Abstract
The DNA double helix undergoes an ‘overstretching’ transition in a narrow force range near 65 pN. Despite numerous studies the basic question of whether the strands are separated or not remains controversial. Here we show that overstretching in fact involves two distinct types of double-helix reorganization: slow hysteretic ‘unpeeling’ of one strand off the other; and a fast, non-hysteretic transition to an elongated double-stranded form. We demonstrate that the relative fraction of these two overstretched forms is sensitive to factors that affect DNA base pair stability, including DNA sequence, salt concentration and temperature. The balance between the two forms shifts near physiological solution conditions. This result, in addition to establishing the existence of an overstretched double-stranded state, also shows that double helix physical properties are tuned so that either unpeeling or overextension can be selected via small changes in molecule environment.
INTRODUCTION
Mechanical properties and base pairing stability of DNA play critical roles in regulating binding of proteins, in particular when protein binding results in deformation of DNA backbones, or disruption of DNA base pairing (1–3). Single-DNA stretching experiments have provided precise measurements of DNA mechanical properties (4) while direct unzipping of two DNA (or RNA) strands by forces ∼15 pN have provided direct measurements of DNA base pair stability at ambient temperatures (5,6). DNA can also be destabilized by direct stretching using forces between 60 and 70 pN; numerous experiments have reported ‘overstretching’ of DNA to a new form about 1.7 times longer than B-DNA, via a sharp (few-pN-wide), cooperative transition near room temperature (7–9).
Given the data for unzipping, a main question has been what role DNA base unpairing plays in overstretching. Many experiments have focused on 48 502 bp λ-DNA; under application of force, a sharp and rapid overstretching transition occurs. However, the reverse transition can be much slower, with appreciable hysteresis, sometimes requiring a long waiting time for the molecule to regain to its original B-DNA force response, suggesting release of one strand from the other (7).
Importantly, this hysteresis behavior has been found to depend on salt concentration and temperature, with the hysteresis absent at elevated salt or at reduced temperature (7,10). From these observations, it was hypothesized that two types of competing structural transitions might occur in an overstretched DNA: one to a double-stranded form where the two strands remain closely associated (S-DNA), and the other to strand separation (7). S-DNA was suggested to be a double helix with severely distorted base pairs (8,11). Atomic force microscopy experiments in which the force–extension curves showed two transition forces under fixed chemical and temperature conditions provided further support for the two-transition hypothesis; the lower (∼65 pN) transition was proposed to be the transition from B-DNA to S-DNA, while the higher (∼100 pN) transition was proposed to be strand separation (12,13). In direct opposition to this conclusion, a series of experiments on λ-DNA showing that changes of solution conditions which favor melting of the double helix reduce the force needed for overstretching (14–17) were used to support the hypothesis that the 65 pN DNA overstretching transition consisted exclusively of force-induced strand separation to form ‘force-melted’ parallel single-stranded DNAs (ssDNA).
Theoretical analysis (18–20) of DNA stretching data from a number of experiments (7,8) led to the conclusion that under physiological solution conditions (pH 7.5 buffered aqueous solution with 150 mM Na+), or at higher NaCl concentrations, or at lower temperatures, the force–extension curves of overstretched DNA could not be explained in terms of two parallel, separated DNA strands. The analysis examined the competition between force-driven formation of the hypothetical S-DNA, the ‘unpeeling’ of one DNA strand from the other starting from a nick or DNA end, and formation of parallel, stretched ssDNAs. It was found that S-DNA and unpeeling required similar free energies and were likely to compete near 65 pN, while parallel-stretched strand separation involved significantly higher free energies and higher forces in physiological buffers near room temperature. The prediction was made that factors that affect DNA base pair stability would determine whether the transition leads to the S-DNA state or to strand unpeeling: in particular higher salt concentration and lower temperature were predicted to favor formation of S-DNA (18). These predictions have not been systematically investigated by experiments.
Recently, elegant experiments on λ-DNA using multicolor, single-molecule fluorescence imaging have verified part of this theoretical prediction, namely that the mode of force-driven DNA melting is unpeeling of one strand from the other starting from a nick (21). However, there was no observation of a rapid, reversible transition to an overstretched double-helix state, which led to the suggestion that in general the 65 pN transition is strand unpeeling (21). The crucial question of the nature of the kinetics of the 65 pN transition as a function of e.g. salt concentration in the >100 mM NaCl concentration range relevant physiologically in Escherichia coli cells was not addressed (22). In addition, the effects of environmental factors, such as temperature and DNA sequences, were not studied.
Although ref. (21) argues that the overstretching transition is exclusively a force-induced strand unpeeling transition, it conflicts with previous experiments reporting a rapid, reversible 65 pN transition to an overstretched base paired double-helix state. We hypothesized that the resolution of these conflicting results is that proposed in (18), namely that there are two, competing modes of overstretching: a slow, hysteretic unpeeling of one strand from a nick as observed in (21), and a rapid, reversible transition to a double-stranded overstretched form of the double helix, i.e. S-DNA. We suspected [in accord with (18)] that which of these two transitions occurs depends sensitively on salt concentration and temperature. Furthermore, we hypothesized that the two transitions might be strongly sequence-dependent, and that λ-DNA (used in most of the studies), which has a marked sequence contrast along its length, might show different modes of overstretching simultaneously. We thus examined the dependence of kinetics of the ∼65 pN transition in the absence of DNA-binding ligands or proteins on salt concentration, temperature and sequence composition.
MATERIALS AND METHODS
DNA constructs
Three DNA constructs were used in the research: the 48 502-bp λ-DNA, the 19 327 bp GC-rich DNA digested from the λ-DNA (1–19 327 bp of the λ-DNA), the AT-rich sequence (33 499–48 502 bp of the λ-DNA). In the experiments, the force was applied to the 3′-ends of the two opposite strands. More details are described in the Supplementary Data.
Magnetic tweezer measurements
Forces were applied to the opposite strands (3′-3′) of the DNA by an improved transverse magnetic tweezer (23) which was previously used to study chromatin assembly in Xenopus egg extracts (24). The digoxygenin end of the DNA was fixed to the side of a #0 cover glass edge, and the biotin end was attached to an 8.2-µ-diameter paramagnetic bead (Bangs Laboratories Inc.) to achieve high force. The DNA was inside a narrow flow channel so experimental buffer could be conveniently replaced. A permanent magnet outside the channel generates controlled forces from 0.04 pN up to 180 pN to the paramagnetic bead in the focal plane, and the extension of DNA was determined to be the distance from the bead to the edge of the cover glass in the force direction. The persistence length of DNA was measured in the force range 1–10 pN by fitting the Marko–Siggia formula (25). The DNA was determined to be a single DNA with persistence length 49 ± 5 nm in pH 7.5 150 mM NaCl buffer (Supplementary Figure S1).
Temperature control
Temperature at below the room temperature was achieved using an ice-water container on the top of the flow channel. Temperature above the room temperature was achieved using a Linkam Warm Stage Controller-MC60. Temperature was measured using the TM-902C Prober Digital Thermometer (Lutron). More details concerning the temperature control are included in the Supplementary Data (Supplementary Figure S2).
RESULTS
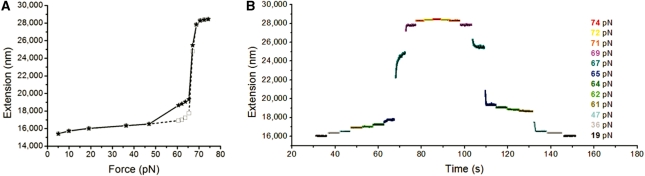
We first reproduced the main features revealed in previous studies of λ-DNA using a high-force, temperature-controlled magnetic tweezer system to eliminate the possibility of sample heating, force-feedback or photochemical effects possible in laser tweezer experiments (Supplementary Figures S1 and S2). Beginning with 10 mM Tris pH 7.5, 150 mM NaCl and 24°C, we slowly increased force through the overstretching transition, pausing for 5 s at each force level; we then decreased force through the same set of force levels. Figure 1A shows average extension during the last 1 s at each force level; open squares show the increasing-force extension scan, solid stars show the subsequent decreasing-force relaxation scan. At the beginning of the transition from B-DNA to overstretched DNA there is hysteresis, i.e. the extension and relaxation scans differ between 50 and 65 pN; note that the DNA extension returned to its original B-form value when force dropped below 50 pN, in reasonable accord with previous studies under similar conditions (7).
Figure 1.
Typical DNA overstretching transition behavior. (A) Force-extension curve representation of the overstretching transition of a λ-DNA in 150 mM NaCl, 10 mM Tris, pH 7.5 at 24°C. Open squares show the increasing-force scan and solid stars show the decreasing-force relaxation scan. (B) The extension change kinetics of the DNA in (A). Each force is denoted by a unique color. Blank gaps correspond to the periods when forces were being changed.
Figure 1B shows the extension change kinetics for Figure 1A. The kinetics reveal that extension of λ-DNA beyond 20 µm is cooperative and rapid: an increase in force by ∼2 pN from 67 pN led to an increase in extension by ∼7 µm and reached a steady state within 5 s. The reverse (retraction) transition was initially also rapid, but the extension did not reach the pre-stretch levels in 5 s. Thus slow dynamics during retraction led to the hysteresis shown in Figure 1A. Figure 1B indicates that two processes are involved in overstretching: a rapid, cooperative initial extension which is partially reversible, plus a slow, hysteretic recovery observed during the end of the retraction process. We hypothesized that the rapid cooperative part of the process was conversion of double helix to a new, double-stranded overstretched form, while the slow, hysteretic behavior was due to the unpeeling of DNA as observed in (21); the slow kinetics are simply explained in terms of the variable free energy barriers that must be crossed during the unpeeling process (18).
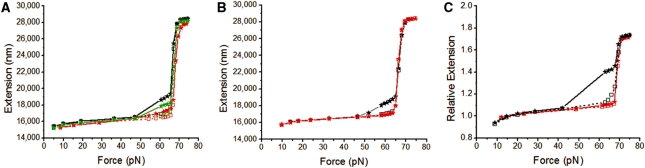
If the hypothesis that the hysteretic behavior in Figure 1 is due to unpeeling is correct, then the hysteresis should be suppressed by stabilization of DNA base pairing. We first tested this by increasing salt concentration; Figure 2A shows overstretching of λ-DNA in 10 mM Tris buffer (pH 7.5) at 24°C for three concentrations of NaCl: 150 mM, 300 mM and 1 M following the same procedure outlined above for Figure 1A. The degree of hysteresis was systematically reduced as salt concentration is increased, and at 1 M, the hysteresis during the 5 s pause experiment was almost entirely absent, in good accord with data of (7,10). We then tested the effect of temperature: Figure 2B compares results for the same 5-s-pause type of experiment in 10 mM Tris buffer (pH 7.5) and 150 mM NaCl for 24°C and 19°C for a λ-DNA. The hysteresis was suppressed at the lower temperature, where DNA strand separation is suppressed.
Figure 2.
Increased stability of base pairs decreases overstretching hysteresis. Open squares show the increasing-force scan and solid stars show the decreasing-force scan. (A) Effects of NaCl concentration for stretching a λ-DNA: 150 mM (black), 300 mM (green, same DNA as black), and 1 M (red, a different DNA from black and green). (B) Effects of temperature for stretching λ-DNA: 24°C (black), 19°C (red). (C) Effects of sequence composition; GC-rich DNA (red) shows less hysteresis than AT-rich DNA (black). Extensions are scaled with their respective contour lengths.
We then examined the role played by DNA sequence heterogeneity in the hysteresis of Figure 1A. The left and the right halves of λ-DNA have markedly different GC content and melting properties. We prepared the left 19 327 bp (57% GC) and the right 15 004 bp (54% AT) of λ-DNA for separate experiments. For 10 mM Tris buffer (pH 7.5), 150 mM NaCl, at 24°C, Figure 2C shows that the GC-rich segment displayed much less hysteresis than does the AT-rich segment. This experiment indicates that a rapid, essentially reversible overstretching transition occurs on the left GC-rich side of λ-DNA, while a slow, hysteretic transition occurs on the right AT-rich side of λ-DNA, for 150 mM NaCl at 24°C.
We thus concluded that the hysteresis observed during relaxation of an overstretched λ-DNA is enhanced by decreased base pairing stability; alternately hysteresis can be suppressed or even eliminated by increasing DNA base pair stability. A rapid, reversible overstretching transition remains under conditions where the hysteresis is suppressed. These results favor the picture that two distinct overstretched forms exist in an overstretched DNA, the selection of which sensitively depends on solution conditions and DNA sequences.
Having demonstrated that the hysteresis in the reverse transition sensitively depends on DNA base pair stability while the forward transition plateau does not, we examined dynamics of the transition further. We quickly cycled between two forces, one below the transition force, and one above. Figure 3A shows cycling of a whole λ-DNA between 61 pN and 74 pN in 10 mM Tris buffer (pH 7.5), 150 mM NaCl, 24°C. Before the transition, the extension was 16 900 nm at 61 pN. In the first cycle, the DNA was held at 74 pN for 0.3 s, during which the DNA extension reached 28 370 nm. When force was dropped back to 61 pN, the extension rapidly dropped to 17 400 nm, showing rapid reverse dynamics. Following this initial fast transition, a slow stepwise re-annealing process occurred. The extension did not return to the B-form extension after >60 s, so we reduced the force to 19 pN and re-annealing occurred. After B-form DNA extension was recovered, the force was increased to 61 pN, and an extension of 16 900 nm was again obtained. In the second force cycle, the DNA was held at 74 pN for 10 s, during which a gradual extension elongation occurred and a final extension of 28 430 nm was reached, about 60 nm longer than the extension obtained in the first cycle. Then, when force was reduced back to 61 pN, the step-wise re-annealing process started from an extension 18 400 nm, about 1000 nm larger compared with that in the previous cycle. We then carried out a third cycle. This time the DNA was held at 74 pN for 30 s, during which the DNA extension reached 28 440 nm. Then, when force was returned to 61 pN the slow step-wise re-annealing dynamics started at an even larger extension of 18 800 nm.
Figure 3.
Force cycling for λ-DNA at different salt concentrations and temperatures. (A–C) Force cycling between 61 pN (blue) and 74 pN (red) for one DNA at different NaCl concentrations: 150 mM (A), 300 mM (B) and 1 M (C). A lower force of 19 pN (orange) was used to facilitate the re-annealing. (D–E) Force cycling between 58 pN (blue) and 75 pN (red) for λ-DNA at different temperatures: 24°C (D) and 19°C (E). A lower force of 46 pN (orange) was used to facilitate the re-annealing.
Figure 3A emphasizes that there are two distinct overstretched DNA structures: the rapid initial elongation indicates formation of a first structure, while the slow elongation at 74 pN indicates a slow conversion to the second, presumably unpeeled structure. When force was reduced to 61 pN, a similar bimodal kinetics was observed; first a rapid contraction occurred, followed by a slow, stepwise contraction.
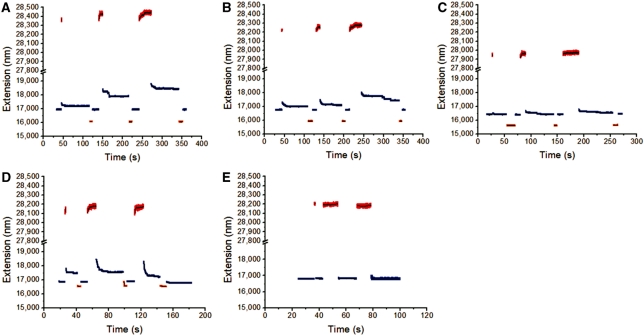
We next investigated how the fast and slow transitions respond to changes in the salt concentration and in temperature. Figure 3B and C show experiments on λ-DNA at 24°C, but at higher salt concentrations of 300 mM and 1 M NaCl, respectively. As salt concentration was increased, the double helix was stabilized, and the slow dynamics after overstretching were suppressed; in the 1 M case there was virtually no hysteresis on the few second time scale. Similarly, Figure 3D and E show experiments in 150 mM NaCl but at 24°C and 19°C respectively for λ-DNA; the 5°C reduction in temperature essentially eliminated the slow opening after overstretching and hysteresis. These experiments show that increased NaCl concentration or reduced temperature, both known to suppress DNA melting, suppress the slow overstretching mode. However, the rapid, reversible overstretching mode remained when melting was suppressed.
The previous experiments indicated that there are two distinct overstretching modes which differ in their kinetics, with the slower mode made less dominant under conditions where double-helix strand separation is suppressed. We next studied the dynamics of overstretching for the AT-rich and GC-rich fragments of λ-DNA which showed different hysteresis of overstretching (Figure 2C). We first considered the AT-rich fragment, which should be more prone to strand-separation than the GC-rich fragment; in 150 mM NaCl at 24°C overstretching the AT-rich fragment showed slow kinetics of overstretching following initial, rapid overstretching, followed by slow relaxation when force is released (Figure 4A). However, the slow overstretching and hysteresis is suppressed by increasing salt concentration (Figure 4B and C, the same DNA as in Figure 4A, at 24°C in 300 mM and 1 M NaCl, respectively) and when temperature is reduced (Figure 4D and E, for the AT-rich DNA in 150 mM NaCl at 24°C and 19°C, respectively).
Figure 4.
Force cycling for the AT-rich segment of λ-DNA at different salt concentrations and temperatures. (A–C) Force cycling between 49 pN (blue) and 75 pN (red) for AT-rich DNA at different NaCl concentrations: 150 mM (A), 300 mM (B) and 1 M (C). A lower force of 31 pN (orange) was used to facilitate the re-annealing. (D–E) Force cycling between 53 pN (blue) and 74 pN (red) for another DNA at different temperatures: 24°C (D) and 19°C (E). A lower force of 39 pN (orange) was used to facilitate the re-annealing.
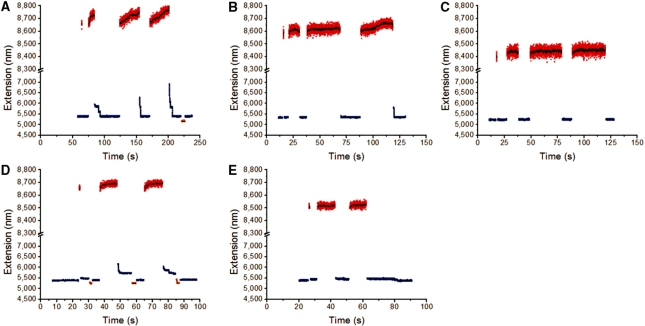
Similar experiments on the GC-rich fragment at 150 mM NaCl and 24°C showed little slow overstretching mode and hysteresis (Figure 5A), in accord with the greater stability of GC-rich DNA against strand separation. If salt concentration was reduced (Figure 5B and C, for the GC-rich DNA at 24°C in 50 mM and 10 mM NaCl, respectively) or if temperature was increased (Figure 5D and E, for the GC-rich DNA in 150 mM NaCl, 24°C and 28°C, respectively), slow overstretching and hysteresis were induced.
Figure 5.
Force cycling for the GC-rich segment of λ-DNA at different salt concentrations and temperatures. (A–C) Force cycling between 47 pN (blue) and 75 pN (red) for GC-rich DNA at NaCl concentrations of 150 mM (A), 50 mM (B) and 10 mM (C). A force of 30 pN (orange) was used to facilitate the re-annealing. (D) Force cycling between 56 pN (blue) and 75 pN (red) for another DNA at 24°C. DNA extension was observed to quickly return to its original value after cycling up and down in force. (E) Same DNA and force cycling as (D), but at 28°C. Slow re-annealing dynamics were observed. Two lower forces of 43 pN (orange) and 10 pN (olive) were used to facilitate the re-annealing.
The experiments shown in Figures 4 and 5 clarify the behavior of the entire λ-DNA molecule at 24°C and 150 mM NaCl (Figure 3A). The overstretching transition on the full λ-DNA involves two different modes on different regions of the molecule: the slow, hysteretic transition occurred on part of the AT-rich region, while the GC-rich left side underwent only the rapid, reversible overstretching transition. We observed a systematic pattern whereby changing solution conditions so as to favor strand separation (decreased NaCl concentration, increased temperature) increased the degree to which the slow, hysteretic transition is observed. On the other hand, changing solution conditions so as to stabilize the double helix (increased NaCl concentration, decreased temperature) eliminated the slow hysteretic overstretching mode. Comparison with the experiments of (21) where overstretching of the λ-DNA was identified as unpeeling from a nick allows us to identify the slow, hysteretic overstretching transition which occurs more prominently on AT-rich DNA as unpeeling. This leaves the fast, reversible transition, which we identify as a transition to an extended DNA state, i.e. to S-DNA. We emphasize that both the AT-rich and the GC-rich sides of λ-DNA display the fast, reversible transition to S-DNA, but that the AT-rich side subsequently undergoes unpeeling for 24°C and 150 mM NaCl indicating that for at least part of the AT-rich side, unpeeling from a nick is more favorable for at least some fraction of the molecule (we estimate that >70% of the λ-DNA overstretched at 24°C and 150 mM NaCl remains in double-stranded form; Supplementary Data).
DISCUSSION
We emphasize that our results indicate that, for a DNA with free ends or nicks, there are two competing modes of overstretching: formation of a stretched double-stranded structure (S-DNA), and alternately unpeeling of one strand from the other starting from one DNA end or a nick, and that this conclusion provides a reconciliation of many apparently conflicting experiments. For example, the data presented in (21), which emphasized that large-scale unpeeling occurs on λ-DNA, are for 50 mM NaCl univalent salt, which favors unpeeling relative to 150 mM NaCl, particularly for AT-rich sequences. We have observed a strong temperature dependence of the overstretching mode; temperatures of the experiments in (21) were not reported and it is conceivable that the laser tweezer system heated the flow chamber to temperatures above ambient (26–28).
One might hypothesize that the two modes of relaxation following overstretching are due to differences in formation of secondary structure in the unpeeled strand. However, this does not explain why the slower relaxation kinetics was greatly suppressed under conditions when the DNA base pair stability was enhanced (high salt concentration, low temperature or high GC percentage), which would presumably also stabilize ssDNA secondary structure. Furthermore, our finding that the re-annealing kinetics could be dramatically sped up by decreasing the relaxation force indicates that there was not significant secondary structure formed in the unpeeled strand. However, since the longest time we were able to hold a DNA at forces >70 pN before it broke was <1 min, we cannot exclude the possibility that significantly more stable secondary structures may form in the unpeeled strand if the DNA could be held for much longer time at forces larger than 70 pN.
Finally, on the basis of theoretical considerations we expect that for solution conditions (salt levels, temperature, pH) where the first transition is from B-DNA to S-DNA, a second, subsequent unpeeling transition should occur at a second, higher force (18). The point at which B-DNA, S-DNA and unpeeling are in ‘coexistence’ is quite close to physiological solution conditions and room temperature (18), and is significantly changed by DNA sequence. On top of this, formation of the unpeeled state is crucially dependent on the presence of a nick along DNA; without a nick in a sufficiently unstable region, no unpeeling will occur. This sensitivity of overstretching mode on solution conditions and DNA topology suggests regulatory function: unnicked DNA under large tension undergoes an overstretching transition to a double-stranded form; on the other hand, placing a nick in an AT-rich region will trigger an unpeeling reaction. The former might be used to facilitate opening of the double helix to allow base access, while the latter might play a role in strand invasion and exchange reactions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Education of Singapore (grants R144000192112 and R144000251112 to J.Y.); Research Centre of Excellence in Mechanobiology at the National University of Singapore (seeding grant to J.Y.); National Science Foundation (grants DMR-0715099 and PHY-0852130) for work at NU; National Institutes of Health (grant U54-CA143869-01); Chicago Biomedical Consortium with support from The Searle Funds at The Chicago Community Trust. Funding for open access charge: Ministry of Education of Singapore (grant R144000251112 to J.Y.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Simona Cocco (Princeton University) for helpful discussions. H.F., H.C. and J.Y. performed the experiments and analyzed the data. J.F.M. and J.Y. conceived the experiments and wrote the article.
REFERENCES
- 1.Xu H, Hoover TR. Transcriptional regulation at a distance in bacteria. Curr. Opin. Microbiol. 2001;4:138–144. doi: 10.1016/s1369-5274(00)00179-x. [DOI] [PubMed] [Google Scholar]
- 2.Rice PA, Yang S, Mizuuchi K, Nash HA. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 3.Wojtuszewski K, Hawkins ME, Cole JL, Mukerji I. HU binding to DNA: evidence for multiple complex formation and DNA bending. Biochemistry. 2001;40:2588–2598. doi: 10.1021/bi002382r. [DOI] [PubMed] [Google Scholar]
- 4.Smith SB, Finzi L, Bustamante C. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science. 1992;258:1122–1126. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- 5.Essevaz-Roulet B, Bockelmann U, Heslot F. Mechanical separation of the complementary strands of DNA. Proc. Natl Acad. Sci. USA. 1997;94:11935–11940. doi: 10.1073/pnas.94.22.11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liphardt J, Onoa B, Smith SB, Tinoco IJ, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 7.Smith SB, Cui Y, Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 8.Cluzel P, Lebrun A, Heller C, Lavery R, Viovy JL, Chatenay D, Caron F. DNA: an extensible molecule. Science. 1996;271:792–794. doi: 10.1126/science.271.5250.792. [DOI] [PubMed] [Google Scholar]
- 9.Baumann CG, Smith SB, Bloomfield VA, Bustamante C. Ionic effects on the elasticity of single DNA molecules. Proc. Natl Acad. Sci. USA. 1997;94:6185–6190. doi: 10.1073/pnas.94.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao H, Arias-Gonzalez JR, Smith SB, Tinoco I., Jr, Bustamante C. Temperature control methods in a laser tweezers system. Biophys. J. 2005;89:1308–1316. doi: 10.1529/biophysj.104.054536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haijun Z, Yang Z, Zhong-can O-Y. Bending and base-stacking interactions in double-stranded DNA. Physical Rev. Lett. 1999;82:4560. [Google Scholar]
- 12.Rief M, Clausen-Schaumann H, Gaub HE. Sequence-dependent mechanics of single DNA molecules. Nat. Struct. Biol. 1999;6:346–349. doi: 10.1038/7582. [DOI] [PubMed] [Google Scholar]
- 13.Calderon CP, Chen WH, Lin KJ, Harris NC, Kiang CH. Quantifying DNA melting transitions using single-molecule force spectroscopy. J. Phys. Condens. Matter. 2009;21:34114. doi: 10.1088/0953-8984/21/3/034114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rouzina I, Bloomfield VA. Force-induced melting of the DNA double helix. 1. Thermodynamic analysis. Biophys. J. 2001;80:882–893. doi: 10.1016/S0006-3495(01)76067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rouzina I, Bloomfield VA. Force-induced melting of the DNA double helix. 2. Effect of solution conditions. Biophys. J. 2001;80:894–900. doi: 10.1016/S0006-3495(01)76068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wenner JR, Williams MC, Rouzina I, Bloomfield VA. Salt dependence of the elasticity and overstretching transition of single DNA molecules. Biophys. J. 2002;82:3160–3169. doi: 10.1016/S0006-3495(02)75658-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams MC, Wenner JR, Rouzina I, Bloomfield VA. Effect of pH on the overstretching transition of double-stranded DNA: evidence of force-induced DNA melting. Biophys. J. 2001;80:874–881. doi: 10.1016/S0006-3495(01)76066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cocco S, Yan J, Leger JF, Chatenay D, Marko JF. Overstretching and force-driven strand separation of double-helix DNA. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2004;70:011910. doi: 10.1103/PhysRevE.70.011910. [DOI] [PubMed] [Google Scholar]
- 19.Chen H, Fu HX, Koh CG. Sequence-dependent unpeeling dynamics of stretched DNA double helix. J. Comput. Theoretical Nanosci. 2008;5:1381–1386. [Google Scholar]
- 20.Whitelam S, Pronk S, Geissler PL. There and (slowly) back again: entropy-driven hysteresis in a model of DNA overstretching. Biophys. J. 2008;94:2452–2469. doi: 10.1529/biophysj.107.117036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Mameren J, Gross P, Farge G, Hooijman P, Modesti M, Falkenberg M, Wuite GJ, Peterman EJ. Unraveling the structure of DNA during overstretching by using multicolor, single-molecule fluorescence imaging. Proc. Natl Acad. Sci. USA. 2009;106:18231–18236. doi: 10.1073/pnas.0904322106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konopka MC, Weisshaar JC, Record MT., Jr Methods of changing biopolymer volume fraction and cytoplasmic solute concentrations for in vivo biophysical studies. Methods Enzymol. 2007;428:487–504. doi: 10.1016/S0076-6879(07)28027-9. [DOI] [PubMed] [Google Scholar]
- 23.Yan J, Skoko D, Marko JF. Near-field-magnetic-tweezer manipulation of single DNA molecules. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2004;70:011905. doi: 10.1103/PhysRevE.70.011905. [DOI] [PubMed] [Google Scholar]
- 24.Yan J, Maresca TJ, Skoko D, Adams CD, Xiao B, Christensen MO, Heald R, Marko JF. Micromanipulation studies of chromatin fibers in Xenopus egg extracts reveal ATP-dependent chromatin assembly dynamics. Mol. Biol. Cell. 2007;18:464–474. doi: 10.1091/mbc.E06-09-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marko JF, Siggia ED. Stretching DNA. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- 26.Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peterman EJ, Gittes F, Schmidt CF. Laser-induced heating in optical traps. Biophys. J. 2003;84:1308–1316. doi: 10.1016/S0006-3495(03)74946-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abbondanzieri EA, Shaevitz JW, Block SM. Picocalorimetry of transcription by RNA polymerase. Biophys. J. 2005;89:L61–L63. doi: 10.1529/biophysj.105.074195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.