Abstract
Endogenous short RNAs (esRNAs) play diverse roles in eukaryotes and usually are produced from double-stranded RNA (dsRNA) by Dicer. esRNAs are grouped into different classes based on biogenesis and function but not all classes are present in all three eukaryotic kingdoms. The esRNA register of fungi is poorly described compared to other eukaryotes and it is not clear what esRNA classes are present in this kingdom and whether they regulate the expression of protein coding genes. However, evidence that some dicer mutant fungi display altered phenotypes suggests that esRNAs play an important role in fungi. Here, we show that the basal fungus Mucor circinelloides produces new classes of esRNAs that map to exons and regulate the expression of many protein coding genes. The largest class of these exonic-siRNAs (ex-siRNAs) are generated by RNA-dependent RNA Polymerase 1 (RdRP1) and dicer-like 2 (DCL2) and target the mRNAs of protein coding genes from which they were produced. Our results expand the range of esRNAs in eukaryotes and reveal a new role for esRNAs in fungi.
INTRODUCTION
The gene silencing pathways using endogenous short RNAs (esRNAs) 20–24 nt in length are very diverse in eukaryotes (1). Most classes of esRNAs are produced from double-stranded RNA usually by a member of the Dicer family and are incorporated into an effector complex containing a member of the Argonaute family (1). esRNAs that are produced from and target transposons seem to be the most ancient because these were found in fungi (2), plants (3) and animals (4,5). These esRNAs act in cis and may lead to DNA methylation and/or histone modifications (2,3). Another class of esRNAs, microRNAs (miRNAs), are produced from hairpin structure non-coding transcripts and target mRNAs in trans (6). miRNAs have been found in plants and animals and play a role in diverse processes including development and adaptation to environmental changes (6). Although miRNAs have not been found in fungi, dicer mutants of several fungi have been reported to be affected in vegetative and developmental processes (7,8) suggesting the existence of esRNAs with regulatory functions in this kingdom. However, information on esRNAs in fungi is very scarce.
Mucor circinelloides, a basal fungus of the clade zygomycete, has been revealed as a model organism in the fungal kingdom for the study of RNA silencing and other processes due to the availability of molecular tools and its evolutionary distance from other fungal model organisms. The existence of a transgene-induced RNA silencing mechanism in M. circinelloides has been previously demonstrated (9) and the pathway has been dissected by identifying genes and proteins involved in silencing. As a result, two dicer genes, dcl1 and dcl2, have been identified (8,10). The DCL2 protein plays a major role in gene silencing induced by sense or hairpin transgenes, being responsible for the production of two different classes of antisense esRNAs, 21 and 25 nt long (9). Null dcl2 mutants (dcl2−) show a significant reduction in the production of asexual spores, which suggests a role for dcl2 in the control of vegetative development (8). Similarly, mutants affected in the dcl1 gene (dcl1−) show defects in vegetative growth and hyphal morphology, even though this gene is not required for efficient gene silencing triggered by transgenes (10). In addition, two genes coding for RNA-dependent RNA polymerases (RdRPs) are involved in the silencing mechanism in M. circinelloides. rdrp1 is required for sense transgene-induced silencing to generate dsRNA molecules but the role of rdrp2 is not understood (S.C., S.T-M., R.M.R-V., our unpublished results).
Relatively little is known about the physiological roles of the fungal RNA-silencing pathways (7). Availability of M. circinelloides mutants affected in the silencing mechanism and the recent completion of its genome sequencing have provided the genetic and molecular tools required to investigate how esRNAs regulate gene expression in this basal fungus. We analyse here the esRNA content of M. circinelloides by deep sequencing of short RNAs (18–25 nt) in the wild type and several mutant strains and identify a new class of esRNAs that target mRNAs of protein-coding genes from which they were produced.
MATERIALS AND METHODS
Strains and growth conditions
The leucine auxotroph R7B, derived from M. circinelloides f. sp. lusitanicus CBS 277.49 (syn. Mucor racemosus ATCC 1216b), was used as the wild-type strain. Strain MU406 is a dcl1 null mutant derived from MU402, a uracil auxotroph derivative of R7B (8). Strain MU410 is a dcl2 null mutant derived from MU402 (10). Strain MU411 is a double dcl1/dcl2 null mutant derived from MU410 (10). Strains MU419 and MU420 correspond to rdrp1 and rdrp2 null mutants derived from MU402, respectively (S.C., S.T-M., R.M.R-V., our unpublished results). Cultures were grown at 26°C in complete YPG medium as described previously (10). The pH was adjusted to 4.5 for mycelial growth.
RNA analysis
Short RNA samples were extracted from mycelia grown for 48 h on YPG plates using the miRVana kit (Ambion), following the instructions of the supplier. cDNA libraries of short RNAs were generated and sequenced as described previously (11). Low and high molecular weight RNA was extracted from frozen mycelia and analysed by northern blot as described previously (9).
Sequence analysis
Raw sRNA reads were processed by removing adaptor sequences with exact matches to the first eight bases of the 3'-adaptor. Annotated exon regions were extracted from the M. circinelloides genome (v 1.0) along with annotated transposons and repeat regions. All remaining segments of the M. circinelloides genome were used as the intergenic set for this analysis.
sRNAs were mapped to exons, transposons/repeats and intergenic regions using PatMaN (12) and distinct sRNA producing loci were predicted in intergenic regions using a previously published method (13). Any locus containing fewer than five reads was discarded for the initial locus analysis. miRNA prediction was carried out both on intergenic regions and the full genome using a modified version of the miRCat pipeline described previously (14). In order to correct for variation in sRNA sample sizes and allow us to perform cross-sample expression analysis, sRNA counts for each of the intergenic, exonic and transposon loci were converted into reads per million genome matching reads. sRNA loci were said to be down-regulated in a given sample if the normalized locus abundance showed at least a 4-fold decrease in comparison to the wild-type sample. To increase the stringency of the analysis and avoid lowly expressed regions, any loci with a normalized abundance count of less than 50 in the wild type were excluded from the analysis.
RESULTS AND DISCUSSION
Short RNAs are primarily generated by DCL2 in Mucor
Two dicer-like genes (dcl1 and dcl2) have been identified in Mucor and a possible function has been hypothesized for dcl2 gene (8). In order to identify bona fide esRNAs we generated cDNA libraries of short RNAs from wild-type (dcl1/dcl2), single mutant (dcl1−/dcl2 or dcl1/dcl2−) and double mutant (dcl1−/dcl2−) strains (8 and 10) and sequenced them on the Solexa platform (Table 1; GEO accession number is GSE18958). Non-redundant reads were mapped to the genome, whose sequence was available from the JGI (http://genome.jgi-psf.org/Mucci1/Mucci1.home.html). In fact, this is the first global analysis reported from the Mucor sequencing project. Reads that mapped to the genome in close proximity (≤200 bp) to each other were grouped together into loci (as previously described in ref. 13). A total of 24 111 loci were identified all over the Mucor genome by this criterion. All esRNA producing loci were analysed by comparing accumulation of esRNAs in the wild-type and dcl mutants. Many loci produced esRNAs only in sense orientation at a similar rate in wild-type and all mutant strains, and accumulation of esRNAs from some of these loci was analysed by northern blot. These esRNAs were either not detectable or the probes detected a smear between 15 and 2000 nt but not bands with distinct sizes between 20 and 25 nt (data not shown). Therefore, we concluded that reads from these loci are most likely to be degradation products of ribosomal, transfer and messenger RNAs and were not further analysed.
Table 1.
Sequencing of short RNAs in different Mucor strains
| WT | dcl1− | dcl2− | dcl1−/2− | rdrp− | rdrp− | |
|---|---|---|---|---|---|---|
| Total reads | 6 421 725 | 5 326 957 | 6 155 439 | 10 899 332 | 11 088 786 | 10 846 130 |
| After adaptor removal (total) | 5 701 786 | 4 604 676 | 5 049 070 | 9 303 440 | 10 178 115 | 9 877 133 |
| After adaptor removal (unique) | 1 362 728 | 1 357 556 | 1 354 177 | 3 027 587 | 1 929 224 | 2 654 222 |
| Mapping to genome (total) | 4 317 874 | 3 619 529 | 3 912 401 | 7 094 985 | 8 347 666 | 7 880 896 |
| Mapping to genome (unique) | 777 409 | 808 650 | 890 709 | 2 121 808 | 1 272 679 | 1 512 498 |
The table shows the read numbers of different categories for each strain we have analysed.
To identify DCL-dependent esRNA loci, we selected those that showed at least a 4-fold decrease in normalized esRNA reads in dcl1− or dcl2− mutants compared to wild type. Eight hundred and forty differentially expressed loci were identified, which were grouped based on the annotation of the locus: intergenic, transposon or exon (Table 2). Seven differentially expressed loci showed a decrease only in dcl1−, 828 were down-regulated in only dcl2− and five were suppressed in both. Next, we compared esRNA accumulation in wild-type and dcl1−/dcl2− double mutant. Most of the 840 loci producing fewer esRNAs in either dcl1− or dcl2− or in both also produced fewer esRNAs in the double mutant. In fact, the read numbers were usually lower in the double mutant than in any of the single mutants, demonstrating some redundancy for the two dcl genes (Supplementary Figure S1). In addition a further 140 loci showed differential expression only in the double mutant making the total number of dcl (dcl1 and/or dcl2) dependent esRNA producing loci 980. Despite the redundancy between dcl1 and dcl2, the sequencing data suggested that DCL2 is the primary protein that generates the majority of esRNAs in Mucor, although from a very small number of loci mainly DCL1 produces esRNAs and there are loci from which both Dicers can make esRNAs. The DCL proteins have similar hierarchy in transgene-induced silencing, where DCL2 also plays a more prominent role in the generation of siRNAs than DCL1 (8).
Table 2.
Number of loci down-regulated at sRNA level in dcl1− and dcl2− mutants
| Type of loci | Down-regulated in |
|||
|---|---|---|---|---|
| dcl1− | dcl2− | dcl1−/dcl2− | All dcl− | |
| Transposons | 0 (0) | 207 (207) | 205 (2) | 209 |
| Intergenic regions | 7 (2) | 401 (396) | 441 (44) | 447 |
| Exons | 5 (5) | 225 (225) | 319 (94) | 324 |
| Total | 12 (7) | 833 (828) | 965 (140) | 980 |
Number of loci (with normalized abundance of more than 50 reads per million) showing a fourfold or higher reduction in the different mutant strains compared to the wild type. Numbers in parentheses in the dcl1− and dcl2− columns show the number of loci that require only that Dicer. The numbers in parentheses for double mutant shows the number of loci that are down-regulated only in the double mutant (the number of loci that can be processed by either Dicer). Please note that the four transposon loci that are not down-regulated in dcl1−/dcl2− but reduced in dcl2−, are just below the 4-fold threshold in the double mutant.
Short RNAs are produced from transposons but there is no evidence for miRNA genes
In order to investigate the distribution of DCL-dependent esRNAs across the different types of loci, we calculated the normalized abundance of esRNAs per 1 kb of exonic, intergenic and transposon regions in the wild-type and dcl mutant strains. Supplementary Figure S2 shows that esRNAs are not formed at random across the genome but they are enriched in exonic regions compared with intergenic regions and transposons. In fact, only 209 out of the 980 esRNA producing loci, down-regulated in at least one of the dcl− strains, were annotated transposons or repeats (Table 2). This is in contrast with other organisms, such as Schizosaccharomyces pombe (15), Saccharomyces castellii and Candida albicans (16), where most esRNAs are produced from repeats and transposons.
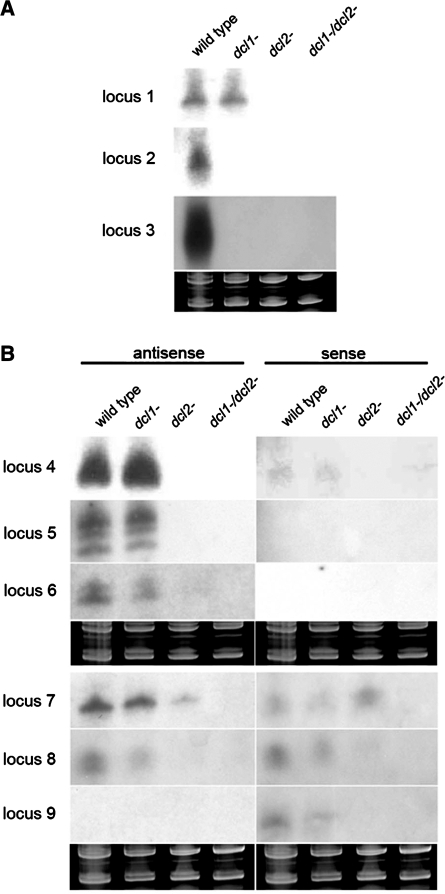
Intergenic and intronic loci are the prime candidates for producing miRNAs. Therefore, we asked whether any of these loci can be folded into a stem–loop structure characteristic of miRNA loci. None of the 447 dicer-dependent intergenic and intronic loci fulfilled the criteria of miRNA loci, although several esRNAs from intergenic regions were confirmed by northern blot (Figure 1A). We also tested all 24 111 esRNA producing loci identified in the initial analysis (including those that are not down-regulated in dcl mutants) but none of them had the features of miRNA genes. It is likely that miRNAs are not present in Mucor, although we cannot exclude the possibility that they are expressed in different growth condition or developmental stages. The apparent lack of miRNAs in fungi and the presence of transposon derived esRNAs in basal fungi, plants and animals suggest that life evolved esRNAs to silence transposons and than each branch of life has utilised this machinery to silence other targets.
Figure 1.
Accumulation of esRNAs in wild type and dcl mutant Mucor strains. Northern blots of esRNAs from intergenic regions (A) and exons of protein coding genes (B). Low-molecular weight RNA was extracted from wild-type, dcl1−, dcl2− and dcl1−/dcl2− double mutant strains, separated on 15% denaturing polyacrylamide gels, transferred to membranes and probed with riboprobes or end-labelled DNA oligonucleotides specific to each locus. For exact probe sequences, see Supplementary Table S1. Ethidium bromide stained images of gels below the radiograms show equal loading of lanes. (B) The accumulation of antisense and sense esRNAs separately. The exon loci correspond to the following proteins: locus 4: ID 80452, serine/threonine kinase; locus 5: ID 82197, no domains found; locus 6: ID 77050, no domains found; locus 7: ID 78553, low similarity to transposase 21 protein; locus 8: ID 85423, Zn-finger CCHC containing protein; locus 9: ID 95230, no domains found. Ten picomoles per lane of 23-mer to 27-mer DNA oligonucleotides in antisense and sense orientation were used as size markers and to control the hybridization specificity. In all cases, the RNA probes hybridized to these controls.
Short RNAs are generated from exons and regulate mRNA accumulation
Next, we focussed on the main class of esRNAs that mapped to exons (Table 2 and Supplementary Figure S2) and we call these exonic-siRNAs (ex-siRNAs) based on their unusual location. In total, 324 exonic loci were identified, which correspond to 276 genes, since some genes contain more than one exon. To validate the sequencing data, accumulation of selected ex-siRNAs was analysed by northern blot (Figure 1B), and in all cases the presence of distinct bands around 20–25 nt was confirmed. Some loci produced almost exclusively antisense ex-siRNAs, whereas others produced ex-siRNAs in both orientations. Results also confirmed the reduced expression of ex-siRNAs in mutant strains compared to wild type, as well as that DCL2 is the primary dicer in Mucor. The northern blots corroborated the redundancy between the two dcl genes because the ex-siRNA signals were often weaker in the double mutant than in the single mutants. Sometimes different sized ex-siRNAs were detected from the same locus, which seems to be a characteristic of the M. circinelloides DCL enzymes. In fact, transgene-induced gene silencing is also associated with two different size classes of siRNAs, 21 and 25 nt long, both in the wild-type and the dcl1− mutant (9,10).
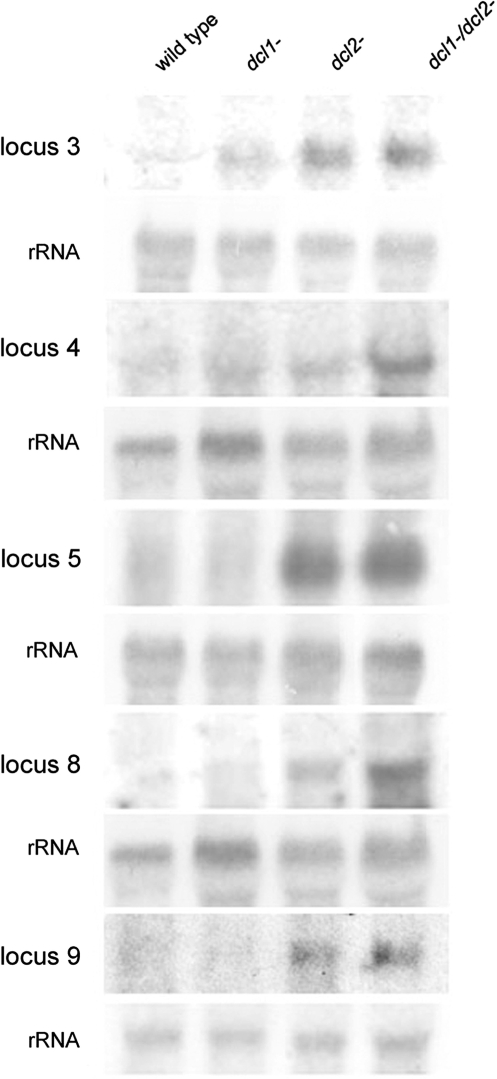
Accumulation of ex-siRNAs does not necessarily mean that they are functional, as for example some non-conserved plant miRNAs do not have target mRNAs (17). To test whether the exon mapping Mucor ex-siRNAs affect gene expression, accumulation of the mRNAs was analysed by northern blot analysis. All the four tested mRNAs that showed reduced ex-siRNA expression in dcl2− accumulated at an increased level in the dcl mutant strains compared to the wild type (Figure 2, loci 4–9) confirming that the Mucor ex-siRNAs are functional. The effect of ex-siRNAs on mRNA accumulation is most likely post-transcriptional, since no significant methylation is associated with gene silencing in Mucor, and siRNAs generated in transgene-induced silencing have been demonstrated to act post-transcriptionally (9). In addition to the four mRNAs, we also tested the accumulation of a potential RNA transcript corresponding to an intergenic region, which produced less esRNA in dcl− strains than in wild type. A transcript was detected from this intergenic locus and it also showed a higher accumulation in the dcl2− single and dcl1−/dcl2− double mutants relative to the wild type (Figure 2, locus 3), supporting that the different esRNAs of Mucor are functional.
Figure 2.
Accumulation of mRNAs in wild type and dcl mutant Mucor strains. Northern blots of high molecular weight RNAs corresponding to an intergenic region (locus 3) or protein coding exons (loci 4, 5, 8 and 9). Total RNA (50 µg) extracted from wild-type and mutant strains were separated in 1.2% denaturing agarose gel, transferred to membranes and hybridized with gene specific or rRNA probes (Supplementary Table S1). The locus numbers correspond to the loci on Figure 1.
Biogenesis of exonic endogenous siRNAs
Next, we wanted to understand the mechanism that produced ex-siRNAs. Since most exons, which generated less ex-siRNAs in dcl2− than in wild type, produced some antisense ex-siRNAs we hypothesised that an RdRP is involved in ex-siRNAs biogenesis. Two different RdRP proteins have been implicated in M. circinelloides transgene-induced gene silencing (S.C., S.T-M., R.M.R-V, our unpublished results), and we tested the involvement of these proteins, RdRP1 and RdRP2, in the biogenesis of ex-siRNAs by deep sequencing of esRNAs from rdrp1− and rdrp2− strains. The normalized reads of ex-siRNAs from the 324 exons that accumulated less ex-siRNA in, at least, one dcl mutant compared to the wild-type strain are shown in Supplementary Table S2, which also shows the strand bias (sense/antisense) of the ex-siRNAs produced by the wild-type and all mutant strains. The accumulation of ex-siRNAs from each exon in the different mutant strains was compared to the wild type and the fold difference is shown in Supplementary Table S3, in which exons are ordered according to the log2 fold change of ex-siRNAs in the dcl2− strain compared to wild type. Four different groups can be easily identified and we call these almost perfectly separated groups class 1–4. Table 3 shows a summary of the properties of the different ex-siRNA classes.
Table 3.
Characteristics of the four classes of ex-siRNA
| sRNA class | Strand bias | Average log2 fold change from WT |
No. of exons | ex-siRNA (%) | 5′ Ua (%) | 3′ penult. Ub (%) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| dcl1− | dcl2− | Dcl1−/2− | rdrp1− | rdrp2− | ||||||
| Class 1 | −0.78 | 0.34 | −12.84 | −8.86 | 3.74 | −1.33 | 9 | 13.42 | 92.18 | 6.11 |
| Class 2 | −0.34 | 0.55 | −3.79 | −8.72 | −5.02 | 1.36 | 222 | 58.74 | 92.12 | 16.63 |
| Class 3 | 0.90 | −0.47 | 0.12 | −3.21 | −4.52 | −3.50 | 88 | 27.36 | 8.39 | 49.57 |
| Class 4 | 0.83 | −2.45 | −0.26 | −1.02 | −2.61 | −2.42 | 5 | 0.48 | 28.28 | 66.61 |
Different characteristics of ex-siRNAs from each classes are shown in the table. Strand bias indicates orientation to mRNAs, where 1 corresponds to all sRNAs in the same orientation as the mRNA, 0 to equal mixture of sRNAs on both strands and −1 to all sRNAs antisense to mRNAs. Numbers in bold indicate a higher than 4-fold down-regulation in the corresponding mutants relative to wild type.
aThe percentage of redundant reads that contain a uracil in the 5′ most position.
bThe last column shows the percentage of redundant reads that contain a uracil in the 3′ penultimate position.
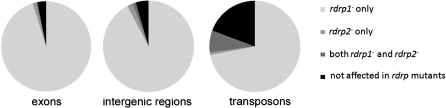
Classes 1 and 2 include all ex-siRNAs that are dcl2-dependent. The first nucleotide of these ex-siRNAs exhibits a strong preference for uracil (92%; Table 3; Supplementary Figure S3). This preference was also shown for Argonaute-bound guide RNAs of animal, plants and other fungi (16). The majority of the DCL2-dependent ex-siRNAs also showed reduced levels in the rdrp1− mutant but not in the rdrp2− strain and define class 2. This is the largest group of ex-siRNAs, with 222 exons that represent 68.5% of the 324 ex-siRNA producing exons. Most likely, these ex-siRNAs act in cis, since mRNAs transcribed from these exons are up-regulated in the absence of the ex-siRNAs (Figure 2). There is not a strong strand bias among these ex-siRNAs, most exons producing a mixed sense and antisense ex-siRNAs, which is expected considering that a large number of ex-siRNAs are produced from each exon (Supplementary Figure S1). The requirement for RdRP1 and DCL2 for the biogenesis of this ex-siRNA class suggests that mRNAs from those loci are turned into dsRNA by RdRP1 and then processed by DCL2. The involvement of these two proteins in the biogenesis of the majority of ex-siRNAs can be extended to esRNAs derived from transposons and intergenic regions because most of the dcl2-dependent esRNAs generated from those loci also require rdrp1 (Figure 3).
Figure 3.
rdrp1 and rdrp2 dependence of DCL2 generated esRNAs. The pie charts show the percentage of DCL2-dependent loci that also show reduced level of esRNAs in rdrp1− and rdrp2− strains.
A very small group of the dcl2-dependent ex-siRNAs, made up by ex-siRNAs from only nine exons belonging to five genes, does not require RdRP1 but most of them depend on RdRP2. These ex-siRNAs (class 1) are the most down-regulated in the dcl2− strain (Supplementary Table S3). Accumulation of these ex-siRNAs is strongly up-regulated in the rdrp1− mutant, which may suggest that both RdRPs can compete for binding the mRNA templates but only RdRP2 can turn them into dsRNA. Consequently, lack of RdRP1 may allow RdRP2 to make more dsRNA that would result in a higher ex-siRNA level.
We have investigated the biological functions of the genes that produce less ex-siRNAs in the dcl2 mutant (classes 1 and 2), trying to link those functions with the phenotype of the dcl2 mutant strain (8). However, the high number of genes affected in the dcl2 mutant makes it difficult to understand their roles and to reveal their biological significance. Nonetheless, it can be emphasized that many of those genes are annotated as encoding for proteins involved in signal transduction and information storage and processing (Supplementary Table S3), which may indicate the involvement of these ex-siRNAs in the regulation of different cellular processes.
A significant group of ex-siRNAs producing exons (88 out of 324) are in class 3 and correspond to ex-siRNAs that are down-regulated only in the double dicer mutant but not in dcl1− or dcl2− single mutants (Supplementary Table S3). This indicates that dsRNA produced from these exons can be processed by either DCL1 or DCL2. However, both RdRP enzymes are involved in the biogenesis of these ex-siRNAs, because they are down-regulated in the absence of either of the two rdrp genes. A peculiar feature of the class 3 ex-siRNAs is that they show a very strong strand bias, almost all of them are exclusively sense to the mRNAs. Besides that, the class 3 ex-siRNA molecules show a random spread of size distribution that is not observed in the dcl2− dependent classes 1 and 2, which produce predominantly 23–24 nt sRNAs (Supplementary Figure S4). This suggests that class 3 ex-siRNAs are not produced by a canonical RNA silencing mechanism. Instead, we can speculate that sequential or combined activity of RdRP1 and RdRP2 generates dsRNA stretches but not dsRNA through the entire length of the mRNA. These discrete dsRNA regions are processed by either DCL1 or DCL2 and after the initial cleavage, the single stranded portions of mRNAs are degraded by non-specific RNases, probably because they lose their cap and/or polyA tail. Alternatively, DCL1 or DCL2 may process these mRNAs beyond the discrete dsRNA regions, since the ability of human Dicer to cleave ssRNA in a partial dsRNA molecule has been described recently (18). We also analysed the nucleotide distribution in each position of class 3 ex-siRNAs and found a very strong bias for uracil in the penultimate position for almost all sizes of short RNAs (20–25 nt; Supplementary Figure S5). This is very surprising considering that uracil is strongly under-represented in all esRNAs (including class 3 ex-siRNAs; Supplementary Table S4). The strong bias for uracil in the penultimate position suggests that the generation of class 3 ex-siRNAs is not random. Another characteristic of this class is that most of these genes are highly expressed, as denoted by the sequencing of high numbers of ESTs. Further experiments are needed to identify what other features, besides high expression, are required for the mRNAs to enter this pathway and exactly how class 3 ex-siRNAs are generated.
Finally, class 4 is a tiny group of ex-siRNAs that derive from only five exons. These are the only ex-siRNAs that are down-regulated in dcl1− but not in dcl2−. Only one of these exons shows a reduced accumulation in the double dcl mutant, although two others are just below the 4-fold threshold in the dcl1−/dcl2− strain (Supplementary Table S3). We cannot rule out the possibility that some of the ex-siRNAs of this class is only an artefact of the sequencing, since it is difficult to explain why the double dcl mutant is different from dcl1−. However, we can point out that one of the exons included in this class (ID: 27711) encodes a conserved protein that in other fungi, such as yeast, co-localize with other proteins in sites of polarized growth (tip of the hypha) (19,20). This, together with the fact that other exons code for proteins involved in mitochondria metabolism and ribosome function may help to understand the phenotype of the dcl1 mutant, that is, abnormal hyphal morphology and a decrease in the growth rate.
The biogenesis and function of the class 2 ex-siRNAs, the largest group of ex-siRNAs we have identified in M. circinelloides, somewhat resemble that of endogenous siRNAs in animals and plants. However, there are clear differences between these ex-siRNAs and previously described esRNAs. The most similar group is the endogenous siRNAs in C. elegans, which regulate the expression of protein coding genes. However, these are mainly generated directly by the RdRP activity without the participation of a Dicer enzyme (21–23). The 24-mer heterochromatin siRNAs in plants are produced by Dicer from an RdRP generated dsRNA, but these siRNAs predominantly target transposons and repeat elements (24). They are therefore similar to the Mucor esRNAs mapping to transposons. Endo-siRNAs found in Drosophila and mouse oocytes are similar to class 2 ex-siRNAs because some of these target protein coding genes, but this class of esRNAs is produced from complementary transcripts without RdRP activity (4). The class 2 ex-siRNA pathway involves elements from all these different pathways. Thus, dsRNA is generated by Mucor RdRP, similarly to plants (heterochromatin-siRNA), the dsRNA is cleaved by Dicer, as occurs in plants (heterochromatin-siRNAs) and higher animals (endo-siRNA) and the generated ex-siRNAs target protein coding genes in cis, which is similar to animals (endo-siRNA and secondary siRNAs). Therefore, the ex-siRNA pathway in Mucor uses all these known elements but in a new and unique combination.
A new class of esRNA, qiRNAs, was recently reported to be involved in DNA-damage response in another filamentous fungus, Neurospora crassa (25). Here, we identified another class of esRNAs that potentially regulate up to 276 mRNAs, suggesting that this layer of regulation is extensive in Mucor and potentially in other fungi. Indeed, the dcl mutants are affected in hyphal morphology and colony growth (dcl1−) (10) and in the production of asexual spores (dcl2−) (8). We identified ex-siRNAs in fungi grown in optimal conditions but qiRNAs are induced by DNA damage (25) and it is also well documented that plant short RNAs are involved in stress responses (26). Therefore, it is possible that expression of other genes is also regulated by ex-siRNAs when the fungus grows in sub-optimal conditions or responds to different signals to execute complex developmental processes, such as asexual sporulation or sexual interaction. Finally, although we cannot rule out that miRNAs will be identified in other classes of fungi, the apparent lack of miRNAs in fungi raises the question whether miRNAs were present in a common eukaryotic ancestor and lost in fungi or appeared independently in plants and animals. The first option is supported by the presence of miRNAs in green alga (13) and the latter is supported by the differences in miRNA biogenesis, degree of complementarity to the target mRNAs and lack of sequence homology between animal and plant miRNAs.
ACCESSION NUMBER
GEO GSE18958.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Commission (FP6 Integrated Project SIROCCO LSHG-CT-2006-037900 to T.D.); Spanish Ministerio de Ciencia e Innovación (BFU2006-02408 and BFU2009-07220 to R.M.R-V.); Office of Science of the US Department of Energy (DE-AC02-05CH11231 to I.V.G.). Funding for open access charge: EU grant, FP6 Integrated Project SIROCCO LSHG-CT-2006-037900.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank for Karim Sorefan for critical reading of the article.
REFERENCES
- 1.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grewal SI, Jia S. Heterochromatin revisited. Nat. Rev. Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 3.Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 2006;20:759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- 4.Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat. Rev. Mol. Cell. Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Nakayashiki H, Nguyen QB. RNA interference: roles in fungal biology. Curr. Opin. Microbiol. 2008;11:494–502. doi: 10.1016/j.mib.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 8.de Haro JP, Calo S, Cervantes M, Nicolás FE, Torres-Martínez S, Ruiz-Vázquez RM. A single dicer gene is required for efficient gene silencing associated with two classes of small antisense RNAs in Mucor circinelloides. Eukaryot. Cell. 2009;8:1486–1497. doi: 10.1128/EC.00191-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicolás FE, Torres-Martínez S, Ruiz-Vázquez RM. Two classes of small antisense RNAs in fungal RNA silencing triggered by non-integrative transgenes. EMBO J. 2003;22:3983–3991. doi: 10.1093/emboj/cdg384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicolás FE, de Haro JP, Torres-Martínez S, Ruiz-Vázquez RM. Mutants defective in a Mucor circinelloides dicer-like gene are not compromised in siRNA silencing but display developmental defects. Fungal Genet. Biol. 2007;44:504–516. doi: 10.1016/j.fgb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Szittya G, Moxon S, Santos DM, Jing R, Fevereiro MP, Moulton V, Dalmay T. High-throughput sequencing of Medicago truncatula short RNAs identifies eight new miRNA families. BMC Genomics. 2008;9:593. doi: 10.1186/1471-2164-9-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prufer K, Stenzel U, Dannemann M, Green RE, Lachmann M, Kelso J. PatMaN: rapid alignment of short sequences to large databases. Bioinformatics. 2008;24:1530–1531. doi: 10.1093/bioinformatics/btn223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molnár A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007;447:1126–1129. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
- 14.Moxon S, Schwach F, Dalmay T, Maclean D, Studholme DJ, Moulton V. A toolkit for analysing large-scale plant small RNA datasets. Bioinformatics. 2008;24:2252–2253. doi: 10.1093/bioinformatics/btn428. [DOI] [PubMed] [Google Scholar]
- 15.Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, Bartel DP. RNAi in budding yeast. Science. 2009;326:544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lima WF, Murray H, Nichols JG, Wu H, Sun H, Prakash TP, Berdeja AR, Gaus HJ, Crooke ST. Human Dicer binds short single-strand and double-strand RNA with high affinity and interacts with different regions of the nucleic acids. J. Biol. Chem. 2009;284:2535–2548. doi: 10.1074/jbc.M803748200. [DOI] [PubMed] [Google Scholar]
- 19.Bi E, Chiavetta JB, Chen H, Chen GC, Chan CSM, Pringle JR. Identification of novel, evolutionarily conserved Cdc42p interacting proteins and of redundant pathways linking Cdc24p and Cdc42p to actin polarization in yeast. Mol. Biol. Cell. 2000;11:773–793. doi: 10.1091/mbc.11.2.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chesneau L, Dupré S, Burdina A, Roger J, Le Panse S, Jacquet M, Cuif MH. Gyp5p and Gyl1p are involved in the control of polarized exocytosis in budding yeast J. Cell Sci. 2004;117:4757–4767. doi: 10.1242/jcs.01349. [DOI] [PubMed] [Google Scholar]
- 21.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 22.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 23.Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 24.Lu C, Kulkarni K, Souret FF, MuthuValliappan R, Tej SS, Poethig RS, Henderson IR, Jacobsen SE, Wang W, Green PJ, et al. MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 2006;16:1276–1288. doi: 10.1101/gr.5530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee HC, Chang SS, Choudhary S, Aalto AP, Maiti M, Bamford DH, Liu Y. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature. 2009;459:274–277. doi: 10.1038/nature08041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phillips JR, Dalmay T, Bartels D. The role of small RNAs in abiotic stress. FEBS Lett. 2007;581:3592–3597. doi: 10.1016/j.febslet.2007.04.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.