Abstract
Glyoxal and methylglyoxal are reactive dicarbonyl metabolites formed and metabolized in physiological systems. Increased exposure to these dicarbonyls is linked to mutagenesis and cytotoxicity and enhanced dicarbonyl metabolism by overexpression of glyoxalase 1 is linked to tumour multidrug resistance in cancer chemotherapy. We report herein that glycation of DNA by glyoxal and methylglyoxal produces a quantitatively important class of nucleotide adduct in physiological systems—imidazopurinones. The adduct derived from methylglyoxal-3-(2′-deoxyribosyl)-6,7-dihydro-6,7-dihydroxy-6/7-methylimidazo-[2,3-b]purine-9(8)one isomers—was the major quantitative adduct detected in mononuclear leukocytes in vivo and tumour cell lines in vitro. It was linked to frequency of DNA strand breaks and increased markedly during apoptosis induced by a cell permeable glyoxalase 1 inhibitor. Unexpectedly, the DNA content of methylglyoxal-derived imidazopurinone and oxidative marker 7,8-dihydro-8-oxo-2′-deoxyguanosine were increased moderately in glyoxalase 1-linked multidrug resistant tumour cell lines. Together these findings suggest that imidazopurinones are a major type of endogenous DNA damage and glyoxalase 1 overexpression in tumour cells strives to counter increased imidazopurinone formation in tumour cells likely linked to their high glycolytic activity.
INTRODUCTION
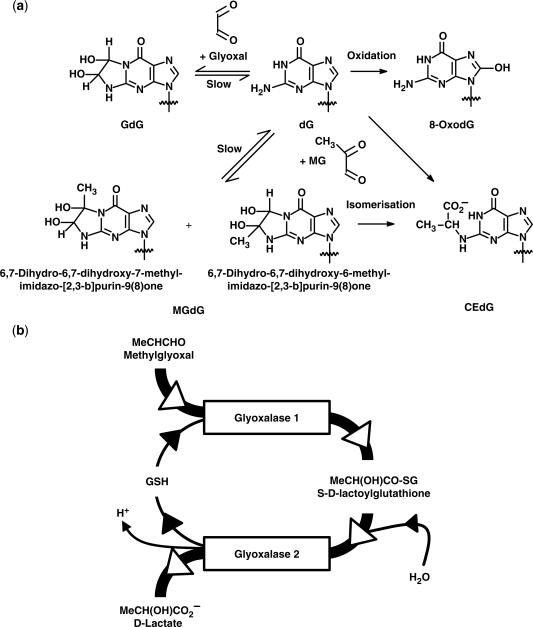
Guanyl bases of nucleotides and nucleosides are susceptible to modification by glyoxal and methylglyoxal (1). Glyoxal and methylglyoxal react with deoxyguanosine under physiological conditions to form mainly imidazopurinone derivatives, 3-(2′-deoxyribosyl)-6,7-dihydro-6,7-dihydroxyimidazo[2,3-b]purin-9(8)one (GdG) and 3-(2′-deoxyribosyl)-6,7-dihydro-6,7-dihydroxy-6/7-methylimidazo-[2,3-b]purine-9(8)one (MGdG)—a 6- and 7-methyl structural isomeric mixture, respectively (2) (Figure 1a). Glyoxal and methylglyoxal also form N2-carboxymethyl-deoxyguanosine (CMdG) (3) and N2-(1,R/S-carboxyethyl)-deoxyguanosine (CEdG)—the latter a stereoisomeric mixture of R/S-epimers at the N2-1-carboxyethyl chiral centre (4). Glyoxal and methylglyoxal are formed in physiological systems: glyoxal is formed by lipid peroxidation and also by degradation of glycated proteins and monosaccharides; methylglyoxal is formed mainly by non-enzymatic degradation of triosephosphates and is also formed by ketone body metabolism and threonine catabolism. Increased methylglyoxal formation occurs in cells with high glycolytic activity. Many tumours have high glycolytic activity which is thought to be a survival adaptation to growth under hypoxic conditions (5,6).
Figure 1.
(a) Formation of glycation and oxidation adducts of deoxyguanosine. The common 2′-deoxyribosyl group has been omitted for clarity. (b) The glyoxalase pathway.
Detection of imidazopurinones derived from glyoxal and methylglyoxal in cellular DNA in vitro and in vivo has proven generally elusive to date. There has been extensive research quantifying the level of the dG-derived oxidative marker 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-OxodG) nucleotides in DNA and related nucleosides released into plasma and excreted in urine (7), as well as other trace endogenous dG adducts (8). There have also been some reports on the minor methylglyoxal-derived nucleotide adduct CEdG (9) and a recent report on the minor glyoxal-derived adduct CMdG (10). Failure to detect imidazopurinones in cell systems may have been due to poor adduct stability and recovery in pre-analytic processing of analytical protocols.
Dicarbonyl adducts of DNA are of likely functional importance because glyoxal and methylglyoxal are both weak mutagens. Diseases associated with high plasma levels of dicarbonyls—diabetes and renal failure—are also associated with increased mutagenicity, cancer risk and vascular cell apoptosis (11). Protection against dicarbonyl mutagenicity and cytotoxicity is provided mainly by the glutathione-dependent cytosolic glyoxalase system. The glyoxalase system is comprised of glyoxalase 1 (Glo1), glyoxalase 2 (Glo2) and a catalytic amount of glutathione. Glo1 catalyses the detoxification of glyoxal and methylglyoxal to S-glycolylglutathione and S-d-lactoylglutathione, respectively, and Glo2 catalyses the hydrolysis of these glutathione thioesters to glycolate and d-lactate, reforming glutathione consumed by Glo1 (Figure 1b) (12). In 2000, Tsuruo and co-workers (13) discovered overexpression of Glo1 as a novel factor producing multidrug resistance (MDR) in tumours. Glo1-linked MDR was found in tumour cells of lung, colorectal, breast and prostate origin, was acquired by experimental Glo1 overexpression and could be countered by the cell permeable Glo1 inhibitor S-p-bromobenzylglutathione cyclopentyl diester (BBGD) (14)—an experimental cancer chemotherapeutic agent (15).
In this report, we describe the concurrent quantitation of imidazopurinones, GdG and MGdG, and CEdG and 8-OxodG by stable isotopic dilution analysis liquid chromatography with tandem mass spectrometric detection (LC-MS/MS) in physiological samples and explore the link of levels of imidazopurinone adducts to DNA strand breaks and Glo1-associated MDR in human tumour cell lines.
MATERIALS AND METHODS
Materials
2′-Deoxyguanosine monohydrate and glyoxal and methylglyoxal solutions (40%), ribonuclease (RNase) A from bovine pancreas, RNase T1 from Aspergillus oryzae, deoxyribonuclease (DNase) II from porcine spleen, phosphodiesterase (PDE) II from bovine spleen, acid phosphatase from potato were purchased from Sigma (Poole, Dorset, UK). Protease was from Qiagen. [13C10,15N5]-2′-deoxyguanosine (all >98% isotopic purity) was purchased from Cambridge Isotope Laboratories (Andover, MA, USA). GdG, MGdG and related stable isotopic standards were prepared as described earlier (16). CEdG was conveniently prepared from the crude product mixture of MGdG by addition of 10 mM phosphate buffer, adjustment of the pH to7.4 and continued incubation at 37°C for 6 days—the MGdG therein degrading in part to CEdG. CEdG was purified by preparative anion exchange HPLC on DEAE Protein-Pak, formate form (2 × 10 cm column; Waters); sample loading 20 mg. The column was equilibrated and eluted isocratically for 10 min with 5 mM ammonium formate buffer, pH 4.0, and then with a linear gradient of 5–100 mM of the same buffer from 10–45 min; the flow rate was 4 ml/min. Fractions were analysed by LC-MS/MS (see below) and those containing CEdG were lyophilized to dryness and analysed by 1H and 13C NMR and mass spectrometry. Analytical characteristics were as reported previously (9,17). The preparation contained an equal mixture of S- and R-epimers.
Isolation of DNA from cell samples
Human mononuclear leukocytes, isolated from venous blood by density gradient centrifugation of the buffy coat (18), or human tumour cells grown in culture (5–30 × 106 cells), were washed three times in phosphate buffered saline and re-suspended in 2 ml lysis buffer (320 mM sucrose, 10 mM Tris/HCl, 5 mM MgCl2, 100 μM desferroxamine [DFOM], 1% Triton X-100, pH 7.5) and kept on ice for 20 min to swell. Cells were then lysed by vigorous agitation (three cycles of 5 min vortexing followed by 5 min on ice). Nuclei were collected by centrifugation (1500g, 10 min, 4°C), washed with 2 ml lysis buffer (1500g, 10 min, 4°C) and stored at −80°C at this step until further processing. The sedimented nuclei were re-suspended in 1.2 ml nuclear lysis buffer (10 mM Tris/HCl, 5 mM EDTA, 150 μM DFOM and 50 µl 10% SDS, pH 8.0) and left on ice for 20 min. The nuclei were lysed by three cycles of vortexing (as described earlier), 40 µl RNase A (1 mg/ml) and 15 µl RNase T1 (1 kU/ml) added and incubated for 15 min at 50°C. Protease (20 mg/ml; 40 µl) was added and samples were incubated for 2 h at 37°C. DNA was precipitated by addition of 3 ml NaI solution (7.6 M NaI, 40 mM Tris/HCl, 20 mM EDTA and 300 μM DFOM, pH 8.0) and 4 ml 2-propanol at 4°C. Precipitation was promoted by inversion of the sample tube 5–10 times. The emergent DNA strand was collected from solution on a spooler, transferred to a microcentrifuge tube and washed twice with 2 ml of 40% 2-propanol and then twice with 2 ml 70% ethanol at 4°C. The resulting DNA pellet was dried under argon to remove the remaining ethanol and re-dissolved in argon-saturated water containing 200 μM DFOM, assisted by sample rotary inversion for 24 h at 4°C in the dark. The concentration of the DNA in the sample was determined by UV nanodrop spectrophotometry at 260 nm by diluting an aliquot (2 μl) 50-fold in buffer (10 mM Tris/HCl, 1 mM EDTA and 200 μM DFOM, pH 7.4). Genomic DNA was stored at 4°C overnight or −80°C for the longer periods.
Digestion of DNA
Genomic DNA was washed into 83 mM ammonium acetate buffer, containing 8.3 mM MgSO4, 20 mM NH4Cl, 500 μM aminoguanidine hydrochloride and 100 μM DFOM, pH 5.0, by three cycles of concentration to 100 μl and dilution to 500 μl with buffer in microspin ultrafilters (10 kDa cut-off, Nanosep, low binding polypropylene) at 4°C. DNA (25 μg in 30 μl buffer) was digested by addition of 2 μl of DNase II (2 U/μg DNA) and 2 μl PDE II (1.5 U/mg DNA). Samples were flushed with argon and incubated at 37°C for a further 30 min. Acid phosphatase (type VII, 10 μl; 35 U/mg DNA) was then added, samples again flushed with argon and incubated at 37°C for a further 4 h. Enzymes were then removed from the sample by ultrafiltration (10 kDa cut-off microspin filter, 4°C). Analytical recoveries were performed by splitting extracted DNA into multiple aliquots and spiking half of the samples with standard analyte prior to digestion and comparing this amount with the analyte increment detected in LC-MS/MS analysis of samples post-DNA digestion. Addition of aminoguanidine (500 μM) during digestion was found to be essential to avoid overestimation of glycation adduct analytes.
Incubation of calf thymus DNA
Calf thymus DNA (30 mg) was suspended in 10 mM Tris/HCl, pH 7.4, containing 1 mM EDTA and 0.2 mM DFOM (10 ml, TED buffer) and dissolved by gentle agitation over 24 h at 4°C. The concentration of DNA was checked by UV absorbance at 260 nm. Aliquots of DNA (final concentration 1.00 mg/ml) were incubated in 50 mM potassium phosphate buffer, pH 7.4 and 37°C, with glyoxal or methylglyoxal (0.01–100 mM), total volume 1.00 ml, for 15 h with gentle shaking. DNA was then precipitated with three volumes of 100% ethanol (1 ml; 4°C), the precipitate sedimented by centrifugation (2000g, 2 min) and washed three times with 80% ethanol (1 ml; cold). Resultant pellets were dissolved in 1 ml of TED buffer and the concentration of DNA determined. Control and modified DNA were digested as described earlier and samples stored at −80°C until LC-MS/MS analysis.
Tumour cell lines with low and high expression of glyoxalase 1
Two human tumour cell lines of relatively low Glo1 expression—NCI-H460 large cell lung carcinoma and A549 alveolar basal epithelial carcinoma, and two human tumour cell lines of relatively high Glo1 expression—MG63 osteosarcoma and NCI-H522 non-small cell lung adenocarcinoma, as defined by Tsuruo and co-workers (14), were cultured as described earlier (14,19,20). The level of Glo1 expression was confirmed by qPCR. Cells were grown to confluence in 150 cm2 flasks, trypsinized, re-suspended in media and centrifuged at 400g for 5 min. The medium was removed and the cells were lysed in 10 ml of Trizol (Invitrogen, Paisley, UK). The RNA was then extracted using the chloroform (0.2 ml/1 ml Trizol), 2-propanol (0.5 ml/1 ml of Trizol) and 75% ethanol purification. The extracted RNA was dissolved in diethyl pyrocarbonate-treated water (Ambion, Huntingdon, UK). cDNA was obtained by two-step reverse transcription using SuperScript III reverse transcriptase kit (Invitrogen) with amplification grade DNAse I (Sigma-Aldrich, Poole, UK) and random nonamer primers (Sigma-Aldrich) as per manufacturer’s instructions. Real time PCR was performed by the comparative CT method (21). The β-actin gene served as an endogenous control. Sequences of the primers used were: β-actin—forward AGAGCTACGAGCTGCCTGAC, reverse TGAAGGTAGTTTCGTGGATGC; Glo1—forward GCGCTCTCCAGAAAAGCTAC, reverse TGCCATTGTGGTAACTCTGG.
Plasma and urine ultrafiltrate
Healthy human subjects (recruited for the VITAGE project) (22) were recruited at the Human Nutrition and Metabolism Research and Training Center, Karl Franzens University of Graz, Austria. The study protocols have been approved by the local Ethics Committee and written informed consent was obtained from all study subjects. Venous blood samples were collected from all study subjects in the fasting state and plasma prepared; 24 h urine collections were made for the VITAGE study. Ultrafiltrates were prepared by microspin ultrafiltration of plasma (10 kDa cut-off, 100 μl) and urine (3 kDa cut-off, 100 μl), collecting ca. 50 μl ultrafiltrate. Assessment of analytical recoveries was made by spiking plasma and urine with 50 and 500 fmol adducts, respectively, prior to ultrafiltration and analysis of spiked and unspiked sample ultrafiltrates. Stable isotopic standards were added to the ultrafiltrates prior to analysis.
LC-MS/MS of nucleotide damage markers
For LC-MS/MS, the DNA digest and ultrafiltrates (40 μl) were spiked with 10 μl isotopic standard mixture containing 0.1 nmol [13C10,15N5]dG, 1 pmol [13C10,15N5]8-OxodG, 0.73 pmol [13C10,15N5]MGdG, 0.09 pmol [13C10,15N5]CEdG and 1.4 pmol [13C10,15N5]GdG. LC-MS/MS was performed using an AcquityTM UPLC-Quattro Premier tandem mass spectrometer with a BEH C18 1.7 µm particle size, 2.1 × 100 mm column. The mobile phase was 0.1% formic acid with a linear gradient of 0–10% acetonitrile from 2 to 10 min and isocratic 10% acetonitrile from 10 to15 min; the flow rate was 0.25 ml/min. After analysis, the column was washed with 50% acetonitrile containing 0.1% formic acid for 10 min and thereafter re-equilibrated with initial mobile phase for 10 min. The column temperature was set to temperatures in the range 4–30°C as required.
Nucleoside adduct stability studies
The stability of GdG, MGdG and CEdG were investigated in buffers used in DNA isolation and digestion, autosampler storage and in urine for a 24 h collection at ambient temperature. For stability in buffers, nucleoside adducts (100 pmol/ml) were incubated at 37°C for 0–36 h in buffers: (i) 10 mM ammonium acetate, pH 5.0, with 100 µM DFOM; (ii) 10 mM Tris/HCl, pH 7.4 with 1 mM EDTA and 100 µM DFOM; and (iii) 10 mM ammonium bicarbonate, pH 9.0. After incubation, 20 µl was mixed with isotopic standards (1 pmol [13C10,15N5]GdG, pmol [13C10,15N5]MGdG and 0.1 pmol CEdG) and analysed by LC-MS/MS. For autosampler stability, 10 pmol GdG, MGdG and CEdG in DNA digestion buffer was analysed with and without storage at 4°C in the Acquity autosampler for 24 h, with addition of isotopic standards immediately prior to analysis. For analyte stability in urine, aliquots of human urine were analysed with and without storage at ambient temperature (18°C) for 24 h, with addition of isotopic standards immediately prior to analysis.
DNA strand break assay
HL60 cells (1 × 105 cells/ml; 20 ml) were incubated with each of the treatments described. The cells were washed twice with cold phosphate buffered saline, pH 7.4, and fixed in 1% (w/v) paraformaldehyde. The fixed cells were then stained using the terminal deoxynucleotidyl transferase-mediated dUTP-FITC nick-end labeling (TUNEL) method (Apo-Direct™ Kit; Calibiochem®) for labeling DNA strand breaks. Cells were also stained with propidium iodide for cell cycle analysis (23). Cells within the normal cell cycle distribution pattern were analysed for DNA strand breaks. Samples were analysed using Becton-Dickinson FACSCalibur twin laser, 4-channel cytometry/cell sorter and the data processed by FlowJo software v7.1.4 (Tree Star Inc.).
Statistical analysis
Limit of detection is analyte amount producing a signal/noise ratio = 3. Data are mean ± SD for parametric data and median (lower–upper quartile) for non-parametric data. Significance of differences of means or medians of two independent groups of parametric or non-parametric data was assessed by Student’s t-test and Mann–Whitney U test, respectively. Significance of difference in median of two related groups was assessed by Wilcoxon signed rank test. Correlation analysis was by the non-parametric Spearmen method. Data in the study of degradation of nucleotide adducts was fitted to a single exponential by least squares non-linear regression and half-life deduced. Statistical analysis was performed by the SPSS software, v15.
RESULTS
Assay of imidazopurinones by stable isotopic dilution analysis LC-MS/MS
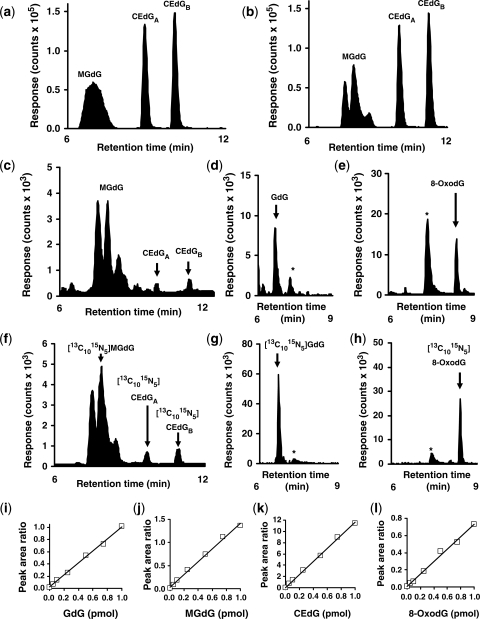
We prepared analytical standards and isotopomer internal standards for GdG, MGdG, CEdG and 8-OxodG. LC-MS/MS reversed phase chromatography with multiple reaction monitoring of the analytes resolved the analyte peaks from dG (Table 1). The chromatographic peak for MGdG was unexpectedly broad with column temperature at 30°C (Figure 2a). On cooling the column to 10°C, the MGdG peak was partially resolved into a three-component peak; no further improvement in peak resolution was achieved at lower column temperatures. This complex peak structure of MGdG is attributed to resolution of structural and stereoisomers; the R/S-epimers of CEdG—CEdGA and CEdGB, were also resolved (Figure 2b). This characteristic chromatographic profile was used in subsequent analysis (Figure 2c–h). Fragment ion scan analysis (collision energy ramp 12–30 eV) showed common fragments of the imidazopurinone base for all three peak components: m/z (neutral fragment loss): 206 (-H2O), 178 (-H2CO2) and 152 (-methylglyoxal); the 178 fragment was also a dominant in the fragment ion scan of CEdGA and CEdGB, as expected for decarboxylation of the N2-(1-carboxyethyl) moiety. The two MGdG component peaks eluting first had similar 178/224 fragment ion intensity ratios that were higher than for final peak component (0.82 and 0.86 versus 0.50). In the 1H NMR spectrum of MGdG in d6-DMSO there were minor peaks duplicating the resonances for 5-NH, 6/7-methyl and 7/6-H protons in integral ratio 86: 14—Supplementary Data, Figure 1. There was no coupling resolved for 5-NH and 6-H protons in GdG and hence resolution of coupling of 5-NH and 6-H protons in the 7-methyl isomer of MGdG was not expected. Shapiro et al. (2) found the 6-methyl isomer is the major product; the major resonances (86%) are therefore assigned to this 6-methyl isomer. The three-component peak of the MGdG chromatogram had two major peaks representing ca. 86% of the peak area—attributed to 6-methyl stereoisomers, and the minor component of longest retention time representing ca. 14% of the peak area—attributed to 7-methyl stereoisomers. Application of LC-MS/MS for quantification of these adducts produced an analytical method of linear response to analyte, high sensitivity and recovery and low variability (Figure 2i–l). For GdG, MGdG, CEdG and 8-OxodG, limits of detection were 0.8, 2.5, 2.2 and 0.7 fmol; analytical recoveries were 104, 97, 98 and 99%, respectively, and coefficients of variation 2–7% in the working analyte range for DNA digests. The high analytical recoveries indicate all four analytes were stable during the DNA hydrolysis procedure. There was no significant loss of analyte during working storage (autosampler at 4°C for 24 h) and over 24 h urine collection at room temperature.
Table 1.
Detection of imidazopurinones and related deoxyguanosine-derived adducts by liquid chromatography-tandem mass spectrometry multiple reaction monitoring
| Analyte | Rt (min) | Molecular > fragment ion transition (Da) | Collision energy (eV) | Cone voltage (V) |
|---|---|---|---|---|
| dG | 7.3 | 268.1 > 152.0 | 37 | 12 |
| GdG | 6.7 | 325.7 > 209.9 | 12 | 12 |
| MGdGa | 7.4, 7.8 and 8.4 | 340.0 > 224.0 | 12 | 11 |
| CEdGa | 9.8 and 11.1 | 340.0 > 224.0 | 12 | 11 |
| 8-OxodG | 8.4 | 283.8 > 168.0 | 12 | 12 |
The neutral fragment loss was 2-dehydro-2-deoxyribose.
aMultiple retention times are shown for adduct structural and stereoisomers isomers.
Figure 2.
Detection of imidazopurinones and related deoxyguanosine-derived adducts by liquid chromatography-tandem mass spectrometry multiple reaction monitoring. (a and b) LC-MS/MS analysis of isobaric MGdG and CEdG adducts (10 pmol) at column temperature 30 and 10°C, respectively. (c–h) Detection of deoxyguanosine-derived adducts in nuclease digests of human mononuclear leukocyte DNA. The asterisk in (d, e, g and h) indicates formation of GdG and 8-OxodG from dG in the sample in the electrospray ionization source of the mass spectrometer. Calibration curves of peak area ratio versus analyte (pmol). (i) GdG: regression equation peak area ratio = (0.98 ± 0.02) × GdG + (0.02 ± 0.01); R2 = 0.996. (j) MGdG: regression equation peak area ratio = (1.38 ± 0.04) × MGdG + (0.04 ± 0.02); R2 = 0.995. (k) CEdG: regression equation peak area ratio = (11.4 ± 0.2) × CEdG + (0.14 ± 0.09); R2 = 0.999. (l) 8-OxodG: regression equation peak area ratio = (0.72 ± 0.03) × 8-OxodG + (0.01 ± 0.01); R2 = 0.994. Amount of stable isotopic standard added was: 1.0 pmol [13C10,15N5]GdG, 0.73 pmol [13C10,15N5]MGdG, 0.09 pmol [13C10,15N5]CEdG and 1.4 pmol [13C10,15N5]8-OxodG. Experimental details are given in the ‘Materials and Methods’ section.
Stability of imidazopurinones
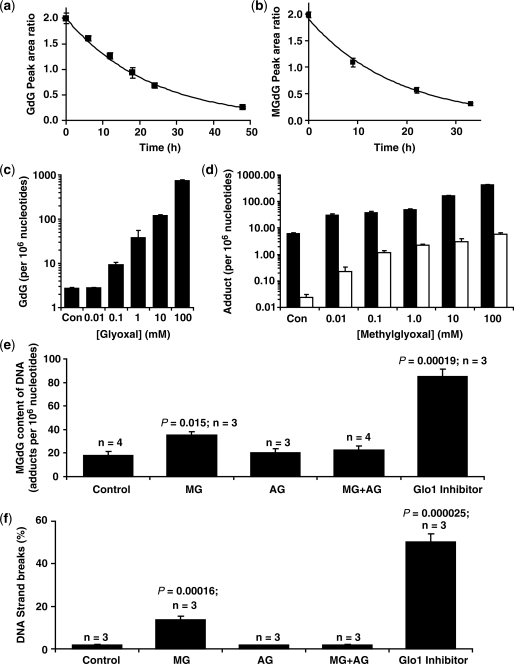
The stability of imidazopurinones at 37°C was investigated in ammonium acetate (pH 5.0) and Tris/HCl (pH 7.4) buffers used in DNA digestion and in ammonium bicarbonate buffer (pH 9.0) used by others for pre-analytic processing. GdG had half-lives at 37°C of 230 ± 6 h at pH 5.0, 16.1 ± 0.4 h at pH 7.4 and <1 h at pH 9. MGdG was less stable than GdG with half-lives at 37°C of 38.1 ± 3.2 h at pH 5.0, 12.0 ± 0.3 h at pH 7.4 and <0.2 h at pH 9 (Figure 3a and b). Addition of the methylglyoxal scavenger aminoguanidine (500 µM) (24) to incubations for the study of MGdG degradation did not prevent the formation of CEdG, suggesting that the MGdG interconversion occurs mainly by intramolecular rearrangement from the 6-methylimidazopurinone isomer. The high instabilities of GdG and MGdG in ammonium bicarbonate buffer at pH 9 explains why GdG and MGdG have been overlooked and markedly underestimated in previous studies employing high pH processing in DNA digests (25,26).
Figure 3.
Imidazopurinone stabilities and adducts of DNA in vitro and in vivo and link to DNA strand breaks. (a and b) Stability of GdG and MGdG in DNA isolation buffer (10 mM Tris/HCl, pH 7.4 with 1 mM EDTA and 100 µM DFOM), pH 7.4 and 37°C. Data are mean ± SD of four determinations with the exponential decay curve of best fit shown. (c and d) Formation of nucleotide AGEs by incubation of calf thymus DNA with glyoxal and methylglyoxal, respectively. Data are mean ± SD (n = 3).Solid bars, imidazopurinone adduct; hollow bars, CEdG. (e and f) Increased MGdG adduct and DNA strand breaks in HL60 cells exposed to increased exogenous and endogenous methylglyoxal. Key: HL60 cells (1 × 105/ml; 3 × 107) incubated for 2 h with no further addition (control), 524 µM methylglyoxal (MG), 500 µM aminoguanidine (AG) and 524 µM methylglyoxal with 500 µM aminoguanidine (MG + AG). A similar incubation for 24 h was with addition of 10 µM BBGD (Glo1 inhibitor). Data are mean ± SD with n and P-values (Student’s t-test) given.
Imidazopurinone adducts formed during the reaction of DNA with glyoxal and methylglyoxal
We studied the formation of imidazopurinone and CEdG adducts in the reaction of calf thymus DNA with glyoxal and methylglyoxal. Calf thymus DNA had ca. 3, 6 and 0.02 adducts of GdG, MGdG and CEdG per 106 nucleotides as supplied. Incubation with 0.01–100 mM glyoxal increased the content of GdG from 3 to 757 adducts per 106 nt. Similar incubation with methylglyoxal increased the MGdG content from 6 to 433 adducts per 106 nt and the CEdG content from 0.02 to 5 adducts per 106 nt. The MGdG/CEdG adduct ratio was 30–300 over the concentration range, indicating that imidazopurinone MGdG was the major adduct formed by modification of DNA with methylglyoxal (Figure 3c and d).
Imidazopurinone adducts in cellular DNA and link to DNA strand breaks
To assess if imidazopurinones were the dominant adduct of physiological DNA damage and had functional links, we determined levels of imidazopurinones, CEdG and 8-OxodG in cellular DNA in vitro and in vivo, and assessed related frequency of DNA strand breaks. For human leukaemia 60 (HL60) cells cultured in vitro, DNA adducts per 106 nt were: GdG 2.67 ± 0.63, MGdG 18.2 ± 6.5, CEdG 1.33 ± 1.12 and 8-OxodG 1.48 ± 0.39. Incubation of HL60 cells with 524 µM methylglyoxal (2 × median growth inhibitory GC50 concentration) (27) for 2 h increased MGdG content of DNA 2-fold without significant increase in other adducts and increased DNA strand breaks by 7-fold. The increase in MGdG and DNA strand breaks was prevented by co-incubation with the dicarbonyl scavenger aminoguanidine. Incubation of HL60 cells with the cell permeable Glo1 inhibitor, BBGD (10 µM—which is twice the median growth inhibitor concentration GC50) (15) increased the DNA content of MGdG by 5-fold and DNA strand breaks by 27-fold. The DNA strand breaks content of HL60 cells correlated with MGdG content of DNA; r = 0.76, P = 0.0015, Spearman (Figure 3e and f).
Imidazopurinone adducts of cellular DNA and related nucleosides in plasma and urine in vivo
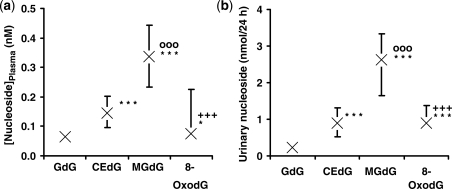
For estimation of imidazopurinone adducts in cellular DNA in vivo, we analysed adducts of DNA extracted from peripheral human mononuclear leukocytes from healthy human subjects. DNA adduct contents were (adducts per 106 nt; n = 3): GdG 2.11 ± 0.74, MGdG 8.73 ± 2.41, CEdG 1.03 ± 0.38 and 8-OxodG 2.79 ± 0.64. To assess the release from DNA and excretion of imidazopurinones and 8-OxodG in human subjects in vivo, we determined the concentrations of related nucleoside adducts in plasma and urine of healthy human subjects. Analytical recoveries for estimation of GdG, CEdG MGdG and 8-OxodG were: in plasma—93 ± 5%, 96 ± 8%, 99 ± 4% and 82 ± 14%, respectively; and in urine were 85 ± 7, 96 ± 5, 98 ± 6 and 87 ± 12 (n = 4). Median plasma concentrations of GdG, CEdG, MGdG and 8-OxodG in healthy human subjects were 0.066, 0.144, 0.338 and 0.076 nM, respectively. Median urinary excretion rates of GdG, CEdG MGdG and 8-OxodG and in healthy human subjects were 0.23, 0.90, 2.63 and 0.90 nmol/24 h, respectively. Significance testing indicated the plasma concentration and urinary excretion of dG adducts was in the order: MGdG> 8-oxodG ≈ CEdG> GdG (Figure 4a and b). Plasma and urinary levels of MGdG were, therefore, 4- and 3-fold higher than 8-OxodG in plasma and urine, respectively. For urinary excretions, GdG correlated positively with urinary MGdG (r = 0.58, P < 0.01) and CEdG correlated positively with GdG (r = 0.64, P < 0.001) and MGdG (r = 0.56, P < 0.01).
Figure 4.
Nucleoside glycation and oxidation adducts in plasma (a) and urine (b) of healthy human subjects. Data are median (upper–lower quartile), n = 28. Characteristics of human subjects: age 61 ± 8 years, gender—male, body mass index 26.1 ± 1.8 kg/m2, fasting plasma glucose 5.6 ± 0.5 mM and glomerular filtrate rate 94 ± 33 ml/min/1.73 m2. Nucleoside adducts were determined as described in the ‘Materials and Methods’ section. *P < 0.05 and ***P < 0.001 with respect to GdG; oooP < 0.001 with respect to CEdG; and +++P < 0.001 with respect to MGdG (Wilcoxon signed rank test).
Imidazopurinone adducts in cellular DNA of tumour cell lines with low and high expression of glyoxalase-1
Imidazopurinone, CEdG and 8-OxodG adduct contents were determined in DNA of two human cell lines of relatively low Glo1 expression—NCI-H460 large cell lung carcinoma and A549 alveolar basal epithelial carcinoma and two human cell lines of relatively high Glo1 expression—MG63 osteosarcoma and NCI-H522 non-small cell lung adenocarcinoma, as defined by Tsuruo and co-workers (14) and confirmed by qPCR. Relative Glo1 expression relative to β-actin and normalized to the expression in NCI-H460 cells was: NCI-H460 cells, 1.0; A549 cells, 6.6; MG63, 10.6 and NCI-H522 cells, 33.2. The median contents of MGdG and 8-oxodG in DNA were increased significantly in cell lines with relatively high Glo1 expression. For all cell lines, MGdG content of DNA correlated positively with GdG adduct content (r = 0.82, P < 0.01) and 8-OxodG content (r = 0.87, P < 0.001) but not with CEdG adduct content of DNA (n = 12) (Table 2).
Table 2.
Imidazopurinone and related DNA adducts of human tumour cells lines with low and high expression of glyoxalase 1
| Glyoxalase 1 expression |
||
|---|---|---|
| Analyte | Low | High |
| GdG | 1.01 (0.23–1.65) | 1.69 (1.56–2.71) |
| MGdG | 1.10 (0.69–1.83) | 10.43 (4.32–13.47)** |
| CEdG | 0.49 (0.43–0.69) | 0.77 (0.19–1.77) |
| 8-OxodG | 1.08 (0.77–1.58) | 8.10 (6.80–15.50)** |
Low Glo1 expression cell lines were NCI-H460 and A549, and high glyoxalase 1 cell lines were MG63 and NCI-H522.
Data are median (upper–lower quartile); adducts per 106 nt.
**P < 0.01 (Mann–Whitney U-test).
DISCUSSION
In this study we found that modification of DNA by physiological dicarbonyls gives rise to quantitatively important steady-state levels of dG-derived adducts in cellular DNA in vitro and in vivo—imidazopurinone derivatives. Imidazopurinone contents of DNA were markedly higher than that of other physiological aldehydes—4HNE, malondialdehyde and others (8). Hence, measurement of imidazopurinone adducts and nucleosides in body fluids may be valuable biomarkers of quantitative and functionally important DNA damage in vivo.
Initial approaches to develop methods to quantify nucleotide adducts formed by glyoxal and methylglyoxal in DNA involved 32P-labelling studies of DNA digests (28) and immunoassay (4). Stable isotopic dilution analysis LC-MS/MS was applied successfully herein for assay of imidazopurinones, related CEdG and 8-OxodG concurrently. The high specificity and sensitivity of LC-MS/MS makes this the preferred method for DNA damage marker quantitative analysis. For relatively unstable nucleotide adducts such as the imidazopurinones, it was necessary to devise a pre-analytic processing protocol to minimize formation, interconversion and degradation of adducts. Our method involves use of acid nuclease and phosphatase, metal ion chelator throughout and the dicarbonyl scavenger aminoguanidine and thereby avoids artifactual formation and minimizes loss of adducts in pre-analytic processing. Investigation of the stabilities of imidazopurinones indicated previous use of high pH and prolonged elevated temperatures in DNA digestion protocols likely led to the degradation of labile GdG and MGdG. Hence, the quantitative importance of imidazopurinone adducts has been overlooked. The stability of GdG in phosphate-buffered saline, pH 7.4 and 37°C, has been reported previously and was ca. 12 h (10). This was repeated herein and the half-life was 11.4 ± 0.4 h—lower than for the determination in Tris/HCl–EDTA–DFOM buffer (16.1 ± 0.4 h). Phosphate buffer—particularly the conjugate base HPO42−—likely de-stabilizes imidazopurinones by deprotonation. The LC-MS/MS method also resolved the adducts from dG chromatographically to avoid interference from formation of adducts by partial degradation of dG during electrospray ionization.
CEdG was determined by immunoassay previously in human urine of normal healthy human subjects with estimates in the range 3.4–344 pmol/mg creatinine and median of ca. 30 pmol/mg creatinine (29). Estimation herein by LC-MS/MS gave median value equivalent to 0.55 pmol/mg creatinine suggesting that immunoassay overestimated urinary CEdG by ca. 50-fold. Similar overestimation with immunoassay of 8-OxodG was reported (30). This is likely caused by interference due to imperfect epitope specificity of the monoclonal antibody used and analyte formation from other sample components during pre-analytic sample processing. A recent independent estimate by Synold et al. (9) of CEdG content of tumours using stable isotopic dilution analysis LC-MS/MS found ca. 1.5 adducts per 106 dG which is similar to the estimate in tumour cell lines with high Glo1 expression herein. The sample processing procedures used in pre-analytic processing, however—such as DNA heated to 95°C, samples incubated at pH 8 and 50°C for 12–18 h and release of adducts from sample clean-up columns with 3% ammonium hydroxide—can be judged as inappropriate for imidazopurinone detection from adduct stability studies herein. Moreover, they risk compromising even CEdG detection by artifactual formation of analyte by sample component degradation to analyte under these conditions of high temperature and pH. Similar compromized analysis of CEdG applies to the study of Yuan et al. (3) where DNA hydrolysates were incubated for 4 h with addition Tris–HCl buffer, pH 8.9 and 37°C to the hydrolysate (final pH ca. 8.4) which is expected to lead to loss of MGdG with formation of CEdG—see above. Incubation of MGdG under these conditions produced complete loss of analyte (data not shown).
Imidazopurinone adducts of dG are formed by reactivity of glyoxal derivatives with the N2-amino group of dG with cyclic attachment to ring N1. Similar reactivity of the exocyclic N2-amino group of dG with aldehydes such as 4-hydroxynonenal (4HNE) has been reported (31). For methylglyoxal, this forms a mixture of 6- and 7-methyl isomers. The major isomer was the 6-methyl stereoisomer. Stereoisomerism of imidazopurinones provided a characteristic chromatographic profile of MGdG and CEdG. Formation of imidazopurinones produces two chiral centres at positions 6 and 7, giving rise to four stereoisomers of GdG (not resolved chromatographically) and eight structural and stereoisomers of MGdG (partially resolved). Chromatographic resolution of isomers of MGdG and CEdG has been reported previously (28,32). Relative detection response peak areas for component peaks for MGdG and CEdG were not significantly different for all test and standards samples, suggesting isomer distribution is constant in standard and test samples and hence quantitation was not compromized. Mechanistically, CEdG is also likely formed by the reaction encounter of methylglyoxal monohydrate with dG. CEdG may be formed by glycating agents other than methylglyoxal—ascorbic acid and glucose, for example, involving complex degradation and fragmentation reactions (33,34). Alternative sources of CEdG as well as different rates of repair of CEdG and MGdG lesions may also contribute to the ratio of CEdG/MGdG being higher in cellular DNA than in DNA modified by methylglyoxal in cell-free system.
Imidazopurinone adducts of DNA were associated with DNA breaks herein. Methylglyoxal modification of DNA is also linked to increased mutation frequencies (35). It may be linked to mutagenesis in ageing, diabetes, renal failure and other disorders associated with increased levels of dicarbonyl metabolites (4,11,36). Increased formation of CEdG was detected in human melanoma WM-266-4 cells incubated with exogenous methylglyoxal and with high glucose concentration—although part may have been formed by degradation of MGdG in pre-analytic processing (3). CEdG was also found to be mutagenic and induced DNA strand breaks (37). This occurred, however, at engineered CEdG contents of DNA over 1000-fold higher than found in cellular DNA physiologically. The effect of CEdG on DNA integrity and mutagenic activity at physiological adduct levels is unknown. MGdG content of DNA was increased by exposure of cells to exogenous methylglyoxal and by increasing endogenous methylglyoxal by inhibition of Glo1 with S-p-bromobenzylglutathione delivered intracellularly by BBGD. The latter was more effective than the former probably because an active Glo1 in the cytosol intercepts effectively methylglyoxal diffusing from the culture medium to the nucleus. BBGD produced a profound increase of MGdG adducts in cellular DNA; it also overcomes MDR in human tumours associated with overexpression of Glo1 (14).
The finding of increased MGdG content of DNA in human tumour cell lines with high relatively expression of Glo1 linked to MDR was counterintuitive. Increased expression and activity of Glo1, given a similar flux of methylglyoxal formation in cells with similar glycolytic activities, is expected to lead to decreased levels of MGdG. The tumour cell lines studied with high Glo1 expression [for example MG63 cells (19)] are known, however, to have high glycolytic activities relative to cell lines with low Glo1 expression and activities [for example A549 cells (20)]. We propose, therefore, that increased Glo1 expression is an imperfect adaptation of tumour cells with relatively high glycolytic activity to suppress increased level of methylglyoxal formed and prevent potential cytotoxicity. The concentration of methylglyoxal in cells is in the range 0.5–2 µM increasing 2–3-fold further in some cells in hyperglycaemia and does not induce acute cytotoxicity (38,39). High concentrations of exogenous methylglyoxal (200–500 µM) induced growth arrest and toxicity in tumour cells (27) but lower concentrations (10–20 µM) induced apoptosis in some cell types (40). Increased flux of formation of methylglyoxal in tumour cells without increased activity of Glo1 or other detoxification activity may induce cytotoxicity. High expression and activity of Glo1 may be thereby permissive of growth for tumours with high glycolytic activity and unavoidable concomitant increased flux of methylglyoxal formation. The correlation of MGdG and 8-OxodG content of DNA in tumour cell lines might be expected since cells exposed to increased endogenous levels of methylglyoxal—as in the nematode Caenorhabditis elegans in old age—show increased modification of mitochondrial proteins by methylglyoxal with related increased formation of reactive oxygen species and oxidative damage (41). This is reported in the tumour cells herein by increased DNA content of 8-OxodG. The lack of correlation of MGdG and CEdG contents of DNA may indicate significant contribution of glycating agents other than methylglyoxal to formation of CEdG—see above.
In conclusion, from their high relative content in DNA and the link to functional effects, imidazopurinones derived from glyoxal and methylglyoxal are of likely pathogenic and diagnostic as well as, potentially, therapeutic significance—particularly in Glo1-linked MDR tumours.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Cancer Research UK (C790/A5147 to P.J.T.); Commonwealth Scholarship Commission—split-site PhD studentship (INCN-2007-34 to S.W./P.J.T.); Fifth Framework Programme, Key Action 1, Food, Nutrition and Health, acronym VITAGE (VITamin A, Vitamin E and Carotenoid Status and Metabolism during AGEing: Functional and Nutritional Consequences) (QLK1-CT-1999-00830 to B.M.W.R.); Michael and Betty Kadoorie Cancer Genetics Research Programme (to T.S.); Wellcome Trust (077012/Z/05/Z to M.R.S.). Funding for open access charge: University of Warwick.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Sandra Wuga, Human Nutrition and Metabolism Research and Training Center, Institute of Molecular Biosciences, Karl Franzens University of Graz, Austria, for clinical technical assistance.
REFERENCES
- 1.Krymkiewicz N. Reactions of methylglyoxal with nucleic acids. FEBS Letts. 1973;29:51–54. doi: 10.1016/0014-5793(73)80013-4. [DOI] [PubMed] [Google Scholar]
- 2.Shapiro R, Cohen BI, Shiuey S-J, Maurer H. On the reaction of guanine with glyoxal, pyruvaldehyde and ketoxal and the structure of the acylguanines. A new synthesis of N2-alkylguanines. Biochemistry. 1969;8:238–245. doi: 10.1021/bi00829a034. [DOI] [PubMed] [Google Scholar]
- 3.Yuan B, Cao H, Jiang Y, Hong H, Wang Y. Efficient and accurate bypass of N2-(1-carboxyethyl)-2′-deoxyguanosine by DinB DNA polymerase in vitro and in vivo. Proc. Natl Acad. Sci. USA. 2008;105:8679–8684. doi: 10.1073/pnas.0711546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, Nakamura S, Miyazaki S, Morita T, Suzuki M, Pischetsrieder M, Niwa T. N2-carboxyethyl-2'-deoxyguanosine, a DNA glycation marker, in kidneys and aortas of diabetic and uremic patients. Kidney Internat. 2006;69:388–392. doi: 10.1038/sj.ki.5000064. [DOI] [PubMed] [Google Scholar]
- 5.Yeluri S, Madhok B, Prasad K, Quirke P, Jayne D. Cancer craving for sugar: an opportunity for clinical exploitation. J. Cancer Res. Clin. Oncol. 2009;135:867–877. doi: 10.1007/s00432-009-0590-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nature Revs. Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 7.Cooke MS, Olinski R, Loft S. and members of the European Standards Committee on Urinary (DNA) Lesion Analysis (ESCULA) (2008) Measurement and meaning of oxidatively modified DNA lesions in urine. Cancer Epidemiol. Biomarkers Prev., 17, 3-14. [DOI] [PubMed] [Google Scholar]
- 8.De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 9.Synold T, Xi B, Wuenschell GE, Tamae D, Figarola JL, Rahbar S, Termini J. Advanced glycation end products of DNA: quantification of N2-(1-Carboxyethyl)-2′-deoxyguanosine in biological samples by liquid chromatography electrospray ionization tandem mass spectrometry. Chem. Res. Toxicol. 2008;21:2148–2155. doi: 10.1021/tx800224y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, Cao H, Wang Y. Quantification of N2-carboxymethyl-2'-deoxyguanosine in calf thymus DNA and cultured human kidney epithelial cells by capillary high-performance liquid chromatography-tandem mass spectrometry coupled with stable isotope dilution method. Chem. Res. Toxicol. 2010;23:74–81. doi: 10.1021/tx900286c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thornalley PJ. Protecting the genome: defence against nucleotide glycation and emerging role of glyoxalase I over expression in multidrug resistance in cancer chemotherapy. Biochem. Soc. Trans. 2003;31:1372–1377. doi: 10.1042/bst0311372. [DOI] [PubMed] [Google Scholar]
- 12.Thornalley PJ. The glyoxalase system in health and disease. Mol. Aspects Med. 1993;14:287–371. doi: 10.1016/0098-2997(93)90002-u. [DOI] [PubMed] [Google Scholar]
- 13.Sakamoto H, Mashima T, Kazaki A, Dan S, Hashimoto Y, Naito M, Tsuruo T. Glyoxalase I is involved in resistance of human leukemia cells to antitumour agent-induced apoptosis. Blood. 2000;95:3214–3218. [PubMed] [Google Scholar]
- 14.Sakamoto H, Mashima T, Sato S, Hashimoto Y, Yamori T, Tsuruo T. Selective activation of apoptosis program by S-p-bromobenzylglutathione cyclopentyl diester in glyoxalase I-overexpressing human lung cancer cells. Clin. Cancer Res. 2001;7:2513–2518. [PubMed] [Google Scholar]
- 15.Thornalley PJ, Edwards LG, Kang Y, Wyatt C, Davies N, Ladan MJ, Double J. Antitumour activity of S-p-bromobenzylglutathione cyclopentyl diester in vitro and in vivo. Inhibition of glyoxalase I and induction of apoptosis. Biochem. Pharmacol. 1996;51:1365–1372. doi: 10.1016/0006-2952(96)00059-7. [DOI] [PubMed] [Google Scholar]
- 16.Fleming T, Rabbani N, Thornalley PJ. Preparation of nucleotide advanced glycation endproducts – imidazopurinone adducts formed by glycation of deoxyguanosine with glyoxal and methylglyoxal. Ann. NY Acad. Sci. 2008;1126:280–282. doi: 10.1196/annals.1433.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao H, Jiang Y, Wang Y. Stereospecific synthesis and characterization of oligodeoxyribonucleotides containing an N2-(1-Carboxyethyl)-2′-deoxyguanosine. J. Am. Chem. Soc. 2007;129:12123–12130. doi: 10.1021/ja072130e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyum A. Separation of lymphocytes, granulocytes, and monocytes from human blood using iodinated density gradient media. Meth. Enzymol. 1984;108:88–109. doi: 10.1016/s0076-6879(84)08076-9. [DOI] [PubMed] [Google Scholar]
- 19.Duewelhenke N, Krut O, Eysel P. Influence on mitochondria and cytotoxicity of different antibiotics administered in high concentrations on primary human osteoblasts and cell lines. Antimicrob. Agents Chemother. 2007;51:54–63. doi: 10.1128/AAC.00729-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo FM, Liu XJ, Yan NH, Li SQ, Cao GQ, Cheng QY, Xia QJ, Wang HJ. Hypoxia-inducible transcription factor-1alpha promotes hypoxia-induced A549 apoptosis via a mechanism that involves the glycolysis pathway. BMC Cancer. 2006;6:26. doi: 10.1186/1471-2407-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST(C)) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rock E, Winklhofer-Roob BM, Ribalta J, Scotter M, Vasson MP, Brtko J, Brigelius-Flohe R, Bronner A, Azais-Braesco V. Vitamin A, vitamin E and carotenoid status and metabolism during ageing: functional and nutritional consequences (VITAGE PROJECT) Nutr. Metab. Cardiovasc. Dis. 2001;11:70–73. [PubMed] [Google Scholar]
- 23.Thornalley PJ, Tisdale MJ. Inhibition of proliferation of human promyelocytic leukaemia HL60 cells by S-D-lactoylglutathione in vitro. Leuk. Res. 1988;12:897–904. doi: 10.1016/0145-2126(88)90016-1. [DOI] [PubMed] [Google Scholar]
- 24.Thornalley PJ. Use of aminoguanidine (Pimagedine) to prevent the formation of advanced glycation endproducts. Arch. Biochem. Biophys. 2003;419:31–40. doi: 10.1016/j.abb.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 25.Frischmann M, Bidmon C, Angerer J, Pischetsrieder M. Identification of DNA adducts of methylglyoxal. Chem. Res. Toxicol. 2005;18:1586–1592. doi: 10.1021/tx0501278. [DOI] [PubMed] [Google Scholar]
- 26.Hou SM, Nori P, Fang JL, Vaca CE. Methylglyoxal induces HPRT mutation and DNA-adducts in human T-lymphocytes in vitro. Environm. Mol. Mutagen. 1995;26:286–291. doi: 10.1002/em.2850260404. [DOI] [PubMed] [Google Scholar]
- 27.Kang Y, Edwards LG, Thornalley PJ. Effect of methylglyoxal on human leukaemia 60 cell growth: modification of DNA, G1 growth arrest and induction of apoptosis. Leuk. Res. 1996;20:397–405. doi: 10.1016/0145-2126(95)00162-x. [DOI] [PubMed] [Google Scholar]
- 28.Vaca CE, Fang J-L, Conradi M, Hou S-M. Development of a 32P-postlabelling technique for the analysis of 2′-deoxyguanosine-3′-monophosphate and DNA of methylglyoxal. Carcinogenesis. 1994;15:1887–1894. doi: 10.1093/carcin/15.9.1887. [DOI] [PubMed] [Google Scholar]
- 29.Schneider M, Thoss G, Hubner-Parajsz C, Kientsch-Engel R, Stahl P, Pischetsrieder M. Determination of glycated nucleobases in human urine by a new monoclonal antibody specific for N2-carboxyethyl-2′-deoxyguanosine. Chem. Res. Toxicol. 2004;17:1385–1390. doi: 10.1021/tx049929d. [DOI] [PubMed] [Google Scholar]
- 30.Cooke MS, Singh R, Hall GK, Mistry V, Duarte TL, Farmer PB, Evans MD. Evaluation of enzyme-linked immunosorbent assay and liquid chromatography-tandem mass spectrometry methodology for the analysis of 8-oxo-7,8-dihydro-2′-deoxyguanosine in saliva and urine. Free Radical Biol. Med. 2006;41:1829–1836. doi: 10.1016/j.freeradbiomed.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 31.Chung FL, Nath RG, Ocando J, Nishikawa A, Zhang L. Deoxyguanosine adducts of 4-hydroxy-2-nonenal are endogenous DNA lesions in rodents and humans: detection and potential sources. Cancer Res. 2000;60:1507–1511. [PubMed] [Google Scholar]
- 32.Cao H, Jiang Y, Wang Y. Stereospecific synthesis and characterization of oligodeoxyribonucleotides containing an N2-(1-carboxyethyl)-2′-deoxyguanosine. J. Am. Chem. Soc. 2007;129:12123–12130. doi: 10.1021/ja072130e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larisch B, Pischetsrieder M, Severin T. Formation of guanosine adducts from L-ascorbic acid under oxidative conditions. Bioorg. Med. Chem. Lett. 1997;7:2681–2686. [Google Scholar]
- 34.Papoulis A, Al-Abed Y, Bucala R. Identification of N2(1-carboxyethyl)guanine (CEG) as a guanine advanced glycosylation endproduct. Biochemistry. 1995;34:648–655. doi: 10.1021/bi00002a032. [DOI] [PubMed] [Google Scholar]
- 35.Seidel W, Pischetsrieder M. DNA glycation leads to depurination by the loss of N2-carboxyethylguanine in vitro. Cell. Mol. Biol. 1998;44:1165–1170. [PubMed] [Google Scholar]
- 36.Baynes JW. The Maillard hypothesis on aging: time to focus on DNA. Ann. NY Acad. Sci. USA. 2002;959:360–367. doi: 10.1111/j.1749-6632.2002.tb02107.x. [DOI] [PubMed] [Google Scholar]
- 37.Pischetsrieder M, Seidel W, Munch G, Schinzel R. N−2(1-Carboxyethyl)deoxyguanosine, a nonenzymatic glycation adduct of DNA, induces single strand breaks and increases mutation frequences. Biochem. Biophys. Res. Commun. 1999;264:544–549. doi: 10.1006/bbrc.1999.1528. [DOI] [PubMed] [Google Scholar]
- 38.Nicolay JP, Schneider J, Niemoeller OM, Artunc F, Portero-Otin M, Haik G, Thornalley PJ, Schleicher E, Wieder T, Lang F. Stimulation of suicidal erythrocyte death by methylglyoxal. Cell. Physiol. Biochem. 2006;18:223–232. doi: 10.1159/000097669. [DOI] [PubMed] [Google Scholar]
- 39.Dobler D, Ahmed N, Song LJ, Eboigbodin KE, Thornalley PJ. Increased dicarbonyl metabolism in endothelial cells in hyperglycemia induces anoikis and impairs angiogenesis by RGD and GFOGER motif modification. Diabetes. 2006;55:1961–1969. doi: 10.2337/db05-1634. [DOI] [PubMed] [Google Scholar]
- 40.Chan WH, Wu HJ, Shiao NH. Apoptotic signaling in methylglyoxal-treated human osteoblasts involves oxidative stress, c-jun N-terminal kinase, caspase-3, and p21-activated kinase 2. J. Cell. Biochem. 2007;100:1056–1069. doi: 10.1002/jcb.21114. [DOI] [PubMed] [Google Scholar]
- 41.Morcos M, Du X, Pfisterer F, Hutter H, Sayed AAR, Thornalley P, Ahmed N, Baynes J, Thorpe S, Kukudov G, et al. Glyoxalase-1 prevents mitochondrial protein modification and enhances lifespan in Caenorhabditis elegans. Aging Cell. 2008;7:260–269. doi: 10.1111/j.1474-9726.2008.00371.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.