, van Galen M, Villerius MP, van Ommen G-JB, den Dunnen JT, Hoen PAC't. CORE_TF: a user-friendly interface to identify evolutionary conserved transcription factor binding sites in sets of co-regulated genes. BMC Bioinformatics. 2008;9:495. doi: 10.1186/1471-2105-9-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sui SJH, Fulton DL, Arenillas DJ, Kwon AT, Wasserman WW. oPOSSUM: integrated tools for analysis of regulatory motif over-representation. Nucleic Acids Res. 2007;35:W245–W252. doi: 10.1093/nar/gkm427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith AD, Xuan Z, Zhang MQ. Using quality scores and longer reads improves accuracy of Solexa read mapping. BMC Bioinformatics. 2008;9:128. doi: 10.1186/1471-2105-9-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faulkner GJ, Forrest ARR, Chalk AM, Schroder K, Hayashizaki Y, Carninci P, Hume DA, Grimmond SM. A rescue strategy for multimapping short sequence tags refines surveys of transcriptional activity by CAGE. Genomics. 2008;91:281–288. doi: 10.1016/j.ygeno.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 21.Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 22.Vencio RZN, Brentani H, Patrao DFC, Pereira CAB. Bayesian model accounting for within-class biological variability in Serial Analysis of Gene Expression (SAGE) BMC Bioinformatics. 2004;5:119. doi: 10.1186/1471-2105-5-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoen PAC't, Ariyurek Y, Thygesen HH, Vreugdenhil E, Vossen RHAM, de Menezes RX, Boer JM, van Ommen G-JB, denDunnen JT. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res. 2008;36:e141. doi: 10.1093/nar/gkn705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huber W, vonHeydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl. 1):S96–104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 25.Smyth G, Yang Y, Speed T. Statistical issues in CDNA microarray data analysis. Methods Mol. Biol. 2003;224:111–136. doi: 10.1385/1-59259-364-X:111. [DOI] [PubMed] [Google Scholar]
- 26.Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. [Google Scholar]
- 27.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J.R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 28.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Camon E, Barrell D, Lee V, Dimmer E, Apweiler R. The Gene Ontology Annotation (GOA) Database–an integrated resource of GO annotations to the UniProt Knowledgebase. In Silico Biol. 2004;4:5–6. [PubMed] [Google Scholar]
- 30.Jelier R, Schuemie MJ, Veldhoven A, Dorssers LCJ, Jenster G, Kors JA. Anni 2.0: a multipurpose text-mining tool for the life sciences. Genome Biol. 2008;9:R96. doi: 10.1186/gb-2008-9-6-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CAM, Taylor MS, Engstrom PG, Frith MC, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 32.Hubbard TJP, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kel AE, Gossling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen CD, Klingenhoff A, Boucherot A, Nitsche A, Henger A, Brunner B, Schmid H, Merkle M, Saleem MA, Koller K-P, et al. Comparative promoter analysis allows de novo identification of specialized cell junction-associated proteins. Proc. Natl Acad. Sci. USA. 2006;103:5682–5687. doi: 10.1073/pnas.0511257103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dohr S, Klingenhoff A, Maier H, deAngelis MH, Werner T, Schneider R. Linking disease-associated genes to regulatory networks via promoter organization. Nucleic Acids Res. 2005;33:864–872. doi: 10.1093/nar/gki230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Velculescu VE, Madden SL, Zhang L, Lash AE, Yu J, Rago C, Lal A, Wang CJ, Beaudry GA, Ciriello KM, et al. Analysis of human transcriptomes. Nat. Genet. 1999;23:387–388. doi: 10.1038/70487. [DOI] [PubMed] [Google Scholar]
- 39.Buskin JN, Hauschka SD. Identification of a myocyte nuclear factor that binds to the muscle-specific enhancer of the mouse muscle creatine kinase gene. Mol. Cell Biol. 1989:2627–2640. doi: 10.1128/mcb.9.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olson EN. MyoD family: a paradigm for development? Genes Dev. 1990;4:1454–1461. doi: 10.1101/gad.4.9.1454. [DOI] [PubMed] [Google Scholar]
- 41.Nishida W, Nakamura M, Mori S, Takahashi M, Ohkawa Y, Tadokoro S, Yoshida K, Hiwada K, Hayashi K, Sobue K. A triad of serum response factor and the GATA and NK families governs the transcription of smooth and cardiac muscle genes. J. Biol. Chem. 2002;277:7308–7317. doi: 10.1074/jbc.M111824200. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki H, Forrest ARR, vanNimwegen E, Daub CO, Balwierz PJ, Irvine KM, Lassmann T, Ravasi T, Hasegawa Y, deHoon MJL, et al. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat. Genet. 2009;41:553–562. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gustincich S, Sandelin A, Plessy C, Katayama S, Simone R, Lazarevic D, Hayashizaki Y, Carninci P. The complexity of the mammalian transcriptome. J. Physiol. 2006;575:321–332. doi: 10.1113/jphysiol.2006.115568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fejes-Toth K, Sotirova V, Sachidanandam R, Assaf G, Hannon GJ, Kapranov P, Foissac S, Willingham AT, Duttagupta R, Dumais E, et al. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]