Abstract
Data presented here extends our previous observations on α-globin transcriptional regulation by the CP2 and PIAS1 proteins. Using RNAi knockdown, we have now shown that CP2b, CP2c and PIAS1 are each necessary for synergistic activation of endogenous α-globin gene expression in differentiating MEL cells. In this system, truncated PIAS1 mutants lacking the ring finger domain recruited CP2c to the nucleus, as did wild-type PIAS1, demonstrating that this is a sumoylation-independent process. In vitro, recombinant CP2c, CP2b and PIAS1 bound DNA as a stable CBP (CP2c/CP2b/PIAS1) complex. Following PIAS1 knockdown in MEL cells, however, the association of endogenous CP2c and CP2b with the α-globin promoter simultaneously decreased. By mapping the CP2b- and CP2c-binding domains on PIAS1, and the PIAS1-binding domains on CP2b and CP2c, we found that two regions of PIAS1 that interact with CP2c/CP2b are required for its co-activator function. We propose that CP2c, CP2b, and PIAS1 form a hexametric complex with two units each of CP2c, CP2b, and PIAS1, in which PIAS1 serves as a clamp between two CP2 proteins, while CP2c binds directly to the target DNA and CP2b mediates strong transactivation.
INTRODUCTION
Transcription factor CP2c, a member of the CP2 gene family, participates in diverse processes, including hematopoiesis, immune response, the cell cycle and neural development, by regulating expression of specific target genes (1). The ubiquitous expression of CP2c has drawn attention to its regulatory mechanisms in various cellular and developmental contexts. Interactions between CP2c and cell-restricted factors may determine the specificity of target gene expression. In erythroid cells, CP2c interacts with an erythroid-specific factor, NF-E4, to form a stage-selector protein complex that binds to and activates the γ-globin promoter (2–4). The zinc finger transcription factor GATA-1, expressed within the hematopoietic system, interacts with CP2c to bind to adjacent CP2/GATA-1 sites within the promoters of GATA-1, NF-E4 and EKLF genes (5). Interaction between CP2c and the neuron-specific protein Fe65 blocks cell cycle progression by downregulating thymidylate synthase gene expression (6,7).
Many transcription factors exist in families of isoforms with distinct activities generated by alternative splicing, or by gene duplication and diversification during evolution. Heteromeric combinations of isoforms exert different effects on target genes, leading to various cellular responses. The CP2 transcription factor family, with six human isoforms (LBP-1a, -1b, -1c, -1d, -9 and -32) and four murine (CP2a/NF2d9, CP2b, CP2c/CP2 and CRTR), exemplifies this pattern (1). CP2c was initially identified as an activator of the murine α-globin gene that binds to the consensus sequence CNRG-N6-CNR(G/C) within the α-globin promoter (8–12). Indeed, overexpression of CP2c strongly activates the α-globin promoter in vitro and in vivo (10,11). Furthermore, experimental CP2c depletion suppresses α- and ß-globin gene transcription, and hemoglobin synthesis during terminal differentiation of murine erythroid leukemia (MEL) cells (13). A novel CP2 isoform, CP2b, is identical to CP2a except for an additional 36 amino acids encoded by an extra exon that gives an additional transcriptional activation domain. A heteromeric complex of CP2b and CP2c activates α-globin expression (14). Investigations with exogenous CP2 proteins reveal that CP2a and CP2b localize exclusively in the cytoplasm and nucleus, respectively (15). Following knockdown of CP2a/CP2b, cells retain CP2c in the cytoplasm. However, in cells that coexpress CP2b, exogenous CP2c localizes predominantly in the nucleus, whereas in cells that coexpress CP2a, CP2c remains in the cytoplasm. Since each CP2 family protein may form homo- and heterodimers and/or oligomers (16,17), it is postulated that CP2c may localize in either the nucleus or the cytoplasm, depending on the relative levels of CP2a and CP2b. In this context, CP2b would potentiate erythroid cell-specific α-globin expression by recruiting CP2c into the nucleus (15). In addition, the heteromeric complex of CP2b and CP2c activates α-globin in an erythroid cell-specific manner; that is, the dimer activates the α-globin promoter in erythroid K562 and MEL cells, but not in non-erythroid 293T cells (14). Finally, we identified PIAS1 (protein inhibitor of activated STAT1), as an interactive partner to CP2 proteins and a potent co-activator for CP2c/CP2b-mediated α-globin expression in erythroid cells (14).
The PIAS protein family includes at least five evolutionarily conserved genes and/or splice variants (PIAS1, PIAS3, PIASxα, PIASxβ and PIASy) (18). Mammalian PIAS proteins were initially identified as negative regulators of STAT signaling (19,20), but may interact with more than 70 proteins. One major function of these proteins is the control of gene transcription (21). PIAS proteins can act as either an activator or a repressor, depending on the target gene or interactions with transcriptional regulators (18,21,22). PIAS proteins can also act as small ubiquitin-like-modifier (SUMO) E3 ligases (23), because they share the central RING-finger domain (RFD, amino acids 309–400) required for SUMO E3 ligase activity (21,24). This finding sheds light on the mechanistic role of sumoylation in transcriptional regulation by the PIAS proteins. Sumoylation affects the stability (25), subcellular localization (26,27) and activity of PIAS target proteins (21,28–31), which include c-Jun, p53, lymphoid enhancer factor 1, Smad, androgen receptor and GATA4. Transcription factors comprise the largest group of proteins subject to sumoylation, although some transcription factors are not coupled to this function (18,21,32,33).
Here, we report that PIAS1 induces the movement of CP2c from the cytoplasm to the nucleus, which may be required to activate CP2c/CP2b-mediated α-globin expression. In addition, we found that PIAS1 stabilizes the CP2c/CP2b complex and strongly enhances its recruitment to consensus binding sites within the α-globin promoter, through interaction with CP2 proteins. Two PIAS1-binding regions were mapped in both CP2c and CP2b. The N-terminal and C-terminal halves of PIAS1 also interact with CP2c and CP2b, and both regions are required for PIAS1 to function as a co-activator. An RFD-deleted PIAS1 mutant interacts with CP2 proteins to enhance α-globin transcription by increasing the DNA binding activity of the CP2c/CP2b complex. These data suggest that PIAS1 functions as a clamp for formation of an active CBP (CP2c/CP2b/PIAS1) complex, conferring DNA-binding activity on the CP2c/CP2b complex.
MATERIALS AND METHODS
Cell culture
The murine erythroid leukemia (MEL) cell line DS19 was cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco BRL) supplemented with 10% fetal bovine serum (Hyclone), 50 U/ml penicillin, and 50 µg/ml streptomycin (Gibco BRL). Terminal differentiation of MEL cells was induced by supplementation with hexametylene-bis-acetamide (HMBA, 5 mM). Human myelogenous leukemia K562 cells were maintained in RPMI 1640 (Gibco BRL) supplemented with 10% fetal bovine serum (Hyclone). The human embryonic kidney cell line 293T was maintained in DMEM plus 10% fetal bovine serum (Hyclone).
Plasmid construction
Full-length cDNAs of mouse CP2a and CP2b were cloned into the pEGFPN1 vector (Clontech) yielding the expression plasmids pEGFPN1-CP2a and pEGFPN1-CP2b, respectively. The pEGFPN1-CP2c, pCMV-HA CP2c, pCMV-HA CP2b and pcDNA3-FLAG-PIAS1 plasmids have been described previously (14). Truncation mutants were generated from pGEX2TK-PIAS1 (a gift from Dr. Guntram Suske), pGEX-4T2 CP2b and pGEX-4T2 CP2c (14). Both pcDNA3-PIAS1 (1-480, c-FLAG) and pcDNA3-PIAS1 (400-651, c-FLAG) were kind gifts from Dr. Wilson Xu. A polymerase chain reaction (PCR) product encoding amino acids 1–320 of PIAS1 was inserted into pcDNA3-FLAG (Invitrogen), to which PCR products corresponding to amino acids 394–540 and 394–651 were ligated in frame to make pcDNA3-Flag-PIAS1 1-540 (ΔRFD) and PIAS1 1-651 (ΔRFD). The pcDNA3-FLAG-PIAS1-pseudo vector was generated by substitution of a full-length cDNA of glutathione-S-transferase for amino acids 135–400 of PIAS1. All DNA sequences used for vector construction were confirmed by sequencing. To generate the small hairpin RNA (shRNA) specific for CP2c, an oligonucleotide (5′-GGG CCC GAA TGC TAG ACA ATA GAA TTC AAG AGA TTC TAT TGT CTA GCA TTC GTT TTT-3′, italics indicate sequences specific for CP2c) was inserted downstream of the U6 promoter of a pGEM-T vector. The PIAS1 oligonucleotide sequences used to construct mouse and human PIAS1 shRNA expression vectors were mPIAS1i, 5′-CAT TCC ACA GCT CAC TTA-3′; and hPIAS1i, 5′-CAT TCC ACA ACT CAC TTA-3′. To generate the shRNA specific for human LBP-1a/b, an oligonucleotide (LBP-1a/bi, 5′-GAT CCC CCA GAC TTC TTG ATT TAG ATT TCA AGA GAA TCT AAA TCA AGA AGT CTG TTT TT-3′, italics indicate the sequence specific for LBP-1a/b) was inserted downstream of the H1 promoter of a pLV-TH vector (a gift from Dr. Didier Trono).
siRNA transfection
The synthetic siRNAs targeting mouse CP2b, CP2c and PIAS1 were purchased from Dharmacon. The siRNA sequences are (sense strand indicated): siCP2a/b, 5′-GAU GCU GGA UAA UCG CAA A-3′; siCP2c, 5′-CGG UGA AGC UCC ACG ACG A-3′; and siPIAS1, 5′-GGA AUA AGG AAU CCG GAU C-3′. MEL DS19 cells were transfected with siRNAs (at a final concentration of 100 nM) in Opti-MEM (Gibco) using Lipofectamine 2000 (Invitrogen) repeatedly at intervals of 17 h, according to the manufacturer’s protocol. We used non-targeting siRNA (Dharmacon, D-001810-01) as a negative control. After the second transfection, cells were treated with 5 mM HMBA for 24 h to induce differentiation and then harvested for RNA or protein preparation.
Transfection and luciferase assays
A luciferase reporter construct containing the mouse α-globin promoter was constructed using a pGL3-TATAA vector, as previously described (14). K562 and MEL cells (2 × 104/well) were plated in 24-well dishes. DNA (400–600 ng), including both the luciferase reporter construct and CP2 expression vectors in various combinations, was transiently transfected using Effectene (Qiagen), according to the manufacturer’s recommendations. 293T cells (5 × 104) were plated in 24-well dishes, and DNA (2 µg), including the reporter and CP2 expression vectors, was transiently transfected using the calcium phosphate method. Cells were harvested 48 h after transfection in 100 μl passive lysis buffer (Promega), and 20 μl of lysate was used for luciferase assays on a Lumat LB9501 luminometer (Berthold). A dual-luciferase assay system (Promega) was used according to the manufacturer’s instructions, and firefly luciferase expression was normalized against renilla luciferase. All experiments were repeated at least three times independently.
Expression of GST-fusion proteins and affinity chromatography
GST-fusion proteins for deletion mutants of CP2 isoforms and PIAS1 were expressed in Escherichia coli BL21. Fusion proteins were purified using glutathione-Sepharose (Amersham-Pharmacia), and their integrity was confirmed by Coomassie blue staining and western blotting. For in vitro protein–protein interaction assays, glutathione-Sepharose beads were incubated with GST or GST-fusion proteins at 4°C for 1 h. After washing, the beads were resuspended in 100 µl binding buffer (20 mM Tris–HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 1% NP-40, 0.1% SDS), including protease inhibitor cocktail (Roche), and then incubated at 4°C for 2 h with whole lysate from 293T cells transfected with pCMV-HA-CP2c, pEGFPN1-CP2b or pcDNA3-FLAG-PIAS1. After extensive washing, retained proteins were eluted by boiling in SDS protein-loading buffer and analyzed by western blotting using anti-HA (Santa Cruz Biotechnology), anti-FLAG (Sigma), anti-EGFP (Clontech) or anti-GST antibodies.
Western blotting and immunocytochemistry
Cell extracts were prepared using lysis buffer (20 mM Tris–HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1 mM β-mercaptoethanol, 10% glycerol, 1% NP40, 0.1% SDS, 0.1 mM phenylmethylsulfonyl fluoride, 1 mM NaF, 1 mM Na3VO4). Whole cell extract proteins (10–30 µg) were electrophoresed on 10% SDS-PAGE gels and transferred to a nitrocellulose membrane (Bio-Rad). The membrane was blocked and incubated with appropriate dilutions of the primary antibody at room temperature for 1 h. After washing, a 1:5000 dilution of the appropriate secondary antibody (horseradish peroxidase-conjugated) was added at room temperature for 1 h. Polyclonal anti-β-tubulin antibody (Santa Cruz Biotechnology) was used as a loading control for immunoblotting. Proteins were visualized by chemiluminescence using an ECL system (Amersham-Pharmacia). For immunocytochemisty, 293T cells were transiently transfected with pEGFPN1-CP2c, pEGFPN1-CP2b and pcDNA3-FLAG-PIAS1, singly or in combinations, and fixed at 48 h after transfection. Subcellular localization of each of the CP2 isoforms was visualized using a fluorescent microscope (Fluoview, Olympus). To analyze the distribution patterns of green fluorescent fusion proteins, 293T cells were seeded onto cover glasses and grown to 50–60% confluence. At 48 h after transfection, cells were fixed in 50% methanol/acetone at room temperature for 5 min and then washed with phosphate-buffered saline (PBS). Over 500 cells showing enhanced green fluorescent protein (EGFP) expression were observed and classified into three groups, i.e. with fluorescent proteins in the nucleus, cytosol, or in both nucleus and cytosol. Nuclei were identified by Hoechst 33258 staining. To analyze the effect of PIAS1 on CP2 isoform localization, full-length FLAG-PIAS1 or FLAG-tagged deletion mutants of PIAS1 were transiently transfected into 293T cells with an expression vector containing the EGFP-fused CP2 isoform. Cells were treated with mouse-anti FLAG (Sigma), followed by Cy5-conjugated anti-mouse antibody (Zymed). To analyze the effect of CP2b and PIAS1 on the localization of endogenous CP2c, MEL and K562 cells transfected with siCP2a/b, siPIAS1 or siControl RNA were analyzed by immunostaining of cells with anti-CP2c antibody (Abcam). Cytospin preparations of MEL and K562 cells were made on poly-L-lysine-coated slide glasses by spinning for 3 min at 113 g under medium acceleration in a Cytospin 4 cytocentrifuge (Shandon).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed, as previously described (34), with some modifications. To initiate protein–DNA cross-linking, MEL DS19 cells (0.6 to 2 × 107 cells) were incubated with formaldehyde at a final concentration of 1% for 7 min at room temperature with gentle agitation. Glycine (0.125 M) was added to quench the reaction. Cells were washed and harvested in cold 1X PBS, and the pellet was lysed in 1 ml cold nuclear lysis buffer [50 mM Tris, pH 8.0, 10 mM EDTA, 1% SDS, 1X protease inhibitor cocktail (Roche)], then diluted to 2 ml with IP dilution buffer [20 mM Tris, pH 8.0, 150 mM NaCl, 2 mM EDTA, 0.01% SDS, 1% Triton X-100, 1X protease inhibitor cocktail (Roche)]. The lysate was sonicated with 10 rounds of 20 s each at 70% of maximum output using an ultrasonic processor H200 (Dr Hielscher, Germany). Soluble chromatin was precleared by addition of 200 μl protein A-Sepharose beads (50% slurry in IP dilution buffer; GE Healthcare). An aliquot of precleared chromatin was removed (input control), and the remainder of the chromatin was incubated with anti-CP2b (custom antibody obtained from Peptron), anti-CP2c (13), and anti-PIAS1-specific antibodies (Abcam) overnight at 4°C. Normal rabbit IgG (Santa Cruz) served as a negative control. Immune complexes were collected by incubation with 30 μl protein A-Sepharose for 2 h at 4°C. The precipitates were washed twice with IP buffer (20 mM Tris, pH 6.8, 150 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% TritonX-100, 0.5X protease inhibitor cocktail), once with LiCl wash buffer (50 mM Tris, pH 8.0, 250 mM LiCl, 1 mM EDTA, 1% NP40, 0.5% deoxycholate, 0.5X protease inhibitor cocktail), and twice with Tris-EDTA buffer. Immune complexes were eluted twice with 200 μl elution buffer (0.1 M NaHCO3, 1% SDS). The eluate and input control were reverse cross-linked at 65°C for 5 h and treated with RNase A and proteinase K individually. DNA was purified by phenol/chloroform extraction and the Phase Lock Gel system (5 Prime). Purified DNA was analyzed by real-time quantitative PCR using primers covering the mouse α-globin promoter (forward: 5′-GGT TTG AGG GAC TTG CTT CT-3′ and reverse: 5′-GGT AGA GCA AGC ACA AAC CAG-3′).
Electrophoretic mobility shift assay
Nuclear extracts were prepared from 293T cells transfected with expression vectors for HA-tagged CP2 isoforms, FLAG-tagged PIAS1 truncation/deletion forms, or a control vector, as described previously (14). Each of the nuclear extracts was incubated individually or in various combinations in 25-µl reactions containing 100 mM KCl, 10 mM Tris–HCl, pH 7.9, 1 mM EDTA, 1 mM DTT, 4% glycerol, 0.1% NP-40, 1 μg poly dI·dC (Pharmacia), and a 32P-labeled oligonucleotide probe. The probe DNA corresponded to the CP2c consensus binding sites in the mouse α-globin promoter: 5′-GAT CCC AAG TTT TAC TCG GTA GAG CAA GCA CAA ACC AGG-3′ (−156 to −124 from the start codon). The reaction mixtures were separated on native 5% polyacrylamide gels, and the dried gels were autoradiographed. The same procedure was used for the purified GST-CP2c, GST-CP2b and GST-PIAS1. For supershift analysis, 3 μg anti-CP2c, anti-CP2b, anti-PIAS1 antibody, or IgG was added to the reaction mixture.
Real-time quantitative reverse transcription (RT)-PCR
Total RNA was isolated using Trizol reagent (Invitrogen). RNA (300 ng) was reverse transcribed by incubation with 10 pmol random hexamer and 100 U M-MLV reverse transcriptase (Invitrogen). cDNA was analyzed by real-time PCR, which was performed using SYBR premix ExTaq (Takara) and the Light Cycler 1.5 system (Roche). For RT-PCR, the following primers were used: mCP2b, forward 5′-GCC AGA GAA TCA CCG TAG TC-3′ and reverse 5′-GCT TGT GCA GCT CCA TCT CC-3′; mCP2c, forward 5′-CGT ACC TCA ATC AAG GAC AG-3′ and reverse 5′-CGG AAT ATG CTC TTC ACC AG-3′; mPIAS1, forward 5′-TGC CTT GAC ACC ACA AC-3′ and reverse 5′-GCT TTG GTT CCA CAC CG-3′; mGAPDH, forward 5′-CGT GCC GCC TGG AGA AAC C-3′ and reverse 5′-TGG AAG AGT GGG AGT TGC TGT TG-3′; and mα-globin, forward 5′-GCT GCC TGG GGG AAG ATT GG-3′ and reverse 5′-GCA GGC AGT GGC TCA GGA GC-3′. All amplifications were performed in triplicate. The expression ratio was calculated using the manufacturer’s software (Lightcycler 4.1) with GAPDH as the internal reference gene.
Statistical analysis
Data are expressed using the mean and the standard deviation when at least two independent experiments were performed. Student’s t-test was used for performing analysis of variance in Excel software. A P-value of 0.05 or less was considered statistically significant.
RESULTS
CP2c, CP2b and PIAS1 are indispensable for erythroid-specific α-globin expression
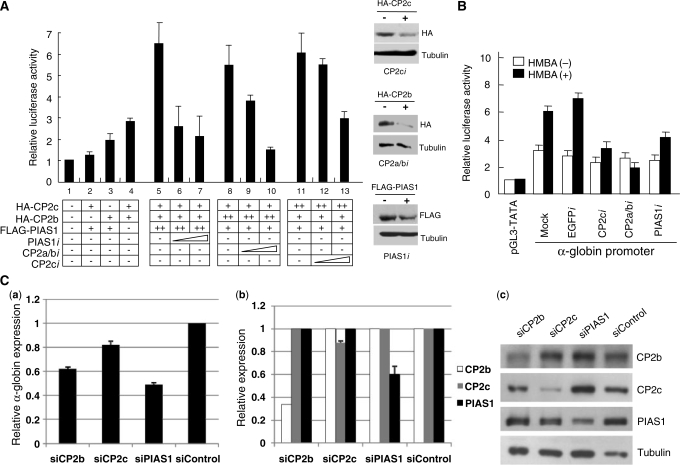
We reported previously that CP2c, CP2b and PIAS1 bind together to the α-globin promoter in vivo and that PIAS1 strongly enhances CP2c/CP2b complex-mediated α-globin gene activation (14). To identify components of the CBP complex that are essential for α-globin gene activation, we tested the effects of knockdown of each protein, using an RNAi-based gene targeting strategy. Expression plasmids for individual CP2 isoforms (HA-CP2c and HA-CP2b) and PIAS1 (FLAG-PIAS1) were transfected into K562 cells, along with a reporter plasmid encoding a luciferase gene controlled by the mouse α-globin promoter (Figure 1A). Coexpression of CP2c, CP2b and PIAS1 in K562 cells induced synergistic increases of α-globin gene expression, as previously described (Figure 1A, lanes 2–5). These transactivation effects were significantly decreased by expression of specific shRNAs for CP2 isoforms (CP2ci or CP2a/bi) or PIAS1 (PIAS1i), in a dose-dependent manner (Figure 1A, lanes 6, 7, 9, 10, 12 and 13). Protein levels of CP2 isoforms and PIAS1 in cells with or without the shRNA constructs were measured by western blotting to ensure that the decreased luciferase activity was actually due to altered CP2 and PIAS1 levels (Figure 1A, right panel).
Figure 1.
CP2c, CP2b and PIAS1 are essential for maximal upregulation of α-globin expression in erythroid cells. (A) K562 cells were transiently transfected with a luciferase reporter vector driven by the mouse α-globin promoter along with expression vectors, as indicated. At 48 h after transfection, whole cell lysates were prepared, and firefly and Renilla luciferase activities were measured simultaneously. Average values and standard deviations were obtained from three independent experiments. One-tenth (20 μg) of each whole cell extract was analyzed on a western blot with anti-FLAG or anti-HA antibody, and with β-tubulin as a control. (B) A reporter vector containing the α-globin promoter was transiently cotransfected with RNAi expression vectors specific for EGFP, CP2c, CP2a/b or PIAS1 into MEL cells in the absence or presence of 5 mM HMBA, as indicated. Each value was measured, as described in (A). (C) Effects of knockdown of endogenous CP2b, CP2c or PIAS1 on α-globin gene induction in differentiating MEL cells. MEL cells were transfected with siRNAs specific for CP2b, CP2c or PIAS1 (siCP2b, siCP2c and siPIAS1) and control siRNA (siControl). After 24 h of HMBA treatment, total RNA was isolated and analyzed by real-time qRT-PCR. (a) Expression of the α-globin gene relative to that of the GAPDH gene is shown. Values were normalized to results obtained from cells transfected with an siControl. Gene-specific decreases in expression corresponding to each siRNA were confirmed at the levels of mRNA and protein, using real-time qRT-PCR (b) and western blotting (c). Expression of CP2b, CP2c and PIAS1 genes relative to that of the GAPDH gene are shown, with values normalized as in (a). (c) Proteins (10 μg) were analyzed on western blots with anti-CP2b, anti-CP2c, anti-PIAS1 and anti-tubulin antibodies (as a control).
To determine which proteins are necessary for α-globin gene expression in erythroid differentiation, the effects of shRNA-induced knockdown of endogenous CP2c, CP2a/b and PIAS1 were examined in differentiating MEL cells by the reporter assays. Knockdown of CP2c, CP2a/b or PIAS1 protein in differentiating MEL cells reduced the α-globin promoter activity to a basal level, whereas transfection of the empty vector (mock) or the EGFP shRNA expression vector did not affect the α-globin promoter activity (Figure 1B).
To confirm the knockdown effect of CP2c, CP2b or PIAS1 on endogenous α-globin expression in erythroid differentiation, we transfected siRNAs specific for each of these genes [CP2c, CP2b, PIAS1 (siCP2c, siCP2a/b, siPIAS1)] or control siRNAs (siControl) into MEL cells. After two rounds of siRNA transfection, erythroid differentiation was induced by incubating the MEL cells in HMBA (5 mM) for 24 h. Total RNA and whole lysates were prepared from the transfected cells for real-time quantitative reverse transcription (qRT-PCR) and western blotting, respectively. We confirmed specific decreases in CP2b, CP2c or PIAS1 expression at the levels of mRNA and protein [Figure 1C, (b) and (c)]. The level of α-globin mRNA in knocked-down MEL cells was compared to that in the control [Figure 1C, (a)]. Knockdown of CP2b, CP2c or PIAS1 decreased α-globin transcription significantly in differentiating MEL cells. Cells transfected with siPIAS1 showed the greatest inhibition of α-globin gene expression (∼50%). These data imply that all three factors are required for α-globin gene expression in erythroid cell differentiation in vivo.
PIAS1 directs CP2c to the nucleus
The subcellular localization of a transcription factor complex responds dynamically to intracellular conditions, with corresponding changes in binding affinity for specific promoters (35–37). We showed previously that nuclear localization of CP2c depends on the relative cellular levels of CP2a and CP2b, with CP2b recruiting CP2c into the nucleus, and CP2a holding CP2c in the cytoplasm by direct protein–protein interaction (15).
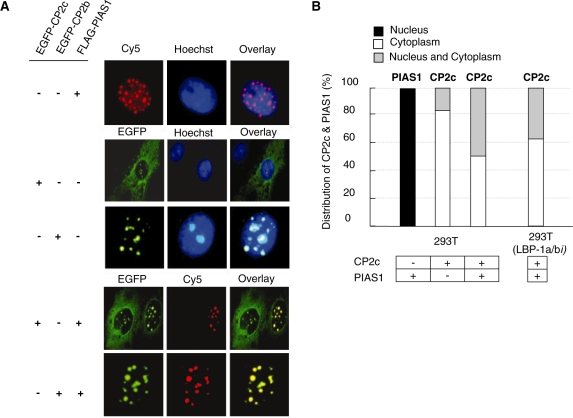
In the presence of CP2c and CP2b, PIAS1 synergistically enhances α-globin expression (see Figure 1) (14), which led us to ask whether PIAS1 also affects the subcellular localization of CP2c. In 293T cells, exogenous PIAS1 displayed a punctuate distribution within the nucleus (Figure 2A, upper panel), as reported for other cell types (29,30), whereas CP2b localized exclusively in nucleus and CP2c in both nucleus and cytoplasm, (Figure 2A, middle panel). Consistent with previous data for the physical interaction between CP2c and PIAS1 (14), immunocytochemical data showed that, when coexpressed with PIAS1, CP2c localized concurrently in both the nucleus and cytoplasm in 50% of cotransfected cells (Figure 2B). We observed that CP2c always colocalized with PIAS1 in the nucleus (Figure 2A, bottom panel). Thus, the nuclear recruitment of CP2c by PIAS1 stands in parallel to the previous finding that CP2b induces the nuclear localization of CP2c (15). In addition, even with depletion of LBP-1a/b (a human homolog of CP2a/b) in 293T cells, PIAS1 still enhanced the movement of CP2c to the nucleus (Figure 2B). These findings indicate that PIAS1 induces the nuclear localization of CP2c in a CP2b-independent way.
Figure 2.
PIAS1 enhances the nuclear translocation of CP2c in a CP2b-independent manner. (A) 293T cells were transfected with pcDNA3-FLAG-PIAS1, pEGFPN1-CP2c or pEGFPN1-CP2b vectors, individually or in combination, as indicated. The FLAG-PIAS1 fusion proteins were detected with FLAG antibody followed by incubation with a Cy5-conjugated secondary antibody (red). (B) 293T or 293T (LBP-1a/bi) cells were transfected with pcDNA3-FLAG-PIAS1, or pEGFPN1-CP2c vectors, individually or in combination, as indicated. The subcellular distribution patterns of PIAS1 or CP2c were analyzed by scoring about 500 cells showing EGFP or FLAG expression and classifying individual cells into one of three groups, depending on its location (nucleus, cytoplasm or nucleus and cytoplasm).
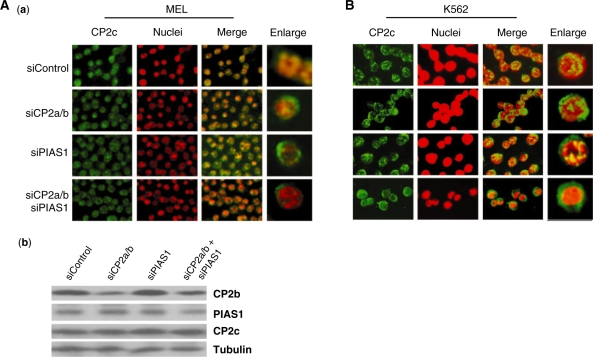
To verify the dependence of CP2c nuclear localization on CP2b and/or PIAS1, we examined the localization of CP2c in MEL cells with endogenous CP2b and/or PIAS1 knocked down using siRNA (Figure 3). MEL cells were transfected with siRNAs specific for CP2a/b, or PIAS1 (siCP2a/b, and siPIAS1) and scrambled siRNA (siControl) and induced to differentiate by treatment with HMBA for 24 h. Western blots showed that endogenous CP2b and PIAS1 proteins were reduced by more than 50% in an siRNA-specific manner, with no effect on the level of CP2c expression [Figure 3A, (b)]. CP2c localized in the nucleus more prominently in siControl-transfected cells, and both in the nucleus and cytoplasm in siCP2a/b- or siPIAS1-transfected cells [Figure 3A, (a)]. CP2c localized more prominently in cytoplasm in the knockdown of both CP2a/b and PIAS1. Similar results were obtained in K562 cells (Figure 3B). These data correlate well with the results in Figure 2 that show that both PIAS1 and CP2b direct CP2c to the nucleus. Furthermore, since the PIAS1 level increased about 4-fold during MEL cell differentiation, as did other CP2 isoforms (Supplementary Figure S1), our data suggest that CP2b and PIAS1 enhance α-globin expression in erythroid differentiation by inducing the movement of CP2c to the nucleus.
Figure 3.
The localization of CP2c in MEL (A) and K562 (B) cells in which the endogenous PIAS1 and/or CP2a/b were knocked down using siRNAs. (A) MEL cells were transfected with siRNAs specific for CP2a/b, or PIAS1 (siCP2a/b, and siPIAS1) and control siRNA (siControl). (a) After treatment with HMBA for 24 h, cells were harvested, attached to glass slides using cytospin and analyzed for CP2c by immunocytochemistry. Nuclei were identified by Hoechst 33258 staining. (b) An aliquot of each cell preparation (a) was analyzed via western blot to confirm protein levels following knockdown. (B) K562 cells were transfected with the same siRNAs shown in (A) and analyzed for CP2c by immunocytochemistry.
PIAS1 forms a complex with CP2c/CP2b and enhances the DNA-binding activity of the CP2c/CP2b complex
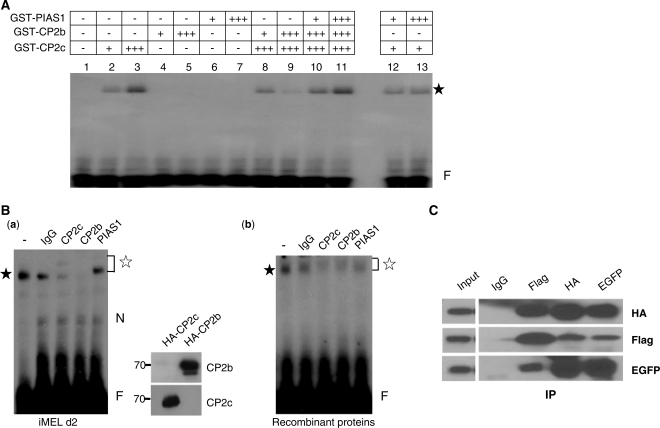
Since PIAS1 acts as an interacting partner with CP2 proteins and as a co-activator for CP2c/CP2b-mediated α-globin expression in erythroid cells (14), we hypothesized that PIAS1 might be required to modulate DNA binding activity of the CP2c/CP2b complex at the α-globin promoter. We tested the effect of PIAS1 on DNA binding of the CP2 complex using recombinant CP2c, CP2b and PIAS1 in electrophoretic mobility shift assay (EMSA) (Figure 4A). PIAS1 strongly enhanced the DNA-binding activity of the CP2 complex, which otherwise would bind negligibly to the α-globin promoter (compare lanes 8 and 9 to lanes 10 and 11). PIAS1 did not affect the DNA-binding activity of CP2c alone (lanes 12 and 13), nor did it bind directly to the α-globin promoter (lanes 6 and 7). However, migration of the protein–DNA complex, including CP2c, CP2b and PIAS1, did not differ from that of CP2c alone (compare lane 3 to lanes 10 and 11). We also observed this phenomenon in the reactions containing nuclear extracts of MEL, K562 or 293T cells (data not shown; see also reference 14). To test whether the protein–DNA complexes in the reaction containing CP2c, CP2b and PIAS1 still retained all three proteins, supershift EMSA analyses were performed using factor-specific antibodies (Figure 4B). The specificities of the anti-CP2b and CP2c antibodies were confirmed by western blotting [Figure 4B, right side of (a)]. In EMSA with nuclear extracts of MEL cells at differentiation Day 2, protein–DNA complexes were shifted upward by the addition of PIAS1 antibody and did not appear with the addition of anti-CP2b or anti-CP2c antibodies, whereas IgG produced no change [Figure 4B, (a)]. These data suggest that the protein–DNA complexes contained at least one each of the CP2c, CP2b and PIAS1 proteins. Similar results were obtained in the reactions using recombinant CP2c, CP2b and PIAS1 proteins [Figure 4B, (b)]. In addition, when nuclear extracts prepared from 293T cells transiently cotransfected with pcDNA3-FLAG-PIAS1, pcDNA3-HA-CP2c and pEGFPN1-CP2b vectors were subjected to Co-IP experiments using specific antibodies, all three proteins co-precipitated (Figure 4C). These data together imply that PIAS1 increases the DNA-binding affinity of the CP2c/CP2b complex for the α-globin promoter through cooperative binding with both CP2c and CP2b proteins. Hereafter, we refer to this protein complex as CBP (CP2c/CP2b/PIAS1).
Figure 4.
PIAS1 enhances the DNA-binding ability of the CP2c/CP2b complex. (A) GST-PIAS1 and GST-CP2 isoforms purified from bacteria were used in EMSA. For EMSA, each protein, individually or in combination, was incubated with the 32P-labeled DNA probe for consensus CP2 binding sites derived from the mouse α-globin promoter, as indicated in ‘Materials and Methods’ section. An asterisk indicates the protein–DNA complex. A plus sign (+) indicates 0.2 µg protein. (B) MEL cell nuclear extracts at differentiation Day 2 (a) or purified bacterial GST-CBP proteins (CP2c/CP2b/PIAS1) (b) were used in supershift EMSA assays. Mock assay (lane 1) and assays in which 3 μg IgG, anti-CP2c, anti-CP2b or anti-PIAS1 antibody was added to the reaction mixtures shown in (A) (lanes 2–5). The closed and open asterisks indicate the specific protein–DNA and supershifted protein–DNA complexes, respectively. N and F represent the nonspecific and free probe bands, respectively. The specificities of anti-CP2b and CP2c antibodies were tested by western blotting [(a), right]. Custom peptide antibody was prepared using CP2b peptide (271-LTEMRLEPIIEDAVEHEQKKSSKRTLPADYGDSLAKRGS-309). The HA-CP2b or HA-CP2c expression vector was transfected into 293T cells, and total lysates were prepared. Proteins (10 μg) were analyzed by western blotting with anti-CP2b or anti-CP2c antibody. (C) 293T cells were transiently cotransfected with pcDNA3-FLAG-PIAS1, pcDNA3-HA-CP2c and pEGFPN1-CP2b vectors. Cell lysates prepared from these cells were immunoprecipitated using IgG, anti-FLAG, anti-HA or anti-EGFP antibody, and precipitates were analyzed in western blot assays using antibodies to EGFP, FLAG or HA.
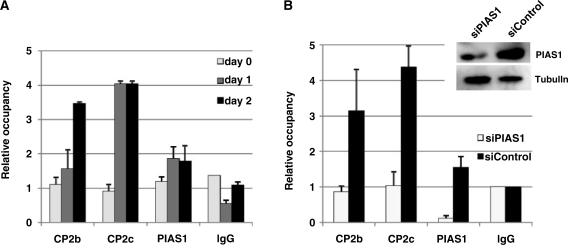
To conclusively demonstrate the effect of PIAS1 on the DNA-binding activity of the CP2c/CP2b complex, we used ChIP to analyze the endogenous target gene promoter in the presence or the absence of PIAS1 during MEL cell differentiation (Figure 5). First, we tested the association of PIAS1, CP2c and CP2b at the endogenous α-globin promoter by ChIP-real time qPCR analysis (Figure 5A). Immunoprecipitation was performed using antibodies against CP2c, CP2b and PIAS1, with normal rabbit IgG as a control. The relative occupancy of CP2c, CP2b and PIAS1 at the α-globin promoter in differentiating MEL cells was determined by comparison to the IgG (control) measurements. Consistent with the previous ChIP observation (14), all three proteins bound to the α-globin promoter, and their associations were enhanced during differentiation of MEL cells. Occupancy of CP2c at the α-globin promoter increased 4-fold from Day 1 of differentiation (Figure 5A). Recruitment of CP2b and PIAS1 to the α-globin promoter was also observed in samples where α-globin expression was active. Notably, the associations of CP2c and PIAS1 with the promoter were saturated at differentiation Day 1, whereas CP2b occupancy continued to increase until Day 2 (Figure 5A), although all of these proteins continued to increase in expression until Day 2 of differentiation [Supplementary Figure S1a]. These results provide strong evidence that all of the CBP complex proteins are associated with the α-globin promoter, and this association correlates with α-globin expression during erythroid differentiation.
Figure 5.
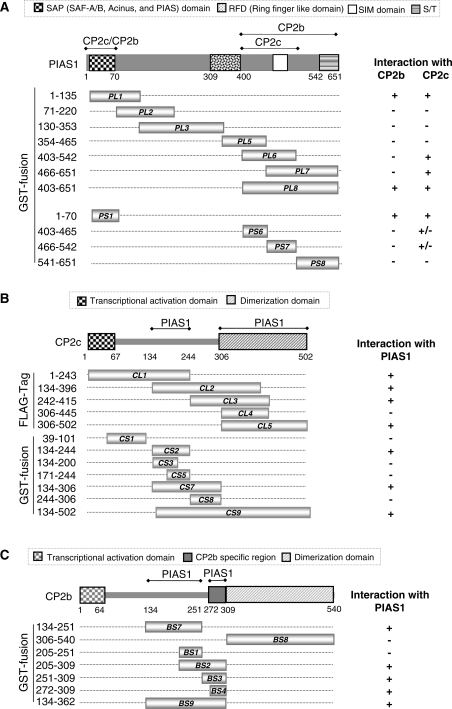
CP2b, CP2c and PIAS1 assemble concurrently at the α-globin promoter, and PIAS1 is essential for CP2b and CP2c to bind to the α-globin promoter. (A) ChIP-real-time qPCR analysis of the endogenous α-globin promoter in differentiating MEL cells. The relative occupancy of CP2b, CP2c and PIAS1 at the α-globin promoter in differentiating MEL cells is shown in comparison to the IgG (control) samples. Values represent the averages of three independent experiments. Mouse Nanog primers were used as a negative control for all real-time qPCR (data not shown). (B) Effects of PIAS1 knockdown on the association of CP2b and CP2c with the α-globin promoter in differentiating MEL cells. PIAS1-specific siRNA (siPIAS1) or scrambled siRNA (siControl) was transfected into MEL cells. The relative occupancy of CP2b, CP2c and PIAS1 at the α-globin promoter was compared between PIAS1-knockdown and control cells. Results represent an average of two independent experiments. The decrease in PIAS1 protein was confirmed in siPIAS1-treated cells using western blotting.
Since PIAS1 co-localized with CP2c/CP2b at the α-globin promoter (Figure 4), and since this association increased during MEL cell differentiation (Figure 5A), we hypothesized that PIAS1 would be involved in forming a stable CBP complex in this setting. Accordingly, we tested whether CP2b or CP2c binds in vivo to the α-globin promoter in MEL cells following knockdown of PIAS1. After transfecting PIAS1-specific siRNA (siPIAS1) or scrambled siRNA (siControl) into MEL cells, we compared the relative occupancy of CP2b, CP2c and PIAS1 at the α-globin promoter between control cells and cells with PIAS1 knocked down. Indeed, knockdown of endogenous PIAS1 prevented recruitment of CP2b and CP2c to the α-globin promoter in differentiating MEL cells (Figure 5B), coincident with suppression of the gene’s induction [Figure 1C, (a)]. This finding suggests that PIAS1 acts as a clamp holding CP2b and CP2c, which stabilizes the CBP complex at the α-globin promoter and activates the transcriptional potential of CP2 proteins.
Two separate regions of each factor in the CBP complex engage in mutual interactions
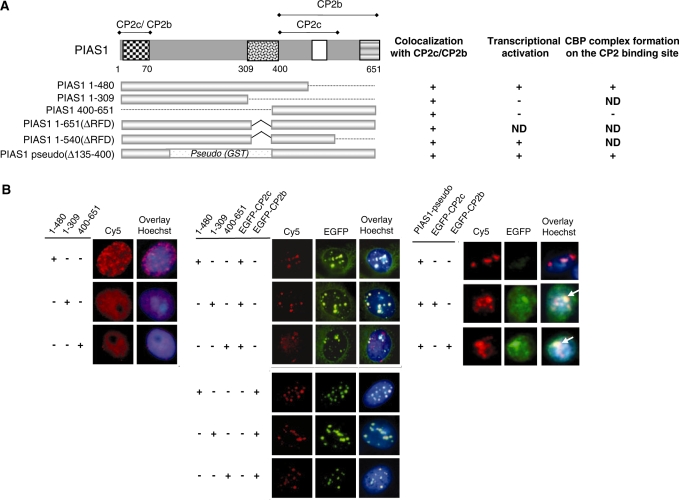
To define the mutual interactions among CP2c, CP2b and PIAS1 in the CBP complex, we mapped the CP2c/CP2b-binding domains on PIAS1, and the PIAS1-binding domains on CP2c and CP2b. We anticipated that there would be two interacting regions in each of the factors if PIAS1 functions as a clamp to hold CP2c and CP2b together. We constructed a series of deletion mutants of PIAS1, CP2b and CP2c and then performed GST pull-down assays (Supplementary Figure S2). Purified recombinant GST-PIAS1 fusion proteins bound to glutathione beads were incubated with cell extracts containing overexpressed CP2c or CP2b proteins. Analyses of truncated PIAS1 proteins showed that either of two domains in the N-terminus and C-terminus was sufficient for the interaction with both CP2c and CP2b (Figure 6A and Supplementary Figure S2A). A small domain in the N-terminus (amino acids 1–70, PS1) is sufficient for binding to both CP2b and CP2c. However, in the C-terminal region, the CP2b-binding domain (PL8) is larger than the CP2c-binding domain (PL6). Similarly, analyses of truncated CP2c proteins revealed that two independent regions of CP2c (CS2, amino acids 134–244; CL5, amino acids 306–502) were required to interact with PIAS1 (Figure 6B and Supplementary Figure S2B). In CP2b, two independent regions (BS7, amino acids 134–251; BS4, amino acids 272–309) were also shown to interact with PIAS1 (Figure 6C and Supplementary Figure S2C). Taken together, these findings suggest that the physical association of factors comprising the CBP complex involve two separate regions of each factor.
Figure 6.
Determination of binding regions joining PIAS1 and CP2c/CP2b. (A) Schematic representation of the PIAS1 deletion mutants and their binding capacities for CP2c and CP2b. The amino acids corresponding to the truncated PIAS1 proteins are indicated. Binding ability of each deletion mutant for CP2b or CP2c is indicated by plus (+) and minus (−) signs. Whole cell extracts from 293T cells expressing the HA-CP2c or EGFP-CP2b protein were mixed with each of the purified GST-tagged PIAS1 deletion mutants or GST and then subjected to pull-down analysis (Supplementary Figure S2A). PL and PS indicate the large and small fragments of PIAS1, respectively. (B) Schematic representation of CP2c deletion mutants and their binding capacities for PIAS1. Total cell extracts from 293T cells expressing the large fragments of FLAG-CP2c (CL1–CL5) or FLAG-PIAS1 were incubated with GST-PIAS1 or GST-CP2c deletion mutants (CS1–CS9), as indicated. GST pull-down proteins were analyzed by immunoblotting against anti-FLAG or GST antibodies (Supplementary Figure S2B). (C) Schematic representation of CP2b deletion mutants and their binding capacities for PIAS1. Whole cell extracts from 293T cells expressing FLAG-PIAS1 were mixed with purified GST-tagged small fragments of CP2b (BS) or GST, as indicated, and binding activities were analyzed in GST pull-down assays, followed by immunoblotting with FLAG or GST antibodies (Supplementary Figure S2C).
PIAS1 functions as a clamp for CP2b and CP2c, to enhance DNA binding and transcriptional activation
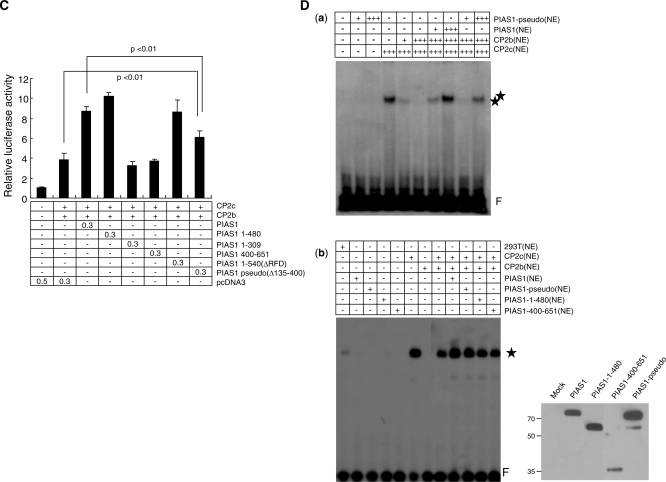
To test whether PIAS1 mutants containing a CP2-binding domain can recruit CP2c into the nucleus, we analyzed the effects of various PIAS1 truncation mutants, including PIAS1 mutants lacking the RFD, on the subcellular localization of CP2 proteins (Figure 7A). Although the RFD of PIAS1 is essential for its sumoylating activity (18,38), it is not involved in the nuclear localization of PIAS1 (32). All of the mutant PIAS1 proteins tested localized in the nucleus in 293T cells (Figure 7B, left panel). When the CP2c or CP2b expression vectors were cotransfected with the PIAS1 mutants, FLAG-PIAS proteins containing the N-terminus (PIAS1 1-480 or PIAS1 1-309) or C-terminus (PIAS1 400-651) induced movement of CP2c into the nucleus, and also co-localized with some of the CP2c and most of the CP2b in the nucleus (Figure 7B, middle panel). FLAG-PIAS1 mutant proteins defective in sumoylation due to the RFD deletions [PIAS1 1-651(ΔRFD) and PIAS1 1-540(ΔRFD)] also co-localized with both CP2c and CP2b in the nucleus (data not shown). In addition, the PIAS1 hybrid [PIAS1 pseudo (Δ135-400)], which contained a GST peptide linker between the two minimal CP2c/CP2b binding regions in both the N-terminus and C-terminus of PIAS1, co-localized with both CP2c and CP2b in the nucleus (Figure 7B, right panel). However, it is noteworthy that the localization phenotype of the full-length protein differs from that of the truncation mutants, which is much more diffuse within the nucleus. The overall data suggest that the PIAS1 mutants containing at least one CP2-binding domain can mobilize CP2c into the nucleus.
Figure 7.
PIAS1 enhances CP2c/CP2b-mediated α-globin gene expression through concerted interaction with both CP2c and CP2b. (A) Schematic representation of PIAS1 deletion mutants and summary for the results of (B), (C) and (D). (B) 293T cells were transfected with FLAG-tagged PIAS1 deletion mutants along with pEGFPN1-CP2c or pEGFPN1-CP2b expression vectors, singly or in combination, as indicated. FLAG-tagged proteins were detected with anti-FLAG antibody followed by incubation with Cy5-conjugated secondary antibody (red). Direct-immunofluorescence analysis was performed using an Olympus confocal laser scanning system. Nuclei were identified by Hoechst 33258 staining. (C) K562 cells were transiently transfected with a reporter vector containing the mouse α-globin promoter along with the indicated expression vectors. Each transfection was performed with 0.5 μg DNA. The three repetitions of each experiment were analyzed using the Student’s t-test. P-values were obtained for the comparison of bar 2 with bar 8 (P < 0.01) and bar 3 with bar 8 (P < 0.01). (D) Nuclear extracts were prepared from 293T cells expressing FLAG-PIAS1, FLAG-PIAS1-pseudo, FLAG-PIAS1 1-480, FLAG-PIAS1 400-651 or HA-CP2 isoforms. EMSA was performed, as described in Figure 4A. In (b), the indicated amounts of nuclear extracts from 293T cells expressing CP2c, CP2b, PIAS1 and/or PIAS1 mutants were added to the reaction. A plus sign (+) indicates 5 µg nuclear extract. Asterisks indicate the specific protein–DNA complex, whereas F indicates the free probe. Expression of wild-type PIAS1 and each of the PIAS1 mutant proteins was confirmed by western blot analysis using the anti-FLAG antibody (right side of (b)).
To test whether these PIAS1 mutant proteins can serve as coactivators of CP2c/CP2b-mediated α-globin gene transcription, we performed reporter assays with CP2b and CP2c on the α-globin promoter using the full-length or deletion mutant PIAS1 (Figure 7C). Interestingly, the PIAS1 mutant containing both the N- and C-terminal CP2-binding regions (PIAS1 1-480) enhanced transcriptional activity of the reporter gene, whereas PIAS1 mutants containing either the N- or C-terminal CP2c/CP2b binding domain (PIAS1 1-309 and PIAS1 400-651) did not. These data suggest that the PIAS1 truncation mutants containing one CP2c/CP2b binding site can direct movement of CP2c into the nucleus or co-localize with CP2b by protein–protein interaction, but cannot join with the CP2 proteins in target gene activation because the mutants cannot perform the clamping function that holds CP2c and CP2b together. In support of this hypothesis, the RFD deletion mutant [PIAS1 1-540(ΔRFD)], containing two CP2c/CP2b binding sites, enhanced reporter gene transcription (Figure 7C). The PIAS1 hybrid [PIAS1 pseudo (Δ135-400)] also significantly enhanced reporter gene transcription, although the level of enhancement did not reach that of wild type PIAS1. To test whether the PIAS1 pseudo (Δ135-400) mutant binds with and activates a CBP complex at the promoter, we performed EMSA using extracts of 293T cells transiently transfected with CP2c, CP2b and PIAS1 pseudo (Δ135-400) expression vectors. As shown in a (a) of Figure 7D, the PIAS1-pseudo (Δ135-400) mutant enhanced DNA-binding activity of the CBP complex, although the level of enhancement was somewhat lower than with the full-length PIAS1. In support of this, the addition of the PIAS1-pseudo (Δ135-400) mutant to the reaction enhanced the protein–DNA complex formation in EMSA [Figure 7D, (b)]. However, other truncation mutants (PIAS1 1-480, 400-651) did not affect the binding activity of CP2c/CP2b complex. This finding is consistent with the requirement for both CP2c/CP2b-interactive regions of PIAS1 in the transactivation function. Thus, the PIAS1 truncation mutants with only one of the CP2c/CP2b binding sites exclude either CP2c or CP2b from the protein–DNA complex formation. Note, on the other hand, that the PIAS1 1-480 mutant shows no clamping function in EMSA, but strong transactivation activity in the reporter assay [compare lane 4 in Figure 7C and lane 11 in (b) of Figure 7D]. This finding may result from a weak clamping ability, sufficient to enhance intracellular transcriptional activity, but not sufficient to hold the protein–DNA complex in EMSA. This mutant had one complete and one incomplete CP2c/CP2b-interacting region (Figure 6A). Thus, our overall findings suggest that PIAS1 acts as a clamp joining CP2c and CP2b to enhance DNA-binding activity and activate transcriptional potential and that this clamping function also requires both of two CP2c/CP2b-interacting regions, located in the N- and C-terminal regions of PIAS1, respectively.
DISCUSSION
Erythroid-specific α-globin expression requires formation of the CBP complex
Previously, we identified CP2c, CP2b and PIAS1 as factors essential for α-globin gene expression in erythroid cells (14). We found that all three proteins bind to the endogenous α-globin promoter and that the extent of binding increased in differentiating MEL cells. Even in non-erythroid 293T cells, co-expression of CP2c, CP2b and PIAS1 activated an exogenous α-globin promoter. In this study, we confirmed that CP2c, CP2b and PIAS1 are indispensable for erythroid-specific α-globin expression (Figure 1). The strong transactivation effects of CP2c, CP2b and PIAS1 decreased in a dose-dependent manner with expression of shRNA specific for CP2 isoforms (CP2ci or CP2a/bi) or PIAS1 (PIAS1i) (Figure 1A). Knockdown of CP2c, CP2b or PIAS1 reduced the transcriptional activity of an exogenous α-globin promoter in differentiating MEL cells to the levels found in undifferentiated cells (Figure 1B), and significantly reduced endogenous α-globin gene expression in differentiating MEL cells (Figure 1C). Thus, CP2c, CP2b and PIAS1 are all required for α-globin gene expression in differentiating MEL cells.
To understand the molecular mechanisms for PIAS1-mediated α-globin gene expression, we first tested the subcellular localization of PIAS1. Exogenous PIAS1 itself localized in the nucleus and could recruit CP2c from the cytosol to the nucleus, as did CP2b (14). In 293T cells, exogenous PIAS1 clearly enhanced nuclear localization of CP2c, whereas exogenous CP2c remained largely in the cytosol (80%) (Figure 2B). In 293T cells with knockdown of the human CP2a/b (LBP-1a/b), exogenous PIAS1 still recruited CP2c to the nucleus. Thus, CP2b and PIAS1 appear to recruit CP2c to the nucleus by separate pathways. Indeed, knockdown of either CP2a/b or PIAS1 with siRNA reduced nuclear localization of CP2c in both MEL and K562 cells, and knockdown of both CP2a/b and PIAS1 reduced nuclear residency of CP2c more severely (Figure 3). These findings indicate that PIAS1 and CP2b recruit CP2c to the nucleus in an additive manner via separate pathways.
PIAS1 revealed the additional function of recruiting the CP2c/CP2b complex to the α-globin promoter. In the EMSA (Figure 4), we found that the mobility shift of the α-globin promoter accounted for the retention of CP2c, CP2b and PIAS1 proteins together. However, the mobility shift with all three recombinant proteins was essentially identical to that seen with CP2c alone (Figure 4A), even though one would expect an additional mobility shift if all three proteins were involved in the protein–DNA complex. One would also argue that PIAS1 is more likely to function by sequestering CP2b away from CP2c so that CP2c itself can restore its DNA-binding activity. However, we confirmed that all three factors coexist in the mobility shift, as demonstrated by supershift assays using factor-specific antibodies (Figure 4B). This result was repeated in the reaction using MEL cell extracts. Furthermore, all factors mutually interact among themselves, as revealed by Co-IP assays (Figure 4C). Thus, we anticipate that the mobility shift with all three proteins represents a stoichiometry different from that with CP2c alone, despite nearly identical gel migration. We found additional evidence that the three proteins associate to form a single complex. First, association of all three factors at the endogenous α-globin promoter increased during MEL cell differentiation (Figure 5A). Second, depletion of endogenous PIAS1 in the differentiating MEL cells by siRNA transfection reduced the association of both CP2c and CP2b with the endogenous α-globin promoter (Figure 5B). These data together suggest that the three factors associate and act together concertedly.
Finally, we found that PIAS1 physically holds CP2c and CP2b together at the promoter. We identified two separable binding regions in each protein component of the CBP complex (Figure 6). Because we just showed pair-wise interactions in this experiment, one may argue that we did not test simultaneous interaction among them. Nevertheless, both of the two CP2-binding regions of PIAS1 were required, though not sufficient, for the clamping function as revealed by EMSA and reporter assays, whereas either CP2 binding region was sufficient to mobilize CP2c to the nucleus and to sequester CP2 proteins from the promoter (Figure 7).
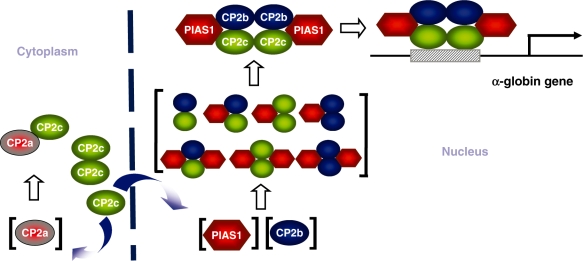
Based on our findings, we propose a model for the role of PIAS1 in transcriptional activation of the α-globin gene in erythroid cells (Figure 8). PIAS1 serves a dual function in erythroid cell-specific globin gene transcription, i.e. nuclear translocation of CP2c and formation of the active CBP complex, by clamping the CBP components together. The action may plausibly proceed, as follows: CP2b and PIAS1, though localized in the nucleus, cannot act by themselves on the α-globin promoter because they cannot bind to the DNA (14). CP2c is recruited into the nucleus by PIAS1 and CP2b in response to a differentiation signal. CP2 isoforms form homo- and/or heterodimers through the dimerization/oligomerization domain in their C-terminal half (16,17). As a dimer or oligomer, CP2c can bind directly to the target DNA using its own DNA-binding domain (14). PIAS1 can interact with both CP2b and CP2c through two regions, located in its N- and C-termini. In the absence of PIAS1, CP2b may interact with CP2c to form an unstable protein–DNA complex (which may explain its ability to exclude CP2c from the mobility shift assay). However, PIAS1 clamps CP2b and CP2c together to form an active CBP complex, thus, stabilizing its binding to the α-globin promoter. Since stable DNA binding requires a CP2c dimer, as well as simultaneous interaction of PIAS1 with two CP2 proteins, the minimal protein complex containing CP2c, CP2b and PIAS1 on the α-globin promoter may be hexametric with two units each of three proteins.
Figure 8.
A model of the CP2c/CP2b/PIAS1 complex activating α-globin gene expression. CP2c and CP2a localize mainly in the cytoplasm, whereas CP2b and PIAS1 co-localize in the nucleus. A specific differentiation cue may increase the amount and/or activity of CP2b and PIAS1, inducing movement of CP2c to the nucleus. Components of the hexametric CP2c/CP2b/PIAS1 complex, through a concerted interaction, strongly enhance binding to the α-globin gene promoter and α-globin transcription in erythroid cells.
Functional significance of the PIAS1-mediated CBP complex formation
We have demonstrated that PIAS1 enhances DNA-binding activity of the CP2c/CP2b complex to the CP2 consensus sequences, in which PIAS1 functions as a clamp to hold together CP2c and CP2b in a sumoylation-independent manner (Figures 4 and 7). Other transcription factors with regulation independent of the sumoylating functions of PIAS include STAT1, STAT3, NF-κB and COUP-TF1 (19,39,40). However, the clamping function of PIAS1 in transcription factor regulation is not yet fully explored. Two separate regions in each factor were involved in the interactions between CP2 isoforms and PIAS1 (Figure 6). The N-terminal CP2 binding region of PIAS1 overlapped with the scaffold attachment factor-A/B/acinus/PIAS (SAP) motif, whereas the C-terminal CP2c binding region includes the SUMO-interacting motif (SIM), and the C-terminal CP2b binding region requires additional Ser/Thr-rich sequences.
We also showed here that PIAS1 mobilizes CP2c from the cytoplasm to the nucleus (Figure 2). Reports indicate that PIAS proteins effect subnuclear localization of targets without sumoylation activity. For instance, PIASy directs the Wnt/β-catenin mediator LEF1 to PML nuclear bodies, which results in repression of LEF1 activity (29). PIASy potently inhibits Oct4-mediated transcriptional activation, sequestering Oct4 protein from the vicinity of Cajal bodies and splicing speckles at the nuclear periphery (41). Other PIAS family members, PIAS1 and PIAS3, also interact with Oct4 in vivo, and target Oct4 to the nuclear periphery in a cell-type specific manner. The homeoprotein Msx1 moves to the nuclear periphery by the action of PIAS1, which allows Msx1 to bind and repress the MyoD and Myf5 gene promoters (42). These examples show that the PIAS proteins, in a manner similar to our findings in this report, can alter the subnuclear localization of target transcription factors in a sumoylation-independent manner through specific protein–protein interactions. However, these examples represent transcriptional repression rather than activation, as observed in this study.
Implications of the CBP protein complex in erythroid cell-specific gene expression
Erythroid differentiation depends vitally on CP2c binding to consensus sequences in both the α- and β-globin gene promoters and consequent activation of the genes (13,43). In addition, CP2c regulation of human globin gene expression correlates with putative CP2-binding sequences identified in the promoters of human α-, ε-, γ- and β-globin genes (43). Mutations in GATA-1 and CP2-binding sites within the promoter of the uroporphyrinogen III synthase gene, the fourth enzyme in the heme biosynthetic pathway, may cause congenital erythropoietic porphyria (44). The CP2c/CP2b complex binds in different patterns depending on the cellular context; for example, CP2b enhances CP2c binding to consensus sites in K562 cells, but inhibits it in 293T cells (14). These findings suggest that nuclear redistribution of CP2c protein by PIAS1 and other CP2 members, and formation of an active CBP complex through cooperative interaction between PIAS1 and CP2 proteins represent key regulatory steps in erythroid cell hemoglobin synthesis. Similar regulatory patterns occur in the expression of marker genes in smooth muscle cell (SMC) differentiation. Serum response factor (SRF) mediates the expression of most SMC differentiation marker genes through cooperative interactions with myocardin, an SMC/cardiomyocyte-specific SRF coactivator and through binding to highly conserved CArG elements in SMC-specific promoters (45–48). Through cooperative interactions with both SRF and class I bHLH proteins, PIAS1 also modulates transcriptional activation of SMC marker genes (49). These findings suggest that PIAS1 and CP2 proteins help to synchronize erythroid marker gene expression, which might be tested by analysis of putative target genes containing CP2 consensus sites.
The regulation of globin gene expression by an active CBP complex presents a novel paradigm for understanding the complex, but highly precise regulation of, tissue-specific mammalian gene expression. Important elements include the diversity of CP2 isoforms involved in transactivation, DNA binding and heteromerization, control of subcellular localization and interaction with specific partner proteins, like PIAS1, in addition to changes in the expression levels (1).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
A Korea Research Foundation Grant funded by the (MOEHRD), Republic of Korea (KRF-2007-313-C00594); the 2002 Research and Developmental Project on Bio-Health of the Ministry of Health and Welfare, Republic of Korea (No. 02-PJ10-PG8-EC01-0012); and the Brain Korea 21 Project (to J.H.C. and J.J.), in part, from the MOEHRD, through the Research Team for Mammalian Development and Differentiation. Funding for open access charge: Hanyang University.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We appreciate the help and advice of our colleagues at the Laboratory of Molecular Genetics, Department of Life Science, Hanyang University. Special thanks go to M.-A. Park and J. J. Lee for their technical support. We also thank Dr Guntram Suske (Philipps-University of Marburg, Germany), Dr Wilson Xu (Memorial Sloan-Kettering Cancer Center and Graduate School of Medical Sciences, Cornell University, USA) and Dr Didier Trono (University of Geneva, Switzerland) for their gifts of plasmids or reagents.
REFERENCES
- 1.Veljkovic J, Hansen U. Lineage-specific and ubiquitous biological roles of the mammalian transcription factor LSF. Gene. 2004;343:23–40. doi: 10.1016/j.gene.2004.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jane SM, Ney PA, Vanin EF, Gumucio DL, Nienhuis AW. Identification of a stage selector element in the human gamma-globin gene promoter that fosters preferential interaction with the 5′ HS2 enhancer when in competition with the beta-promoter. EMBO J. 1992;11:2961–2969. doi: 10.1002/j.1460-2075.1992.tb05366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou W, Clouston DR, Wang X, Cerruti L, Cunningham JM, Jane SM. Induction of human fetal globin gene expression by a novel erythroid factor, NF-E4. Mol. Cell. Biol. 2000;20:7662–7672. doi: 10.1128/mcb.20.20.7662-7672.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou W, Zhao Q, Sutton R, Cumming H, Wang X, Cerruti L, Hall M, Wu R, Cunningham JM, Jane SM. The role of p22 NF-E4 in human globin gene switching. J. Biol. Chem. 2004;279:26227–26232. doi: 10.1074/jbc.M402191200. [DOI] [PubMed] [Google Scholar]
- 5.Bosè F, Fugazza C, Casalgrandi M, Capelli A, Cunningham JM, Zhao Q, Jane SM, Ottolenghi S, Ronchi A. Functional interaction of CP2 with GATA-1 in the regulation of erythroid promoters. Mol. Cell. Biol. 2006;26:3942–3954. doi: 10.1128/MCB.26.10.3942-3954.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruni P, Minopoli G, Brancaccio T, Napolitano M, Faraonio R, Zambrano N, Hansen U, Russo T. Fe65, a ligand of the Alzheimer's beta-amyloid precursor protein, blocks cell cycle progression by down-regulating thymidylate synthase expression. J. Biol. Chem. 2002;277:35481–35488. doi: 10.1074/jbc.M205227200. [DOI] [PubMed] [Google Scholar]
- 7.Zambrano N, Minopoli G, Candia P, Russo T. The Fe65 adaptor protein interacts through its PID1 domain with the transcription factor CP2/LSF/LBP1. J. Biol. Chem. 1998;273:20128–20133. doi: 10.1074/jbc.273.32.20128. [DOI] [PubMed] [Google Scholar]
- 8.Barnhart KM, Kim CG, Banerji SS, Sheffery M. Identification and characterization of multiple erythroid cell proteins that interact with the promoter of the murine alpha-globin gene. Mol. Cell. Biol. 1988;8:3215–3226. doi: 10.1128/mcb.8.8.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim CG, Barnhart KM, Sheffery M. Purification of multiple erythroid cell proteins that bind the promoter of the α-globin gene. Mol. Cell. Biol. 1988;8:4270–4281. doi: 10.1128/mcb.8.10.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim CG, Swendeman SL, Barnhart KM, Sheffery M. Promoter-elements and erythroid cell nuclear factors that regulate α-globin gene transcription in vitro. Mol. Cell. Biol. 1990;10:5958–5966. doi: 10.1128/mcb.10.11.5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim LC, Fang L, Swendeman SL, Sheffery M. Characterization of the molecularly cloned murine alpha-globin transcription factor CP2. J. Biol. Chem. 1993;268:18008–18017. [PubMed] [Google Scholar]
- 12.Lim LC, Swendeman SL, Sheffery M. Molecular cloning of the alpha-globin transcription factor CP2. Mol. Cell. Biol. 1992;12:828–835. doi: 10.1128/mcb.12.2.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chae JH, Lee YH, Kim CG. Transcription factor CP2 is crucial in hemoglobin synthesis during erythroid terminal differentiation in vitro. Biochem. Biophys. Res. Commun. 1999;263:580–583. doi: 10.1006/bbrc.1999.1408. [DOI] [PubMed] [Google Scholar]
- 14.Kang HC, Chae JH, Lee YH, Park MA, Shin JH, Kim SH, Ye SK, Cho YS, Fiering S, Kim CG. Erythroid cell-specific alpha-globin gene regulation by the CP2 transcription factor family. Mol. Cell. Biol. 2005;25:6005–6020. doi: 10.1128/MCB.25.14.6005-6020.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chae JH, Kang HC, Kim CG. The relative cellular levels of CP2a and CP2b potentiates erythroid cell-specific expression of the α-globin gene by regulating the nuclear localization of CP2c. Biochem. Biophys. Res. Commun. 2009;380:813–817. doi: 10.1016/j.bbrc.2009.01.172. [DOI] [PubMed] [Google Scholar]
- 16.Uv AE, Thompson CR, Bray SJ. The Drosophila tissue-specific factor Grainyhead contains novel DNA-binding and dimerization domains which are conserved in the human protein CP2. Mol Cell Biol. 1994;14:4020–4031. doi: 10.1128/mcb.14.6.4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shirra MK, Zhu Q, Huang HC, Pallas D, Hansen U. One exon of the human LSF gene includes conserved regions involved in novel DNA-binding and dimerization motifs. Mol. Cell Biol. 1994;14:5076–5087. doi: 10.1128/mcb.14.8.5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shuai K, Liu B. Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat. Rev. Immunol. 2005;5:593–605. doi: 10.1038/nri1667. [DOI] [PubMed] [Google Scholar]
- 19.Chung CD, Liao J, Liu B, Rao X, Jay P, Berta P, Shuai K. Specific inhibition of Stat3 signal transduction by PIAS3. Science. 1997;278:1803–1805. doi: 10.1126/science.278.5344.1803. [DOI] [PubMed] [Google Scholar]
- 20.Liu B, Liao J, Rao X, Kushner SA, Chung CD, Chang DD, Shuai K. Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl. Acad. Sci. USA. 1998;95:10626–1031. doi: 10.1073/pnas.95.18.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt D, Müller S. PIAS/SUMO: new partners in transcriptional regulation. Cell. Mol. Life Sci. 2003;60:2561–2574. doi: 10.1007/s00018-003-3129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharrocks AD. PIAS proteins and transcriptional regulation–more than just SUMO E3 ligases? Genes Dev. 2006;20:754–758. doi: 10.1101/gad.1421006. [DOI] [PubMed] [Google Scholar]
- 23.O'Shea JJ, Watford W. A peek at PIAS. Nat. Immunol. 2004;5:875–876. doi: 10.1038/ni0904-875. [DOI] [PubMed] [Google Scholar]
- 24.Jackson PK. A new RING for SUMO: wrestling transcriptional responses into nuclear bodies with PIAS family E3 SUMO ligases. Genes Dev. 2001;15:3053–3058. doi: 10.1101/gad.955501. [DOI] [PubMed] [Google Scholar]
- 25.Weger S, Hammer E, Heilbronn R. SUMO-1 modification regulates the protein stability of the large regulatory protein Rep78 of adeno associated virus type 2 (AAV-2) Virology. 2004;330:284–294. doi: 10.1016/j.virol.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 26.Besnault-Mascard L, Leprince C, Auffredou MT, Meunier B, Bourgeade MF, Camonis J, Lorenzo HK, Vazquez A. Caspase-8 sumoylation is associated with nuclear localization. Oncogene. 2005;24:3268–3273. doi: 10.1038/sj.onc.1208448. [DOI] [PubMed] [Google Scholar]
- 27.Pichler A, Melchior F. Ubiquitin-related modifier SUMO1 and nucleocytoplasmic transport. Traffic. 2002;3:381–387. doi: 10.1034/j.1600-0854.2002.30601.x. [DOI] [PubMed] [Google Scholar]
- 28.Kahyo T, Nishida T, Yasuda H. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell. 2001;8:713–718. doi: 10.1016/s1097-2765(01)00349-5. [DOI] [PubMed] [Google Scholar]
- 29.Sachdev S, Bruhn L, Sieber H, Pichler A, Melchior F, Grosschedl R. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 2001;15:3088–3103. doi: 10.1101/gad.944801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kotaja N, Karvonen U, Jänne OA, Palvimo JJ. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol. 2002;22:5222–5234. doi: 10.1128/MCB.22.14.5222-5234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Girdwood DW, Tatham MH, Hay RT. SUMO and transcriptional regulation. Semin. Cell Dev. Biol. 2004;15:201–210. doi: 10.1016/j.semcdb.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 32.Ilmarinen T, Kangas H, Kytömaa T, Eskelin P, Saharinen J, Seeler JS, Tanhuanpää K, Chan FY, Slattery RM, Alakurtti K, et al. Functional interaction of AIRE with PIAS1 in transcriptional regulation. Mol. Immunol. 2008;45:1847–1862. doi: 10.1016/j.molimm.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 33.Diao Y, Wang X, Wu Z. SOCS1, SOCS3, and PIAS1 promote myogenic differentiation by inhibiting the leukemia inhibitory factor-induced JAK1/STAT1/STAT3 pathway. Mol. Cell Biol. 2009;29:5084–5093. doi: 10.1128/MCB.00267-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forsberg EC, Downs KM, Christensen HM, Im H, Nuzzi PA, Bresnick EH. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc. Natl Acad. Sci. USA. 2000;97:14494–14499. doi: 10.1073/pnas.97.26.14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen LF, Greene WC. Regulation of distinct biological activities of the NF-kappaB transcription factor complex by acetylation. J. Mol. Med. 2003;81:549–557. doi: 10.1007/s00109-003-0469-0. [DOI] [PubMed] [Google Scholar]
- 36.Duma D, Jewell CM, Cidlowski JA. Multiple glucocorticoid receptor isoforms and mechanisms of post-translational modification. J. Steroid Biochem. Mol. Biol. 2006;102:11–21. doi: 10.1016/j.jsbmb.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 37.Schwoebel ED, Moore MS. The control of gene expression by regulated nuclear transport. Essays Biochem. 2000;36:105–113. doi: 10.1042/bse0360105. [DOI] [PubMed] [Google Scholar]
- 38.Kotaja N, Karvonen U, Jänne OA, Palvimo JJ. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol Cell Biol. 2002;22:5222–5234. doi: 10.1128/MCB.22.14.5222-5234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu B, Yang R, Wong KA, Getman C, Stein N, Teitell MA, Cheng G, Wu H, Shuai K. Negative regulation of NF-kappaB signaling by PIAS1. Mol. Cell. Biol. 2005;25:1113–1123. doi: 10.1128/MCB.25.3.1113-1123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurihara I, Shibata H, Kobayashi S, Suda N, Ikeda Y, Yokota K, Murai A, Saito I, Rainey WE, Saruta T. Ubc9 and protein inhibitor of activated STAT 1 activate chicken ovalbumin upstream promoter-transcription factor I-mediated human CYP11B2 gene transcription. J. Biol. Chem. 2005;280:6721–6730. doi: 10.1074/jbc.M411820200. [DOI] [PubMed] [Google Scholar]
- 41.Tolkunova E, Malashicheva A, Parfenov VN, Sustmann C, Grosschedl R, Tomilin A. PIAS proteins as repressors of Oct4 function. J. Mol. Biol. 2007;374:1200–1212. doi: 10.1016/j.jmb.2007.09.081. [DOI] [PubMed] [Google Scholar]
- 42.Lee H, Quinn JC, Prasanth KV, Swiss VA, Economides KD, Camacho MM, Spector DL, Abate-Shen C. PIAS1 confers DNA-binding specificity on the Msx1 homeoprotein. Genes Dev. 2006;20:784–794. doi: 10.1101/gad.1392006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chae JH, Kim CG. CP2 binding to the promoter is essential for the enhanced transcription of globin genes in erythroid cells. Mol. Cells. 2003;15:40–47. [PubMed] [Google Scholar]
- 44.Solis C, Aizencang GI, Astrin KH, Bishop DE, Desnick R. Uroporphyrinogen III synthase erythroid promoter mutations in adjacent GATA1 and CP2 elements cause congenital erythropoietic porphyria. J. Clin. Invest. 2001;107:753–762. doi: 10.1172/JCI10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim S, Ip HS, Lu MM, Clendenin C, Parmacek MS. A serum response factor-dependent transcriptional regulatory program identifies distinct smooth muscle cell sublineages. Mol. Cell. Biol. 1997;17:2266–2278. doi: 10.1128/mcb.17.4.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li L, Miano JM, Mercer B, Olson EN. Expression of the SM22 promoter in transgenic mice provides evidence for distinct transcriptional regulatory programs in vascular and visceral smooth muscle cells. J. Cell Biol. 1996;132:849–859. doi: 10.1083/jcb.132.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mack CP, Owens GK. Regulation of smooth muscle α-actin expression in vivo is dependent on CArG elements within the 5′ and first intron promoter regions. Circ. Res. 1999;84:852–861. doi: 10.1161/01.res.84.7.852. [DOI] [PubMed] [Google Scholar]
- 48.Yoshida T, Kawai-Kowase K, Owens GK. Forced expression of myocardin is not sufficient for induction of smooth muscle differentiation in multipotential embryonic cells. Arterioscler. Thromb. Vasc. Biol. 2004;24:1596–1601. doi: 10.1161/01.ATV.0000137190.63214.c5. [DOI] [PubMed] [Google Scholar]
- 49.Kawai-Kowase K, Kumar MS, Hoofnagle MH, Yoshida T, Owens GK. PIAS1 activates the expression of smooth muscle cell differentiation marker genes by interacting with serum response factor and class I basic helix-loop-helix proteins. Mol. Cell. Biol. 2005;25:8009–8023. doi: 10.1128/MCB.25.18.8009-8023.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.