Abstract
The RNA binding protein Larp1 was originally shown to be involved in spermatogenesis, embryogenesis and cell-cycle progression in Drosophila. Our data show that mammalian Larp1 is found in a complex with poly A binding protein and eukaryote initiation factor 4E and is associated with 60S and 80S ribosomal subunits. A reduction in Larp1 expression by siRNA inhibits global protein synthesis rates and results in mitotic arrest and delayed cell migration. Consistent with these data we show that Larp1 protein is present at the leading edge of migrating cells and interacts directly with cytoskeletal components. Taken together, these data suggest a role for Larp1 in facilitating the synthesis of proteins required for cellular remodelling and migration.
INTRODUCTION
Normal epithelial cells display a regular structure, are joined to neighbouring cells, have a characteristic apico-basal polarity and minimal ability to migrate (1). In physiological states such as embryonic development, wound healing and immunity, epithelial cells undergo cytoskeletal and cell signalling changes to become motile (2–4). This also occurs in pathological processes such as cancer invasion and metastasis where cells acquire migratory or mesenchymal characteristics. The complex alterations in cell signalling required for this process [reviewed in ref. (5)] result in a series of cellular changes including detachment from neighbouring cells, loss of apico-basal polarity, formation of a spindle-shaped morphology and the ability to migrate through the extracellular matrix (ECM) (6). Migration requires the activation of localized actin polymerization at the ‘leading edge’ of the cell to form protrusive structures beneath the plasma membrane, such as flat lamellipodia and finger-like filopodia (7,8). In addition, actin-based stress fibres and focal adhesions are formed, providing attachment and traction between the cell and the ECM. Forward gliding in the direction of the leading edge is achieved through interactions between these actin filaments and contractile proteins such as myosin II (9). Disruption of intercellular junctions allows detachment from neighbouring cells, and digestion of the surrounding ECM by proteolytic enzymes permits 2D (amoeboid) migration (10,11). Additionally, dynamic protrusive structures called ‘invasive pseudopodia’ form, which facilitate 3D (mesenchymal) movement (12).
The acquisition of a migratory phenotype is triggered by extracellular stimulants such as hepatocyte growth factor and epithelial growth factor (13). The process linking these signals to co-ordinated formation of invasive components remains poorly understood. The major downstream regulators of migration are the Rho GTPases which primarily regulate lamellipodia and filopodia formation via WAVE and formin (mDia/Drf) pathways, respectively (14). Actin-binding and capping proteins regulate actin filament assembly and disassembly (15,16). Cell types vary in their proportions of lamellipodia and filopodia, and some studies suggest they are generated independently (17).
Many of these processes are controlled at the level of messenger RNA (mRNA) translation. This occurs in three steps. The first or ‘initiation’ step is rate-limiting and requires the binding of the eukaryotic initiation factor (eIF)4F complex to the 5′-end of the mRNA. The complex contains the cap-binding protein eIF4E, a scaffold protein, eIF4G and a helicase, eIF4A, which unwinds the RNA (18). Assembly of the cap complex and a second pre-initiation complex facilitates recruitment of the 40S ribosomal subunit and scanning to the first AUG or ‘start’ codon. Global regulation of mRNA translation is mainly achieved by directly altering the phosphorylation of eIFs or their binding partners. As well as global regulation, transcript-specific regulation of translation occurs, mediated by interactions between trans-acting factors and sequences within the 5′ and/or 3′ mRNA untranslated regions (UTRs) (19). Inactive mRNA transcripts can also be shuttled to specific cellular destinations before protein synthesis is activated; this spatio-temporal regulation of mRNA translation has been demonstrated in embryogenesis and patterning in Drosophila (20) and in meiotic spindle assembly and chromosome condensation in Xenopus (21). In mammalian axons, mRNAs encoding β-actin are stored in an inactive state and transported to distal neuronal processes following axonal injury to facilitate regeneration, achieved through the binding of CPE-proteins to cis-acting sequences within their 3′ or 5′ UTR (22). In addition to a role in development and axonal recovery, site-specific protein synthesis is likely to be required for other asymmetric cellular processes.
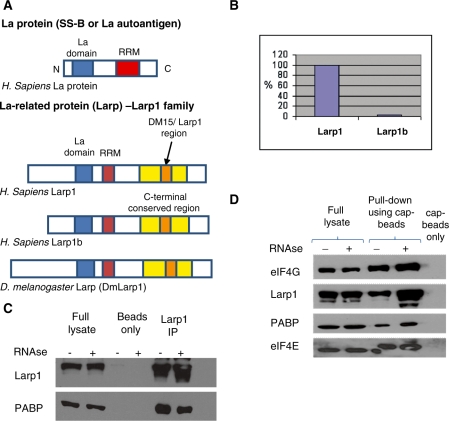
Larp1 was first described in Drosophila and shown to be required for spermatogenesis, embryogenesis and cell cycle progression (23–25). Drosophila Larp1 interacts with poly A binding protein (PABP) (25) suggesting the phenotype observed in Larp1 mutants may be due to defective mRNA translation or regulation, supported by the finding in Caenorhabditis elegans that LARP-1 is an RNA-binding protein (26). Although five Larp proteins exist in the human genome, only two carry a combination of a La domain, an RNA recognition motif and a DM15/Larp1 region (Figure 1A); Larp1 and Larp1b (previously named Larp2) (27). Larp 1 (positioned at 5q34) encodes a 1097 amino-acid protein with 50% identity to Drosophila Larp1, and Larp1b (at 4q28) encodes a 915 amino acid protein with 46% identity to Drosophila Larp1. Here we demonstrate that Larp1 exists in complexes with both PABP and eIF4E, and is required for ordered mitosis, cell survival and migration.
Figure 1.
(A) Larp proteins are conserved in metazoans and members of the Larp1 family contain an N-terminal La domain (similar to La proteins) and a C-terminal conserved or Larp1 region containing DM15 tandem repeat regions (33). There is a single ‘Larp’ gene in D. melanogaster [now termed DmLarp1 (27)] and two homologues in humans, Larp1 and Larp1b (previously termed Larp2) encoded by genes located at 5q33.2 and 4q28.2, respectively. Larp1 and Larp1b share 50 and 59% identity with Dm Larp1, respectively. There are two isoforms of Larp1 protein of 1019 and 1096 amino acids, respectively, and three isoforms of Larp1b protein of 914, 512 and 358 amino acids. (B) Of the two human Larp proteins, Larp1 is more abundant than Larp1b as shown by qPCR in HeLa cells (after normalization to housekeeping genes). The relative expression level of Larp1b (when standardized to 100% Larp1) was 2.5%, therefore Larp1 is 40-fold more abundant. (C) Immunoprecipitation, using antibodies to Larp1 and PABP and protein G sepharose beads showed an interaction between Larp1 and PABP, as demonstrated using western blotting of the immunoprecipitate. This interaction was not observed using beads alone (control) but was not disrupted following treatment with RNAse A. (D) Western blot of protein pull-down using 7-Methyl-GTP cap-binding sepharose beads demonstrates that Larp1 exists in complex with eIF4E. As expected, PABP and eIF4G were also ‘pulled down’ from the lysate using the cap beads in an association that was not disrupted after RNAse treatment. Additionally, loss of Larp1 (by RNAi) did not remove the interaction between either PABP and eIF4E or eIF4G and eIF4E (data not shown), suggesting these interactions are non-Larp1 dependent.
MATERIALS AND METHODS
Cell culture
HeLa cells were maintained in DMEM supplemented with l-glutamine (Gibco, 2mM), FCS (10%, First Link UK Ltd.) and PenStrep (Gibco, 50 U/ml). PE01 and PE04 cells were a kind gift from Dr Simon Langdon (CRUK, Edinburgh) and maintained in RPMI supplemented as earlier. The cell lines were kept at 37°C at 5% CO2.
Qualitative real time PCR
RNA isolated from samples underwent reverse transcription. Total RNA (1 µg) was made up to a volume of 12.7 µl with diethylpyrocarbonate (DEPC) water. The samples were then incubated at 65°C for 5 min followed by incubation at 37°C for 2 min. RT-PCR Mix (7.3 µl) [4 µl of 5× MMLV RT buffer, 2 µl dNTP’s (4 mM), 1 µl oligo dT15 (10 µg/ml) and 0.3 µl MMLV reverse transcriptase (5 U/µl)] was then added to the RNA solution and mixed by pipetting. This was incubated at 37°C for 1 h followed by incubation at 95°C for 5 min. cDNA was stored at −20°C. cDNA from untransfected HeLa cells was used to make a set of standards ranging from 0.2 to 0.000064. The sample cDNA was diluted 1: 50 and 2 µl of either sample cDNA or the standards was added to each well of a 96-well plate along with 8 µl of master mix [1.8 µl DEPC water (Bioline, BIO-38031), 5 µl Syber green (Invitrogen, 11733-038), 0.2 µl ROX dye (Invitrogen, 11733-038) and 0.5 µl of each the forward and reverse primer (stock 100 µM) for each gene of interest]. The plate was sealed with a clear plastic film and centrifuged for 2 min at 1200 r.p.m. The plate was then placed in an Applied biosystems 7900ht thermal cycler using the following settings: 50°C for 2 min, 95°C for 2 min, then 40 cycles of 95°C for 3 s and 60°C for 30 s as well as a dissociation step. Standard curves were created for all genes. Sample RNA levels were normalized against results obtained for the housekeeping genes. For this experiment glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and the 18S ribosomal subunit (18S) were used as housekeeping genes.
Larp1 primers: For (CAAGACACAGTTCAAACCCA), Rev (GTTTCCGCTCATTAAGGCAG);
Larp2 primers: For (AGACAUUCCUCUACUUCUG), Rev (GAACCAGAACAAGAAGAAC);
GAPDH primers: For (CATGGCCTCCAAAGGAGTAAGAC), Rev (TCTCTTCCTCTTGTGCTCTTGCT);
18S primers: For (CACGCCAGTACAAGATCCCA), Rev (CAGTCGCTCCAGGTCTTCAC).
Immunoprecipitation
HeLa cells were grown to 70–80% confluency in 10-cm dishes. The cells were then washed once with PBS followed by the addition of 1 ml of lysis buffer [1% Triton X-100, 150 mM NaCl, 50 mM Tris–HCl (pH 7.2), 0.2 mM Na3VO4, 50 mM NaF, 2 mM EDTA, 1 mM PMSF, 40 µl/ml 25× protease inhibitor cocktail (Roche, 11697498001) and 10 µl/ml 100× phosphatase inhibitor cocktail (Calbiochem, 524625)] ± RNAse A 2 µg/ml (Sigma, R6148) for 30 min at 4°C. The lysates were then centrifuged for 20 min at 13–14 000 r.p.m. at 4°C. The supernatant was removed and aliquoted as follows: 20 µl was used for direct loading control and 400 µl each was placed in a tube for IP sample and extract only. Lysis buffer (400 µl) was placed in a tube for antibody only control. Larp1 antibody (3 µl) was added to the IP sample and antibody-only control tubes and incubated rotated overnight at 4°C. Following this, 20–30 µl of pre-washed protein G sepharose beads was added to the IP sample, extract only and antibody only control tubes and rotated for 1 h at 4°C. The samples were then centrifuged for 2 min at 10 000 r.p.m. at 4°C and washed twice with lysis buffer centrifuging as above, followed twice with PBS. Sample buffer was added to direct loading control, IP sample, extract only and antibody only control and boiled for 5 min at 95°C followed by centrifugation at 10 000 r.p.m. for 2 min. Samples were resolved by SDS–PAGE and visualized by immunoblotting.
7-methyl GTP sepharose beads pull-down
HeLa cells were grown to 70–80% confluency in 10 cm dishes. The cells were then washed once with PBS followed by the addition of 600 µl lysis buffer [1% Triton X-100, 150 mM NaCl, 50 mM Tris–HCl (pH 7.2), 0.2 mM Na3VO4, 50 mM NaF, 2 mM EDTA, 1 mM PMSF, 40 µl/ml 25× protease inhibitor cocktail (Roche, 11697498001) and 10 µl/ml 100× phosphatase inhibitor cocktail (Calbiochem, 524625)] ± RNAse A 2 µg/ml (Sigma, R6148) for 30 min at 4°C. The lysates were then centrifuged for 20 min at 13–14 000 r.p.m. at 4°C. The supernatant was removed and aliquoted as follows: 20 µl was used for a direct loading control and 500 µl was placed in a clean labelled tube for the pull-down. Pre-washed (30 µl) 7-methyl GTP sepharose beads (GE Healthcare, 27-5025-01) was added to the 500 µl of lysate and rotated overnight at 4°C. The samples were then centrifuged for 2 min at 10 000 r.p.m. at 4°C and washed twice with lysis buffer centrifuging as above, followed twice with PBS. Sample buffer was then added to all the tubes and boiled for 5 min at 95°C followed by centrifugation at 10 000 r.p.m. for 2 min. Samples were then resolved by SDS–PAGE and visualized by immunoblotting.
RNAi
HeLa cells were seeded in either six-well plates (for protein lysates) or 10 cm dishes (for RNA lysates) and allowed to reach 70–80% confluency. A transfection mixture was made as described: 100 nM of a control scramble siRNA (Eurofins: sense 5′-GGUCCGGCUCCCCCAAAUG-3′) or 100 nM of a mixture of two Larp1 siRNA sequences (Eurofins: first, sense 5′-GAAUGGAGAUGAGGAUUGCTT-3′; second, sense 5′-AGACUCAAGCCAGACAUCATT-3′) was diluted 1: 10 with siRNA buffer (Eurofins) and then diluted 1: 2 with optimem (Gibco, 51985-026). This was left for 5 min during which time transfection reagent was diluted 1: 40 with optimem and also left for 5 min. Equal volumes of siRNA mixture and transfection reagent mixture were combined and left for 20 mins at room temperature before being diluted 1: 12.5 with cell media and added to each well. Cells were left for 24 h before a repeat transfection was performed as described earlier. Cells were then left for a further 24 h. To create cell lysates the cells were washed with PBS before being treated with 200 µl lysis buffer [1% Triton X-100, 150 mM NaCl, 50 mM Tris–HCl (pH 7.2), 0.2 mM Na3VO4, 50 mM NaF, 2 mM EDTA and 1 mM PMSF] scraped and collected. RNA was extracted using the Absolutely RNA Miniprep kit (Stratagene, 400800) as per manufacturer’s instructions.
Antibodies and western blotting
Proteins were separated by SDS–PAGE using Amersham MINIVE kit and then transferred onto a nitrocellulose membrane followed by immunoblotting. Non-specific binding sites on the nitrocellulose membranes were blocked overnight with 5% milk in PBS-T (PBS:1% Tween 20). Antibodies used were as follows: Larp 1 (SDI, 1 : 5000), PABP (Abcam, 1 : 2000), eIF4E, 4E-BP1, p-eIF2α and eIF2α (cell signalling, 1 : 1000). Additional antibodies to p-eIF4E, eIF4E, eIF4G and actin were donated by Simon Morley (University of Sussex; used at 1 : 1000). Binding of the horseradish peroxidase conjugated secondary antibodies was visualized by enhanced chemiluminescence according to the manufacturer’s instructions and exposed to X-ray film.
Mass spectrometry
Immunoprecipitates were generated as described earlier and separated by SDS–PAGE gel electrophoresis. Protein bands were excised from Coomassie-stained gels, digested with trypsin and resulting peptides analysed by LC-MS/MS using a nanoLC (Waters) coupled to a Q-TOF2 mass spectrometer (Waters). Uninterpreted fragmentation data was used to search the NCBI database using the MASCOT search engine (Matrix Science) allowing identification of the protein(s) within a gel band.
Methionine incorporation
The overall rate of protein synthesis in intact cells was measured by the incorporation of [35S] methionine into trichloroacetic acid-insoluble material. After pulse-labelling for 1 h with up to 15 mCi/ml of the radioactive amino acid (in the presence of the normal level of methionine in the cell culture medium), cells were briefly centrifuged, washed once in cold phosphate-buffered saline containing 100 mM unlabelled methionine, dissolved in 0.3 M NaOH and precipitated with 10% trichloroacetic acid in the presence of 0.5 mg bovine serum albumin carrier protein. Precipitates were washed with 5% trichloroacetic acid and industrial methylated spirit and the radioactivity determined by scintillation counting.
Flow cytometry
HeLa cells were incubated for 3 days at 37°C, the medium was then removed from each well and placed in a 15 ml tube. Remaining cells were then trypsinized to detach them from the well. This was achieved by washing the cells first with 0.5 ml 1× trypsin and then adding 1 ml of 1× trypsin to each well, and incubating for 5 min at 37°C. When the cells became detached they were removed and added to the media in the 15 ml tube. This was then centrifuged at 1000 r.p.m. for 5 min. After centrifugation, the cell pellet was washed in 1× PBS solution and fixed by resuspending in 1 ml 70% ice-cold ethanol, added drop-wise, while vortexing. Samples were left on ice for at least 30 min, and then centrifuged at 1000 r.p.m. for 5 min. The ethanol solution was then removed and the pellet resuspended in 500 µl PBS with 200 µg/ml RNase A (Qiagen), followed by 500 µl PBS with 200 µg/ml of propidium iodide (Sigma). Samples were then incubated in a 37°C waterbath for 30 min and resuspended using a syringe and 25G needle prior to FACS analysis.
Cell staining
HeLa cells were grown to ∼50% confluency on coverslips in six-well plates. Cells were then transfected with Larp1 RNAi or control RNAi (as stated earlier) 48 and 24 h before staining. FLAG-Larp plasmid for Larp1 overexpression was introduced to the cells using Mega Trans 1.0 transfection reagent (Origene) according to the manufacturer’s protocol 24 h before staining. On the day of staining cells were rinsed with PBS and incubated with PHEM fixative (4% paraformaldehyde, 10 mM EGTA pH 6.8, 4 mM MgSO4, 60 mM PIPES pH 6.8, 30 mM HEPES pH 7.0) for 10 min. Cells were then washed in PBS twice before being blocked and permeabilized in PBSTB (PBS, 0.1% Triton-X, 1% BSA) for 45 min. Primary antibodies were diluted as detailed below in PBSTB and applied to cells and left overnight at 4°C. Following overnight incubation, cells were washed several times in PBS before being incubated with a secondary antibody diluted in PBSTB. After 1 h incubation, cells were washed three times with PBS and mounted on slides with 25 µl Vectashield mounting medium containing 49,6-diamidino-2-phenylindole (Vector Laboratories). Actin was visualized with the aid of Phalloidin-TRITC (Fluka) diluted in PBSTB at 1: 500 and added at the same time as the secondary antibodies.
For the mitotic staining the cell media was removed and cells fixed for 15 min with 4% formalin in PBS. Cells were washed three times for 5 min with PBST (PBS + 0.1% Triton X-100) followed by 1 h of blocking with PBNT (PBST + 4% normal goat serum). Primary antibody against Cyclin B was applied over night at 4°C followed by three washes of 5 min. Anti-rabbit Alexa594 secondary antibody was applied for 2 h at room temperature followed by incubation with Hoechst 33258 (1 : 1000 dilution) for 10 min. Cells were washed three times for 5 min with PBST before being analysed.
| Target | Primary antibody | Secondary Antibody |
|---|---|---|
| Larp1 | Larp1 (Strategic Diagnostic) 1: 100 | Anti-Rabbit Cy5, (Invitrogen) 1: 500 |
| Microtubules | YL1/2 αTubulin antibody (Oxford Biotechnologies) 1: 50 | Anti-rat Alexa 488, (Molecular Probes) 1: 400 |
| Centrosomes | GTU88 γ-Tubulin (Sigma) 1: 200 | Anti-mouse Rhodamine, (JIR) 1: 100 |
| Cyclin B | Cyclin B (Sc-752 Santa Cruz) 1: 100 | Anti-rabbit Alexa 594, (Molecular Probes) 1: 500 |
| Total actin | Alexa Fluor Phalloidin (A22283-Invitrogen) 1: 500 | – |
| β-actin | β-actin (Ab8227-AbCam) 1: 100 | Anti-rabbit Alexa 594, (Molecular Probes) 1: 500 |
| γ-actin | γ-actin (A8481- Sigma) 1: 100 | Anti-mouse Alexa 488 (Molecular Probes) 1: 500 |
Migration assay
HeLa cells were seeded in six-well plates and allowed to reach ∼70% confluency. The cells were then treated with RNAi against Larp1 or control RNAi as mentioned above. After two transfections of 24 h each, plate was washed once in PBS which was removed and replaced with low serum media DMEM without phenol red (Gibco) supplemented with 1% FCS and 25 mM sodium pyruvate for 6 h. Wounds were made by scratching cells using a sterile pipette tip and evaluated under phase contrast microscopy at 0, 12, 15 and 18 h. To generate confocal images, cells were seeded and treated with control and Larp1 RNAi as above. At 12 h following scratch, cells were fixed, permeabilized and immunostained as described earlier.
Statistics
Statistical analysis was used to compare cell characteristics between treatment pairs, i.e. FLAG Larp1 versus transfection control and Larp1 RNAi versus control RNAi. The median (inter-quartile range) score was calculated for each variable. The P-value, a measure of difference between the variables was obtained using the non-parametric Mann–Whitney U test.
RESULTS
Human Larp1 interacts with PABP and eIF4E
The relative mRNA levels of Larp1 and Larp1b were assayed by Qualitative real time PCR (qPCR) (Figure 1B). We demonstrated that Larp1 was 40× more abundant than Larp1b and that siRNA to Larp1b did not cause a quantifiable phenotypic change in HeLa cells (data not shown). Thus the work described in this article is focused on Larp1. As Drosophila Larp1 has already been demonstrated to interact with Drosophila PABP, we investigated whether this interaction was also conserved in human cells. Larp1 interacting proteins were assessed in HeLa cells lysates by immunoprecipitation using protein-G sepharose beads coated with anti-Larp1 or anti-PABP antibodies (Figure 1C). Immunoprecipitated proteins were separated by SDS–PAGE and immunoblotted. This showed that PABP and Larp1 co-immunoprecipitated in an interaction that was not perturbed following RNAse treatment. This excludes an indirect interaction between the proteins via mRNA. To determine whether cap-binding proteins were present in complex with Larp1 and PABP, 7-methyl GTP sepharose beads were used. HeLa cell lysates were incubated with the 7-methyl GTP sepharose beads and proteins pulled down in this interaction were separated by SDS–PAGE and immunoblotted. As expected, eIF4E was bound to the 7-methyl GTP beads, but PABP, eIF4G and Larp1 were also present (Figure 1D). Neither the interactions between eIF4E and PABP or eIF4E and eIF4G were abolished after Larp1 RNAi (data not shown), suggesting these associations are non-Larp1 dependent.
Larp1 may have a role in translation initiation
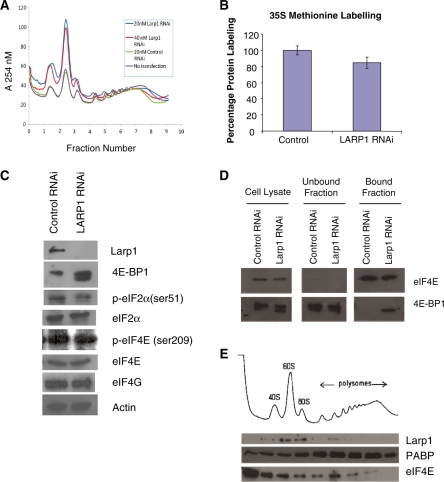
PABP is known to stimulate translation initiation by indirectly interacting with eIF4E (via an interaction with the cap-complex scaffold protein, eIF4G) and circularising the mRNA (28) and, in addition, PABP has been found to interact with polysomally-associated mRNAs (29) and it has a direct role in 60S subunit joining (30). To determine whether depletion of Larp1 might have an effect upon polysomal association of mRNAs, HeLa cells were transfected with Larp1 siRNA and the subsequent ribosome sedimentation profiles were examined two days following transfection (Figure 2A). Compared to controls (cells untransfected or transfected with ‘control’ siRNA) the profiles demonstrated an increase in the amount of subpolysome components (40S, 60S and 80S) and a relative decrease in polysomes after Larp1 RNAi (using 20 and 40 nM siRNA) treatment, suggesting that Larp1 may be involved either directly or indirectly in an early stage of translation initiation. To assess the extent of inhibition, global protein synthesis rates in cells treated with Larp1 siRNA and non-sense siRNA were compared using 35S methionine incorporation (Figure 2B). This showed a 15% reduction in total protein synthesis in cells which lacked Larp1. To determine whether levels of Larp1 had an effect on the expression or phosphorylation status of canonical initiation factors, western blotting analysis was performed on protein lysate samples obtained from HeLa cells treated with either Larp1 siRNA or non-sense siRNA. The only significant changes observed after loss of Larp1 was an increase in the hypophosphorylated form of 4E-BP1 (Figure 2C) and an increase in the binding of this protein to eIF4E (Figure 2D). This would have the net effect of reducing the amount of eIF4E available for eIF4F complex formation, in agreement with the observed reduction in total protein synthesis. To determine whether Larp1 may be directly involved in binding to the translational machinery sucrose density gradients were fractionated and western blotting was performed to determine the location of Larp 1 protein (Figure 2E). Larp 1 was found to be associated primarily with pre-polysomal subunits.
Figure 2.
(A) Ribosome sedimentation profiles using HeLa cells treated with either Larp1 siRNA (20 and 40 nM), control siRNA (20 nM) or untreated. Compared to controls, the profiles demonstrated a considerable increase in the ratio of the subpolysome components (40S, 60S and 80S) compared to polysomes after Larp1 siRNA (20 and 40 nM) treatment, indicating that loss of Larp1 results in defective translation initiation. (B) Global protein synthesis rates in cells treated with Larp1 siRNA and non-sense siRNA were compared using 35S methionine incorporation and show that there is a 15% reduction in total protein synthesis in cells which lack Larp 1. (C) Western blotting of HeLa cell lysates pre and post Larp1 RNAi probed with antibodies for translation initiation components. Loss of Larp1 did not alter levels of eIF4G, eIF4E or eIF2α, but was associated with increase in levels of hypophosphorylated 4E-BP1. (D) Western blotting following cap pull-down of HeLa lysates pre- and post-Larp1 RNAi showed increased association between eIF4E and 4E-BP1. (E) Western blotting of proteins precipitated from elute fractions obtained during ribosome sedimentation profiling showed a predominant association between Larp1 and the pre-polysome components, the 40S, 60S and 80S ribosome fractions.
Loss of Larp1 results in a cell cycle arrest and apoptotic phenotype
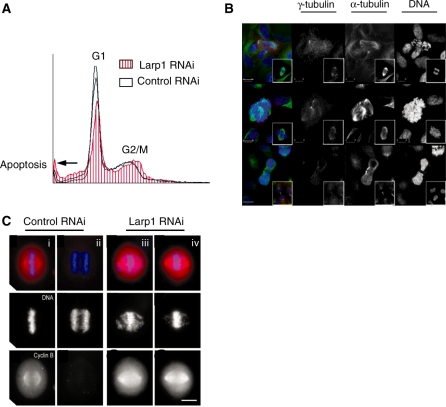
FACS profiling in HeLa cells treated with Larp1 or control siRNA demonstrated that Larp1 knockdown caused an accumulation of 4C cells (relative to 2C cells) consistent with G2/M arrest along with a sub G1 peak signifying cell death (Figure 3A). Immunostaining (here in U2OS cells) following Larp1 RNAi demonstrated (as previously observed in Drosophila Larp mutants) abnormal chromatin condensation, failure of bipolar mitotic spindle formation, abnormal centrosome migration, irregular nuclear morphology and defective cytokinesis (Figure 3B). This was quantified (Table 1) showing increased apoptosis, a greater number of chromosome defects and a mitotic arrest in cells treated with Larp1 siRNA when compared to controls. In addition, in control cells, chromosomes were aligned in a tight metaphase configuration [Figure 3C (i)] at which point Cyclin B levels were high. As they progressed into anaphase, Cyclin B was abruptly degraded [Figure 3C (ii)]. In contrast, the majority of Larp1 depleted mitotic cells showed chromosomes scattered on the mitotic spindle in the presence of elevated levels of Cyclin B [Figure 3C (iii)], suggesting defective chromosome congression or segregation and failure to exit mitosis via activation of the spindle assembly checkpoint.
Figure 3.
(A) Flow cytometry profile of HeLa cells following Larp1 RNAi treatment (red hatched) compared to control RNAi (black line). In cells treated with Larp1 siRNA, an increase in 4C relative to 2C cells is observed along with an increase in cell death (arrow). RNAi was performed using 20 nM Larp1 siRNA and 20 nM control siRNA. (B) Immunofluorescence showing U2OS cells during mitosis compared to controls (inset). Following knockdown of Larp1 after siRNA treatment, cells have a distinct ‘scattered chromosome’ appearance. Alpha-tubulin (green), γ-tubulin (red) and DNA (blue) (scale bar = 10 µm) compared to controls. (C) In control treated HeLa cells Cyclin B levels are elevated in metaphase (i) but are decreased following anaphase (ii). Following Larp1 RNAi, levels of Cyclin B remain high, showing the characteristic ‘scattered chromosome’ phenotype and without transition to anaphase (iii and iv) (scale bar = 10 µm).
Table 1.
Quantification of defects in HeLa cells following RNAi to Larp1, control dsRNA and transfection reagent alone
| Transfection reagent only control | Control (non-sense RNAi) | Larp1 RNAi | |
|---|---|---|---|
| Apoptosis (preG1) | 7.02% ± 0.84 | 7.8% ± 1.04 | 14.38% ± 1.93 |
| Mitotic index | 100% ± 0 | 100.8% ± 4.7 | 174.92% ± 20.18 |
| Cell density | 100% ± 0 | 75.8% ± 9.04 | 41.99% ± 4.35 |
| Chromosome defects | 2.14% ± 1.08 | 1.83% ± 0.67 | 6.33% ± 1.44 |
Following Larp1 RNAi, there is a greater percentage of sub G1 cells (indicative of apoptosis) and a higher number of cells with chromosome defects compared to controls. The mitotic index was also elevated following Larp1 RNAi.
Larp1 is localized to cell protrusions in HeLa cells and Larp1 over-expression causes multiple lamellopodia while loss of Larp1 results in an inhibition of cell migration
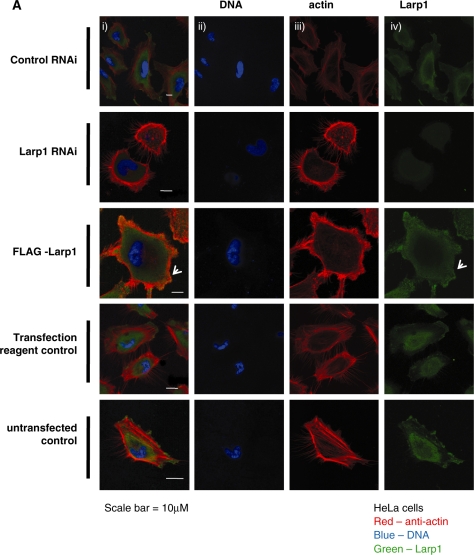
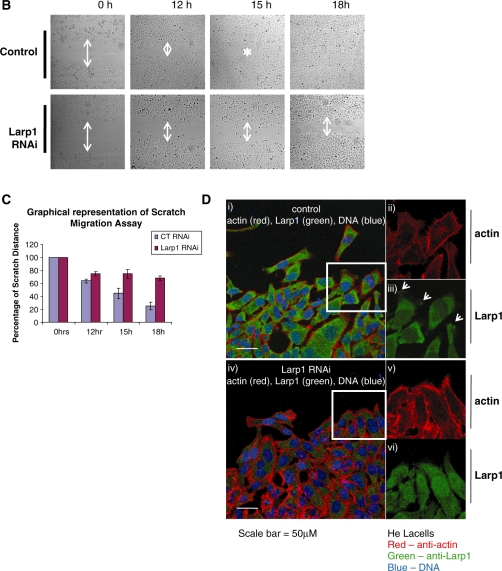
By immunostaining HeLa cells using anti-Larp1 antibody, we demonstrated the presence of Larp1 within the cytoplasm and at the leading edge of cells (Figure 4A). The presence of Larp1 at the leading edge was most pronounced in cells over-expressing Larp1 (FLAG-Larp1) and, when compared to controls, these cells demonstrated a greater number of lamellopodia containing co-localized actin and Larp1. To quantify, cells were measured in two dimensions (area) and the number of filopodia and lamellipodia were counted and data was analysed statistically (Table 2). Cells over-expressing Larp1 (FLAG-Larp1) were significantly smaller by area, had fewer filopodia (even when number was controlled for cell size) but a greater number of lamellipodia compared to transfection reagent-alone controls. In contrast, cells treated with Larp1 RNAi were larger by area, had a greater number of filopodia and fewer lamellipodia compared to cells treated with control RNAi. As lamellipodia are a feature of actively migrating cells, we explored the effect of Larp1 on cell motility using a scratch assay. A monolayer of HeLa cells was grown to confluence on a glass coverslip, scored to produce an open furrow using a pipette tip and then examined at a series of time points. In controls the wound gap was refilled by 18 h, but in cells treated with Larp1 RNAi there was little or no filling of the gap after 18 h (Figure 4B). This is demonstrated graphically (Figure 4C).This assay was repeated but cells at the scratch margin were fixed after 12 h and stained with antibodies to Larp1 and actin (total). Cells at the scratch furrow edge were then examined using confocal microscopy. This demonstrated a concentration of Larp1 at the cellular leading edge and within protrusions in the direction of cell travel i.e. towards the furrow (arrowheads, Figure 4D).
Figure 4.
(A) Immunofluorescence of HeLa cells following treatment with control RNAi, Larp1 RNAi and in cells transfected with Larp1 over-expressing construct (FLAG-Larp1) examined using confocal microscopy. Panels show images in combined colour channel (i), blue channel (DNA) (ii), red channel (actin) (iii) and green channel (Larp1) (iv). Larp1 over-expression (FLAG-Larp1) causes a mesenchymal-like phenotype, with cells displaying multiple lamellipodia (white arrow). Cells treated with Larp1 RNAi were larger and had more filopodia when compared to controls. (B) Scratch migration assay in HeLa cells treated with Larp1 siRNA or control siRNA control. An open furrow was generated by scratching confluent cells using a pipette tip. Confluency was restored in controls between 15 and 18 h. However, in cells treated with Larp1 siRNA, confluency was not restored after 18 h. The distance between furrow edges in the control and Larp1 siRNA treated cells was measured and is presented graphically in (C). In (D), cells were fixed and immunostained 12 h after scratch. Cells at the furrow edge were examined using confocal microscopy. This showed, in cells treated with control RNAi (top panel), a concentration of Larp1 within their leading edge (small arrows in magnified figures) in the direction of cell travel. This was not observed after Larp1 RNAi (bottom panel), where cells were closely adherent to each other without a clear direction of travel.
Table 2.
Phenotype quantification of cell size, filopodia number and number of projections in HeLa cells either over-expressing Larp1 (FLAG-Larp1), after treatment using transfection reagent alone, cells treated with Larp1 RNAi or control RNAi
| Control RNAi median (IQR) n = 192 | Larp1 RNAi median (IQR) n = 178 | P-value | FLAG Control median (IQR) n = 202 | FLAG Larp1 median (IQR) n = 234 | P-value | |
|---|---|---|---|---|---|---|
| Filopodia | 35 (23–53) | 61 (43–75) | <0.001 | 40 (25–55) | 35 (22–47) | 0.04 |
| Area | 2000 (1440–2800) | 2320 (1750–3444) | 0.006 | 2400 (1600–3400) | 1500 (1000–2275) | <0.001 |
| Protrusions | 0 (0–1) | 0 (0–0) | 0.005 | 0 (0–2) | 2 (0–3) | <0.001 |
| Filopodia / area | 0.016 (0.011–0.029) | 0.026 (0.015–0.038) | <0.001 | 0.018 (0.009–0.026) | 0.023 (0.013–0.036) | 0.001 |
| Protrusions / area | 0.000 (0.000–0.001) | 0.000 (0.000–0.000) | 0.004 | 0.001 (0.000–0.002) | 0 (0–6) | <0.001 |
Data analysed using Mann–Whitney U test.
Larp1 interacts with cytoskeletal proteins
To determine with which other proteins Larp1 interacted, immunoprecipitation experiments were performed using cell lysates incubated with protein-G sepharose beads coated with anti-Larp1 antibody. The subsequent immunoprecipitates were analysed by mass spectrometry (Table 3). To control for PABP-interacting proteins, a second set of immunoprecipitates were analysed using beads coated with anti-PABP antibody, and overlapping candidates identified by mass spectrometry were excluded. This showed that Larp1 was in a complex with cytoskeletal proteins, as well as proteins required for transcription, protein synthesis, cell signalling and metabolism. Cytoskeletal proteins identified as being in complex with Larp1 included tubulin, keratins, actin and myosin as well as actin-binding proteins: plectin, spectrin, actinin and tropomyosin and actin-capping proteins CapZA and CapZB. The tight junction protein, zona occludens 1 (ZO-1) was also identified along with other cell–cell junction proteins such as tropomyosin and spectrin.
Table 3.
Proteins identified by mass spectrometry as interacting with Larp1 using lysates of two isogenic epithelial ovarian cancer cell lines, PE01 and PE04
| Cytoskeleton | Protein synthesis |
|---|---|
| (β actin | hnRNPE, G, Q2 |
| α2 (smooth muscle) actin | 40S ribosome proteins S14, S25 |
| actin prepeptide | 60S ribosome proteins L22, L23A |
| α-1, α-4 actinin | nucleolin |
| CAPZA and CAPZB | PABP |
| α-tubulin | RNA (DEAD box) helicase |
| keratins 1, 2e, 4, 5, 6B, 7, 8, 9, 10, 18, 19 | |
| LIM-domain containing protein | Transcription / chromatin |
| myosins 1, 2, 6, 9 | histones 1B, 2A, 4, testicular H1 |
| plectin 1 | Thymopoietin α (LAP2 alpha) |
| α-2 and β-spectrin | YB1 |
| Tropomysin | |
| zona occludens 1 | |
| Chaperone / cell cycle / cell signalling | |
| Metabolism | Hsp70 and 90 |
| Lactate dehydrogenase A | Protein phosphatase 1 |
| Phenylalanine hydroxylase | Ig κ light chain |
| mitotic spindle associated protein | |
| annexin A2 | |
| EF hand domain family D2 | |
| 14-3-3 σ and θ | |
| ubiquitin | |
| SUMO | |
| Clathrin | |
| SLC25A4 |
Candidate proteins identified using PABP controls were excluded. Proteins were grouped by function. This showed that Larp1 predominantly interacts with cytoskeletal proteins, but also proteins required for protein synthesis, chaperones, cell signalling, transcription and metabolism.
Larp1 affects the distribution of β- and γ-actin
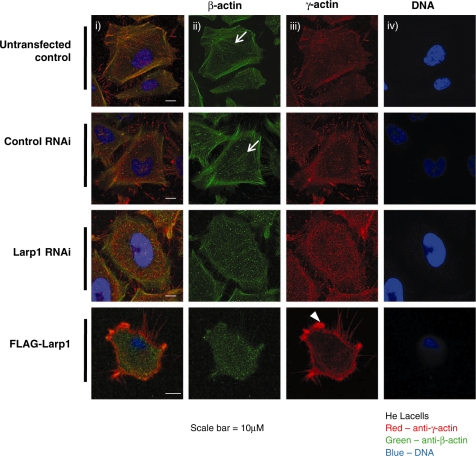
To address the effect of Larp1 on cytoskeletal organization, we observed the distribution of two isoforms of actin (β and γ) in HeLa cells following Larp1 RNAi and over-expression, compared to controls (Figure 5). In untreated cells or cells transfected with non-sense RNAi, β-actin was observed within stress fibres and co-localized with γ-actin in cortical fibres. After Larp1 RNAi, stress fibres were reduced and both β and γ actin were diffusely distributed throughout the cytoplasm. In cells in which Larp was over-expressed (FLAG-Larp1), γ-actin was concentrated at the leading edge and within lamellipodia. Stress fibres were absent in these cells and β-actin was diffusely distributed throughout the cytoplasm.
Figure 5.
Immunofluorescence of HeLa cells following treatment with control RNAi, Larp1 RNAi and in cells transfected with Larp1 over-expressing construct (FLAG-Larp1) examined using confocal microscopy. Panels show images in combined colour channel (i), green channel (β-actin) (ii), red channel (γ-actin) (iii) and blue channel (DNA) (iv). Stress fibres containing β-actin are present in control cells (white arrows) along with β- and γ-actin colocalization in cortical fibres. After Larp1 RNAi, stress fibres are no longer present and β-actin has a diffusely cytoplasmic localization. Following Larp1 over-expression (FLAG-Larp1) cells again demonstrate a mesenchymal morphology with absent stress fibres but a concentration of γ-actin within lamellipodia (arrowhead).
DISCUSSION
To summarize our findings, we have showed that, like its Drosophila counterpart, Larp1 interacts with PABP but also that Larp1 is in complex with the mRNA cap-binding protein eIF4E. Phenotypically, loss of Larp1 causes a mitotic arrest, apoptosis and changes in cell morphology such as a reduction in formation of migratory lamellipodia. The mitotic phenotype observed in HeLa cells following a reduction of Larp1 expression by siRNA is consistent with that observed in Drosophila Larp mutants, namely failure of centrosome separation and abnormal migration around the nuclear periphery, as well as defective chromosome condensation, nuclear morphology and cytokinesis. The mitotic arrest phenotype observed after Larp1 RNAi (and in Drosophila Larp mutants) could be as a result of reduced synthesis of cytoskeletal proteins, such as tubulin and keratins, which are required for ordered cell-cycle progression as well as for migration, although whether Larp1 has a direct or indirect effect in this process and the exact candidates responsible for each characteristic have yet to be resolved.
When over-expressed, Larp1 causes an increase in number of lamellipodia and a switch to a mesenchymal-like or migratory phenotype while loss of Larp1 in HeLa cells caused defective ‘wound healing’ as evidenced in our scratch assay. In addition, Larp1 appears to alter the balance between β and γ-actin expression and stress fibre formation. Although these two isoforms of cytoplasmic actin differ by only four amino acids, they are known to have functional diversity in the migrating cell, and it is possible that their expression is post-transcriptionally regulated by Larp1 (31). After immunostaining, we demonstrated dense Larp1 staining within lamellipodia and at the leading edge of HeLa cells, which was more pronounced in cells over-expressing Larp1. Loss of Larp1 by RNAi caused an increase in number of filopodia per cell, while filopodia numbers were decreased in cells over-expressing Larp1. We showed by immunofluorescence that, although Larp1 was present in lamellipodia, it did not localize to filopodia.
Although the data suggest that Larp1 has a role in the regulation of protein synthesis, it is unclear as to whether the effect of Larp1 is direct or indirect. In C. elegans, LARP-1 was observed in P-bodies and it is possible that Larp1 may have a role in trafficking a selection of mRNA transcripts to or from stress granules within the cytoplasm. Both eIF4E and PABP have been identified within cytoplasmic granules (32) and this mechanism may provide a means to protect mRNA transcripts until their protein products are required within the cell. We show here that Larp1 interacts with cytoskeletal components which could alternatively suggest a model whereby Larp1, perhaps in complex with eIF4E, journeys around the cytoskeleton to activate translation at specific sites, such as at the leading edge. Our finding of Larp1 at the leading edge in HeLa cells provides, albeit circumstantial, evidence to support this. These data raise exciting questions about the role of Larp1 in mRNA translation and cell migration, and suggest Larp1 may have relevance in a number of conditions in which these processes are disrupted, such as in tissue regeneration and cancer.
FUNDING
Biotechnology and Biological Sciences Research Council (BBSRC) (to K.S. studentship, to A.E.W. Professorial Fellowship); Cancer Research UK (to D.M.G.); Wellbeing of Women (to S.-J.L. Entry Level Scholarship) and Experimental Cancer Medicine Centre (ECMC) (to C.B.). Funding for open access charge: Charitable research fund.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Paul Bassett from Stats Consultancy Ltd for statistical advice and analysis. Dr Ernesto Yague and Dr Justin Sturge (both Imperial College) for informative discussions and manuscript advice.
REFERENCES
- 1.Schock F, Perrimon N. Molecular mechanisms of epithelial morphogenesis. Annu. Rev. Cell Dev. Biol. 2002;18:463–493. doi: 10.1146/annurev.cellbio.18.022602.131838. [DOI] [PubMed] [Google Scholar]
- 2.Aman A, Piotrowski T. Cell migration during morphogenesis. Dev. Biol. 2010;341:20–33. doi: 10.1016/j.ydbio.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 3.Lambrechts A, Van Troys M, Ampe C. The actin cytoskeleton in normal and pathological cell motility. Int. J. Biochem. Cell Biol. 2004;36:1890–1909. doi: 10.1016/j.biocel.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 4.Insall RH, Machesky LM. Actin dynamics at the leading edge: from simple machinery to complex networks. Dev Cell. 2009;17:310–322. doi: 10.1016/j.devcel.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Cannito S, Novo E, Valfrè di Bonzo L, Busletta C, Colombatto S, Parola M. Epithelial-mesenchymal transition: from molecular mechanisms, redox regulation to implications in human health and disease. Antioxid. Redox Signal. 2010 doi: 10.1089/ars.2009.2737. [17 February 2010, Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 6.Friedl P. Prespecification and plasticity: shifting mechanisms of cell migration. Curr. Opin. Cell. Biol. 2004;16:14–23. doi: 10.1016/j.ceb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Caldieri G, Ayala I, Attanasio F, Buccione R. Cell and molecular biology of invadopodia. Int. Rev. Cell Mol. Biol. 2009;275:1–34. doi: 10.1016/S1937-6448(09)75001-4. (Review) [DOI] [PubMed] [Google Scholar]
- 8.Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- 9.Naumanen P, Lappalainen P, Hotulainen P. Mechanisms of actin stress fibre assembly. J. Microsc. 2008;231:446–454. doi: 10.1111/j.1365-2818.2008.02057.x. [DOI] [PubMed] [Google Scholar]
- 10.Etienne-Manneville S. Polarity proteins in migration and invasion. Oncogene. 2008;27:6970–6980. doi: 10.1038/onc.2008.347. [DOI] [PubMed] [Google Scholar]
- 11.Friedl P, Wolf K. Proteolytic interstitial cell migration: a five-step process. Cancer Metastasis Rev. 2009;28:129–135. doi: 10.1007/s10555-008-9174-3. [DOI] [PubMed] [Google Scholar]
- 12.Machesky LM. Lamellipodia and filopodia in metastasis and invasion. FEBS Lett. 2008;582:2102–2111. doi: 10.1016/j.febslet.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 13.Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J. Clin. Invest. 2009;119:1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ladwein M, Rottner K. On the Rho’d: the regulation of membrane protrusions by Rho-GTPases. FEBS Lett. 2008;582:2066–2074. doi: 10.1016/j.febslet.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 15.Le Clainche C, Carlier MF. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev. 2008;88:489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- 16.Akin O, Mullins RD. Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell. 2008;133:841–851. doi: 10.1016/j.cell.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steffen A, Faix J, Resch GP, Linkner J, Wehland J, Small JV, Rottner K, Stradal TE. Filopodia formation in the absence of functional WAVE- and Arp2/3-complexes. Mol. Biol. Cell. 2006;17:2581–2591. doi: 10.1091/mbc.E05-11-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Quesne JP, Spriggs KA, Bushell M, Willis AE. Dysregulation of protein synthesis and disease. J. Pathol. 2010;220:140–151. doi: 10.1002/path.2627. [DOI] [PubMed] [Google Scholar]
- 20.Mendez R, Richter JD. Translational control by CPEB: a means to the end. Nat. Rev. Mol. Cell Biol. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- 21.Eliscovich C, Peset I, Vernos I, Méndez R. Spindle-localized CPE-mediated translation controls meiotic chromosome segregation. Nat. Cell. Biol. 2008;10:858–865. doi: 10.1038/ncb1746. [DOI] [PubMed] [Google Scholar]
- 22.Yoo S, van Niekerk EA, Merianda TT, Twiss JL. Dynamics of axonal mRNA transport and implications for peripheral nerve regeneration. Exp. Neurol. 2010;223:19–27. doi: 10.1016/j.expneurol.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ichihara K, Shimizu H, Taguchi O, Yamaguchi M, Inoue YH. A Drosophila orthologue of larp protein family is required for multiple processes in male meiosis. Cell. Struct. Funct. 2007;32:89–100. doi: 10.1247/csf.07027. [DOI] [PubMed] [Google Scholar]
- 24.Chauvet S, Maurel-Zaffran C, Miassod R, Jullien N, Pradel J, Aragnol D. dlarp, a new candidate Hox target in Drosophila whose orthologue in mouse is expressed at sites of epithelium/mesenchymal interactions. Dev. Dyn. 2000;218:401–413. doi: 10.1002/1097-0177(200007)218:3<401::AID-DVDY1009>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 25.Blagden SP, Gatt MK, Archambault V, Lada K, Ichihara K, Lilley KS, Inoue YH, Glover DM. Drosophila Larp associates with poly(A)-binding protein and is required for male fertility and syncytial embryo development. Dev. Biol. 2009;334:186–197. doi: 10.1016/j.ydbio.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 26.Nykamp K, Lee MH, Kimble J. C. elegans La-related protein, LARP-1, localizes to germline P bodies and attenuates Ras-MAPK signaling during oogenesis. RNA. 2008;14:1378–1389. doi: 10.1261/rna.1066008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bousquet-Antonelli C, Deragon JM. A comprehensive analysis of the La-motif protein superfamily. RNA. 2009;15:750–764. doi: 10.1261/rna.1478709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otero LJ, Ashe MP, Sachs AB. The yeast poly(A)-binding protein Pab1p stimulates in vitro poly(A)-dependent and cap-dependent translation by distinct mechanisms. EMBO J. 1999;18:3153–3163. doi: 10.1093/emboj/18.11.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu W, Kwon Y, Oko R, Hermo L, Hecht NB. Poly (A) binding protein is bound to both stored and polysomal mRNAs in the mammalian testis. Mol. Reprod. Dev. 1995;40:273–285. doi: 10.1002/mrd.1080400303. [DOI] [PubMed] [Google Scholar]
- 30.Kahvejian A, Svitkin YV, Sukarieh R, M'Boutchou MN, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19:104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dugina V, Zwaenepoel I, Gabbiani G, Clément S, Chaponnier C. Beta and gamma-cytoplasmic actins display distinct distribution and functional diversity. J. Cell Sci. 2009;122:2980–2988. doi: 10.1242/jcs.041970. [DOI] [PubMed] [Google Scholar]
- 32.Kedersha N, Tisdale S, Hickman T, Anderson P. Real-time and quantitative imaging of mammalian stress granules and processing bodies. Methods Enzymol. 2008;448:521–552. doi: 10.1016/S0076-6879(08)02626-8. [DOI] [PubMed] [Google Scholar]
- 33.Ponting CP, Mott R, Bork P, Copley RR. Novel protein domains and repeats in Drosophila melanogaster: insights into structure, function, and evolution. Genome Res. 2001;11:1996–2008. doi: 10.1101/gr.198701. [DOI] [PubMed] [Google Scholar]