Abstract
Borrelia burgdorferi produces Erp outer surface proteins throughout mammalian infection, but represses their synthesis during colonization of vector ticks. A DNA region 5′ of the start of erp transcription, Operator 2, was previously shown to be essential for regulation of expression. We now report identification and characterization of a novel erp Operator 2-binding protein, which we named BpaB. erp operons are located on episomal cp32 prophages, and a single bacterium may contain as many as 10 different cp32s. Each cp32 family member encodes a unique BpaB protein, yet the three tested cp32-encoded BpaB alleles all bound to the same DNA sequence. A 20-bp region of erp Operator 2 was determined to be essential for BpaB binding, and initial protein binding to that site was required for binding of additional BpaB molecules. A 36-residue region near the BpaB carboxy terminus was found to be essential for high-affinity DNA-binding. BpaB competed for binding to erp Operator 2 with a second B. burgdorferi DNA-binding protein, EbfC. Thus, cellular levels of free BpaB and EbfC could potentially control erp transcription levels.
INTRODUCTION
The Lyme disease spirochete, Borrelia burgdorferi, persists in nature through infectious cycles between vertebrate hosts and Ixodes spp. tick vectors. Human infection is generally accompanied by an expanding ‘bull’s-eye’-shaped rash and nonspecific symptoms such as fever, aches and malaise. Untreated, B. burgdorferi infections may manifest as significant inflammatory arthritis, carditis and/or neurological disorders (1,2).
Borrelia burgdorferi produces proteins that are specific for each stage of its vertebrate-arthropod infectious cycle. Among these are the Erp proteins, a polymorphic family of surface-exposed outer membrane lipoproteins that are highly expressed during all stages of mammalian infection but largely repressed during tick colonization (3,4). Among the known functions of Erp proteins are adherence to host laminin and the binding of host plasminogen, complement regulator factor H and the factor H-related proteins FHR-1, FHR-2 and FHR-5 (5–15). These infection-associated functions make Erp proteins attractive targets for developing new protective and curative therapies for Lyme disease.
All Lyme disease spirochetes naturally contain multiple, distinct 32-kb circular prophages, collectively called cp32s (3,16). Each cp32 contains a mono- or bicistronic erp locus. Nine different erp-containing cp32s are present in the B. burgdorferi type strain, B31 (16–18). A 10th erp locus is found in strain B31 on a mutant cp32 known as lp56, an ∼56-kb linear plasmid that consists of an entire cp32 integrated into an unrelated linear replicon (17–19). Another natural plasmid of strain B31, named cp9-1, evolved from a cp32 through a series of deletion and rearrangement mutations, and no longer carries an erp locus (18).
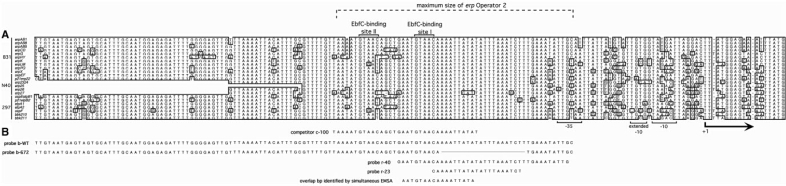
All erp loci contain highly conserved DNA sequences 5′ of the open reading frames (Figure 1A). Genetic and biochemical studies revealed that two small regions 5′ of erp loci contain binding sites for B. burgdorferi cytoplasmic proteins (20). The DNA region closest to the transcriptional start site, Operator 2, appears to play a role in transcriptional regulation, as deletion of that sequence resulted in constitutive, high-level transcription (20). DNA-affinity chromatography indicated that at least three proteins specifically bind to erp 5′ DNA (21). We previously identified one of these as EbfC, which is a novel type of DNA-binding protein that is encoded on the borrelial main chromosome. EbfC preferentially binds the palindromic sequence GTnAC, where ‘n’ can be any base, with all erp Operator 2 regions containing two adjacent EbfC-binding sites (21,22). We now report identification and characterization of a family of closely related borrelial proteins named BpaB (Borrelial ParB analog) that also bind erp Operator 2 DNA. All cp32 family members encode a BpaB protein, which appears to play a role in replication and segregation, possibly analogous to the ParB DNA-binding proteins of low copy-number plasmids in other bacterial species (18,23–29). Predicted protein sequences of BpaB proteins are distinct from any known plasmid partition-segregation protein, and may represent a novel type of such proteins (18,24,28). Some species’ ParB proteins are known to also be involved in plasmid gene regulation, by binding specific DNA sites and occluding promoter elements (23,30–33). We therefore analyzed the DNA-binding characteristics of BpaB proteins encoded by cp32 family members.
Figure 1.
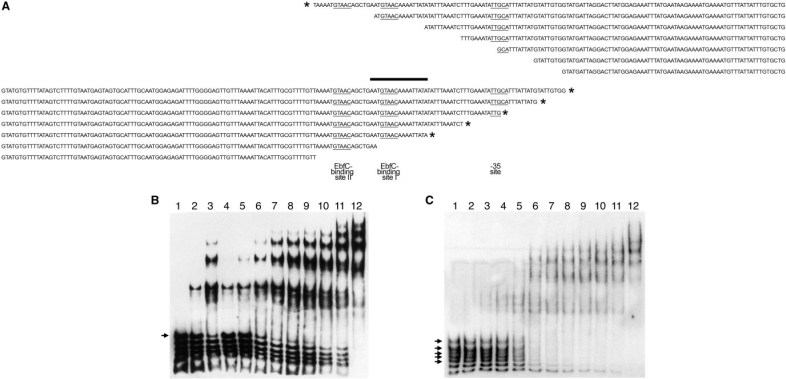
(A) DNA sequences 5′ of all known erp loci of B. burgdorferi strains B31, N40 and 297, indicating transcriptional promoter motifs, start of transcription (+1), the maximal borders of Operator 2 and two EbfC-binding sequences. Identical nucleotides found in the majority of known erp loci are boxed and shaded. For some erp loci of strain N40, only parts of some 5′ sequences are known, as indicated by blanks on this figure. These three strains, plus B. burgdorferi strain BL206 and Sh-2-82, are the only Lyme disease spirochetes for which the complete erp locus sequences have been published. The erp locus sequences of strain BL206 are identical to those of strain B31, and those of Sh-2-82 are identical to the erp loci of strain 297, and so were omitted from this figure (our unpublished data and references 18, 40, 43 and 44). (B) DNA sequences of biotin- and radiolabeled DNA probes used in EMSA studies. Base pairs present in the wild-type probe (b-WT) but absent from probe b-672 are indicated by a line. The DNA sequence identified as including the initial BpaB-binding site through studies illustrated in Figure 5 are shown on the lower line. All EMSA probes and competitors were double-stranded DNAs; for simplicity, only one strand (5′ to 3′) is illustrated.
MATERIALS AND METHODS
Bacteria
The B. burgdorferi type strain, B31, was the original source for all DNAs and recombinant proteins used in these studies. The complete genome sequence of a subculture of strain B31, known as B31-MI, has been determined (18,34). Strain B31-MI contains nine cp32 family members: cp32-1, cp32-3, cp32-4, cp32-6, cp32-7, cp32-8, cp32-9, cp9-1 and lp56. Two additional cp32 replicons, cp32-2 and cp32-5, have been found in other subcultures of the original B31 isolate, and have been partially sequenced (16,17). A 12th cp32 family member, cp9-2, was identified in another B31 derivative, but its probable bpaB locus has yet to be sequenced (35).
Modeling analyses
Primary amino acid sequences of proteins were compared with all known sequences in the nonredundant protein sequence database of GenBank using BLAST-P and Psi-BLAST (http://www.ncbi.nlm.nih.gov/BLAST). Protein folding probabilities were determined using Protein Homology/analogY Recognition Engine (PHYRE) (http://www.sbg.bio.ic.ac.uk/∼phyre). Predictions of disorder within proteins were determined using Predictor of Naturally Disordered Regions (PONDR, www.pondr.com) using VL-XT. Access to PONDR® was provided by Molecular Kinetics (Indianapolis, IN, USA). VL-XT is copyright©1999 by the WSU Research Foundation, all rights reserved, and PONDR® is copyright©2004 by Molecular Kinetics, all rights reserved. Probabilities of coiled-coil formation by proteins were determined using COILS (http://www.ch.embnet.org/software/COILS_form.html) and PCOILS (http://toolkit.tuebingen.mpg.de/pcoils).
Recombinant proteins
All recombinant proteins contained amino-terminal polyhistidine tags. Alleles of bpaB were named according to the replicon on which the gene is located, e.g. cp32-1 contains bpaB321, lp56 contains bpaB56, cp9-1 contains bpaB9, lp38 carries bpaB38, etc. For production of full-length recombinant BpaB321, BpaB56, BpaB9 and BpaB38, their respective genes were polymerase chain reaction (PCR)-amplified and cloned into pET200-TOPO (Invitrogen). Plasmids encoding truncated BpaB321 proteins were produced by sequence overlap extension PCR mutagenesis (36) of the full-length BpaB321 expression plasmid. Inserts of all recombinant plasmids were completely sequenced on both strands to confirm expected results. Recombinant EbfC was produced using a previously described plasmid (21). Escherichia coli strain Rosetta(DE3)(pLysS) (Invitrogen) was individually transformed with each plasmid. Production of recombinant proteins was induced by addition of isopropyl-β-d-galactopyranose (IPTG, final concentration 1 mM) to mid-exponential phase 37°C cultures, then incubated overnight with aeration at room temperature. Alternatively, E. coli were grown overnight at 37°C in 100 ml expansion broth (Zymo Research), inoculated into 500 ml over-expression broth (Zymo Research) the following day, then cultured for another 24 h at 37°C. Bacteria were harvested by centrifugation. Cells were resuspended in binding buffer (20 mM NaPO4, 1 M NaCl, 30 mM imidazole, pH 7.4), and bacteria lysed by sonication. Lysates were cleared by centrifugation at 15 000g, then injected onto a pre-equilibrated 5 ml HisTrap-HP column on an AKTA-FPLC with a UPC-900 absorbance monitor and a Frac920 fraction collector (all GE Healthcare). The column was washed with 15 column volumes of binding buffer, followed by linear gradient elution with imidazole concentrations steadily increasing from 30 to 750 mM. Aliquots of eluted fractions were analyzed for purity by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie brilliant blue staining. Proteins were then dialyzed against 50 mM Tris–HCl, pH = 7.5, 1 mM phenylmethanesulfonyl fluoride (PMSF), 1 mM dithiothreitol (DTT) and 10% (vol/vol) glycerol. Final protein concentrations were determined by bicinchoninic acid (BCA) protein assay (Pierce). Purified, dialyzed proteins were snap frozen in liquid nitrogen and stored at −80°C.
Electrophoretic mobility shift assays
A previously constructed recombinant plasmid containing B31 erpAB1 5′ non-coding DNA (including Operator 2) served as template for sequence overlap extension PCR mutagenesis of erp Operator 2 DNA (21,36), deleting 20 bp from erp Operator 2 to yield pBLS672. The erp 5′ noncoding sequence of that mutant plasmid was completely sequenced on both strands to confirm expected results.
The wild-type erp Operator 2-containing plasmid was used as template for producing wild-type electrophoretic mobility shift assay (EMSA) probe b-WT and the labeled probes used in simultaneous EMSA studies which initially identified the high-affinity, initial binding site of BpaB competitor DNAs. Plasmid pBLS672 served as template for production of probe b-672. For each of these labeled probes, the upstream primer used had been modified with a biotin moiety on the 5′ end. For unlabeled DNA competitors, unmodified oligonucleotides were used. Amplification products were separated by agarose gel electrophoresis, the appropriate band excised, and DNA cleaned using Wizard SV gel purification systems (Promega). Prior to use in EMSA, the concentration of each probe was determined spectrophotometrically (A260 of 1.0 = 50 µg DNA/ml), then diluted to a concentration of 1 nM. EMSA reactions were performed in 20 µl total volumes, containing 2 fmol biotinylated DNA probe in 50 mM Tris–HCl (pH = 7.5), 1 mM dithiothreitol, 8 µl/ml protease inhibitor (Sigma, St. Louis, MO, USA), 2 µl/ml phosphatase inhibitor cocktail II (Sigma), 50 µg/ml bovine serum albumin, 10% glycerol, 40 ng/µl poly(dI–dC) (Sigma), and various concentrations of recombinant protein. Reaction mixes were incubated for 20 min at room temperature, followed by electrophoresis in pre-cast 6% acrylamide gels (Invitrogen) for 90 min at 100 V. DNAs were transferred to nylon membranes (Pierce), and cross-linked by exposure to ultraviolet (UV) light. Labeled DNAs were detected using chemiluminescent nucleic acid detection kits (Pierce) and exposure to Kodak Biomax film.
Radiolabeled EMSA probes r-40 and r-23 were produced from pairs of complimentary oligonucleotides either 40 or 23 bp in length, respectively (Figure 1). Following are details on production of probe r-40. Probe r-23 was produced in exactly the same manner, except with use of oligonucleotides 23-F and 23-R. For probe r-40, two complimentary 40 base sequences, 40-F and 40-R, were synthesized. Oligonucleotide 40-F was 5′-end labeled using γ-32P-ATP (Perkin-Elmer) and T4 Kinase (New England Biolabs). The radiolabeled DNA was ethanol precipitated, resuspended in H20, passed through a Sephadex 10 (Sigma) column and dialyzed in TE buffer, pH = 8. A 1.2-fold excess of the complementary oligomer, 40-R, was added to the radiolabeled 40-F oligomer. The DNAs were annealed by first incubating for 5 min in a boiling water bath, then the heat turned off and allowed to cool to ∼37°C (3–4 h). The resulting dsDNA was dialyzed with EMSA buffer. Varying concentrations of purified BpaB321 protein were added to 0.2 µM 32P-labeled r-40, and mixtures were equilibrated at 4°C for 4 h. Electrophoresis was carried out in an 8% polyacrylamide gel using low salt TAE buffer 0.8 M Tris and 0.02 M EDTA pH 7.6. Autoradiographic images were captured on storage phosphor screens (type GP, GE Healthcare) detected with a Storm phosphorimager and quantitated with Image-Quant software (GE Healthcare).
For studies to determine binding constants, a luminescent form of wild-type erpAB Operator 2 probe b-WT was produced. The upstream primer had been modified with an IRDye 700 fluorescent infrared dye at its 5′-end (LI-COR). Amplification products were separated by agarose gel electrophoresis, the appropriate DNA band excised, then cleaned as above. EMSAs were carried out as described previously. Luminescent DNA bands were detected using an Odyssey Infrared Imaging System (LI-COR). Disassociation constants (Kd) for BpaB–DNA interactions were determined by comparing the signal strengths of bound and unbound DNA in each reaction. Signal strengths were normalized for each lane. Ratios of bound : free DNA were graphed in relation to the known concentrations of free protein [P], using Microsoft Excel. Since the ratios of free protein to DNA were extremely high, the binding of protein to DNA did not appreciably change the concentration of free protein (37) The disassociation constant was therefore calculated as the concentration of BpaB where the ratio of bound to free DNA = 1 (22,37–39).
Analyses of DNA conformation
Five 150-bp DNA fragments, overlapping stepwise by 30-bp increments over a total of 250 bp, were produced by PCR, using the cloned erpAB insert of pBLS434a as template (16,22). The upstream oligonucleotide primer for each DNA fragment was modified with a biotin moiety at the 5′ end. Natural curvature of the DNAs was analyzed by electrophoresis through 15% polyacrylamide gels and staining with ethidium bromide. EMSA using purified recombinant BpaB321 was performed as described previously.
RESULTS
BpaB binds to erp Operator 2 DNA
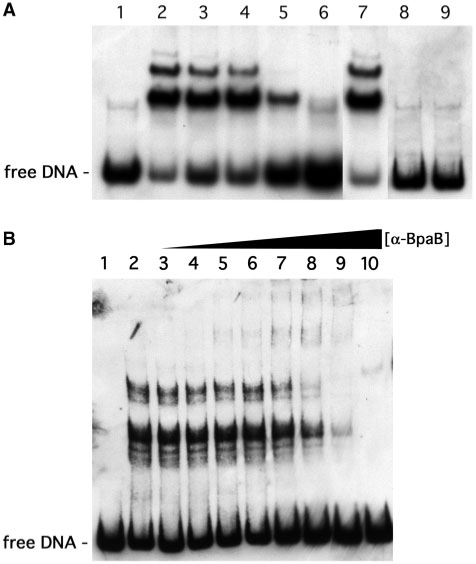
We previously reported that DNA-affinity chromatography using erp 5′ noncoding DNA and B. burgdorferi cytoplasmic extracts led to isolation of three different proteins (21). Subsequent mass spectrometric analyses of tryptic fragments of the largest, ∼22-kDa, erp-binding protein suggested it to be encoded by a gene located on the B. burgdorferi cp32 prophages (data not shown). These genes have been implicated in plasmid maintenance, and have been variously referred to as ORF-3, ORF-D, paralog family 49 or pfam01672 (18,19,24,26,28,40,41). Due to apparent functional similarities between the ∼22-kDa proteins encoded by these borrelial genes and the ParB/SopB proteins of other bacterial species, we named the borrelial proteins ‘BpaB’ (Borrelial ParB analog). The validity of this proteomic identification of BpaB as an erp-binding protein was investigated by EMSA using labeled DNA probe b-WT, derived from the wild-type erpAB1 locus (Figure 1B), and recombinant BpaB allele 321, based on the bpaB gene sequence of B. burgdorferi strain B31 cp32-1. Incubation of probe b-WT with BpaB321 protein yielded slowly migrating EMSA bands, consistent with DNA-protein complexes (Figure 2A). Preincubation of recombinant protein and DNA at room temperature, 4°C or 37°C made no significant differences in protein binding, so all subsequent EMSAs were performed at room temperature (Figure 2A, lanes 2–4). BpaB specificity for erp Operator 2 DNA was assessed by competition EMSA using excess quantities of unlabeled DNAs derived from either erp 5′ DNA or from an unrelated source. Competitor C-100, which consists of part of erp Operator 2, partially inhibited formation of the protein–DNA complexes (Figure 2A, lane 5). A 100-fold excess of unlabeled DNA identical in sequence to the labeled probe almost completely eliminated the protein–DNA complex bands (Figure 2A, lane 6). However, addition of a 100-fold excess of unlabeled DNA derived from the promoter region of the unrelated B. burgdorferi flaB gene did not have any detectable effects on DNA–protein complex formation (Figure 2A, lane 7). As additional controls, heating recombinant BpaB321 in a boiling water bath for 10 min prior to incubation with DNA, or addition of SDS (final concentration 10%) to the DNA–protein mixture prevented formation of the upper bands, consistent with compositions that include protein (Figure 2A, lanes 8 and 9).
Figure 2.
Demonstration that BpaB binds specifically to erp Operator 2 DNA. (A) EMSA with labeled erp probe b-WT and purified recombinant protein based on the sequence of bpaB from B. burgdorferi strain B31 cp32-1 (BpaB321). Lane 1 lacked added BpaB321, while 0.12 µM recombinant protein was added to the other lanes. (Lanes 2–4) Pre-incubation of DNA and protein at room temperature, 4°C and 37°C, respectively. (Lanes 5–7) DNA and protein mixtures were preincubated with 100-fold excesses of unlabeled competitor DNAs C-100, unlabeled-WT, or flaB, respectively. (Lane 8) BpaB321 was heated in a boiling water bath for 10 min prior to mixture with DNA. (Lane 9) 10% SDS, final concentration, was added to the DNA–protein mixture prior to EMSA. (B) Supershift EMSA using BpaB-directed antibodies. Lane 1 contained only DNA, lane 2 contained DNA plus 0.19 µM BpaB321, and lanes 3–10 contained the same concentrations of DNA and BpaB321 plus increasing concentrations of anti-BpaB321 polyclonal antiserum.
Confirmation that the slowly migrating bands consisted of DNA–BpaB complexes was obtained by antibody super-shift studies. Addition of increasing concentrations of polyclonal rabbit antiserum raised against and specific for BpaB led to the appearance of EMSA bands with even slower mobilities, consistent with DNA–BpaB–antibody complexes (Figure 2B). Antibodies in the absence of BpaB did not affect levels of free DNA (data not shown).
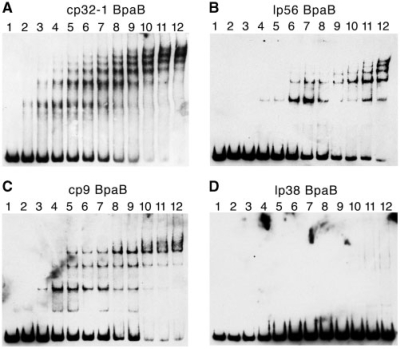
Initial EMSAs, such as those shown in Figure 2, indicated multiple BpaB–DNA complexes. The natures of these multiple complexes were investigated by additional EMSA, using a constant concentration of erpAB1 Operator 2 DNA and increasing amounts of BpaB321. These analyses revealed formation of a single protein–DNA complex at low concentrations of BpaB321, with additional, more slowly migrating complexes forming as protein concentrations were increased (Figure 3A). At the higher concentrations of BpaB321, the more rapidly migrating DNA–protein complexes disappeared. Additional studies described below revealed that this pattern was due to initial binding of a BpaB protein to a specific site on the DNA, with that bound protein facilitating the binding of additional BpaB molecules.
Figure 3.
EMSA using labeled probe b-WT and increasing concentrations of recombinant BpaB proteins based on sequences of B31 plasmids cp32-1, lp56, cp9-1 and lp38. For all EMSAs, lane 1 contained DNA without protein. (A) Lanes 2–12 contained in addition 0.033, 0.066, 0.099, 0.13, 0.17, 0.20, 0.23, 0.26, 0.30, 0.33 and 0.40 µM recombinant BpaB321 protein, respectively. (B) Lanes 2–12 contained 0.0048, 0.012, 0.024, 0.048, 0.097, 0.15, 0.19, 0.24, 0.29, 0.34 and 0.39 µM recombinant BpaB56 protein, respectively. (C) Lanes 2–12 contained 0.084, 0.17, 0.25, 0.34, 0.42, 0.50, 0.59, 0.67, 0.75, 0.84 and 1.00 µM recombinant BpaB9 protein, respectively. (D) Lanes 2–12 contained 0.069, 0.14, 0.21, 0.28, 0.35, 0.42, 0.48, 0.55, 0.62, 0.69 and 0.83 µM recombinant BpaB38 protein, respectively. Note that some of the higher-order DNA–protein complexes do not always resolve crisply, and appear faint or as smears in EMSAs [e.g. the upper EMSA bands in lane 12 of (C)].
Based upon the disappearance of free probe with increasing concentrations of BpaB321, a Kd value of 190 nM was calculated for the binding of protein to free erp Operator 2 DNA. This is obviously an underestimate of the protein’s affinity for free DNA, since analyses necessarily included BpaB binding to DNA to which other BpaB molecules had previously attached.
BpaB proteins from different cp32 family members bind the same DNA sequence
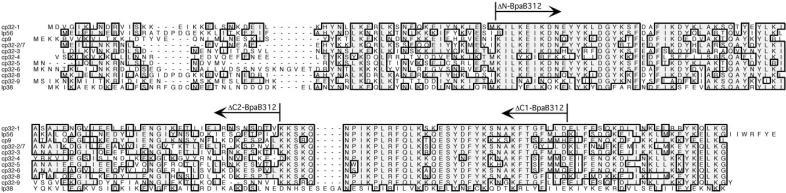
The BpaB proteins encoded by the different naturally occurring cp32 family members share ∼ 35–70% amino acid sequence identities (Figure 4). As discussed previously and illustrated in Figure 1A, the Operator 2 sequences of all known erp loci are highly similar. This suggested that a protein which can bind a sequence within one erp locus’ Operator 2 would probably be able to bind the Operator 2 regions of other erp loci, as is the case with EbfC (21,22). This hypothesis predicts that since BpaB from cp32-1 binds erp Operator 2 DNA, then BpaB proteins encoded by other cp32 family members will also do so.
Figure 4.
Alignments of the predicted amino acid sequences of BpaB proteins encoded by B. burgdorferi B31 cp32 family members, and by the strain B31 plasmid lp38. Identical residues found in the majority of proteins are boxed and shaded. Gaps introduced to maximize homology are indicated by dashes. The amino terminus of protein ΔN-BpaB321 and the carboxy-termini of proteins ΔC1-BpaB321 and ΔC2-BpaB321 are indicated.
This question was first addressed by producing and purifying a recombinant BpaB protein derived from the bpaB allele encoded by the cp32 family member lp56 (BpaB56). EMSA using cp32-1-derived DNA and BpaB56 yielded banding patterns similar to those obtained when using BpaB321 (Figure 3B). The Kd value of recombinant BpaB56 binding to this DNA was calculated to be 220 nM.
The small cp9-1 plasmid of B. burgdorferi strain B31 is a naturally occurring cp32-derivative that had undergone extensive mutations prior to isolation of the bacterium from nature (18). cp9-1 lacks an erp locus, but does encode a BpaB paralog. This protein’s predicted sequence is <50% identical to the other cp32 family members’ BpaB paralogs (Figure 4). It is, however, well conserved in the carboxy-terminal region that we identified as being involved with binding to erp Operator 2 DNA (see below). We therefore produced a recombinant protein based on the cp9-1 sequence, BpaB9. This recombinant protein also yielded a laddered banding pattern with cp32-1-derived erp Operator 2 DNA (Figure 3C). The Kd value of recombinant BpaB9 binding to this DNA fragment was calculated to be 530 nM.
All but two of the naturally occurring plasmids of strain B31 contain a bpaB gene (18,29). Having found that the BpaB proteins encoded by cp32-1, lp56 and cp9-1 all bind erpAB1 Operator 2 DNA, the obvious next step was to examine the BpaB of a borrelial plasmid that is not related to the cp32 family (18). To that end, a recombinant protein was produced based on the bpaB allele of the 38-kb linear plasmid lp38, BpaB38 (Figure 4). Unlike the cp32 family member BpaB proteins, BpaB38 was not able to bind erp Operator 2 DNA (Figure 3D). The recombinant BpaB38 was produced and purified two different times, from separately transformed E. coli colonies, to control for possible denaturation during purification, yet the same result was obtained from both preparations.
Mapping the initial BpaB-binding site within erp Operator 2
To narrow down the high-affinity BpaB-binding site, additional, simultaneous EMSAs were performed. In one set of studies, seven different biotin-labeled DNA probes were produced from erpAB1 5′ DNA, each using the same downstream oligonucleotide primer paired with one of seven distinct upstream primers (Figure 5A, upper 7 sequences). Being of different sizes, each DNA migrated at a different rate under PAGE. Equimolar amounts of all seven DNA probes were simultaneously incubated with various concentrations of BpaB321, then separated by PAGE. BpaB321 bound to the largest DNA probe at much lower protein concentrations than it did to the six shorter probes, indicating that the largest probe contained sequences necessary for initial high-affinity binding of BpaB, while the shorter probes lacked those bases (Figure 5B). The same procedure was performed using another set of seven labeled probes, this time produced with the same upstream primer and different downstream primers (Figure 5A, lower 7 sequences). The longest five of these probes bound BpaB321, indicating that they contained sequences for BpaB-binding that were lacking from the two smaller probes (Figure 5C). Comparisons of BpaB-binding activities and the various sizes of probes used in these studies indicated that DNA probe size did not itself have an appreciable effect upon the results. These analyses demonstrated the presence of a high-affinity binding-site for BpaB within a region of erp Operator 2 DNA that overlaps EbfC-binding site I (Figures 1 and 5A). The identified DNA region is consistent with additional analyses described below.
Figure 5.
Identification of the high-affinity BpaB-binding site in erp Operator 2, by use of simultaneous titration of multiple probes with increasing concentrations of BpaB321. In this method, several DNAs that overlap in sequence were simultaneously incubated with protein, then subjected to EMSA. These DNAs which contain the high-affinity protein binding site were preferentially bound by lower concentrations of protein, and signals corresponding with those free DNA disappeared soonest. (A) Sequences of labeled DNA probes used in simultaneous EMSAs. Asterisks indicate DNAs preferentially bound by BpaB. The –35 sequence of the erpAB promoter and the two consensus EbfC-binding sites are indicated by underlining. The upper seven nested DNAs were anchored 51 bases downstream of the erpAB transcription start site, and EMSA data are illustrated in (B). The lower seven nested DNAs were anchored at bp –174 relative to the start of erpAB transcription, and EMSA data are illustrated in (C). The sequence contained in all DNAs that preferentially bound BpaB but absent from lower-affinity DNAs is indicated by a thick horizontal bar. (B) Simultaneous EMSA of recombinant BpaB321 with the upper seven DNAs of (A). Lane 1 contains only the DNAs. Lanes 2–12 contain DNAs plus increasing concentrations of recombinant BpaB321. Quantification of signal strengths indicated that BpaB bound preferentially to the largest DNA (marked with an arrowhead): in lanes 9, 10 and 12, the proportion of free DNA for this probe was 21, 0.10 and 0% of lane 1, respectively, while that of the next largest probe was 52, 13 and 10% of lane 1, respectively. (C) Simultaneous EMSA of recombinant BpaB321 with the lower seven DNAs of (A). Lane 1 contains only the DNAs. Lanes 2–12 contain DNAs plus increasing concentrations of recombinant BpaB321. Quantification of signal strengths indicated that BpaB bound preferentially to the five largest DNAs (marked with arrowheads): in lanes 7, 8 and 9, the proportion of free DNA for the largest probe was 16, 10 and 0% of lane 1, respectively, while that of the smallest probe was 75, 55 and 43% of lane 1, respectively.
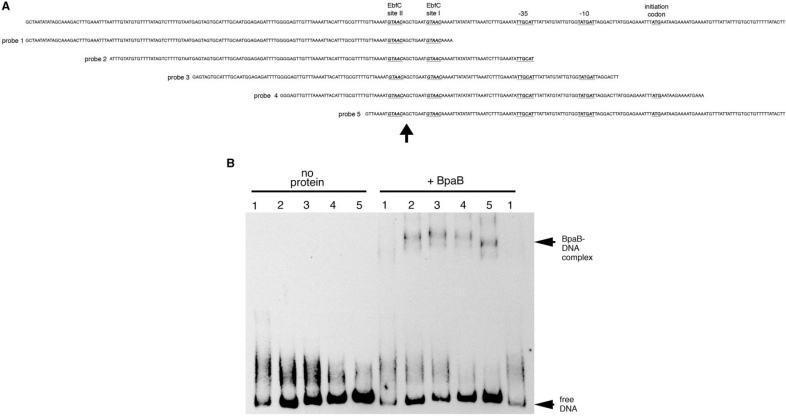
Binding of a protein often alters the conformation of DNA, and protein-induced bending may be detected by EMSA. To explore whether BpaB-binding distorts DNA conformation, and to further define the location of initial BpaB-binding in erp Operator 2 DNA, we utilized a series of five identically sized DNA fragments of erpAB1 5′ DNA that overlapped each other by 30-bp increments (Figure 6A). PAGE of the free DNAs indicate that erpAB1 5′ DNA contains a naturally occurring bend near the −10 site of the promoter (Figure 6 and reference 22). Addition of BpaB, at a concentration sufficient to produce a single protein–DNA complex EMSA band, yielded signals indicative of bending centered within DNA fragment 3 (Figure 6B). The center of that DNA fragment lies near EbfC-binding site I (Figure 6A), indicating a location for the high-affinity, initial BpaB-binding site consistent with the results obtained from the simultaneous EMSA studies described earlier.
Figure 6.
Binding of BpaB changes DNA conformation. (A) Sequence of the 5′ end of the erpAB1 locus (top line) and five biotinylated 150-bp fragments of that DNA (probes 1 to 5). Arrowhead under the panel indicates the center of probe 3. (B) Comparisons of mobilities of free probes 1–5 and those probes bound by BpaB321. Numbers above each lane indicate the labeled probe being analyzed. Note that free probe 5 migrates the slowest, indicative of a naturally occurring bend near the center of that DNA (22). Binding of BpaB321 resulted in probe 3 migrating the slowest, indicating an induced change in DNA conformation.
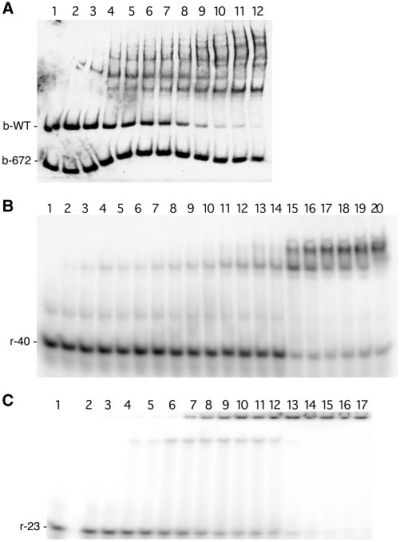
Additional EMSA studies further indicated this region of DNA to be critical for initial BpaB-binding to erp Operator 2. Site-directed mutagenesis was utilized to produce a labeled probe, b-672, that lacked 20 bp of erpAB1 Operator 2 but was otherwise identical to the previously used probe b-WT (Figure 1B). Simultaneous EMSA using probes b-672 and b-WT demonstrated appreciable BpaB binding to the wild-type DNA, but not to the mutant DNA (Figure 7A). In the illustrated EMSA, the highest concentration of BpaB assayed bound almost all the free b-WT probe, but did not bind even half of the free mutant b-672 probe. EMSA using probe b-672 alone, with concentrations of BpaB similar to those above, detected very faint shifted bands upon extended film exposures, but due to the weakness of interactions, a binding affinity could not be calculated (data not shown). These results indicate that the 20 bp deleted from b-672 contain nucleotides necessary for high-affinity binding of BpaB. Noting that the deletion of these 20 bp prevented formation of any readily detectable BpaB–DNA complexes, these experiments also demonstrated that formation of the other BpaB–DNA complexes observed with wild-type erp DNAs require formation of the initial DNA–protein complex.
Figure 7.
Confirmation of the initial BpaB-binding site in erp Operator 2 (A) Simultaneous EMSA using wild-type recombinant BpaB321 protein and both biotin-labeled wild-type erpAB1 probe b-WT and b-672. Probe b-672 is identical to b-WT except for deletion of 20 bp (Figure 1). Lane 1 contained only DNA. Lanes 2–12 contained BpaB321 at concentrations of 0.086, 0.18, 0.26, 0.35, 0.44, 0.53, 0.61, 0.70, 0.79, 0.86 and 1.1 µM, respectively. (B) EMSA using the radioactively labeled 40 bp probe r-40 (Figure 1). Lane 1 contained only DNA. Lanes 2–20 contained BpaB321 at concentrations of 0.40, 0.80, 1.0, 1.2, 1.5, 2.0, 2.5, 3.0, 3.5, 4.0, 5.0, 7.0, 10, 15, 20, 25, 30, 40 and 50 µM, respectively. (C) EMSA using the radioactively labeled 23-bp probe r-23, which encompasses the 20-bp sequence present in b-WT but absent from b-672 (see Figure 1). Lane 1 contained only DNA. Lanes 2–17 contained BpaB321 at concentrations of 0.40, 0.80, 1.0, 1.2, 1.5, 2.0, 2.5, 3.0, 3.5, 4.0, 5.0, 7.0, 10, 15, 20 and 30 µM, respectively.
A second series of EMSAs utilized a 40-bp probe, r-40, which includes the DNA regions identified by the above-described experiments (Figure 7B). The Kd of recombinant BpaB321 binding to probe r-40 was calculated to be 20 µM. At most, three protein–DNA species were detected by EMSA when using probe r-40, in contrast with the six complexes observed when using the 125-bp probe b-WT (Figure 3A). These data support the hypothesis that the multiple complexes observed in BpaB–DNA EMSAs are due to multiple protein–DNA interactions.
The 23-bp probe, r-23, which incorporates the sequence deleted from probe b-672, was used for additional EMSAs. Recombinant BpaB321 bound to this small piece of DNA with a Kd of 3.2 µM (Figure 7C). Thus, this 23-bp segment of erp Operator 2 contains a nucleotide sequence sufficient for high-affinity binding of BpaB. This result is consistent with the above-described studies with probe b-672 (Figure 7A) and with the simultaneous EMSAs using overlapping DNA probes (Figure 5).
Identification of the DNA-binding domain of BpaB
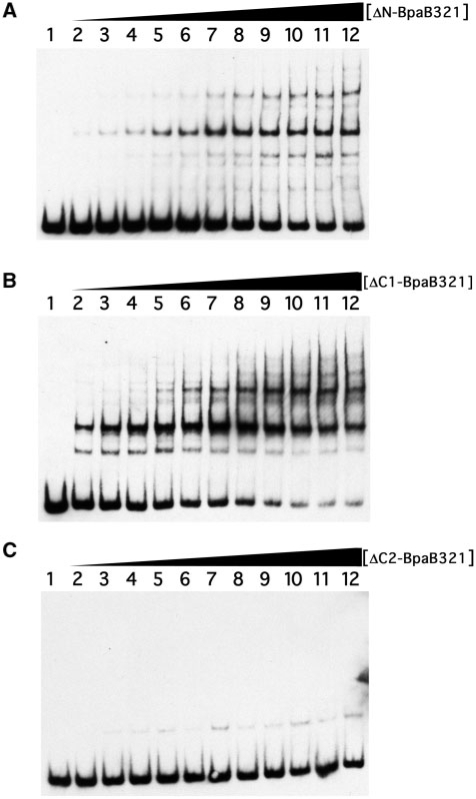
Comparisons of the BpaB proteins encoded by cp32 family members show limited identities in the amino-terminal third (Figure 4). Site-directed mutagenesis of the cp32-1-encoded BpaB was employed to produce truncated recombinant protein ΔN-BpaB321, which lacks the first 53 residues. EMSA using the truncated protein did not show significantly different results from those found with the full-length protein (Figure 8A). Addition of increased concentrations of ΔN-BpaB321 to EMSA reactions yielded reduced amounts of free probe DNA and increased laddering of protein–DNA bands. These results indicate that the amino-terminal third of BpaB is not required for either initial or subsequent binding of DNA.
Figure 8.
EMSAs using wild-type erpAB1 DNA and truncated BpaB321 proteins. For all panels, lane 1 contained DNA without protein. (A) Lanes 2–12 contained 0.013, 0.026, 0.039, 0.052, 0.065, 0.078, 0.090, 0.10, 0.12, 0.13 and 0.16 µM recombinant ΔN-BpaB321, respectively. (B) Lanes 2–12 contained 0.078, 0.016, 0.023, 0.031, 0.039, 0.047, 0.055, 0.062, 0.070, 0.078 and 0.094 µM recombinant ΔC1-BpaB321 respectively. (C) Lanes 2–12 contained 0.0075, 0.015, 0.023, 0.030, 0.038, 0.045, 0.053, 0.060, 0.068, 0.075 and 0.090 µM recombinant ΔC2-BpaB321, respectively.
Truncated protein ΔC1-BpaB321 was next produced, lacking the carboxy-terminal 22 amino acids (Figure 4). EMSAs using that recombinant protein were not appreciably different from those using full-length BpaB321 (Figure 8B).
A third truncated protein was produced, ΔC2-BpaB321, which lacks the carboxy-terminal 58 amino acids of BpaB321 (Figure 4). EMSA using this truncated protein revealed significantly weakened DNA binding (Figure 8C). Laddering was not seen, even at high protein concentrations. A single, weak, slowly migrating signal was observed, but diminution of the free DNA was not detected and so the binding affinity of ΔC2-BpaB321 was too weak to be determined. Protein ΔC2-BpaB321 was induced and purified two separate times, with both preparations giving the same results, indicating that failure to bind DNA was unlikely to have been due to denaturation during purification. Hence, the 36 amino acid region absent from ΔC2-BpaB321 but present in ΔC1-BpaB321 is necessary for high-affinity DNA binding. Modeling analyses of BpaB molecules predicted these proteins to be comprised of multiple α-helices, with the 36-residue region involved in high-affinity binding predicted to be a disordered region bridging two α-helices (data not shown). Additional analyses to identify specific residues of BpaB involved with binding to erp Operator 2 and with protein–protein interactions are ongoing.
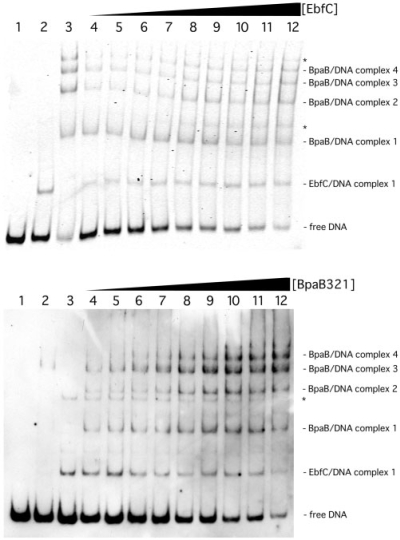
BpaB and EbfC compete for binding to erp Operator 2 DNA
The above-described results, combined with previous data (21,22), indicate that both BpaB and EbfC bind to adjacent or overlapping DNA sequences of erp Operator 2 (Figure 1). To determine the effects each protein has upon DNA-binding by the other, EMSAs were performed in which labeled erp Operator 2 DNA was preincubated with a fixed amount of one protein, then increasing concentrations of the other protein were added.
When the concentration of BpaB321 was held constant, increasing concentrations of EbfC led to reduced levels of free DNA and increased levels of EbfC–DNA complexes (Figure 9A). There were not any detectable changes observed in band intensities of any of the BpaB–DNA complexes. Measured ratios of free DNA signal to BpaB–DNA complex 1 signal diminished from 3.9 : 1 in lane 4 to 0.57 : 1 in lane 12 of Figure 9A. Thus, EbfC preferentially bound to free erp Operator 2 DNA, as opposed to previously formed BpaB–DNA complexes.
Figure 9.
EbfC and BpaB compete for binding to erp Operator 2. (A) EMSAs in which the concentration of BpaB321 was held constant while increasing concentrations of EbfC were added. (Lane 1) Labeled probe b-WT alone. (Lanes 2 and 3) Probe b-WT preincubated with 2.05 and 2.89 µM recombinant EbfC or BpaB321, respectively. (Lanes 4–12) Probe b-WT preincubated with 0.97 µM recombinant BpaB321, plus EbfC at concentrations of 0.27, 0.51, 1.03, 1.54, 2.05, 2.56, 3.08, 3.59 and 4.10 µM, respectively. Compositions of EMSA bands are indicated to the right of the panel. Asterisks indicate nonspecific DNA binding by EbfC observed at high concentrations of that protein (our unpublished data). Ratios of free DNA to BpaB–DNA complex 1 in lanes 4 through 12 were 3.9 : 1, 3.5 : 1, 2.7 : 1, 2.1 : 1, 1.4 : 1, 1.1 : 1, 1.1 : 1, 0.86 : 1 and 0.57 : 1, respectively, with the BpaB–DNA complex signals remaining constant, indicating that EbfC preferentially bound to free DNA. Quantification of signal intensities indicated that the proportion of EbfC–DNA complexes steadily increased with addition of EbfC protein, and that total of signal strengths remained constant across the EMSA gel. (B) EMSAs in which the concentration of EbfC was held constant while increasing concentrations of BpaB321 were added. (Lane 1) Labeled probe b-WT alone. (Lanes 2 and 3) Probe b-WT pre-incubated with 0.2 and 0.3 µM recombinant EbfC or BpaB321, respectively. (Lanes 4–12) Probe b-WT preincubated with 0.3 µM recombinant BpaB321, plus EbfC at concentrations of 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9 and 1 µM, respectively. Compositions of EMSA bands are indicated to the right of the panel. The asterisk indicates nonspecific DNA binding by EbfC observed at high concentrations of that protein (our unpublished data). Quantification of signal strengths indicated that proportions of free DNA and DNA bound by EbfC decreased as additional BpaB was added, while proportions of BpaB–DNA complexes increased.
In other studies, the DNA probe was preincubated with a constant level of EbfC, then BpaB concentrations were increased. The signal strengths of EbfC–DNA complexes and free DNA all decreased, while signal strengths of BpaB–DNA complexes increased (Figure 9B). Thus, under these experimental conditions, recombinant BpaB321 can drive recombinant EbfC off Operator 2 DNA. Together, these competition EMSAs indicate that BpaB and EbfC compete for erp Operator 2 DNA.
DISCUSSION
Previous studies by our laboratory indicated that a conserved region immediately 5′ of erp promoters, Operator 2, is critical for repression of erp transcription. We have now identified two different B. burgdorferi proteins that bind specific DNA sequences within erp Operator 2. The high-affinity BpaB-binding sequence is adjacent to and possibly overlaps the high-affinity EbfC-binding sites. Dual protein binding assays indicated that both BpaB and EbfC preferentially bind to free DNA, with no evidence of both proteins binding DNA simultaneously. Thus, it is possible that either BpaB or EbfC is a repressor of erp transcription, while the other protein functions as an activator/anti-repressor by competing for erp Operator 2 DNA. This hypothesis is currently being tested through additional studies to correlate cellular BpaB and EbfC levels with erp transcription.
Addition of low concentrations of BpaB to EMSA reactions produced a single DNA–protein complex, while addition of increasing concentrations of BpaB yielded increasing numbers of BpaB–DNA complexes. Deletion of 20 bp within erp Operator 2 significantly inhibited formation of any BpaB–DNA complex, indicating that this small region must contain sequences critical for all of the BpaB–DNA complexes formed with wild-type erp Operator 2 DNA. Thus, we conclude that BpaB initially forms a high-affinity complex with a specific sequence within erp Operator 2, and that subsequent binding of BpaB to erp DNA is dependent upon that initial binding. Presumably, protein–protein interactions between BpaB molecules stabilize binding to non-preferred DNA sequences. Without the specific DNA sequence, initial BpaB binding was impaired, and so was the binding of additional molecules. The binding of subsequent BpaB proteins from a nucleation site is similar to what has been demonstrated for several other bacterial DNA-binding proteins, such as the spreading phenomena of some ParB proteins and E. coli H-NS (23,30–33,42).
The current studies demonstrated that BpaB proteins encoded by the cp32 family members cp32-1, lp56 and cp9-1 all bound to erp Operator 2 DNA, and all produced multiple DNA–protein complexes. Analyses of mutant proteins indicated that a 36 amino acid region in the carboxy-terminal half of BpaB contains residues necessary for specific DNA binding. The region, between 22 and 58 residues from the carboxy-terminus, is highly conserved among all cp32 family BpaB proteins. These data strongly suggest that all BpaB proteins encoded by cp32 family members can bind erp Operator 2 DNA.
In contrast, the BpaB protein encoded by the B. burgdorferi B31 lp38 replicon did not bind erp Operator DNA. This result is consistent with the low degrees of similarities in the carboxy-terminal regions of lp38 BpaB and the cp32-encoded proteins. These data also indicate that non-cp32 replicons such as lp38 are unlikely to directly influence erp transcription.
BpaB plays a role(s) in B. burgdorferi replicon maintenance (24–27). Almost all borrelial replicons contain a bpaB adjacent to gene bearing strong similarity to the parA/sopA genes of other bacterial species’ replicons. Each replicon within an individual B. burgdorferi cell carries a distinctively different pair of parA homolog and bpaB genes, and those pairings appear to coincide with incompatibility groups (18,24,28). These data suggest that, even though BpaB sequences are dissimilar from characterized ParB proteins of other bacterial plasmids, BpaB may perform segregation functions similar to those of typical ParBs. Bacterial ParB proteins are site-specific DNA-binding proteins with affinities for parS DNA sequences on their replicons, and interactions between ParB, ParA and parS play roles in replicon segregation. It is possible that erp Operator 2 regions contain the parS homologue of cp32 elements, although we feel this to be unlikely due to ability of all tested cp32 family-encoded BpaB proteins to bind that DNA. Alternatively, parS may be located elsewhere on these replicons, while the DNA sequence within erp Operator 2 directs binding of additional BpaB molecules for another reason, such as regulation of erp transcription. Studies are ongoing to determine the consensus high-affinity binding sequence of cp32-encoded BpaB proteins, and where else these proteins bind on the replicons.
In conclusion, a new DNA-binding protein of B. burgdorferi was discovered, and was demonstrated to bind with high affinity to a DNA sequence within erp Operator 2. Binding of BpaB at that site led to DNA-bending and to the binding of additional BpaB molecules. The high-affinity BpaB-binding site is adjacent to or overlaps the high-affinity EbfC-binding sites, and those two proteins were found to compete for binding to erp Operator 2 DNA. Since that region of DNA is required for control of erp operon transcription (20), the present studies suggest that cellular levels of free BpaB and EbfC may contribute to regulation of erp gene expression.
FUNDING
Funding for open access charge: US National Institutes of Health (grant R01-AI044254 to B. S. and R01-GM070662 to M.F.); NIH Training Grant in Microbial Pathogenesis T32-AI49795 (to L.B. and S.R.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Tomasz Bykowski, Catherine Brissette, Ashutosh Verma, and Michael Woodman for assistance during this work.
REFERENCES
- 1.Stanek G, Strle F. Lyme borreliosis. Lancet. 2003;362:1639–1647. doi: 10.1016/S0140-6736(03)14798-8. [DOI] [PubMed] [Google Scholar]
- 2.Steere AC. Lyme disease. New Engl. J. Med. 2001;345:115–125. doi: 10.1056/NEJM200107123450207. [DOI] [PubMed] [Google Scholar]
- 3.Stevenson B, Zückert WR, Akins DR. In: The Spirochetes: Molecular and Cellular Biology. Saier MH, García-Lara J, editors. Oxford: Horizon Press; 2001. pp. 87–100. [Google Scholar]
- 4.Stevenson B, Bykowski T, Cooley AE, Babb K, Miller JC, Woodman ME, von Lackum K, Riley SP. In: Molecular Biology of Spirochetes. Cabello FC, Godfrey HP, Hulinska D, editors. Amsterdam: IOS Press; 2006. pp. 354–372. [Google Scholar]
- 5.Brissette CA, Verma A, Bowman A, Cooley AE, Stevenson B. The Borrelia burgdorferi outer-surface protein ErpX binds mammalian laminin. Microbiology. 2009;155:863–872. doi: 10.1099/mic.0.024604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brissette CA, Haupt K, Barthel D, Cooley AE, Bowman A, Skerka C, Wallich R, Zipfel PF, Kraiczy P, Stevenson B. Borrelia burgdorferi infection-associated surface proteins ErpP, ErpA, and ErpC bind human plasminogen. Infect. Immun. 2009;77:300–306. doi: 10.1128/IAI.01133-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stevenson B, El-Hage N, Hines MA, Miller JC, Babb K. Differential binding of host complement inhibitor factor H by Borrelia burgdorferi Erp surface proteins: a possible mechanism underlying the expansive host range of Lyme disease spirochetes. Infect. Immun. 2002;70:491–497. doi: 10.1128/IAI.70.2.491-497.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haupt K, Kraiczy P, Wallich R, Brade V, Skerka C, Zipfel PF. FHR-1, an additional human plasma protein, binds to complement regulator-acquiring surface proteins of Borrelia burgdorferi. Int. J. Med. Microbiol. 2008;298, S1:287–291. [Google Scholar]
- 9.Hellwage J, Meri T, Heikkilä T, Alitalo A, Panelius J, Lahdenne P, Seppälä IJT, Meri S. The complement regulatory factor H binds to the surface protein OspE of Borrelia burgdorferi. J. Biol. Chem. 2001;276:8427–8435. doi: 10.1074/jbc.M007994200. [DOI] [PubMed] [Google Scholar]
- 10.Kraiczy P, Skerka C, Brade V, Zipfel PF. Further characterization of complement regulator-acquiring surface proteins of Borrelia burgdorferi. Infect. Immun. 2001;69:7800–7809. doi: 10.1128/IAI.69.12.7800-7809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kraiczy P, Hellwage J, Skerka C, Kirschfink M, Brade V, Zipfel PF, Wallich R. Immune evasion of Borrelia burgdorferi: mapping of a complement inhibitor factor H-binding site of BbCRASP-3, a novel member of the Erp protein family. Eur. J. Immunol. 2003;33:697–707. doi: 10.1002/eji.200323571. [DOI] [PubMed] [Google Scholar]
- 12.Kraiczy P, Hartmann K, Hellwage J, Skerka C, Brade V, Zipfel PF, Wallich R, Stevenson B. Immunological characterization of the complement regulator factor H-binding CRASP and Erp proteins of Borrelia burgdorferi. Int. J. Med. Microbiol. 2004;293 S37:152–157. doi: 10.1016/s1433-1128(04)80029-9. [DOI] [PubMed] [Google Scholar]
- 13.Metts MS, McDowell JV, Theisen M, Hansen PR, Marconi RT. Analysis of the OspE determinants involved in binding of factor H and OspE-targeting antibodies elicited during Borrelia burgdorferi infection. Infect. Immun. 2003;71:3587–3596. doi: 10.1128/IAI.71.6.3587-3596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alitalo A, Meri T, Lankinen H, Seppälä I, Lahdenne P, Hefty PS, Akins D, Meri S. Complement inhibitor factor H binding to Lyme disease spirochetes is mediated by inducible expression of multiple plasmid-encoded outer surface protein E paralogs. J. Immunol. 2002;169:3847–3853. doi: 10.4049/jimmunol.169.7.3847. [DOI] [PubMed] [Google Scholar]
- 15.Alitalo A, Meri T, Chen T, Lankinen H, Cheng Z-Z, Jokiranta TS, Seppälä IJT, Lahdenne P, Hefty PS, Akins DR, et al. Lysine-dependent multipoint binding of the Borrelia burgdorferi virulence factor outer surface protein E to the C terminus of factor H. J. Immunol. 2004;172:6195–6201. doi: 10.4049/jimmunol.172.10.6195. [DOI] [PubMed] [Google Scholar]
- 16.Stevenson B, Tilly K, Rosa PA. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J. Bacteriol. 1996;178:3508–3516. doi: 10.1128/jb.178.12.3508-3516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casjens S, van Vugt R, Tilly K, Rosa PA, Stevenson B. Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J. Bacteriol. 1997;179:217–227. doi: 10.1128/jb.179.1.217-227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson RJ, et al. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs of an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- 19.Zückert WR, Meyer J. Circular and linear plasmids of Lyme disease spirochetes have extensive homology: characterization of a repeated DNA element. J. Bacteriol. 1996;178:2287–2298. doi: 10.1128/jb.178.8.2287-2298.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Babb K, McAlister JD, Miller JC, Stevenson B. Molecular characterization of Borrelia burgdorferi erp promoter/operator elements. J. Bacteriol. 2004;186:2745–2756. doi: 10.1128/JB.186.9.2745-2756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Babb K, Bykowski T, Riley SP, Miller MC, DeMoll E, Stevenson B. Borrelia burgdorferi EbfC, a novel, chromosomally-encoded protein, binds specific DNA sequences adjacent to erp loci on the spirochete’s resident cp32 prophages. J. Bacteriol. 2006;188:4331–4339. doi: 10.1128/JB.00005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riley SP, Bykowski T, Cooley AE, Burns LH, Babb K, Brissette CA, Bowman A, Rotondi M, Miller MC, DeMoll E, et al. Borrelia burgdorferi EbfC defines a newly-identified, widespread family of bacterial DNA-binding proteins. Nucleic Acids Res. 2009;37:1973–1983. doi: 10.1093/nar/gkp027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ebersbach G, Gerdes K. Plasmid segregation mechanisms. Annu. Rev. Genet. 2005;39:4530479. doi: 10.1146/annurev.genet.38.072902.091252. [DOI] [PubMed] [Google Scholar]
- 24.Eggers CH, Caimano MJ, Clawson ML, Miller WG, Samuels DS, Radolf JD. Identification of loci critical for replication and compatibility of a Borrelia burgdorferi cp32 plasmid and use of a cp32-based shuttle vector for the expression of fluorescent reporters in the Lyme disease spirochaete. Mol. Microbiol. 2002;43:281–295. doi: 10.1046/j.1365-2958.2002.02758.x. [DOI] [PubMed] [Google Scholar]
- 25.Beaurepaire C, Chaconas G. Mapping of essential replication functions of the linear plasmid lp17 of B. burgdorferi by targeted deletion walking. Mol. Microbiol. 2005;57:132–142. doi: 10.1111/j.1365-2958.2005.04688.x. [DOI] [PubMed] [Google Scholar]
- 26.Stewart PE, Thalken R, Bono JL, Rosa P. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol. Microbiol. 2001;39:714–721. doi: 10.1046/j.1365-2958.2001.02256.x. [DOI] [PubMed] [Google Scholar]
- 27.Stewart PE, Chaconas G, Rosa P. Conservation of plasmid maintenance functions between linear and circular plasmids in Borrelia burgdorferi. J. Bacteriol. 2003;185:3202–3209. doi: 10.1128/JB.185.10.3202-3209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stevenson B, Casjens S, Rosa P. Evidence of past recombination events among the genes encoding the Erp antigens of Borrelia burgdorferi. Microbiology. 1998;144:1869–1879. doi: 10.1099/00221287-144-7-1869. [DOI] [PubMed] [Google Scholar]
- 29.Casjens SR, Huang WM, Gilcrease EB, Qiu W, McCaig WD, Luft BJ, Schutzer SE, Fraser CM. In: Molecular Biology of Spirochetes. Cabello FC, Hulinska D, Godfrey HP, editors. Amsterdam: IOS Press; 2006. pp. 79–95. [Google Scholar]
- 30.Sawitzke JA, Li Y, Sergueev K, Youngren B, Brendler T, Jones K, Austin S. Transcriptional interference by a complex formed at the centromere-like partition site of plasmid P1. J. Bacteriol. 2002;184:2447–2454. doi: 10.1128/JB.184.9.2447-2454.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalnin K, Stegalkina S, Yarmolinsky M. pTAR-encoded proteins in plasmid partitioning. J. Bacteriol. 2000;182:1889–1894. doi: 10.1128/jb.182.7.1889-1894.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartosik AA, Lasocki K, Mierzejewska J, Thomas CM, Jagura-Burdzy G. ParB of Pseudomonas aeruginosa: interactions with its partner ParA and its target parS and specific effects on bacterial growth. J. Bacteriol. 2004;186:6983–6998. doi: 10.1128/JB.186.20.6983-6998.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodionov O, Lobocka M, Yarmolinsky M. Silencing of genes flanking the P1 plasmid centromere. Science. 1999;283:546–549. doi: 10.1126/science.283.5401.546. [DOI] [PubMed] [Google Scholar]
- 34.Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 35.Miller JC, Bono JL, Babb K, El-Hage N, Casjens S, Stevenson B. A second allele of eppA in Borrelia burgdorferi strain B31 is located on the previously undetected circular plasmid cp9-2. J. Bacteriol. 2000;182:6254–6258. doi: 10.1128/jb.182.21.6254-6258.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagensis by overlap extension using polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 37.Adams CA, Fried MG. In: Protein Interactions: Biophysical Approaches for the Study of Complex Reversible Systems. Schuck P, editor. New York: Academic Press; 2007. pp. 417–446. [Google Scholar]
- 38.Fried MG, Crothers DM. Equilibria and kinetics of Lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fried MG, Kanugula S, Bromberg JL, Pegg AE. DNA binding mechanism of O6-alkylguanine-DNA alkyltransferase: stoichiometry and effects of DNA base composition and secondary structure on complex stability. Biochemistry. 1996;35:15295–15301. doi: 10.1021/bi960971k. [DOI] [PubMed] [Google Scholar]
- 40.Stevenson B, Miller JC. Intra- and interbacterial genetic exchange of Lyme disease spirochete erp genes generates sequence identity amidst diversity. J. Mol. Evol. 2003;57:309–324. doi: 10.1007/s00239-003-2482-x. [DOI] [PubMed] [Google Scholar]
- 41.Dunn JJ, Buchstein SR, Butler L-L, Fisenne S, Polin DS, Lade BN, Luft BJ. Complete nucleotide sequence of a circular plasmid from the Lyme disease spirochete, Borrelia burgdorferi. J. Bacteriol. 1994;176:2706–2717. doi: 10.1128/jb.176.9.2706-2717.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lang B, Blot N, Bouffartigues E, Buckle M, Geertz M, Gualerzi CO, Mavathur R, Muskhelishvili G, Pon CL, Rimsky S, et al. High-affinity DNA binding sites for H-NS provide a molecular basis for selective silencing within proteobacterial genomes. Nucleic Acids Res. 2007;35:6330–6337. doi: 10.1093/nar/gkm712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akins DR, Caimano MJ, Yang X, Cerna F, Norgard MV, Radolf JD. Molecular and evolutionary analysis of Borrelia burgdorferi 297 circular plasmid-encoded lipoproteins with OspE- and OspF-like leader peptides. Infect. Immun. 1999;67:1526–1532. doi: 10.1128/iai.67.3.1526-1532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stevenson B, Bono JL, Schwan TG, Rosa P. Borrelia burgdorferi Erp proteins are immunogenic in mammals infected by tick bite, and their synthesis is inducible in cultured bacteria. Infect. Immun. 1998;66:2648–2654. doi: 10.1128/iai.66.6.2648-2654.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]