Abstract
Translation termination in eukaryotes typically requires the decoding of one of three stop codons UAA, UAG or UGA by the eukaryotic release factor eRF1. The molecular mechanisms that allow eRF1 to decode either A or G in the second nucleotide, but to exclude UGG as a stop codon, are currently not well understood. Several models of stop codon recognition have been developed on the basis of evidence from mutagenesis studies, as well as studies on the evolutionary sequence conservation of eRF1. We show here that point mutants of Saccharomyces cerevisiae eRF1 display significant variability in their stop codon read-through phenotypes depending on the background genotype of the strain used, and that evolutionary conservation of amino acids in eRF1 is only a poor indicator of the functional importance of individual residues in translation termination. We further show that many phenotypes associated with eRF1 mutants are quantitatively unlinked with translation termination defects, suggesting that the evolutionary history of eRF1 was shaped by a complex set of molecular functions in addition to translation termination. We reassess current models of stop-codon recognition by eRF1 in the light of these new data.
INTRODUCTION
Eukaryotic translation termination is mediated by two interacting polypeptides, the eukaryotic release factors eRF1 and eRF3 (1–3). eRF1 interacts with the ribosomal A site and, upon decoding of a stop codon, induces hydrolysis of the peptidyl-tRNA:peptide bond. The exact molecular details underlying stop codon decoding by eRF1 are unclear, but several competing models have been proposed that differ substantially in the predicted stop codon binding sites on eRF1. One of these models, termed the ‘cavity model’, posits that specific nucleotide binding pockets in the N-terminal domain of eRF1 physically accommodate the 3 nt of the stop codon. This model has received strong support from several recent studies, and it was proposed that eRF1 adapts to the different stop codons via eRF3-controlled conformational changes that alter the relative arrangement of the respective pockets (4–6).
eRF3 assists the decoding process in a manner dependent on its physical interaction with eRF1 and on the hydrolysis of GTP (7,8). eRF3 also links translation termination to the control of mRNA turnover. The interplay of contacts between this protein, the poly(A) binding protein and factors involved in the non-sense-mediated decay (NMD) pathway are important for the rapid destruction of mRNAs containing premature stop codons (9–11). In addition, contacts between eRF3, the poly(A) binding protein and several other factors control the rate of normal co-translational deadenylation, and the translational life-time of mRNAs (12–14).
Stop codon read-through (i.e. the incorporation of an amino acid upon decoding of a stop codon by a near-cognate tRNA) leads to the production of C-terminally extended proteins while also altering the abundance of the corresponding mRNA. Translation termination therefore has the potential to be a major modulator of the cellular proteome. Consistent with this notion, mutations in the yeast Saccharomyces cerevisiae that affect the fidelity of translation termination display many pleiotropic phenotypes, including sensitivity to osmotic stress (15) and the microtubule-destabilizing drug benomyl (16), chromosome instability (17), respiratory deficiency (18), and cytoskeletal and cell-cycle defects (19). However, pleiotropic phenotypes may not exclusively occur as secondary defects resulting from primary translation termination defects. A non-translational function was recently suggested for eRF1 during cytokinesis, based on the suppression of cytokinesis defects in two eRF1 mutants by overexpression of the myosin light chain Mlc1p, and on the demonstration that eRF1 and Mlc1p interact physically (20). Together, these observations suggest that there are at least two routes by which alterations in translation termination factor activity can modulate quantitative traits, namely via changes in the physical properties and abundance of individual proteins resulting from stop-codon read-through, and via changes in translation-independent functions of the eRFs.
This study describes a quantitative trait analysis of a collection of point mutants in yeast eRF1. The quantitative relationship between termination defects and other phenotypes in these mutants strongly indicates that many pleiotropic phenotypes do not quantitatively correlate with translation termination defects in this organism, and are thus most likely due to defects in non-translational functions of eRF1. Our findings have implications for current molecular models of stop codon recognition.
MATERIALS AND METHODS
Plasmids and yeast strains
Plasmids used in this study are summarized in Table 1, and yeast strains in Table 2.
Table 1.
Plasmids used in this study
| Plasmid name | Description | Reference |
|---|---|---|
| 228 | CEN/ARS LEU2 sup45-S74F | (4) |
| 222 | CEN/ARS LEU2 sup45-M48I | (4) |
| 242 | CEN/ARS LEU2 sup45-D110G | (4) |
| 707 | CEN/ARS LEU2 sup45-H129R | (4) |
| 708 | CEN/ARS LEU2 sup45-I32F | (4) |
| 718 | CEN/ARS LEU2 sup45-P38L | (4) |
| 721 | CEN/ARS LEU2 sup45-L123V | (4) |
| 731 | CEN/ARS LEU2 sup45-V68A | (4) |
| pTH353 and mutant derivatives | CEN/ARS LEU2 SUP45 or sup45 | This study |
| pTH400 | CEN/ARS URA3 PGal-SUP45 | This study |
| pTH422 | 2 µ URA3 SUP35 | This study |
| Dual luciferase readthrough reporter plasmids | (32) |
Table 2.
Yeast strains used in this study
| ΔMT604 | MATa/α SUQ5/SUQ5 ade2-1/ade2-1 his3-11,15/his3-11,15 ura3-1/ura3-1 leu2-1/ leu2-1 trp1-1/trp1-1 can1-100/can1-100 sup45::HIS3/SUP45 [PIN+] [psi−] | (21) |
| YTH82 | MATα SUQ5 ade2-1 his3-11,15 ura3-1 leu2-1 trp1-1 can1-100 sup45::HIS3 [PIN+] [psi−] [pTH400] | This study |
| YTH87 | =YTH82 upf1::KanMX4 | This study |
| LJ1, 74D-694 | MATa/α ade1-14/ade1-14 trp1-2889/trp1-2889 his3Δ200/his3Δ200 ura3-52/ura3-52 leu2-3,122/leu2-3,112 | (23,31) |
| YTH88 | MATa ade1-14 trp1-2889 his3Δ200 ura3-52 leu2-3,112 sup45::KanMX4 [pTH400] | This study |
| BY4740 | MATa/α leu2Δ0/ leu2Δ0 lys2Δ0/ lys2Δ0 ura3Δ0/ ura3Δ0 his3Δ1/ his3Δ1 | (22) |
| YTH90 | MATα leu2Δ0 lys2Δ0 ura3Δ0 his3Δ1 sup45::KanMX4 [pTH400] | This study |
Shuffling strains
The shuffling strain used throughout most of this study (except for experiments in Figure 2) was created from diploid strain ΔMT604 (21), which is heterozygous for the SUP45 locus (MATa/α SUQ5/SUQ5 ade2-1/ade2-1 his3-11,15/his3-11,15 ura3-1/ura3-1 leu2-1/leu2-1 trp1-1/trp1-1 can1-100/can1-100 sup45::HIS3/SUP45 [PIN+] [psi−]). ΔMT604 was transformed with pTH400, a centromeric URA3 marker plasmid containing the SUP45 gene under control of the GAL1-promoter, sporulated, and a spore able to grow on medium containing galactose and lacking histidine, but unable to grow on glucose-containing medium, was selected as final shuffling strain YTH82.
Figure 2.
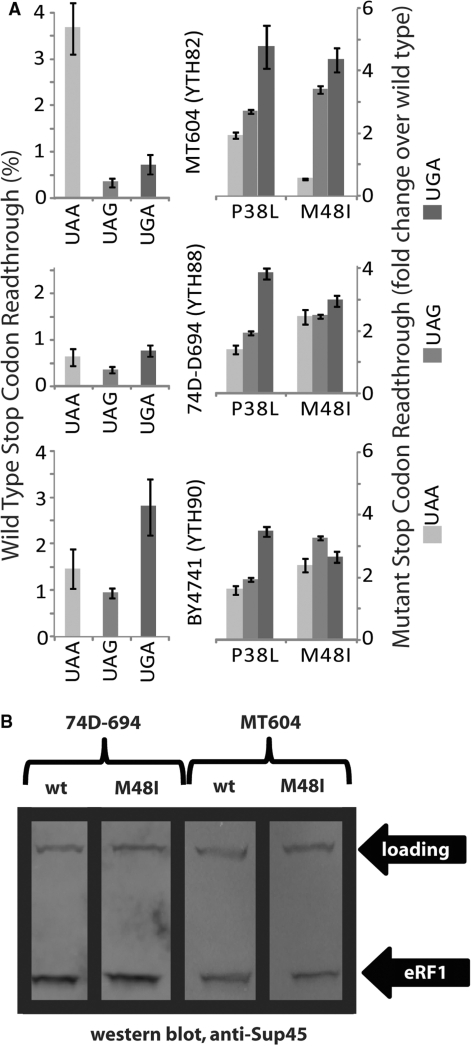
Strain specificity of stop codon read-through. (A) Bar graphs on the left show basal stop codon read-through levels for three SUP45 shuffling strains. Bar graphs on the right show changes in stop codon read-through compared to wild-type for two different mutants. Error bars indicate the variability in data from four independent transformants. Strains used are indicated. See text for discussion. (B) Western blots indicate that there are no differential stability effects that could give rise to the strain specific effect of the M48I mutant observed in panel A.
Shuffling strains based on the BY4743 (22) and 74D-694 (23) backgrounds (used for experiments in Figure 2) were created by first disrupting one of the SUP45 loci in the diploid parent strains using PCR-generated KanMX4 cassettes (24), then transforming with pTH400 and sporulation as described for ΔMT604 above.
Plasmid shuffling was performed as previously described (25), by first introducing LEU2 plasmids that express Sup45 under control of the wild-type SUP45 promoter, and then using 5-FOA as selection medium for cells that have lost the original URA3 plasmids.
Plasmids containing wild-type or mutant alleles of SUP45 were constructed by excising an XbaI–SalI fragment containing the SUP45 gene with the adjacent regulatory sequences from plasmid UKC803 (26) and insertion into pRS315 (27), a centromeric LEU2 marker plasmid, to yield pTH353. sup45 mutants were generated by PCR, either by using existing templates containing the relevant mutations or by using site-directed mutagenesis (28). Mutant DNA sequences were then introduced into pTH353 by digesting the PCR products with BglII and SpeI (enzymes that cut near the extreme 5′- and 3′-ends of the SUP45 sequence), and cloning of the resulting fragments into similarly digested pTH353. A subset of the mutants (4) was provided by Dr Ian Stansfield (Aberdeen, UK) in an identical plasmid background. A 2 µ URA3 marker plasmid for the overexpression of Sup35 was generated by excising an XhoI–NotI fragment containing the SUP35 ORF and regulatory sequences from plasmid UKC1620, and ligation into similarly digested pRS426 (27).
Western blotting was performed using an alkaline lysis method as described in ref. (29). Five ODs of cells were lysed in 100 µl standard buffer without urea for blots with anti-eRF1 antibodies (MT44), or in buffer containing urea for blots with anti-eRF3 antibodies (MT30). Antibodies were raised in rabbit and have been described before (30,31).
Dual-luciferase assays of termination efficiency
Termination efficiency of strains transformed with the luciferase-based reporter constructs was determined in 96-well microtitre plates as follows, using a commercial dual luciferase assay (Dual-Glo Assay, Promega, UK). Individual transformants were inoculated into 150 µl of SC medium lacking uracil in the wells of a 96-well plate and grown with rapid shaking (1000 r.p.m.) at 30°C in a microplate thermoshaker (Grant Bio). After overnight growth, 10 µl of these cultures were transferred into 140 µl of fresh medium in a new 96-well microplate, and grown for an additional 4 h. Immediately prior to the luciferase measurements, 40 µl of passive lysis buffer (PLB, Promega, UK) was added per well. Culture/ PLB mixture (25 µl) were then mixed with 25 µl of the Firefly luciferase substrate in the wells of an opaque 96-well plate, incubated for 20 min at room temperature, and Firefly luciferase activity was measured in a BMG Fluostar microplate reader. Stop-and-Glo reagent (25 µl) was then added per well, and the Renilla luciferase activity was measured after another 20 min of incubation.
Read-through values were calculated as follows (32): F/R ratios were calculated by dividing firefly and Renilla values from each well. Percent read-through was then calculated by dividing the F/R ratio of a construct containing the relevant stop codon by the F/R ratio of a control construct without any stop codon. Values presented in this study are the average of at least four biologically independent replicates.
Quantitative colony colour determination
Cells (5 µl) diluted to an OD of 0.1 in water were spotted onto agar plates and incubated at 30°C for 48 h. Plates were then removed from the incubator, allowed to cool to room temperature and scanned at 360 dpi and 24 bit colour using an Epson Perfection 2580 Photo scanner. The acquired images were analysed using the box tools and histogram function of ImageJ (v1.33u, http://rsbweb.nih.gov/ij/), which returns the distribution and mean of the RGB values in the analysed image section.
Quantitative analyses of phenotypes
Growth assays were conducted in minimal medium lacking uracil, using strains that had been transformed to URA3 with the dual-Luciferase reporter plasmids. Logarithmic growth rates of three independent transformants were determined by following the OD600 over time.
For other phenotypic analyses, 5 µl of yeast cultures were spotted onto plates at serial 10-fold dilutions (final ODs of 0.1, 0.01 and 0.001) and incubated at the respective growth temperatures and media for 48 h. Plates were then scanned and the diameter of colonies that had grown without touching adjacent colonies was determined using the program ImageJ (version 1.33, http://rsb.info.nih.gov/ij/). Clustering analyses and multidimensional scaling were performed using the program ClustanGraphics (ver. 8.04, http://www.clustan.com). Phenotypic traits were scaled between the wild-type and worst observed phenotype, and clustered using Pearson’s rho.
Evolutionary conservation scores
eRF1 sequences from a published set of organisms with standard code stop codon usage (33) were used to calculate average Gonnet scores for every amino acid position in yeast eRF1.
RESULTS
Generation of an eRF1 mutant collection
As a starting point to our study, we established an extensive collection of isogenic yeast strains containing single amino acid exchanges in eRF1. Plasmid-borne mutant alleles of the SUP45 gene (which encodes eRF1 in S. cerevisiae) were collated from a wide variety of sources including alleles previously described in other strain backgrounds (20,34–37), alleles that displayed functional defects in in vitro models of stop codon decoding (38–40) but that had to date not yet been tested in vivo, fortuitous mutants arising during cloning procedures in our lab, and mutants identified in an in vivo suppressor screen (4). A complete list of eRF1 mutants used in this study is given in Table 3.
Table 3.
SUP45 alleles used in this study
| SUP45 mutant | Amino acid distance scorea | Published allele name | Readthrough frequency (%) |
Growth rate (% of wt.) | eRF1 expression level (% of wt.) | Ref. | ||
|---|---|---|---|---|---|---|---|---|
|
UAAC |
UAGC |
UGAC |
||||||
| wt [psi−] | – | 3.7 | 0.3 | 0.7 | 100 | 100 | This study | |
| wt [PSI+]s | – | 12.7 | 2.0 | 5.8 | 101 | nd | This study | |
| wt [PSI+]m | – | 9.8 | 1.1 | 3.4 | 99 | nd | This study | |
| wt [PSI+]w | – | 5.6 | 0.7 | 1.5 | 97 | nd | This study | |
| S30P | 0.4 | sl23ts | 5.0 | 0.6 | 1.3 | 99 | 101 | (20) |
| I32F | 1.0 | sup45-708 | 6.4 | 0.2 | 5.8 | 87 | 30 | (4) |
| L34S | −2.1 | sup45-36ts | Unstable | (34) | ||||
| P38L | −2.3 | sup45-718 | 7.1 | 0.9 | 3.5 | 80 | 96 | (4) |
| M48I | 2.5 | sup45-222 | 2.1 | 1.2 | 3.2 | 75 | 129 | (4) |
| L49A | −1.2 | 3.0 | 0.3 | 0.7 | 100 | nd | This study | |
| E52A | 0.0 | 3.4 | 0.3 | 0.7 | 100 | nd | This study | |
| G54D | 0.1 | Not viable | This study | |||||
| T55A | 0.6 | 5.4 | 0.4 | 0.6 | 97 | 86 | This study | |
| N58A | −0.3 | 22.4 | 1.9 | 3.8 | 71 | 140 | This study | |
| K60A | −0.4 | Not viable | This study | |||||
| S61A | 1.1 | 3.8 | 0.4 | 0.8 | 100 | nd | This study | |
| R62A | −0.6 | 5.1 | 0.4 | 1.1 | 92 | 87 | This study | |
| R65C | −2.2 | sup45-3 | Unstable | (35) | ||||
| V68A | 0.1 | sup45-731 | 8.9 | 0.3 | 4.6 | 87 | 85 | (4) |
| S74F | −2.8 | sup45-228 | 13.2 | 1.6 | 2.0 | 80 | 96 | (4) |
| V107D | −2.9 | 10.2 | 0.8 | 4.7 | 100 | 29 | This study | |
| D110G | 0.1 | sup45-242 | 12.2 | 0.9 | 2.8 | 98 | 119 | (4) |
| L123V | 1.8 | sup45-721 | 4.4 | 0.3 | 10.0 | 101 | 118 | (4) |
| H129R | 0.6 | sup45-707 | 4.3 | 0.3 | 5.4 | 98 | 96 | (4) |
| P174Q | −0.2 | 7.0 | 0.7 | 2.5 | 89 | 82 | This study | |
| K175A | −0.4 | Not viable | This study | |||||
| K176A | −0.4 | Not viable | This study | |||||
| G180A | 0.5 | Not viable | This study | |||||
| G181A | 0.5 | Not viable | This study | |||||
| Q182E | 1.7 | Unstable | (37) | |||||
| Q182N | 0.7 | Unstable | (37) | |||||
| I222S | −1.8 | sup45-2 | 9.1 | 1.4 | 3.9 | 88 | 85 | (36) |
| T295A | 0.6 | 3.7 | 0.4 | 0.7 | 100 | nd | This study | |
| T305A | 0.6 | 4.3 | 0.4 | 0.9 | 100 | nd | This study | |
| T357A | 0.6 | 3.9 | 0.4 | 0.8 | 100 | nd | This study | |
| T388A | 0.6 | 4.3 | 0.3 | 0.9 | 100 | nd | This study | |
| F401Y | 5.1 | 4.1 | 0.3 | 0.6 | 85 | nd | This study | |
| Y410F | 5.1 | 5.0 | 0.4 | 1.6 | 87 | 74 | This study | |
Statistically significant differences to wild-type are indicated in bold (P <0.05) or bold italic (P <0.01). All data for sup45 mutants were generated in a [psi−] background (strain YTH82/MT604).
aGiven as the log odds score for observing this amino acid substitution in natural protein alignments, based on the PAM250-normalized Gonnet matrix (42). Lower numbers indicate less frequently observed substitutions.
Nd, not determined.
Mutants were introduced into yeast strain YTH82/MT604, which contains a deletion of the chromosomal SUP45 gene, by using a standard plasmid shuffling strategy (25). To ensure that any observed termination defect was solely the result of the introduced eRF1 mutations, we utilized the ade2-1 non-sense allele present in our shuffling strain. This allele leads to accumulation of a red intermediate in the adenine synthesis pathway (33). Defects in the termination machinery lead to partial suppression of the ade2-1 allele and restoration of white colony colour, thus allowing an easy visual assessment of termination defects. We patched 10 or more independently shuffled colonies per mutant onto one-fourth Yeast extract/Peptose/Dextrose medium (YPD) plates, and individual colonies were only subjected to further analysis if the colour change on one-fourth YPD was consistent with the majority of shuffled samples. For each shuffled mutant, we also ensured that colony colour reverted to the non-suppressed colour when a plasmid-borne wild-type SUP45 allele was re-introduced into the strains.
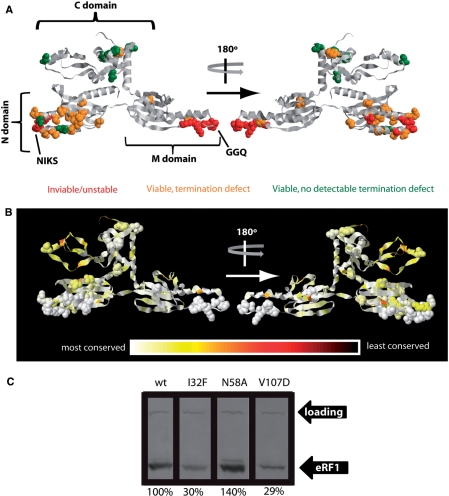
Out of the 34 mutant sup45 alleles tested, 28 conferred viability when present as the sole source of eRF1. Four of these 28 alleles initially yielded identical termination defects in independently shuffled strains, but these defects reverted to the wild-type phenotype within short periods of sub-culturing the cells (days to weeks). Because of the difficulties in analysing such phenotypically unstable mutants, these four mutants were discounted from further analyses. Figure 1A summarizes the location of all inviable and unstable mutations. All of the respective amino acids are located within or directly adjacent to the highly conserved TASNIKS and GGQ motifs. Our in vivo results are thus consistent with the previous identification of these motifs in vitro as sites of particular functional importance for release factor activity and translation termination (38,39).
Figure 1.
eRF1 point mutants affect viability and termination efficiency in yeast. Yeast strain YTH82 was used to generate data for this figure. (A) Amino acids mutated in this study are shown in spacefill in a ribbon model of yeast eRF1. The yeast model was generated by homology modelling based on the published structure of human eRF1 (48). Phenotypes of the respective mutants are indicated in red (inviable or genetically unstable), orange (viable but detectable termination defect on at least one of the three stop codons) or green (viable and no significant termination defect on any of the analysed stop codons). (B) The relative conservation of amino acids in the eRF1 sequence is indicated by colour. Amino acids mutated in this study are shown in spacefill as in panel A. (C) Some eRF1 mutants alter the intracellular abundance of the protein. A representative western blot is shown for the three mutants that showed significantly different changes in abundance compared to wild-type. Numbers under the blots give the abundance relative to wild-type, as average of three independent experiments. ‘Loading’ indicates a high molecular weight band arising from a cross reaction of the anti-eRF1 antibody, which was used as loading control.
Termination defects in eRF1 mutants
A dual-luciferase based reporter system (41) was used to accurately measure termination defects associated with the 24 phenotypically stable alleles. Stop codon read-through at all three stop codons was measured, in each case using cytosine as the base 3′ of the stop codon since this context is known to confer the highest levels of basal read-through (7). This analysis revealed widespread translation termination defects for mutations in the N-terminal and middle domains of eRF1 (amino acids 1–276, Table 3 and Figure 1). In contrast, only one of the mutations introduced into the C-terminal domain (encompassing amino acids 277–437) produced a significant increase in stop codon read-through. In order to assess whether the observed termination defects were the result of functional changes in eRF1 or of changes in abundance, we determined relative levels of eRF1 in the mutants via a quantitative protein extraction and western blotting procedure (29). Two of the mutants (I32F and V107D) showed statistically significant reductions in abundance, while the N58A mutant showed a significant increase (Figure 1C). This indicates that, in most mutants, termination defects were the result of functional changes in eRF1.
To facilitate comparisons between the different mutants, we quantitatively analysed both the relative conservation of the mutated amino acids during evolution, and the severity of the introduced amino acid exchange, by using substitution frequencies from the Gonnet matrix as a reference (42). Consistent with previous reports, eRF1 is generally highly conserved, with a slightly lower degree of conservation evident in the C-terminal domain compared with the N- and M-domains. There are only small windows of variable amino acids dotted throughout its primary sequence (Figure 1B). Since none of the mutations described here affect any of these less conserved residues, all would be expected to display similarly strong phenotypes if they involved similarly severe amino acid changes.
The amino acid changes in the mutant alleles varied widely in their impact on translation termination (Table 3), although surprisingly there was no obvious correlation between the severity of the amino acid exchange and the severity of the observed termination defect. This lack of correlation was particularly obvious in the C-domain, where the only mutation resulting in significant stop-codon read-through was Y410F, the most conservative amino acid exchange in our collection. On the other hand, some relatively severe changes in highly conserved residues (e.g. S61A, R62A) displayed no or only very mild termination defects.
For seven mutants from the Stansfield collection where stop codon read-through levels were previously reported using a different reporter assay to the one employed here (4), our results agree only partially with the original report. There is excellent agreement for the L123V and H129R mutants, which confer strong UGA-specific read-through in both our studies. In contrast, for the P38L, M48I, V68A, S74F and D110G mutants, we observe significant differences in the specificity and levels of reported stop-codon read-through. Such differences might either arise from the different yeast strains used in our respective studies or from the different reporter systems employed to measure stop-codon read-through. To address this question, we constructed SUP45 shuffling strains in two additional genetic backgrounds, namely YTH90 which is based on the S288c-derived BY4741 background (22), and YTH88 which is based on 74D-694 (23), a strain background that is not genetically related to S288c. Our existing MT604-based shuffling strain (YTH82) has a genetic make-up that is largely—but not exclusively—derived from S288c (B.S. Cox, personal communication). MT604 also contains the serine-inserting SUQ5 ochre suppressor allele which leads to high UAA read-through levels (43).
Stop codon read-though was measured in all three strains for the wild-type SUP45 allele, as well as the P38L and M48I mutants, for which we had obtained results that differed significantly from the original study (Figure 2). The P38L allele gave highly reproducible results in all three strain backgrounds, in that it increased read-through on UGA codons ∼4-fold, and on UAA and UAG codons ∼2-fold. This is very different from the original data reported for this allele, which showed moderate UAA-specific read-through. In contrast to our reproducible data for the P38L allele, we observed significant strain-specific differences for the M48I allele. While this allele produced a weak omnipotent suppressor phenotype in the BY4741 and 47D-694 derived strains, it increased read-through on UAA and UAG codons more strongly in MT604, but actually decreased stop codon read-through on UAA codons. Repeat measurements in freshly constructed mutant strains confirmed these results. Interestingly, the original study described decreased read-through on UGA codons for this SUP45 allele. From the differences observed in our three strain backgrounds, as well as differences between the study conducted by Bertram et al. (4) and this study, we conclude that the effects of individual termination factor alleles on termination efficiency can be strongly modulated by undefined differences in the host genotype. It is also noteworthy that Bertram et al. (4) discovered the I32F allele as a chromosomal mutation, but were unable to shuffle this allele into their shuffling strain (which differs from the strains used here), while we had no difficulties in shuffling the same I32F mutant in our study. On the other hand, we were unable to shuffle the L34S allele into YTH82/MT604, but this allele was originally discovered as a ts mutation that was viable at low temperatures [sup45-36ts, ref. (34)]. We were unable to shuffle this mutant in multiple attempts, including the use of transformation procedures that avoided the use of high temperatures (44). The differing viability of mutants in different strains supports the notion that the same mutation can have substantially different effects in different strain backgrounds.
The relationship between growth defects and termination efficiency
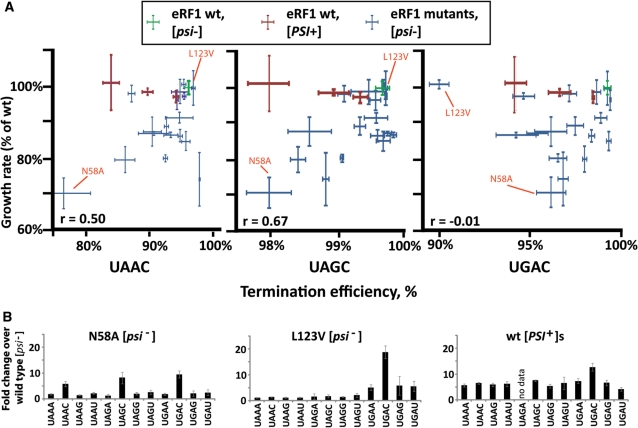
In order to gain insights into the relationship between stop codon read-through and growth defects, we measured growth rates for all viable sup45 mutants in SC medium lacking uracil, i.e. under the same conditions used for measuring termination efficiency. Strains were transformed to URA3 using the stop-codon read-through plasmids for this purpose. As a means of control, we also generated a series of [PSI+] variants with different levels of stop-codon read-through that were isogenic to the eRF1 mutant collection. These [PSI+] variants covered a similar range of stop-codon read-through values for the three codons in a C context as the eRF1 mutants, with the exceptions of the N58A mutant which showed higher UAA read-through levels than any of the [PSI+] variants, and L123V, which showed higher UGA read-through (Table 3). In order to evaluate how the eRF1 mutants compared to the [PSI+] variants for other stop codon contexts, we determined termination efficiency on all possible tetranucleotide stop signals for the strongest [PSI+] variant, and the N58A and L123V mutants (i.e. the strongest omnipotent suppressor and the strongest stop-codon specific suppressor in our collection, Figure 3B). The results indicate that the eRF1 mutants do not show significantly worse termination defects when compared to the [PSI+] variants.
Figure 3.
Relationship between stop codon read-through and growth in yeast eRF1 mutants. Yeast strain YTH82 was used to generate data for this figure. (A) plots of growth rates versus termination efficiency for individual strains for three stop codons with ‘C’ as fourth base context. Growth data are from the same experiment in all three graphs. Error bars (x-axis) indicate the standard error for the growth rate of three independent transformants, y-axis error bars indicate the standard error for termination efficiency of four independent transformants. Pearson product moment correlation coefficients for the correlation between termination defects and growth are given. Data points for eRF1 mutants analysed in panel B are indicated. (B) The change in stop codon read-through was measured for all possible tetranucleotide stop signals in two eRF1 mutants and [PSI+] cells. Error bars indicate the standard deviation of data from six independent transformants. These data indicate that loss of growth in the N58A mutant (panel A) is unlikely to stem from general termination defects, since the [PSI+] strain shows more severe termination defects for most stop signals.
None of the [PSI+] variants showed a statistically significant change in growth rates when compared to the [psi−] wild-type strain. In contrast, many of the [psi−] strains containing eRF1 mutants grew significantly slower than the wild-type [psi−] strain. A moderate correlation was observed between the termination efficiency and growth rate when data for the UAG codon were analysed (Figure 3, pearson product moment correlation coefficient = 0.67), with lower correlation for the other two codons. In summary, while none of the [PSI+] strains showed any reduction in growth rate, eRF1 mutants showed significant reductions when compared to wild-type eRF1 cells. Because eRF1 mutants do not show significantly worse termination defects in any context than the [PSI+] strains, growth rate reductions in these mutants appear to be independent of the termination defects.
Pleiotropic phenotypes caused by mutations in eRF1
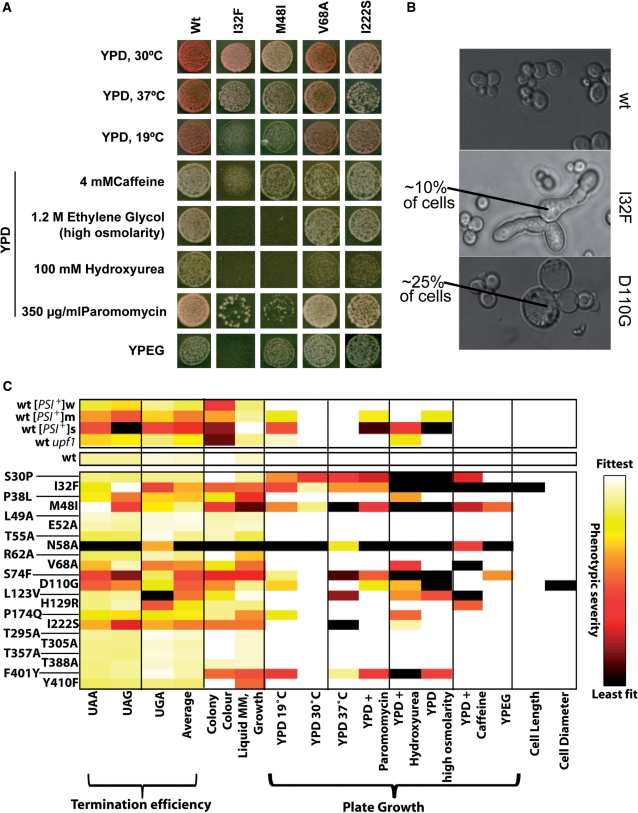
To further characterize potential non-translational defects in eRF1 mutants that might be causally linked to the reduced growth rates, we investigated phenotypic traits associated with the sup45 alleles. An illustrative selection of mutants is shown in Figure 4A. We studied previously published phenotypes of eRF1 mutants including growth at high and low temperatures, on non-fermentable carbon sources, under high osmolarity conditions and in the presence of paromomycin. We also uncovered two phenotypes not previously reported for eRF1 mutants, namely impaired growth on media containing 100 mM hydroxyurea or 4 mM caffeine. The mutants also displayed very specific cell morphology defects (Figure 4B), with sub-populations of I32F mutant cells showing an elongated shape reminiscent of the shmooing process that yeast cells undergo during mating, while cultures of D110G strains contained enlarged, rounded cells. All of the observed phenotypes could be suppressed by re-introducing wild-type SUP45 genes (data not shown), confirming that these phenotypes are tied to the eRF1 alleles in the respective strains.
Figure 4.
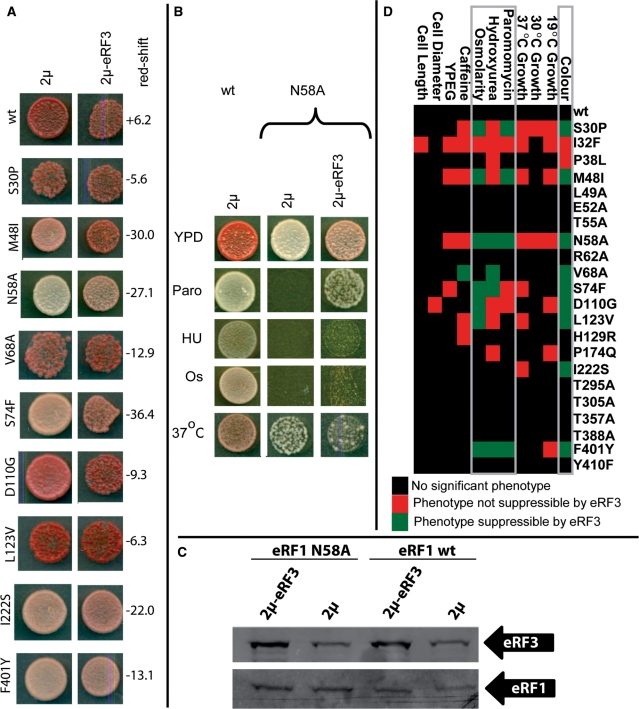
Pleiotropic phenotypes in sup45 mutants. Yeast strain YTH82 was used to generate data for this figure. (A) Observed growth phenotypes of a selection of eRF1 mutants. (B) Cell shape phenotypes observed in I32F and D110G eRF1 mutants. (C) A heat map summarizing all observed phenotypes for all mutants and control strains. Growth phenotypes were quantified by measuring colony diameters after 48 h of growth under the respective conditions, and normalized to values between one (fittest observed phenotype) and zero (worst observed phenotype).
All of the mutant strains were also plated onto the different media at lower cell densities than those shown in Figure 4, and the diameter of individual colonies was measured following 48 h of growth. This parameter was taken to be a quantitative indicator of the severity of the observed phenotypes (Figure 4C). The same analysis was also conducted for a series of control strains, namely the [PSI+] strain series described earlier, and an additional strain containing wild-type SUP45 but a deletion of the NAM7/UPF1 gene, which generates weak termination defects as well as a complete block of the termination-related non-sense-mediated mRNA decay or NMD pathway (9). Two main conclusions emerge from these results: first, four of the phenotypes observed for eRF1 mutants are also observed in the different [PSI+] variants, namely sensitivities to paromomycin, hydroxyurea, high osmolarity and to reduced incubation temperatures. In the [PSI+] variant series, there was a quantitative correlation between these phenotypes and the observed termination defects, whereas these phenotypes did not show an immediately apparent correlation with termination defects for the eRF1 mutant collection. Other phenotypes specific to eRF1 mutants but not observed for the [PSI+] strains were sensitivity to growth at 37°C and to caffeine, inability to utilize non-fermentable carbon sources, and the two cell-shape defects depicted in Figure 4B. Since these phenotypes are also absent in the upf1 deletion strain, they are not the result of specific defects in the NMD pathway that might be associated with SUP45 mutants, but not with the [PSI+] state. Thus, a subset of phenotypes observed in both [PSI+] strains and eRF1 mutants may result from a translation termination defect, whereas those phenotypes observed only in eRF1 mutants are more likely to be result from defects in non-translational functions of eRF1 or in highly mRNA-specific termination defects that are not observed when steady-state levels of functional eRF3 are reduced, as is the case in [PSI+] strains.
Suppression of pleiotropic phenotypes by eRF3 overexpression
For many genes encoding components of macromolecular complexes, defects can be compensated by overexpression of genes encoding physical binding partners [i.e. gene dosage suppression, ref. (45)]. To gain additional insights into the phenotypes associated with the sup45 mutants, we investigated whether increased levels of eRF3 would affect any of the observed phenotypes. We introduced the full-length SUP35 gene encoding eRF3 on a high-copy number plasmid into the different sup45 mutant strains, which resulted in a ca. 3-fold increase in intracellular levels of eRF3 (Figure 5C). In these experiments, colony colour of the transformants was carefully monitored, and occasional colonies which had become [PSI+] due to the high eRF3 levels (46) excluded from analyses. In many of the mutants, eRF3 overexpression led to a detectable shift in colony colour towards red (Figure 5A), which was often but not always accompanied by a partial suppression of the paromomycin sensitivity, sensitivity to hydroxyurea and osmotic sensitivity. This is illustrated for the N58A mutant in Figure 5B, and summarized for all mutants in Figure 5D. In the N58A mutant, we further observed that eRF3 overexpression exacerbated a temperature-sensitivity defect (Figure 5B). For all other phenotypes, only a single case was found, namely caffeine sensitivity in the V68A mutant, where eRF3 overexpression led to amelioration of the phenotype. The observed co-suppression of several mutant-associated traits thus clearly establishes two groups of phenotypes, namely those suppressible by increased eRF3 levels and those that do not respond to this condition.
Figure 5.
eRF3 is a dosage suppressor of eRF1-related phentoypes. Yeast strain YTH82 was used to generate data for this figure. (A) eRF3 overexpression partially suppresses the colony colour change observed in eRF1 mutants. Patched colonies of cells containing either an empty 2µ plasmid, or a 2µ plasmid also containing the eRF3 gene, are shown. Numbers next to the panels indicate the red-shift in the presence of high-copy eRF3, as evaluated from computerized scans of the plates. Positive numbers indicate a shift towards white colour, negative numbers a shift towards red. (B) Effects of high copy expression of eRF3 in the eRF1 N58A mutant. High copy number expression of eRF3 partially suppresses the colony colour and paromomycin phenotypes. It also very weakly suppresses the hydroxyurea and high osmolarity phenotpyes, while exacerbating the temperature sensitivity observed in this mutant. (C) Western blots demonstrate that eRF3 is overexpressed 3-fold in the presence of high copy plasmids encoding its gene, and that neither the stability of wild-type eRF1 nor of N58A eRF1 are affected by this overexpression. (D) Summary of suppression by high copy number eRF3. Paromomycin, hydroxyurea and high osmolarity phenotypes can be co-suppressed together with colony colour in many sup45 (eRF1) mutants.
Correlation analysis of termination and pleiotropic defects in eRF1 mutants
A minimal framework for the quantitative connection between primary termination defects and secondary pleiotropic defects is that if a series of strains is ranked first in order of the severity of the termination defect and then in order of the severity of the pleiotropic defect, both rank orders should be identical. This precise relationship was for example given for the termination defect and paromomycin, hydroxyurea, osmolarity and cold-sensitivity phenotypes in the three [PSI+] strains, which gave a ranking of wild-type <[PSI+]w<[PSI+]m< [PSI+]s for all five phenotypes (Figure 4C). In contrast, no strong correlation between any two phenotypes was observed in the sup45 mutants. This may reflect the fact that, unlike in the [PSI+] strain series, termination defects in the sup45 mutants show varying codon-specificity. If secondary phenotypes were the result of compound termination defects at the three different stop codons, correlations might be masked by this codon specificity.
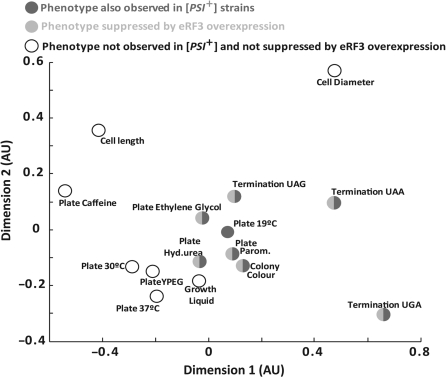
To further analyse potential correlations between termination defects and other phenotypes in the sup45 mutants Spearman’s rank correlation coefficient or rho was determined for all possible pair wise comparisons of phenotypes (Figure 6). The relative degree of similarity in terms of rho is represented by the relative distance between two parameter points. Those phenotypes that were observable in [PSI+] strains and suppressible by eRF3 overexpression, showed a higher degree of rank correlation with termination defects, in particular on UAG and UAA codons, than those phenotypes to which these criteria did not apply. This correlation analysis confirmed the division of the observed phenotypes into two groups, one that is likely to be dependent on primary termination defects, and another group that is likely to be independent of such defects.
Figure 6.
Correlation between eRF1-related phenotypes. Yeast strain YTH82 was used to generate data for this figure. The linear distances between individual data points approximate the degree of correlation between the severities of the two phentoypes, i.e. data points located more closely together show stronger correlation in the rank order of the phenotypes. See text for discussion.
DISCUSSION
Our quantitative analysis of a collection of point mutants in yeast eRF1 has revealed a surprising strain-specificity of eRF1-associated phenotypes, and an apparent lack of correlation between evolutionary conservation and termination-related functionality in eRF1 residues. These findings have important implications for current models of functional epitopes on eRF1.
Strain-specific variability of termination defects in eRF1 mutants
The apparent differences we observed between termination defects published for some sup45 alleles and defects observed for the same alleles in our genetic background prompted us to evaluate how far phenotypes may be strain specific. For example, the M48I allele produced significantly different phenotypes in different genetic backgrounds, even though all experiments were conducted using the same reporter system and identical experimental conditions. Moreover, most of the alleles from the study by Bertram et al. (4) that we measured in another genetic background also gave different results in our studies. There were also differences in the viability of the same allele in different strains [e.g. I32F which could be shuffled into our YTH82 strain but not into the shuffling strain used by Bertram et al. (4), and L34S which could not be shuffled into our strain but was viable in a chromosomal background in another strain (34)]. Taken together, these findings suggest that differences in the molecular details of translation termination exist in strains of different genetic backgrounds, and that such differences are widespread. Although we currently do not know the molecular details of these differences, we anticipate that they may be linked to differences in the abundance or activity of trans-acting modulators of translation termination. Interestingly, quantitative differences in different genetic backgrounds have previously been documented for the efficiency of NMD-mediated mRNA decay, which is functionally linked to translation termination (47).
Our study includes a number of alleles corresponding to human eRF1 mutants previously only examined in in vitro termination assays. Some of the relevant mutations produced effects in vitro that mirror the results from our in vivo study closely, namely the N58A, K60A and S61A alleles [N61A, K63A and S64A in human numbering (38)]. Other alleles show significant defects in in vitro assays but had no effect in our strains in vivo [yeast R62A and E52A, corresponding to human R65A and E55A (38,40)], although it should be noted that the in vitro assays were relatively unphysiological in their set-up and lacked eRF3, which may render the release reaction much more sensitive to amino acid changes in eRF1. In summary, we observe differences in translation termination efficiency for the same mutant in different in vivo studies, for the same mutant in different strains in the same study, and between in vivo and in vitro experiments: these findings highlight the difficulty in constructing reliable structural models of translation termination based on mutagenesis results alone.
Evolutionary conservation and functionality in termination
Analyses of evolutionary conservation have been used to infer details of the molecular functions of eRF1, and to model potential stop-codon binding sites (48,49). Such studies are based on the rationale that the sole function of eRF1 is in translation termination; and that conservation of a residue therefore indicates some form of a functional requirement during termination with respect to this residue. Surprisingly, the peptide 54-GTASNIKSR-62 is one of the highest conserved stretches in eRF1 sequences from a wide variety of organisms (48), yet yeast tolerated amino acid substitutions in some of these residues much better than in others (Figure 1 and Table 3). Although all our exchanges were of very similar severity, the G54D and K60A mutants were inviable, the N58A mutant were viable but caused a severe termination defect, while the T55A, S61A and R62A mutants only caused mild or no defects.
This apparent independence of the mechanism of translation termination on very highly conserved residues suggests that the evolutionary constraint on eRF1 residues may not solely be exerted by the requirements of translation termination. If eRF1 has roles in the cell other than translation, and if these roles contribute to achieving maximum fitness, then evolutionary constraints on sequence changes in eRF1 must reflect such non-translational roles as well as translational ones.
Our quantitative trait analyses reveal several eRF1-linked phenotypes that are unlinked to translation termination as established by three independent criteria: first, they occur in eRF1 mutants, but not when translational efficiency is reduced by prion aggregation of the second essential translation factor, eRF3 (Figure 4). Second, they are not co-suppressed when colony colour, paromomycin sensitivity and sensitivity to high osmotic pressure, three phenotypes known to be linked to translation termination, are suppressed by an extragenic suppressor (Figure 5). Lastly, their quantitative correlation with the termination defects in eRF1 is poor, making it unlikely that they are secondary consequences of a primary termination defect. These unlinked phenotypes indicate that eRF1 has multiple functions in eukaryotic cells, which have collectively shaped the evolutionary history of this essential translation factor.
The molecular mechanism of stop codon decoding
Current models of stop-codon decoding involve variations on two central themes, namely, linear models positing that stop-codons physically bind to the TASNIKS motif in the eRF1 N-domain (49–51), and ‘cavity’ models which propose that the body of the eRF1 N-domain contains several cavities that may physically accommodate the 3 nt of the stop codon (4–6). Evidence for or against these models has included data derived from evolutionary, mutagenesis and biochemical studies.
The various linear models proposed for stop codon recognition differ in the exact amino acids contacting the stop codon nucleotides. Proposed binding sites include TASNIKS (51) and GTASNIKS (49), both of which are not consistent with the absence of strong phenotypes in a T55A mutant (Table 3). The only physical evidence linking stop codons to individual amino acids in eRF1 comes from the observation that in in vitro termination assays, s4U-label can cross-link the first base of the stop codon to either K60, S61 or R62 [TASNIKSR, (50)]. The latter two residues are unlikely to act as primary sites for stop-codon interactions since their replacement by alanines does not lead to significant termination defects (Table 3).
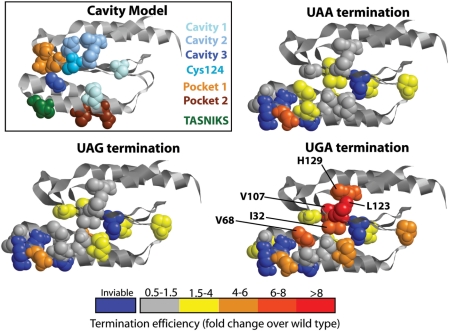
A non-linear model of stop-codon recognition, the so-called cavity model, was first proposed by Bertram et al. (4) based on the results of a screen for unipotent stop-codon suppressors. The eRF1 N-domain contains three cavities in its surface (Figure 7), and based on the stop-codon specificity of suppressor mutants and on the best fit of docking models between the stop codon and the eRF1 surface, it was proposed that these three cavities directly bind to the 3 nt of the stop codon. Additional evidence for an important role of residues in cavity two for UGA decoding was provided in another study (5), which identified C124 as a critical residue for UGA decoding. This study extended the cavity model by suggesting that conformational changes hinging around the TASNIKS motif optimize the relative orientation of the cavities for decoding of UAA/UAG on one hand and UGA on the other. Lastly, based on the results of an anti-suppressor screen, Hatin et al. (6) identified two new regions important for stop codon decoding they termed pocket one and two (Figure 7), which were proposed to be involved in maintaining an overall structural frame-work of the N-domain that is optimal for eRF1 function in translation termination.
Figure 7.
Stop codon specificity of read-through in eRF1 N-domain mutants. The boxed panel shows as reference a summary of residues identified in previous studies. Cavities 1–3 refers to residues identified by Bertram et al. (4), pocket 1/2 to residues identified by Hatin et al. (6). The K and S residues of the TASNIKS motif, and the C124 residue identified by Fan-Minogue et al. (5), are also shown. The other three panels show the same view of the eRF1 N-domain, with residues mutated in this study shown in spacefill and colour coded according to the level of stop codon read-through observed. Residues of mutants conferring strong UGA-specific read-through are indicated.
Our study strongly confirms the importance of residues around cavity two for decoding of the UGA stop codon. Both re-examination of the L123V and H129R mutants already measured by Bertram et al., (4) and new measurements for the I32F and V107D mutants which are located directly adjacent to L123 and H129 (Figure 7), show strong UGA-specific stop-codon read-through when residues of this epitope are mutated. Both the I32F and V107D mutant show reduced levels of eRF1 (Figure 1C), however, the strong stop-codon specificity is unlikely to be a result of eRF1 depletion, and is more likely related to stop-codon specific termination defects. The C124 residue identified by Fan-Minogue et al. (5) is continuous with this epitope, and all of these residues are part of or near cavity two which was proposed to be directly involved in binding to the middle nucleotide of the stop codon, and crucial for UGA recognition but less crucial for UAA/UAG recognition.
In contrast to the strong support for an important role of cavity two, we find that mutant eRF1 alleles used as evidence for the assignment of pockets one and three are poorly reproducible in different genetic backgrounds. While our new data do not conclusively exclude the three cavities as direct sites for stop codon binding, the evident variability of termination defects in eRF1 mutants means that evidence supporting roles for cavities one and three are weaker than for cavity two. The one aspect that appears to be consistently supported by the available evidence is the existence of an extensive epitope critical for UGA decoding centring around the C124 residue. However, the ability to decode UAA, UAG and UGA but exclude UGG codons is likely to be a complex result of properties distributed more widely throughout the eRF1 N-domain.
FUNDING
Wellcome Trust, UK, through a Research Career Development Fellowship (075438 to T.v.d.H.); Biotechnology and Biological Sciences Research Council (BBSRC), UK, through a project grant (96/G06869 to M.F.T.). Funding for open access charge: Wellcome Trust, UK.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Dr Ian Stansfield (University of Aberdeen, UK) for generously providing his collection of eRF1 mutants, and Prof. David M. Bedwell (University of Alabama, Birmingham, Alabama) for the dual-luciferase-based reporter constructs.
REFERENCES
- 1.Kisselev L, Ehrenberg M, Frolova L. Termination of translation: interplay of mRNA, rRNAs and release factors? EMBO J. 2003;22:175–182. doi: 10.1093/emboj/cdg017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rospert S, Rakwalska M, Dubaquie Y. Polypeptide chain termination and stop codon readthrough on eukaryotic ribosomes. Rev. Physiol. Biochem. Pharmacol. 2005;155:1–30. doi: 10.1007/3-540-28217-3_1. [DOI] [PubMed] [Google Scholar]
- 3.von der Haar T, Tuite MF. Regulated translational bypass of stop codons in yeast. Trends Microbiol. 2007;15:78–86. doi: 10.1016/j.tim.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Bertram G, Bell HA, Ritchie DW, Fullerton G, Stansfield I. Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA. 2000;6:1236–1247. doi: 10.1017/s1355838200000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fan-Minogue H, Du M, Pisarev AV, Kallmeyer AK, Salas-Marco J, Keeling KM, Thompson SR, Pestova TV, Bedwell DM. Distinct eRF3 requirements suggest alternate eRF1 conformations mediate peptide release during eukaryotic translation termination. Mol. Cell. 2008;30:599–609. doi: 10.1016/j.molcel.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatin I, Fabret C, Rousset JP, Namy O. Molecular dissection of translation termination mechanism identifies two new critical regions in eRF1. Nucleic Acids Res. 2009;37:1789–1798. doi: 10.1093/nar/gkp012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salas-Marco J, Bedwell DM. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol. Cell Biol. 2004;24:7769–7778. doi: 10.1128/MCB.24.17.7769-7778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stansfield I, Jones KM, Kushnirov VV, Dagkesamankaya AR, Poznyakovski AI, Paushkin SV, Nierras CR, Cox BS, Ter-Avanesyan MD, Tuite MF. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akimitsu N. Messenger RNA surveillance systems monitoring proper translation termination. J. Biochem. 2008;143:1–8. doi: 10.1093/jb/mvm204. [DOI] [PubMed] [Google Scholar]
- 10.Amrani N, Sachs MS, Jacobson A. Early nonsense: mRNA decay solves a translational problem. Nat. Rev. Mol. Cell Biol. 2006;7:415–425. doi: 10.1038/nrm1942. [DOI] [PubMed] [Google Scholar]
- 11.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 12.Funakoshi Y, Doi Y, Hosoda N, Uchida N, Osawa M, Shimada I, Tsujimoto M, Suzuki T, Katada T, Hoshino S. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev. 2007;21:3135–3148. doi: 10.1101/gad.1597707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosoda N, Kobayashi T, Uchida N, Funakoshi Y, Kikuchi Y, Hoshino S, Katada T. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J. Biol. Chem. 2003;278:38287–38291. doi: 10.1074/jbc.C300300200. [DOI] [PubMed] [Google Scholar]
- 14.Keeling KM, Salas-Marco J, Osherovich LZ, Bedwell DM. Tpa1p is part of an mRNP complex that influences translation termination, mRNA deadenylation, and mRNA turnover in Saccharomyces cerevisiae. Mol. Cell Biol. 2006;26:5237–5248. doi: 10.1128/MCB.02448-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh A. Nonsense suppressors of yeast cause osmotic-sensitive growth. Proc. Natl Acad. Sci. USA. 1977;74:305–309. doi: 10.1073/pnas.74.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tikhomirova VL, Inge-Vechtomov SG. Sensitivity of sup35 and sup45 suppressor mutants in Saccharomyces cerevisiae to the anti-microtubule drug benomyl. Curr. Genet. 1996;30:44–49. doi: 10.1007/s002940050098. [DOI] [PubMed] [Google Scholar]
- 17.Borchsenius AS, Tchourikova AA, Inge-Vechtomov SG. Recessive mutations in SUP35 and SUP45 genes coding for translation release factors affect chromosome stability in Saccharomyces cerevisiae. Curr. Genet. 2000;37:285–291. doi: 10.1007/s002940050529. [DOI] [PubMed] [Google Scholar]
- 18.Ter-Avanesyan MD, Zimmermann J, Inge-Vechtomov SG, Sudarikov AB, Smirnov VN, Surguchov AP. Ribosomal recessive suppressors cause a respiratory deficiency in yeast Saccharomyces cerevisiae. Mol. Gen Genet. 1982;185:319–323. doi: 10.1007/BF00330805. [DOI] [PubMed] [Google Scholar]
- 19.Valouev IA, Kushnirov VV, Ter-Avanesyan MD. Yeast polypeptide chain release factors eRF1 and eRF3 are involved in cytoskeleton organization and cell cycle regulation. Cell Motil. Cytoskeleton. 2002;52:161–173. doi: 10.1002/cm.10040. [DOI] [PubMed] [Google Scholar]
- 20.Valouev IA, Urakov VN, Kochneva-Pervukhova NV, Smirnov VN, Ter-Avanesyan MD. Translation termination factors function outside of translation: yeast eRF1 interacts with myosin light chain, Mlc1p, to effect cytokinesis. Mol. Microbiol. 2004;53:687–696. doi: 10.1111/j.1365-2958.2004.04157.x. [DOI] [PubMed] [Google Scholar]
- 21.Eurwilaichitr L, Graves FM, Stansfield I, Tuite MF. The C-terminus of eRF1 defines a functionally important domain for translation termination in Saccharomyces cerevisiae. Mol. Microbiol. 1999;32:485–496. doi: 10.1046/j.1365-2958.1999.01346.x. [DOI] [PubMed] [Google Scholar]
- 22.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 23.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 24.Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 26.Stansfield I, Eurwilaichitr L, Akhmaloka, Tuite MF. Depletion in the levels of the release factor eRF1 causes a reduction in the efficiency of translation termination in yeast. Mol. Microbiol. 1996;20:1135–1143. doi: 10.1111/j.1365-2958.1996.tb02634.x. [DOI] [PubMed] [Google Scholar]
- 27.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mikaelian I, Sergeant A. A general and fast method to generate multiple site directed mutations. Nucleic Acids Res. 1992;20:376. doi: 10.1093/nar/20.2.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von der Haar T. Optimized protein extraction for quantitative proteomics of yeasts. PLoS ONE. 2007;2:e1078. doi: 10.1371/journal.pone.0001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stansfield I, Grant GM, Akhmaloka, Tuite MF. Ribosomal association of the yeast SAL4 (SUP45) gene product: implications for its role in translation fidelity and termination. Mol. Microbiol. 1992;6:3469–3478. doi: 10.1111/j.1365-2958.1992.tb01782.x. [DOI] [PubMed] [Google Scholar]
- 31.von der Haar T, Jossé L, Wright P, Zenthon J, Tuite MF. Development of a novel yeast cell-based system for studying the aggregation of Alzheimer's disease-associated Abeta peptides in vivo. Neurodegener. Dis. 2007;4:136–147. doi: 10.1159/000101838. [DOI] [PubMed] [Google Scholar]
- 32.Keeling KM, Lanier J, Du M, Salas-Marco J, Gao L, Kaenjak-Angeletti A, Bedwell DM. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA. 2004;10:691–703. doi: 10.1261/rna.5147804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von der Haar T, Josse L, Byrne LJ. In: Yeast Gene Analysis. 2nd edn. Stansfield I, Stark MJ, editors. Vol. 36. San Diego: Elsevier Academic Press; 2007. pp. 165–188. [Google Scholar]
- 34.Breining P, Piepersberg W. Yeast omnipotent supressor SUP1 (SUP45): nucleotide sequence of the wildtype and a mutant gene. Nucleic Acids Res. 1986;14:5187–5197. doi: 10.1093/nar/14.13.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mironova LN, Samsonova MG, Zhouravleva GA, Kulikov VN, Soom MJ. Reversions to respiratory competence of omnipotent sup45 suppressor mutants may be caused by secondary sup45 mutations. Curr. Genet. 1995;27:195–200. doi: 10.1007/BF00326148. [DOI] [PubMed] [Google Scholar]
- 36.Stansfield I, Kushnirov VV, Jones KM, Tuite MF. A conditional-lethal translation termination defect in a sup45 mutant of the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 1997;245:557–563. doi: 10.1111/j.1432-1033.1997.00557.x. [DOI] [PubMed] [Google Scholar]
- 37.Studte P, Zink S, Jablonowski D, Bar C, von der Haar T, Tuite MF, Schaffrath R. tRNA and protein methylase complexes mediate zymocin toxicity in yeast. Mol. Microbiol. 2008;69:1266–1277. doi: 10.1111/j.1365-2958.2008.06358.x. [DOI] [PubMed] [Google Scholar]
- 38.Frolova L, Seit-Nebi A, Kisselev L. Highly conserved NIKS tetrapeptide is functionally essential in eukaryotic translation termination factor eRF1. RNA. 2002;8:129–136. doi: 10.1017/s1355838202013262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frolova LY, Tsivkovskii RY, Sivolobova GF, Oparina NY, Serpinsky OI, Blinov VM, Tatkov SI, Kisselev LL. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kolosov P, Frolova L, Seit-Nebi A, Dubovaya V, Kononenko A, Oparina N, Justesen J, Efimov A, Kisselev L. Invariant amino acids essential for decoding function of polypeptide release factor eRF1. Nucleic Acids Res. 2005;33:6418–6425. doi: 10.1093/nar/gki927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salas-Marco J, Bedwell DM. Discrimination between defects in elongation fidelity and termination efficiency provides mechanistic insights into translational readthrough. J. Mol. Biol. 2005;348:801–815. doi: 10.1016/j.jmb.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 42.Gonnet GH, Cohen MA, Benner SA. Exhaustive matching of the entire protein sequence database. Science. 1992;256:1443–1445. doi: 10.1126/science.1604319. [DOI] [PubMed] [Google Scholar]
- 43.Waldron C, Cox BS, Wills N, Gesteland RF, Piper PW, Colby D, Guthrie C. Yeast ochre suppressor SUQ5-ol is an altered tRNA Ser UCA. Nucleic Acids Res. 1981;9:3077–3088. doi: 10.1093/nar/9.13.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007;2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- 45.Hodgkin J. Genetic suppression. In: The C. elegans Research Community, editor. WormBook. 2005. doi/10.1895/wormbook.1.59.1. [Google Scholar]
- 46.Chernoff YO, Derkach IL, Inge-Vechtomov SG. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr. Genet. 1993;24:268–270. doi: 10.1007/BF00351802. [DOI] [PubMed] [Google Scholar]
- 47.Kebaara B, Nazarenus T, Taylor R, Atkin AL. Genetic background affects relative nonsense mRNA accumulation in wild-type and upf mutant yeast strains. Curr. Genet. 2003;43:171–177. doi: 10.1007/s00294-003-0386-3. [DOI] [PubMed] [Google Scholar]
- 48.Inagaki Y, Blouin C, Doolittle WF, Roger AJ. Convergence and constraint in eukaryotic release factor 1 (eRF1) domain 1: the evolution of stop codon specificity. Nucleic Acids Res. 2002;30:532–544. doi: 10.1093/nar/30.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muramatsu T, Heckmann K, Kitanaka C, Kuchino Y. Molecular mechanism of stop codon recognition by eRF1: a wobble hypothesis for peptide anticodons. FEBS Lett. 2001;488:105–109. doi: 10.1016/s0014-5793(00)02391-7. [DOI] [PubMed] [Google Scholar]
- 50.Chavatte L, Seit-Nebi A, Dubovaya V, Favre A. The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. Embo J. 2002;21:5302–5311. doi: 10.1093/emboj/cdf484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakamura Y, Ito K, Ehrenberg M. Mimicry grasps reality in translation termination. Cell. 2000;101:349–352. doi: 10.1016/s0092-8674(00)80845-4. [DOI] [PubMed] [Google Scholar]