Abstract
In this work, we discuss the active or passive character of helicases. In the past years, several studies have used the theoretical framework proposed by Betterton and Julicher [Betterton, M.D. and Julicher, F. (2005) Opening of nucleic-acid double strands by helicases: active versus passive opening. Phys. Rev. E, 71, 11904–11911.] to analyse the unwinding data and assess the mechanism of the helicase under study (active versus passive). However, this procedure has given rise to apparently contradictory interpretations: helicases exhibiting similar behaviour have been classified as both active and passive enzymes [Johnson, D.S., Bai, L. Smith, B.Y., Patel, S.S. and Wang, M.D. (2007) Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell, 129, 1299–1309; Lionnet, T., Spiering, M.M., Benkovic, S.J., Bensimon, D. and Croquette, V. (2007) Real-time observation of bacteriophage T4 gp41 helicase reveals an unwinding mechanism Proc. Natl Acid. Sci., 104, 19790–19795]. In this work, we show that when the helicase under study has not been previously well characterized (namely, if its step size and rate of slippage are unknown) a multi-parameter fit to the afore-mentioned model can indeed lead to contradictory interpretations. We thus propose to differentiate between active and passive helicases on the basis of the comparison between their observed translocation velocity on single-stranded nucleic acid and their unwinding rate of double-stranded nucleic acid (with various GC content and under different tensions). A threshold separating active from passive behaviour is proposed following an analysis of the reported activities of different helicases. We study and contrast the mechanism of two helicases that exemplify these two behaviours: active for the RecQ helicase and passive for the gp41 helicase.
INTRODUCTION
Nucleic acid (NA., i.e. DNA or RNA) helicases are enzymes capable of unwinding double-stranded NA (dsNA) to provide the single-stranded NA (ssNA) template required in many biological events, such as replication, recombination and repair (1–3). In that process, helicases hydrolyse ATP to translocate along one strand of the double helix displacing the other. How this translocation is coupled to unwinding is a much debated issue. In particular two models confront each other. A passive model whereby the translocation of the helicase simply traps transient unwinding fluctuations of the upstream dsNA and an active mechanism whereby the interaction of the enzyme with the dsNA is sufficient to destabilise it and eliminate its possible action as a ‘road block’ on the progression of the enzyme.
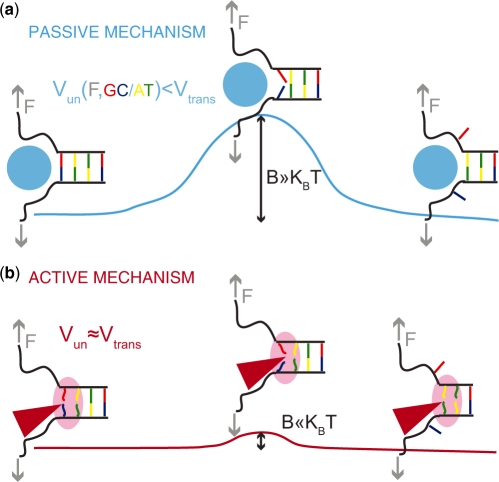
Helicases are enzymes and as such they act by lowering the activation barrier of the reaction they catalyse, i.e. NA unwinding. From that point of view the difference between an active and a passive helicase rests on the size of the activation energy 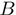 (Figure 1). If
(Figure 1). If 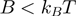 (where
(where 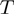 is the temperature and
is the temperature and 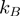 the Boltzman's constant) then the helicase is active (the barrier is negligible). In contrast, if
the Boltzman's constant) then the helicase is active (the barrier is negligible). In contrast, if 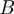 is a few
is a few 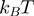 then hoping over the activation barrier may be the rate-limiting step in the reaction and the enzyme is passive (Figure 1).
then hoping over the activation barrier may be the rate-limiting step in the reaction and the enzyme is passive (Figure 1).
Figure 1.
Passive versus active mechanisms: a passive enzyme (a) has to overcome a significant barrier 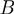 (few
(few 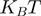 ) to proceed with NA unwinding. Consequently, its rate of progression is significantly reduced by the presence of the NA fork, i.e.
) to proceed with NA unwinding. Consequently, its rate of progression is significantly reduced by the presence of the NA fork, i.e. 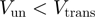 , where
, where 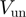 and
and 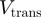 are the helicase rates of unwinding of dsNA and translocation on ssNA. Moreover, its unwinding rate will depend on factors that affect the height of the kinetic barrier such as the NA sequence [AT(U) versus GC base pair shown in different colors] or the presence of a tension (
are the helicase rates of unwinding of dsNA and translocation on ssNA. Moreover, its unwinding rate will depend on factors that affect the height of the kinetic barrier such as the NA sequence [AT(U) versus GC base pair shown in different colors] or the presence of a tension (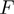 ) that destabilizes the fork. In contrast, an active enzyme (b) is able to destabilize the dsNA and effectively reduce the kinetic barrier to fork melting (
) that destabilizes the fork. In contrast, an active enzyme (b) is able to destabilize the dsNA and effectively reduce the kinetic barrier to fork melting (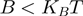 ). An active enzyme will thus unwind at a constant rate (independent of the underlying sequence) and as fast as it translocates along ssNA (
). An active enzyme will thus unwind at a constant rate (independent of the underlying sequence) and as fast as it translocates along ssNA (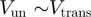 ).
).
Factors that control the height of the activation barrier 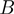 , such as the NA sequence or the tension on the fork (i.e. the force acting to unzip the two NA strands), may affect the unwinding behaviour of helicases. In general, one can write the various contributions to the activation energy
, such as the NA sequence or the tension on the fork (i.e. the force acting to unzip the two NA strands), may affect the unwinding behaviour of helicases. In general, one can write the various contributions to the activation energy 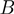 as
as
| (1) |
where  is the number of unwound base pairs (bps) in the transition state,
is the number of unwound base pairs (bps) in the transition state, 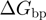 the free energy of base pair formation and
the free energy of base pair formation and 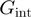 and
and 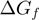 the reduction in that free energy due to the helicase and the unzipping force, respectively. On naked DNA (free of proteins), the free energy of formation of an AT base pair is on average half than that of a GC base pair [at 37°C, 100 mM NaCl and 5 mM Mg
the reduction in that free energy due to the helicase and the unzipping force, respectively. On naked DNA (free of proteins), the free energy of formation of an AT base pair is on average half than that of a GC base pair [at 37°C, 100 mM NaCl and 5 mM Mg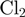
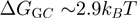 whereas
whereas 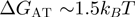 , values obtained by averaging among the different possible neighbouring base pairs (4)]. Similar differences exist between AU and GC RNA base pairs (4). Consequently, a passive enzyme whose unwinding is controlled by the activation barrier to fork melting (Equation 1) will display an unwinding rate that decreases with the NA's GC content. By contrast, the unwinding activity of an active enzyme will be insensitive to the NA sequence,
, values obtained by averaging among the different possible neighbouring base pairs (4)]. Similar differences exist between AU and GC RNA base pairs (4). Consequently, a passive enzyme whose unwinding is controlled by the activation barrier to fork melting (Equation 1) will display an unwinding rate that decreases with the NA's GC content. By contrast, the unwinding activity of an active enzyme will be insensitive to the NA sequence,  . On the other hand, the application of unzipping forces lowers the height of the activation barrier to a point where
. On the other hand, the application of unzipping forces lowers the height of the activation barrier to a point where 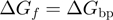 (
(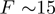 pN for DNA) and the dsNA may spontaneously unwind within the experimental time-scale (5,6). For a passive enzyme, the presence of destabilizing forces will then lead to higher unwinding rates. When the enzyme, is active, the effect of the force may be negligible or counter-productive, i.e. slowing down the enzyme [as expected for an active ‘rolling’ model (1)]. Consequently, the unwinding rate dependence on force and sequence can be used to reveal the active or passive character of the enzyme under study. Notice that, a priori, the NA sequence and the applied force might also affect the translocation activity of helicases along ssNA. In other words, the interaction between the helicase and ssNA that govern the enzyme translocation activity might be altered by the presence of an stretching force or specific NA sequences. For the helicases studied in this work no significant sequence or force dependence was detected in their ssNA translocation activity (see below), but force sensitivity has been reported for other translocases [such as polymerases (7,8)] and hypothesized for the Uvrd helicase (9).
pN for DNA) and the dsNA may spontaneously unwind within the experimental time-scale (5,6). For a passive enzyme, the presence of destabilizing forces will then lead to higher unwinding rates. When the enzyme, is active, the effect of the force may be negligible or counter-productive, i.e. slowing down the enzyme [as expected for an active ‘rolling’ model (1)]. Consequently, the unwinding rate dependence on force and sequence can be used to reveal the active or passive character of the enzyme under study. Notice that, a priori, the NA sequence and the applied force might also affect the translocation activity of helicases along ssNA. In other words, the interaction between the helicase and ssNA that govern the enzyme translocation activity might be altered by the presence of an stretching force or specific NA sequences. For the helicases studied in this work no significant sequence or force dependence was detected in their ssNA translocation activity (see below), but force sensitivity has been reported for other translocases [such as polymerases (7,8)] and hypothesized for the Uvrd helicase (9).
The qualification of the helicase mechanism has typically relied on estimations of the free energy of interaction between the helicase and the NA fork, 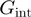 , through a theoretical analysis of the dependence of the unwinding activity on the NA sequence or the unzipping force (10–13). However, as discussed below, reliable estimates of
, through a theoretical analysis of the dependence of the unwinding activity on the NA sequence or the unzipping force (10–13). However, as discussed below, reliable estimates of 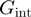 are difficult to obtain. Here, we propose a simpler approach that consists in using the ratio between the translocation and unwinding rates and their dependence on force and NA sequence to assess how active the helicase is. A threshold that separate passive from active behaviour is proposed on the basis of the reported activities of different helicases and some theoretical considerations. The approach is illustrated by investigating the unwinding and translocation activities as a function of force and DNA sequence of two different DNA helicases that exemplify the two extremes of helicase behaviour (active and passive): the Escherichia coli RecQ, an helicase involved in DNA repair and recombination, and the T4 gp41 hexameric replicative helicase.
are difficult to obtain. Here, we propose a simpler approach that consists in using the ratio between the translocation and unwinding rates and their dependence on force and NA sequence to assess how active the helicase is. A threshold that separate passive from active behaviour is proposed on the basis of the reported activities of different helicases and some theoretical considerations. The approach is illustrated by investigating the unwinding and translocation activities as a function of force and DNA sequence of two different DNA helicases that exemplify the two extremes of helicase behaviour (active and passive): the Escherichia coli RecQ, an helicase involved in DNA repair and recombination, and the T4 gp41 hexameric replicative helicase.
MATERIALS AND METHODS
Proteins
The helicases T4 gp41 and E. coli RecQ were prepared as previously described (14,15).
DNA hairpin substrate
The fork structure was formed by two partially annealed oligos of which the flap oligo was 5′-biotinylated to allow for attachment to the magnetic bead. This fork structure and a short hairpin oligo were annealed and ligated to either end of a 1.1 kb fragment based on their compatible ends. Details of the substrate preparation are given elsewhere (16). Experiments with the E. coli RecQ were performed with the full hairpin (FH) containing 1239 bp, whereas experiments with gp41 were performed with the half-hairpin (HH) substrate, containing 604 bp and long ∼600 nt tails [(16) and Supplementary Figure S1a and b].
Single-molecule assay
The glass surface was treated with anti-digoxigenin antibody and passivated with bovine serum albumin (BSA). The magnetic beads (Dynal from Invitrogen) were ∼1  m in diameter and coated with streptavidin. Bead images were acquired at 60 Hz using a PicoTwist inverted microscope and the DNA extension was measured by tracking the bead position in real time (17). The force was measured by using the fluctuation-dissipation theorem and recording the fluctuations of a bead attached to the surface by a long DNA molecule (typically 48 kb DNA molecule made from
m in diameter and coated with streptavidin. Bead images were acquired at 60 Hz using a PicoTwist inverted microscope and the DNA extension was measured by tracking the bead position in real time (17). The force was measured by using the fluctuation-dissipation theorem and recording the fluctuations of a bead attached to the surface by a long DNA molecule (typically 48 kb DNA molecule made from  DNA). A calibration for the force versus magnet vertical position was then obtained. The calibration curve was used to estimate the applied force on the DNA hairpin with 10% error. The mechanical stability of the DNA hairpin was characterized by measuring the extension of the substrate as a function of the pulling force along a force-cycle in which the force is first increased and then relaxed (Supplementary Figure S1c). Experiments with the gp41 helicase were performed at 29°C in 25 mM Tris–Ac (pH 7.5), 150 mM KOAc, 10 mM Mg(OAc)2, 1 mM dithiothreitol (DTT) and 5 mM ATP. The protein concentration was 50 nM gp41 (monomeric concentration). Experiments with the E. coli RecQ helicase were performed at 29°C in 20 mM Tris–HCl (pH 7.5), 25 mM NaCl, 3 mM Mg(Cl)2, 1 mM DTT and 1 mM ATP. The protein concentration was 30 pM RecQ.
DNA). A calibration for the force versus magnet vertical position was then obtained. The calibration curve was used to estimate the applied force on the DNA hairpin with 10% error. The mechanical stability of the DNA hairpin was characterized by measuring the extension of the substrate as a function of the pulling force along a force-cycle in which the force is first increased and then relaxed (Supplementary Figure S1c). Experiments with the gp41 helicase were performed at 29°C in 25 mM Tris–Ac (pH 7.5), 150 mM KOAc, 10 mM Mg(OAc)2, 1 mM dithiothreitol (DTT) and 5 mM ATP. The protein concentration was 50 nM gp41 (monomeric concentration). Experiments with the E. coli RecQ helicase were performed at 29°C in 20 mM Tris–HCl (pH 7.5), 25 mM NaCl, 3 mM Mg(Cl)2, 1 mM DTT and 1 mM ATP. The protein concentration was 30 pM RecQ.
Single-molecule data analysis
Raw data, corresponding to the real-time evolution of the DNA extension 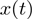 in
in 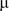 m, was converted into the number of base-pairs unwound
m, was converted into the number of base-pairs unwound 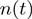 as a function of time using either the equation
as a function of time using either the equation 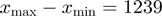 bp unwound for the FH substrate (RecQ experiments) or the equation
bp unwound for the FH substrate (RecQ experiments) or the equation 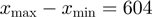 bp unwound for the HH substrate (gp41 experiments). The unwinding or rezipping velocity
bp unwound for the HH substrate (gp41 experiments). The unwinding or rezipping velocity 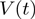 at position
at position 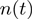 was computed as the slope of the best linear fit to a 30-point (corresponding to 0.5 s) segment
was computed as the slope of the best linear fit to a 30-point (corresponding to 0.5 s) segment 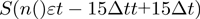 , where
, where  t is 1/60 s, the time interval between two data points. In the force analysis, the velocity was averaged along the hairpin position
t is 1/60 s, the time interval between two data points. In the force analysis, the velocity was averaged along the hairpin position 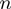 ,
, 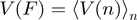 , where
, where 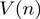 is the mean velocity at position
is the mean velocity at position 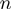 obtained from the average between different trajectories and the brackets represent the average along
obtained from the average between different trajectories and the brackets represent the average along  . Each force point
. Each force point 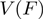 (Figure 3a) was obtained from ∼20–40 trajectories corresponding to ∼3–10 molecules. Values reported correspond to the mean between different molecules. To analyse the dependence of unwinding velocity on the DNA sequence, we used a moving window
(Figure 3a) was obtained from ∼20–40 trajectories corresponding to ∼3–10 molecules. Values reported correspond to the mean between different molecules. To analyse the dependence of unwinding velocity on the DNA sequence, we used a moving window 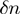 of 15 bp along which the velocity was averaged,
of 15 bp along which the velocity was averaged, 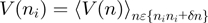 , where
, where 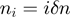 with
with  being an integer, and the brackets represent the average along
being an integer, and the brackets represent the average along 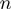 verifying
verifying 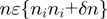 . We next computed the average GC content of each window
. We next computed the average GC content of each window 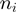 ,
, 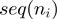 , and obtained the velocity as a function of the GC percentage,
, and obtained the velocity as a function of the GC percentage, 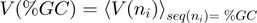 , where the brakets represent the average along
, where the brakets represent the average along 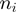 verifying
verifying  . Each velocity point
. Each velocity point 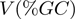 (Figure 3b) was obtained from ∼100–200 trajectories corresponding to ∼50–100 molecules. Values reported correspond to the mean between different molecules as a function of the GC percentage of the
(Figure 3b) was obtained from ∼100–200 trajectories corresponding to ∼50–100 molecules. Values reported correspond to the mean between different molecules as a function of the GC percentage of the 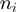 window.
window.
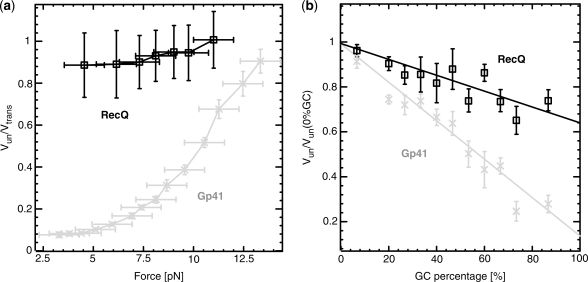
Figure 3.
Comparison between the unwinding rates of RecQ (black) and gp41 (grey) as the force (a) and GC content (b) are increased. The rates reported in the force data (a) are averaged along the sequence and normalized to the translocation rate on ssDNA. Based on the translocation rate measurements presented in Supplementary Figure S4) a constant force-independent translocation rate has been estimated for both gp41 and RecQ. The rates reported in the sequence data (b) are measured at 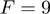 pN and are normalized by the maximal rate (at the 0% GC content). For both helicases, the data are measured at ATP-saturating conditions (5 mM for gp41 and 1 mM for RecQ). Values reported are the mean between different molecules and the error bars represent the standard error of the mean.
pN and are normalized by the maximal rate (at the 0% GC content). For both helicases, the data are measured at ATP-saturating conditions (5 mM for gp41 and 1 mM for RecQ). Values reported are the mean between different molecules and the error bars represent the standard error of the mean.
Model for helicase unwinding
Betterton and Julicher (BJ) (18) proposed a framework to describe the NA unwinding by helicases that has been used to interpret several experimental results (10–13). In this model, the kinetics of the NA fork is governed by the rates 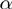 and
and 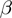 at which the base pair at the NA fork opens and closes. In addition, the helicase movement is governed by the forward and backward translocation rates,
at which the base pair at the NA fork opens and closes. In addition, the helicase movement is governed by the forward and backward translocation rates, 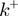 and
and 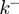 , along ssNA (Figure 4a). The coupling between the helicase movement and the NA opening comes from the fact that these rates depend on the relative position between the helicase and the NA fork, e.g. the fork cannot move back when the helicase is adjacent. Using the index
, along ssNA (Figure 4a). The coupling between the helicase movement and the NA opening comes from the fact that these rates depend on the relative position between the helicase and the NA fork, e.g. the fork cannot move back when the helicase is adjacent. Using the index 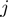 for the discrete position of the helicase relative to NA fork along the NA lattice (Figure 4a) one can write the following equations for the NA opening
for the discrete position of the helicase relative to NA fork along the NA lattice (Figure 4a) one can write the following equations for the NA opening 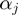 and closing
and closing 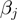 rates
rates
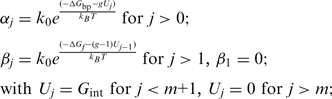 |
(2) |
where 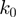 is the attempt frequency,
is the attempt frequency, 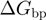 the base pair free energy,
the base pair free energy, 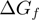 and
and 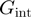 the reduction in that free energy due to the external force and the helicase, respectively,
the reduction in that free energy due to the external force and the helicase, respectively, 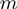 the range of the helicase–NA fork potential interaction and
the range of the helicase–NA fork potential interaction and 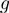 defines how the interaction potential affects the base pair opening and closing kinetics (18). The choice of such rates (Equation 2) is based on the description of the folding/unfolding kinetics of NA molecules under the force given elsewhere (19). Note that we have chosen a simple one-step potential of range
defines how the interaction potential affects the base pair opening and closing kinetics (18). The choice of such rates (Equation 2) is based on the description of the folding/unfolding kinetics of NA molecules under the force given elsewhere (19). Note that we have chosen a simple one-step potential of range 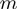 and amplitude
and amplitude 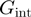 (Supplementary Figure S2a and b). The particular case where
(Supplementary Figure S2a and b). The particular case where 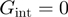 corresponds to the description for a passive helicase. Following the BJ's description one can write the helicase forward
corresponds to the description for a passive helicase. Following the BJ's description one can write the helicase forward 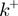 and backward
and backward  rates as
rates as
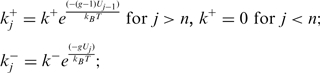 |
(3) |
where 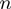 corresponds to the step size of the helicase and
corresponds to the step size of the helicase and 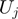 has been defined in Equation 2. The dynamics of the helicase/NA fork is then governed by the master equation for the probability
has been defined in Equation 2. The dynamics of the helicase/NA fork is then governed by the master equation for the probability 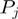 that the helicase is j bases away from the NA fork (
that the helicase is j bases away from the NA fork (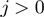 ) (18)
) (18)
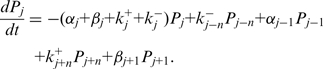 |
(4) |
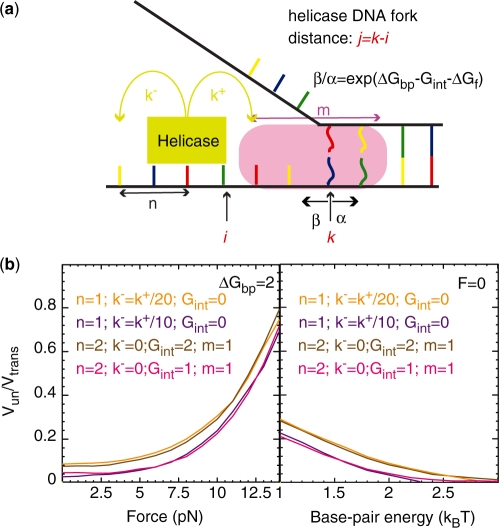
Figure 4.
(a) Schematic figure showing the different parameters characterizing the BJ's model: the ssNA translocation forward 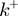 and backward
and backward 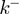 rates; the helicase step size
rates; the helicase step size 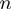 ; the base pair opening
; the base pair opening 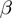 and closing
and closing 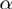 rates; and the range of the helicase–NA fork interaction
rates; and the range of the helicase–NA fork interaction  . The rates
. The rates 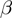 and
and 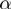 verify detailed balance condition:
verify detailed balance condition: 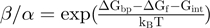 , where
, where 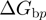 is the base pair free energy and
is the base pair free energy and 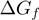 and
and 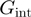 are the reduction due to the external force and the helicase interaction, respectively. The position of the helicase and the fork along the NA lattice is represented by the indexes i and k, respectively. The distance between both is given by the index
are the reduction due to the external force and the helicase interaction, respectively. The position of the helicase and the fork along the NA lattice is represented by the indexes i and k, respectively. The distance between both is given by the index 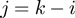 . (b)
. (b) 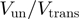 obtained from simulations of the BJ's model (see ‘Materials and Methods’ section) as a function of the unzipping force (left) and the NA stability (right) for different combinations of parameters describing the helicase–NA system giving rise to similar unwinding behaviour. For all cases the parameter
obtained from simulations of the BJ's model (see ‘Materials and Methods’ section) as a function of the unzipping force (left) and the NA stability (right) for different combinations of parameters describing the helicase–NA system giving rise to similar unwinding behaviour. For all cases the parameter 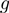 , which defines how the base pair kinetics is affected by the interaction potential, was chosen to be
, which defines how the base pair kinetics is affected by the interaction potential, was chosen to be 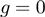 .
.
We performed Monte-Carlo simulations of such dynamical system for different parameters of the system, such as the step size of the helicase (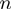 ), the presence of slippage (
), the presence of slippage (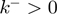 ) and the interaction potential (
) and the interaction potential (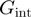 ,
, 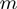 ). The unwinding rates normalized to the helicase translocation rate (given by
). The unwinding rates normalized to the helicase translocation rate (given by 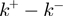 ) as a function of force and NA sequence for different conditions tested are shown in Supplementary Figure S2c. Note that the base-pair stability and the unzipping force enter in the model through
) as a function of force and NA sequence for different conditions tested are shown in Supplementary Figure S2c. Note that the base-pair stability and the unzipping force enter in the model through 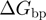 and
and 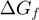 , respectively. The latter is estimated from the ssNA elasticity as
, respectively. The latter is estimated from the ssNA elasticity as 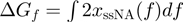 , where
, where 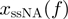 corresponds to the extension of single nucleotide at the given force
corresponds to the extension of single nucleotide at the given force 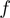 (Supplementary Figure S3). In all simulations, the attempt frequency was chosen to be
(Supplementary Figure S3). In all simulations, the attempt frequency was chosen to be 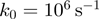 [a value consistent with the nuclear magnetic resonance (NMR) measurements of the base-pair kinetics (20)], the helicase forward translocation rate
[a value consistent with the nuclear magnetic resonance (NMR) measurements of the base-pair kinetics (20)], the helicase forward translocation rate 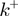 was fixed to 300 bp/s (a value close to the translocation rates reported for different helicases; see Table 1), and the parameter
was fixed to 300 bp/s (a value close to the translocation rates reported for different helicases; see Table 1), and the parameter 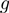 was fixed to 0.
was fixed to 0.
Table 1.
Unwinding and translocation rates
| Helicase | 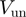 |
 |
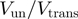 |
|---|---|---|---|
| (bp/s) | (nt/s) | ||
| T7 gp4 | 15–30a (10,27) | 130–300a (10,28) | ∼0.09–0.1 |
| T4 gp41 | 30a–40b (11) | 350–500b (11) | ∼0.08 |
| DnaB | 35 (29) |
 1000c 1000c
|
∼0.035–0.1* |
Rep B B |
226 (30) | 530 (30) | ∼0.43 |
| Uvrd | ∼40c,e−70d (9,31) | ∼40a,d−200e (9,31) | ∼1d |
| RecQb | 80 | 90 | ∼0.9 |
aValues reported at low forces (3–5 pN).
bRates obtained in this work at low forces (∼3 pN).
cReplication rates of 1000 nt/s has been reported in E. coli replisome (32), showing that the DNA-B helicase is able to translocate along ssDNA at a rate  1000 nt/s
1000 nt/s
dValues given correspond to a dimeric Uvrd (9).
eValues given correspond to a monomeric Uvrd (31).
*Values reported in the table for the unwinding and translocation rate were measured at different temperatures. Recent single molecule studies at 25°C measure a ratio Vun/Vtrans of 0.1 (O. Saleh personal communication).
RESULTS
Single-molecule studies of T4 gp41 and E. coli RecQ
To monitor the activity of a single helicase we use a magnetic trap (8,11,16,21). Briefly, a DNA hairpin (with duplex length of 600 or 1200 bps) was tethered by its extremities to a glass surface and a magnetic bead (Supplementary Figure S1). Small magnets positioned above the sample applied a constant force on the molecule. Unwinding the hairpin results in an increase in its end-to-end distance observed as a change in the distance between the bead and the surface (Figure 2a). The measured change in extension can be converted into the number of unwound base pairs, since the maximal change in extension is proportional to the (known) full length of the hairpin (Figure 2c). We first characterized the mechanical unfolding of the hairpin construct in absence of helicase (Supplementary Figure S1). At forces below 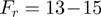 pN (depending on buffer conditions), the hairpin was observed to be stable for the duration of the experiment, while above a critical force
pN (depending on buffer conditions), the hairpin was observed to be stable for the duration of the experiment, while above a critical force 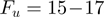 pN the hairpin unfolded rapidly. Therefore, at forces
pN the hairpin unfolded rapidly. Therefore, at forces 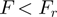 , any unfolding observed in the presence of helicase (and ATP) results from its enzymatic activity. Indeed in absence of helicase, the extension of the DNA molecule remains constant at the level corresponding to the folded hairpin.
, any unfolding observed in the presence of helicase (and ATP) results from its enzymatic activity. Indeed in absence of helicase, the extension of the DNA molecule remains constant at the level corresponding to the folded hairpin.
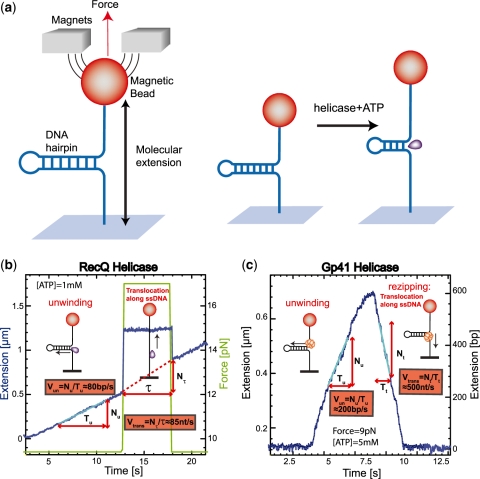
Figure 2.
(a) Schematic representation of the experimental configuration. (b) Experimental trace corresponding to the RecQ helicase activity in a ∼1200 bp hairpin (FH substrate, Supplementary Figure S1a). Molecular extension is shown in blue and the applied force in green. The force is transiently increased during unwinding (at times 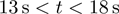 ) to measure the translocation activity on ssDNA. Experiment performed at 1 mM ATP (buffer conditions given in ‘Materials and Methods’ section). (c) Experimental trace corresponding to the gp41 helicase activity in a ∼600 bp hairpin (HH substrate, Supplementary Figure S1b). Extension in
) to measure the translocation activity on ssDNA. Experiment performed at 1 mM ATP (buffer conditions given in ‘Materials and Methods’ section). (c) Experimental trace corresponding to the gp41 helicase activity in a ∼600 bp hairpin (HH substrate, Supplementary Figure S1b). Extension in  m is converted to number of base pairs unwound (right axis) by assigning to the maximal length of the unwinding events the full length of the hairpin. Experiment performed at 5 mM ATP and 9 pN of applied force (buffer conditions given in ‘Materials and Methods’ section). The trace shows the unwinding phase (rising edge) and the rezipping phase (falling edge) along which the enzyme translocates on ssDNA, while the hairpin reanneals in its wake.
m is converted to number of base pairs unwound (right axis) by assigning to the maximal length of the unwinding events the full length of the hairpin. Experiment performed at 5 mM ATP and 9 pN of applied force (buffer conditions given in ‘Materials and Methods’ section). The trace shows the unwinding phase (rising edge) and the rezipping phase (falling edge) along which the enzyme translocates on ssDNA, while the hairpin reanneals in its wake.
Bursts of helicase activity observed in single molecule trajectories allow to directly measure the unwinding and translocation rates (see ‘Materials and Methods’ section). The unwinding rate 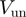 is computed from the slope of the unwinding phase, as shown in Figure 2b and c for the two helicases studied here (see below). On the other hand, the translocation rate on ssDNA,
is computed from the slope of the unwinding phase, as shown in Figure 2b and c for the two helicases studied here (see below). On the other hand, the translocation rate on ssDNA,  , can be deduced from an experiment where the force is transiently increased (to a value
, can be deduced from an experiment where the force is transiently increased (to a value  ) to unfold the hairpin during an unwinding event (Figure 2b). During the time lapse
) to unfold the hairpin during an unwinding event (Figure 2b). During the time lapse  when the hairpin is mechanically unfolded the enzyme translocates on ssDNA. Upon lowering back the force to its initial value the hairpin reanneals up to the position of the helicase. The detectable change in extension is thus due to translocation on ssDNA and the translocation rate at force
when the hairpin is mechanically unfolded the enzyme translocates on ssDNA. Upon lowering back the force to its initial value the hairpin reanneals up to the position of the helicase. The detectable change in extension is thus due to translocation on ssDNA and the translocation rate at force 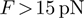 can be obtained by dividing the number of translocated nucleotides
can be obtained by dividing the number of translocated nucleotides 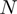 by
by  . The translocation rate at lower force (
. The translocation rate at lower force (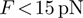 ) can also be measured once the enzyme has passed the hairpin apex and translocates along ssDNA with the hairpin reannealing in its wake [(11) and Figure 2c]. For the two helicases studied, the translocation rates estimated for the two different protocols described are consistent (Supplementary Figure S4), showing that the force does not affect their translocation activity.
) can also be measured once the enzyme has passed the hairpin apex and translocates along ssDNA with the hairpin reannealing in its wake [(11) and Figure 2c]. For the two helicases studied, the translocation rates estimated for the two different protocols described are consistent (Supplementary Figure S4), showing that the force does not affect their translocation activity.
Using the protocols described above we studied RecQ, a 3′-5′ helicase belonging to the SFII superfamily of helicases (such as NS3, PriA and RecG). Adding RecQ in a solution with hairpin molecules under moderate tension (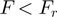 ) results in a processive unwinding of the hairpin at a constant rate
) results in a processive unwinding of the hairpin at a constant rate 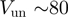 bps/s (Figure 2b) [RecQ activity results in two different types of unwinding events: one displaying a fast and uniform unwinding rate and the other showing a more complex non-uniform unwinding (unpublished results). Here we have only focused on the analysis of the fast and processive regime]. This rate is very close to the translocation rate
bps/s (Figure 2b) [RecQ activity results in two different types of unwinding events: one displaying a fast and uniform unwinding rate and the other showing a more complex non-uniform unwinding (unpublished results). Here we have only focused on the analysis of the fast and processive regime]. This rate is very close to the translocation rate 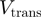 measured during transient mechanical unfolding of the hairpin (Figure 2b) or during hairpin reanneling (Supplementary Figure S4). We also find that
measured during transient mechanical unfolding of the hairpin (Figure 2b) or during hairpin reanneling (Supplementary Figure S4). We also find that 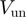 does not significantly increase with the applied force—
does not significantly increase with the applied force— at all forces, Figure 3a—and depends weakly on the GC content of the molecule—the ratio of unwinding rates on purely GC to purely AT rich regions is estimated to be
at all forces, Figure 3a—and depends weakly on the GC content of the molecule—the ratio of unwinding rates on purely GC to purely AT rich regions is estimated to be 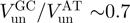 , see ‘Materials and Methods’ section and Figure 3b.
, see ‘Materials and Methods’ section and Figure 3b.
This behaviour is in stark contrast with the 5′-3′ hexameric helicase of the T4 replication complex, gp41 (11). At low forces, gp41 unwinds DNA much slower than it translocates along ssDNA (Figure 2c). The unwinding rate strongly increases with the applied force [(11) and Figure 3a] and is very sensitive to the GC content of the molecule: 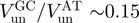 (Figure 3b). These results clearly show that the unwinding mechanism of gp41 is qualitatively different from that of RecQ. More specifically, they suggest that the mechanism of gp41, in contrast to that of RecQ, relies to some degree on thermal fraying of the DNA fork.
(Figure 3b). These results clearly show that the unwinding mechanism of gp41 is qualitatively different from that of RecQ. More specifically, they suggest that the mechanism of gp41, in contrast to that of RecQ, relies to some degree on thermal fraying of the DNA fork.
Characterizing the activeness and passiveness of helicases
The BJ's framework (18) is often used to describe helicase unwinding. In that description, the helicase movement depends on (Figure 4a and ‘Materials and Methods’ section):
the ssNA forward and backward translocation rates,
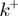 and
and 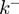 (note that the total ssNA translocation rate corresponds to
(note that the total ssNA translocation rate corresponds to 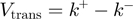 );
);the helicase step size
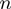 ;
;the base pair free energy
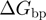 and its reduction due to the external forces
and its reduction due to the external forces  ;
;the interaction potential between the helicase and the fork (defined itself by a set of parameters such as the amplitude
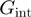 and the interaction range
and the interaction range 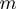 ).
).
Studies on different helicases have used this model to fit the measured variation of the unwinding rate with force or sequence in order to deduce the helicase–NA fork interaction free energy, 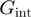 (10–13). These estimates of
(10–13). These estimates of 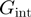 have then been used to qualify the helicase mechanism as active or passive. For an active enzyme
have then been used to qualify the helicase mechanism as active or passive. For an active enzyme 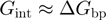 , whereas for a passive one
, whereas for a passive one 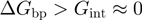 .
.
This procedure has, however, lead to contradictory interpretations. In particular, while very similar results for the unwinding rate variation with applied force have been reported for the T4 and T7 replicative helicases, using a fit to the BJ’s model different conclusions were reached. The T4 and T7 helicases have been proposed, respectively, as examples of a passive (11) and a partially active enzyme (10).
To assess the applicability of the BJ’s model to describe helicase unwinding, we performed Monte-Carlo simulations of the model (‘Materials and Methods’ section) and investigated how the unwinding rates predicted by the model depended on various parameters of the helicase–NA system. As shown in Supplementary Figure S2 the unwinding rate dependence on force and sequence is extremely sensitive to the value of the step size of the helicase 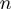 , to the presence of slippage (
, to the presence of slippage (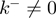 ) and to the specific type of potential interaction between helicase and NA fork, properties that are very difficult to directly measure. These properties are usually estimated by making some assumptions about the translocations and unwinding mechanisms, such as considering that the helicase cannot move backwards (
) and to the specific type of potential interaction between helicase and NA fork, properties that are very difficult to directly measure. These properties are usually estimated by making some assumptions about the translocations and unwinding mechanisms, such as considering that the helicase cannot move backwards (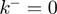 ), or using model-based kinetic schemes to extract the helicase step size (22,23). Visualization of the individual steps or the backward motion of helicases requires using a high spatial resolution technique (typically in the subnanometer range). Direct measurements of the helicase stepping has only been obtained for few slow helicases, such as NS3, where 1 bp has been reported (24,25). On the other hand, backsliding has been observed for RecBCD in presence of stretching forces (26), but estimations of
), or using model-based kinetic schemes to extract the helicase step size (22,23). Visualization of the individual steps or the backward motion of helicases requires using a high spatial resolution technique (typically in the subnanometer range). Direct measurements of the helicase stepping has only been obtained for few slow helicases, such as NS3, where 1 bp has been reported (24,25). On the other hand, backsliding has been observed for RecBCD in presence of stretching forces (26), but estimations of 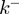 in absence of force have never been reported. Nevertheless, the well-defined polarity along ssDNA that display the majority of helicases imply
in absence of force have never been reported. Nevertheless, the well-defined polarity along ssDNA that display the majority of helicases imply 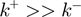 .
.
From the simulations results, we noticed (Figure 4b) that different combinations of parameters in the BJ’s model gave rise to similar unwinding behaviour: a passive helicase (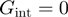 ) of 1 bp step size presenting some slippage behaves as a partially active helicase (
) of 1 bp step size presenting some slippage behaves as a partially active helicase (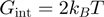 ) with step size of 2 bp. These results imply that when the step size and the slippage rate of the helicase under study are not known a multi-parameter fit to the BJ's model can lead to non-robust results.
) with step size of 2 bp. These results imply that when the step size and the slippage rate of the helicase under study are not known a multi-parameter fit to the BJ's model can lead to non-robust results.
We hereby propose to cut this Gordian knot by simply using the ratio between the translocation and the unwinding rate in absence of force, 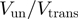 , to qualify the mechanism of helicases. A passive enzyme has to overcome a significant barrier for unwinding dsNA
, to qualify the mechanism of helicases. A passive enzyme has to overcome a significant barrier for unwinding dsNA 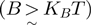 ; the helicase is thus slowed down by the fork and displays an unwinding rate significantly lower than its translocation rate on ssNA,
; the helicase is thus slowed down by the fork and displays an unwinding rate significantly lower than its translocation rate on ssNA, 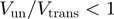 (as observed for gp41 helicase). Indeed, for a passive helicase of 1 bp kinetic step size unwinding a dsNA with average bp energy of 2
(as observed for gp41 helicase). Indeed, for a passive helicase of 1 bp kinetic step size unwinding a dsNA with average bp energy of 2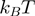 , the BJ's model predicts
, the BJ's model predicts 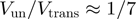 at zero force, a value that decreases with the kinetic step size and rate of helicase slippage. In contrast, for an active enzyme that is able to effectively destabilize the dsNA fork (
at zero force, a value that decreases with the kinetic step size and rate of helicase slippage. In contrast, for an active enzyme that is able to effectively destabilize the dsNA fork (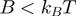 ) unwinding can proceed as fast as ssNA translocation,
) unwinding can proceed as fast as ssNA translocation, 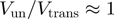 (as observed for the RecQ helicase).
(as observed for the RecQ helicase).
DISCUSSION AND CONCLUSIONS
Helicases are enzymes involved in many processes related to NA metabolism. They used the energy of ATP hydrolysis to translocate along ssNA and unwind dsNA. Depending on the mechanism of coupling translocation to unwinding they are considered as passive or active enzymes. A passive helicase relies on the transient fraying of the dsNA base pairs at the ssNA–dsNA junction in order to displace the fork, whereas an active helicase directly interacts with the fork destabilizing it and promoting NA unwinding. These two behaviours are the two extremes of a continuum spectra. In other words, most helicases will display a behaviour between an ideally active and totally passive behaviour. A common approach to quantify the degree of activeness of helicases has consisted in using estimations of the helicase–NA fork interaction free energy. Since a direct measurement of this free energy is difficult to obtain, several studies have used the theoretical framework proposed by BJ to describe the helicase motion (18) in order to fit the helicase unwinding data and obtain an estimate of the helicase–NA fork interaction free energy.
Here, we performed Monte-Carlo simulations of the BJ’s model for different NA/helicase parameters. Our results show that helicases characterized by different parameters and in particular different interaction energies 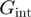 can give rise to similar unwinding behaviour (Figure 4). This could explain the apparent contradictory interpretations reported in different studies (10,11). From this analysis we conclude that a multi-parameter fit to the BJ’s model can lead to an ambiguous characterization of the helicase mechanism, when the helicase step-size and slippage rate have not been previously measured. Since they are both difficult to determine experimentally, we propose to use a less sophisticated experimental (rather than theoretical) criteria to assess how active a helicase is. This approach consists in qualifying the helicase mechanism (active or passive) by comparing the efficiency of dsNA unwinding with the ssNA translocation rate at zero force. An active helicase effectively destabilizes the fork and its unwinding and translocation rate are similar:
can give rise to similar unwinding behaviour (Figure 4). This could explain the apparent contradictory interpretations reported in different studies (10,11). From this analysis we conclude that a multi-parameter fit to the BJ’s model can lead to an ambiguous characterization of the helicase mechanism, when the helicase step-size and slippage rate have not been previously measured. Since they are both difficult to determine experimentally, we propose to use a less sophisticated experimental (rather than theoretical) criteria to assess how active a helicase is. This approach consists in qualifying the helicase mechanism (active or passive) by comparing the efficiency of dsNA unwinding with the ssNA translocation rate at zero force. An active helicase effectively destabilizes the fork and its unwinding and translocation rate are similar: 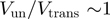 . In contrast, a passive enzyme is slowed down by the presence of the dsNA fork and unwinds it at a much slower rate than it translocates along ssNA:
. In contrast, a passive enzyme is slowed down by the presence of the dsNA fork and unwinds it at a much slower rate than it translocates along ssNA: 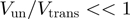 . Moreover, since the activation energy barrier to fork melting is altered by the dsNA sequence and the tension on the fork, a passive helicase unwinding rate will be affected by these factors. In contrast, the unwinding rate of an active helicase will depend weakly (if at all) on tension or sequence. Indeed, results on different helicases have shown that their unwinding rates exhibit widely different sensitivities to both NA sequence and applied force (9–13).
. Moreover, since the activation energy barrier to fork melting is altered by the dsNA sequence and the tension on the fork, a passive helicase unwinding rate will be affected by these factors. In contrast, the unwinding rate of an active helicase will depend weakly (if at all) on tension or sequence. Indeed, results on different helicases have shown that their unwinding rates exhibit widely different sensitivities to both NA sequence and applied force (9–13).
Here, we studied the behaviour of two DNA helicases that exemplify these different mechanisms: active for the E. coli RecQ helicase as opposed to passive for the T4 gp41 helicase. For both helicases, we have examined the ratio between the unwinding and translocation rates 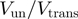 and its dependence on force and DNA sequence.
and its dependence on force and DNA sequence. 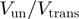 for RecQ is >0.7 for all the conditions tested (forces from 3 to 11 pN and CG content from 0% to 80%), demonstrating the active character of this helicase. In contrast, for gp41
for RecQ is >0.7 for all the conditions tested (forces from 3 to 11 pN and CG content from 0% to 80%), demonstrating the active character of this helicase. In contrast, for gp41 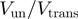 reaches values as low as 0.1 (at low forces or high CG content) and its unwinding activity displays strong force and sequence dependence. We conclude that RecQ is close to an optimally active helicase that unwinds DNA at its maximal ssDNA translocation rate almost independently of both applied force and DNA sequence. In contrast, gp41 exemplify the behaviour of a passive helicase that reaches its maximal unwinding rate when it is assisted by a force sufficiently large to almost unzip the molecule and which unwinding activity is very sensitive to the underlying DNA sequence.
reaches values as low as 0.1 (at low forces or high CG content) and its unwinding activity displays strong force and sequence dependence. We conclude that RecQ is close to an optimally active helicase that unwinds DNA at its maximal ssDNA translocation rate almost independently of both applied force and DNA sequence. In contrast, gp41 exemplify the behaviour of a passive helicase that reaches its maximal unwinding rate when it is assisted by a force sufficiently large to almost unzip the molecule and which unwinding activity is very sensitive to the underlying DNA sequence.
In Table 1, we show the unwinding and translocation rates, 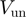 and
and 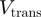 , reported for different helicases. To our knowledge these are the only helicases for which both
, reported for different helicases. To our knowledge these are the only helicases for which both 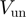 and
and 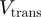 have been measured in absence of accessory proteins. Most of the rates reported in Table 1 were measured in presence of a stretching force. We have then reported the rate at the minimum force measured (typically around 3–5pN). Notice that the helicases listed in Table 1 can be easily divided in two groups: some of them (Uvrd, RecQ and Rep2
have been measured in absence of accessory proteins. Most of the rates reported in Table 1 were measured in presence of a stretching force. We have then reported the rate at the minimum force measured (typically around 3–5pN). Notice that the helicases listed in Table 1 can be easily divided in two groups: some of them (Uvrd, RecQ and Rep2 B) present unwinding rates that are similar to their ssNA translocation rates (
B) present unwinding rates that are similar to their ssNA translocation rates (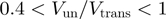 ), whereas others (hexameric T4 gp41, T7 gp4 and DNaB) unwind much slower than they translocate along ssNA (
), whereas others (hexameric T4 gp41, T7 gp4 and DNaB) unwind much slower than they translocate along ssNA (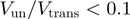 ). We can use the BJ's model to establish a criterion separating active from passive helicases, by setting the helicase-induced reduction in the fork free energy at
). We can use the BJ's model to establish a criterion separating active from passive helicases, by setting the helicase-induced reduction in the fork free energy at 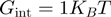 , i.e. about half the average base-pair energy. For a helicase with a one base pair step size and no slippage, the BJ's model then yields a value
, i.e. about half the average base-pair energy. For a helicase with a one base pair step size and no slippage, the BJ's model then yields a value 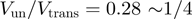 . Based on these theoretical considerations and the observations reported in Table 1, we propose to define as mainly active a helicase for which
. Based on these theoretical considerations and the observations reported in Table 1, we propose to define as mainly active a helicase for which 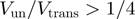 and as mainly passive one for which this ratio is smaller than one-fourth.
and as mainly passive one for which this ratio is smaller than one-fourth.
As discussed above, the unwinding rate of a passive helicase displays a strong force sensitivity (Figures 3a and 4b and Supplementary Figure S2). Since the unwinding rate at 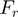 (i.e. close to mechanical unfolding of the fork) is equal to the translocation rate on ssNA (irrespective of the nature of the helicase mechanism), the ratio
(i.e. close to mechanical unfolding of the fork) is equal to the translocation rate on ssNA (irrespective of the nature of the helicase mechanism), the ratio 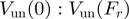 can also be used to qualify the nature of the helicase mechanism (passive if the ratio is below one-fourth and active if above one-fourth). Moreover, the unwinding behaviour of a passive helicase is also altered by its NA content (Figure 3b, Figure 4b and Supplementary Figure S2). Following the BJ’s model, an helicase displaying a behaviour half-way between an active and a passive enzyme (characterized by
can also be used to qualify the nature of the helicase mechanism (passive if the ratio is below one-fourth and active if above one-fourth). Moreover, the unwinding behaviour of a passive helicase is also altered by its NA content (Figure 3b, Figure 4b and Supplementary Figure S2). Following the BJ’s model, an helicase displaying a behaviour half-way between an active and a passive enzyme (characterized by 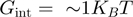 , a step size
, a step size  and no-slippage
and no-slippage 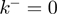 , see above) will advance ∼4 times slower when unwinding a GC-rich region with (
, see above) will advance ∼4 times slower when unwinding a GC-rich region with (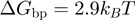 ) than an AT-rich one (
) than an AT-rich one (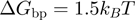 ):
): 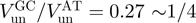 . Consequently active and passive helicases might also be qualified by their unwinding rate on AT- and GC-rich regions. An active helicase will display an unwinding ratio
. Consequently active and passive helicases might also be qualified by their unwinding rate on AT- and GC-rich regions. An active helicase will display an unwinding ratio 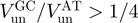 , while a passive helicase will have an unwinding ratio on GC- and AT-rich regions smaller than one-fourth. Table 2 shows a list of the values of that ratio for different helicases. Notice that for the few helicases for which that ratio and the ssNA translocation rate (Table 1) has been measured (RecQ, T7 gp4 and T4 gp41) both ways of classifying the enzymes agree.
, while a passive helicase will have an unwinding ratio on GC- and AT-rich regions smaller than one-fourth. Table 2 shows a list of the values of that ratio for different helicases. Notice that for the few helicases for which that ratio and the ssNA translocation rate (Table 1) has been measured (RecQ, T7 gp4 and T4 gp41) both ways of classifying the enzymes agree.
Table 2.
Unwinding rates for AT- and GC-rich sequences
| Helicase | 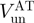 |
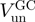 |
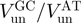 |
|---|---|---|---|
| (bp/s) | (bp/s) | ||
| T7 gp4 | ∼60 (13) | ∼10 (13) | ∼0.17 |
| T4 gp41a | 190 | 24 | ∼0.15 |
| NS3b | 62 (12) | 22 (12) | ∼0.35 |
| RecQa | 90 | 60 | ∼0.7 |
aRates obtained in this work at 9 pN of applied force.
bValues reported at ∼10 pN of applied force.
The existence of different helicase mechanisms might be a strategy that has evolved to satisfy the different functional requirements of these enzymes. Remarkably, studies on different replicative DNA helicases [such as T7 gp4 (10), T4 gp41 (11) or E. coli DNA-B (29)] find that these helicases are essentially passive: they unwind much slower than they translocate on ssDNA and display unwinding rates that are strongly force- and sequence-dependent. However, at the replication fork, when coupled to the full replisome machinery, these helicases move at much higher rates (32–34). The presence of the other replisomal proteins must then affect their unwinding behaviour increasing their unwinding rate. Indeed, in the T7 system the presence of a polymerase synthesizing a new strand in the wake of the gp4 helicase allows gp4 to reach its maximal rate given by  (33). This increase in helicase rate might be due to a change in the helicase–DNA fork interaction potential or to the trapping of the ssDNA bases by the polymerase preventing helicase slippage. Similar helicase–polymerase coupling has also been observed in other systems (29,35,36). The ability of other replisomal proteins to increase the helicase rate of unwinding might be a strategy evolved to co-ordinate leading and lagging strand synthesis and to provide the tight coupling that prevents replisome disassembly.
(33). This increase in helicase rate might be due to a change in the helicase–DNA fork interaction potential or to the trapping of the ssDNA bases by the polymerase preventing helicase slippage. Similar helicase–polymerase coupling has also been observed in other systems (29,35,36). The ability of other replisomal proteins to increase the helicase rate of unwinding might be a strategy evolved to co-ordinate leading and lagging strand synthesis and to provide the tight coupling that prevents replisome disassembly.
On the other hand, results reported on various DNA repair helicases [such as UvrD (9) and RecQ] show that these helicases are essentially active ones: they unwind DNA at their maximal rate irrespective of force or sequence. The unwinding activity of these enzymes is co-ordinated to the activity of other proteins (such as UvrA, UvrB and UvrC for Uvrd or Topo III for RecQ). However, in contrast to replicative helicases, DNA repair helicases mostly work physically uncoupled from their macromolecular partners. In absence of accessory proteins that may act as processivity factors (36) an active helicase mechanism appears optimal for efficient unwinding.
SUPPLIMENTARY DATA
Supplimentary Data are available at NAR Online.
FUNDING
Agence Nationale de Recherche; Human Frontiers Science Program RGP0003/2007-C; EU (BioNanoSwitch); NanoSci-Era contract MolMachines. Funding for open access charge: Human Frontiers Science Program.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank M.M. Spiering and S. Benkovic for preparing the DNA substrate and the gp41 protein and T. Lionnet for initial discussions on the work. We also would like to thank S. Kowalczykowski and O. Saleh for reading the manuscript.
REFERENCES
- 1.Lohman TM, Bjornson KP. Mechanisms of helicase-catalysed unwinding. Annu. Rev. Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 2.West SC. DNA helicases: new breeds of translocating motors and molecular pumps. Cell. 1996;86:177–180. doi: 10.1016/s0092-8674(00)80088-4. [DOI] [PubMed] [Google Scholar]
- 3.Soultanas P, Wigley DB. Unwinding the ‘‘gordian knot’’ of helicase action Trends Biochem. Sci. 2001;26:47–54. doi: 10.1016/s0968-0004(00)01734-5. [DOI] [PubMed] [Google Scholar]
- 4.Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–W581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bockelmann U, Essevaz-Roulet B, Heslot F. Molecular Stick-Slip Motion Revealed by Opening DNA with Piconewton Forces. Phys. Rev. Lett. 1997;79:4489–4492. [Google Scholar]
- 6.Liphardt J, Onoa B, Smith SB, Tinoco I, Jr, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 7.Wuite GJL, Smith SB, Young M, Keller D, Bustamante C. Single-molecule studies of the effect of template tension on T7 DNA polymerase activity. Nature. 2000;404:103–106. doi: 10.1038/35003614. [DOI] [PubMed] [Google Scholar]
- 8.Maier B, Bensimon D, Croquette V. Replication by a single DNA polymerase of a stretched single-stranded DNA. Proc. Natl Acad. Sci. USA. 2000;97:12002–12007. doi: 10.1073/pnas.97.22.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun B, Wei KJ, Zhang B, Zhang XH, Dou SX, Li M, Xi XG. Impediment of E. coli UvrD by DNA-destabilizing force reveals a strained-inchworm mechanism of DNA unwinding. EMBO. 2008;27:3279–3287. doi: 10.1038/emboj.2008.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson DS, Bai L, Smith BY, Patel SS, Wang MD. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell. 2007;129:1299–1309. doi: 10.1016/j.cell.2007.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lionnet T, Spiering MM, Benkovic SJ, Bensimon D, Croquette V. Real-time observation of bacteriophage T4 gp41 helicase reveals an unwinding mechanism. Proc. Natl Acad. Sci. USA. 2007;104:19790–19795. doi: 10.1073/pnas.0709793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng W, Dumont S, Tinoco IJ, Bustamante C. NS3 helicase actively separates RNA strands and senses sequence barriers ahead of the opening fork. Proc. Natl Acad. Sci. USA. 2007;104:13954–13959. doi: 10.1073/pnas.0702315104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donmez I, Rajagopal V, Jeong Y, Patel SS. Nucleic acid unwinding by hepatitis C virus and bacteriophage T7 helicases is sensitive to base pair stability. Nucleic Acids Res. 2007;282:21116–21123. doi: 10.1074/jbc.M702136200. [DOI] [PubMed] [Google Scholar]
- 14.Valentine AM, Ishmael FT, Shier VK, Benkovic SJ. A zinc ribbon protein in DNA replication: primer synthesis and macromolecular interactions by the bacteriophage T4 primase. Biochemistry. 2001;40:15013–15085. doi: 10.1021/bi0108554. [DOI] [PubMed] [Google Scholar]
- 15.Xu HQ, Deprez E, Zhang AH, Tauc P, Ladjimi MM, Brochon JC, Auclair C, Xi XG. The Escherichia coli RecQ helicase functions as a monomer. J. Biol. Chem. 2003;278:34925–34933. doi: 10.1074/jbc.M303581200. [DOI] [PubMed] [Google Scholar]
- 16.Manosas M, Spiering MM, Zhuang Z, Benkovic SJ, Croquette V. Coupling DNA unwinding activity with primer synthesis in the bacteriophage T4 primosome Nat. Chem. Biol. 2009;5:904–912. doi: 10.1038/nchembio.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gosse C, Croquette V. Magnetic tweezers: micromanipulation and force measurement at the molecular level. Biophys. J. 2002;82:3314–3329. doi: 10.1016/S0006-3495(02)75672-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Betterton MD, Julicher F. Opening of nucleic-acid double strands by helicases: active versus passive opening. Phys. Rev. E. 2005;71:11904–11911. doi: 10.1103/PhysRevE.71.011904. [DOI] [PubMed] [Google Scholar]
- 19.Cocco S, Marko JF, Monasson R. Slow nucleic acid unzipping kinetics from sequence-defined barriers Eur. Phys. J. E. 2003;10:153–161. doi: 10.1140/epje/e2003-00019-8. [DOI] [PubMed] [Google Scholar]
- 20.Gueron M, Leroy JL. Studies of basepair kinetics by NMR measurement of proton exchange Meth. Enzymol. 1995;261:383–413. doi: 10.1016/s0076-6879(95)61018-9. [DOI] [PubMed] [Google Scholar]
- 21.Strick TR, Allemand J-F, Bensimon D, Bensimon A, Croquette V. The elasticity of a Single Supercoiled DNA Molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 22.Tomko EJ, Fischer CJ, Niedziela-Majka A, Lohman TM. A nonuniform steppingmechanism for E. coli UvrD monomer translocation along single-stranded DNA. Mol. Cell. 2007;26:335–347. doi: 10.1016/j.molcel.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dillingham JMS, Wigley DB, Webbs MR. Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry. 2004;39:205–212. doi: 10.1021/bi992105o. [DOI] [PubMed] [Google Scholar]
- 24.Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng W, Tinoco I, Jr, Bustamante C. Direct observation of NS3 substeps at single base pair resolution. Biophys.J. 2009;96:367a. [Google Scholar]
- 26.Perkins TT, Li HW, Dalal RV, Gelles J, Block SM. Forward and reverse motion of single RecBCD molecules on DNA. Science. 2004;86:1640–1648. doi: 10.1016/S0006-3495(04)74232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeong Y-J, Levin MK, Patel SS. The DNA-unwinding mechanism of the ring helicase of bacteriophage T7. Proc. Natl Acad. Sci. USA. 2004;101:7264–7269. doi: 10.1073/pnas.0400372101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim DE, Narayan M, Patel SS. T7 DNA Helicase: A molecular motor that processively and unidirectionally translocates along single-stranded DNA. J. Mol. Biol. 2002;321:807–819. doi: 10.1016/s0022-2836(02)00733-7. [DOI] [PubMed] [Google Scholar]
- 29.Kim S, Dallmann HG, McHenry CS, Marians KJ. Coupling of a replicative polymerase and helicase: a τ-Dna-B interaction mediates rapid replication fork movement. Cell. 1996;84:643–650. doi: 10.1016/s0092-8674(00)81039-9. [DOI] [PubMed] [Google Scholar]
- 30.Brendza KM, Wei C, Fischer CJ, Chesnik MA, Niedziela-Majka A, Lohman TM. Autoinhibition of Escherichia coli Rep monomer helicase activity by its 2B subdomain. Proc. Natl Acad. Sci. USA. 2005;102:10076–10081. doi: 10.1073/pnas.0502886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischer CJ, Maluf NK, Lohman TM. Mechanism of ATP-dependent Translocation of E. coli UvrD Monomers Along Single-stranded DNA. J.Mol.Biol. 2004;344:1287–1309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 32.Chandler M, Bird R, Caro L. The replication time of the Escherichia coli K12 chromosome as a function of cell doubling time. J. Mol. Biol. 1975;94:127–131. doi: 10.1016/0022-2836(75)90410-6. [DOI] [PubMed] [Google Scholar]
- 33.Stano NM, Jeong Y-J, Donmez I, Tummalapalli P, Levin MK, Patel SS. DNA synthesis by a polymerase provides the driving force to accelerate DNA unwinding by a helicase. Nature. 2005;435:370–373. doi: 10.1038/nature03615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinha NK, Morris CF, Alberts BM. Efficient in vitro replication of double-stranded DNA templates by a purified T4 bacteriophage replication system. J. Biol. Chem. 1980;255:4290–4293. [PubMed] [Google Scholar]
- 35.Delagoutte E, Von Hippel PH. Molecular mechanisms of the functional coupling of the helicase (gp41) and polymerase (gp43) of bacteriophage T4 within the DNA replication fork. Biochemistry. 2002;40(14:4459–4477. doi: 10.1021/bi001306l. [DOI] [PubMed] [Google Scholar]
- 36.Von Hippel PH, Delagoutte E. A general model for nucleic acid helicases and their coupling within macromolecular machines. Cell. 2002;104:177–190. doi: 10.1016/s0092-8674(01)00203-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.