Abstract
The alternation/deficiency in activation-3 (ADA3) is an essential component of the human p300/CBP-associated factor (PCAF) and yeast Spt-Ada-Gcn5-acetyltransferase (SAGA) histone acetyltransferase complexes. These complexes facilitate transactivation of target genes by association with transcription factors and modification of local chromatin structure. It is known that the yeast ADA3 is required for nuclear receptor (NR)-mediated transactivation in yeast cells; however, the role of mammalian ADA3 in NR signaling remains elusive. In this study, we have investigated how the human (h) ADA3 regulates retinoic acid receptor (RAR) α-mediated transactivation. We show that hADA3 interacts directly with RARα in a hormone-dependent manner and this interaction contributes to RARα transactivation. Intriguingly, this interaction involves classical LxxLL motifs in hADA3, as demonstrated by both ‘loss’ and ‘gain’ of function mutations, as well as a functional coactivator pocket of the receptor. Additionally, we show that hADA3 associates with RARα target gene promoter in a hormone-dependent manner and ADA3 knockdown impairs RARβ2 expression. Furthermore, a structural model was established to illustrate an interaction network within the ADA3/RARα complex. These results suggest that hADA3 is a bona fide transcriptional coactivator for RARα, acting through a conserved mechanism involving direct contacts between NR boxes and the receptor’s co-activator pocket.
INTRODUCTION
Retinoic acids (RAs), the oxidized forms of vitamin A, possess important physiological and pharmacological actions as they determine vertebrae development, promote cell differentiation and affect cancer cell proliferation (1). The genomic actions of RAs are mediated by RA receptors (RARs), members of the nuclear receptor (NR) superfamily. NRs are ligand-dependent transcription factors that control gene expression in target cells. To date, many NRs are known as targets for therapeutic drugs in treating various human disorders, including metabolic diseases and cancers (2). Structurally, NRs share a common domain organization that consists of a variable N-terminal activation function domain (AF-1), a central DNA-binding domain (DBD) followed by a short hinge/D region, and a C-terminal ligand-binding and ligand-dependent transactivation function domain (LBD/AF-2) (3). The highly conserved DBD contains two zinc fingers and is responsible for recognition of hormone responsive elements (HREs). The LBD contributes to a dimerization interface of the receptor, in addition to binding co-activators and corepressors.
The transcriptional activity of NRs is thought to be regulated by a dynamic exchange of co-activators and corepressors (4). Crystallographic studies have revealed that ligand-binding induces a conformational change of the LBD and causes a positional shift of the carboxyl-terminal AF-2 helix (helix 12 or H12) (5). This alternation changes the cofactor-binding surface, triggers the dissociation of corepressors such as silencing mediator for retinoid and thyroid hormone receptors (SMRT) and N-CoR (6,7), and subsequently recruits co-activators like p160/SRC and CBP/p300 (8). The p160 co-activator family includes SRC1/NCoA-1 (9), TIF2/GRIP1 (10,11) and pCIP/AIB1/RAC3/ACTR (12,13). These three conserved co-activators interact with receptors in a hormone- and AF2-dependent manner and facilitate transcriptional activation by NRs. In contrast to the corepressors, co-activators contain intrinsic histone acetyltransferase activity (HAT) and remodel chromatin structure in an ATP-dependent manner, as well as causing covalent modification of histone tails (14). Both corepressors and co-activators interact with the same cofactor-binding pocket on the receptor through their α-helical LxxLL or related motifs (L represents leucine, x represents any amino acids). These LxxLL motifs are also known as CoRNR boxes in corepressors and as NR boxes in co-activators, respectively (15,16). In general, the LxxLL motif adopts a short α-helical structure and docks into a hydrophobic co-activator-binding pocket surrounded by helices H3, H4, H5 and H12 of the LBD (17). Mutational analyses of the co-activators NR boxes have also uncovered a receptor specific code of interaction, suggesting that NR box-mediated differential contacts may determine specific modes of NR action (5).
The alternation/deficiency in activation-3 (ADA3) is an essential component of the human PCAF and yeast SAGA or ADA HAT complexes. These complexes are known to stimulate transcription by association with DNA-binding factors and by modifying local chromatin structure (18,19). Through direct interactions, ADA3 participates in diverse physiological processes, and regulates the functions of several important proteins such as the tumor suppressor p53 (20), HPV E6 (21), several NRs (22) and ANCO-1 (23). In particular, the yeast ADA3 (yADA3) directly interacts with hRXRα and hERα in a ligand-dependent manner to augment their transactivation in yeast cells (24). In contrast, other studies have reported that the mouse ADA3 (mADA3) does not interact with these NRs (25) and the yADA3 and hADA3 reportedly fail to interact with hRARα (24,26). Hence, it is assumed that the hADA3 plays an indirect role in regulating RAR’s transcriptional activity through interaction with either RXRs or p160 co-activators (27).
Previously, we have determined the molecular mechanisms of the assembly of RXRα with corepressor and co-activator complexes (28,29) complexes. To extend our understanding of the mechanisms of retinoid signaling, we have characterized the potential physical and functional interactions between hADA3 and hRARα. Our results suggest that hADA3 is essential for hRARα transactivation and that hADA3 interacts directly with hRARα. A favorable structural model of the ADA3 LxxLL motif/RARα complex is established and the underlying molecular interactions are postulated.
MATERIALS AND METHODS
Chemicals and reagents
All-trans RA (atRA), rifampicin (Rif), 1,25-dihydroxyvitamin D3, 17β-estradiol (E2), and 3,5,3′-triiodothyronine (T3) were purchased from Sigma (St. Louis, MO, USA). The anti-ADA3 polyclonal antibody was a generous gift from Dr Pierre Chambon (IGBMC, Illkirch, France). The anti-Actin monoclonal antibody was purchased from Sigma.
Plasmids
The pCMX-hRARα wild-type and E412K mutant, and the pDR5-tk-luc reporter have been described previously (29). The hRARα K399E, ME400/1AA, I402A, PG403/4AA, SM405/6AA, PP407/8DT, LI409/10AA, L409A, I410A, Q411A, ML413/4AA, M413A, L414A, E415K, NS416/7AA, EG418/9AA and LD420/1AA mutants were created on the pCMXHA-hRARα template using the Quick Change Site-directed Mutagenesis kit (Stratagene, La Jolla, CA, USA). The series of GAL4 DBD-hRARα H12 mutants for expression in yeast cells were constructed in the pGBT9 vector at NcoI and BamHI cloning sites. The pACT-hADA3, pACT-hADA2α, pACT-hADA2β, pGBT-ANCO-1C (aa 2369–2663) and pGEX-hADA3 are as described previously (23). The NR box motifs of hADA3 were mutated as indicated and the primer sequences (5′ to 3′) for each NR box mutants are: mNR1, CTG CAG CTG GAG GCG GAG ACC GCG GCG TCT TCT GCC; mNR2, AGG TCC GCA CAG CTG AGG AGG CAG CGA AGC CCC CA; mNR3, GAC ACT AAA GAT GCG GCT GCC GCG GCG AAG AAG TCT GAG; mNR4, CTG ACG CAG CGC GCC GCG CAG GCC GCG GCG GAG GAA AAT; mNR5, GAG CGT GAG AGC GCC GCG AAG GCG GCG GAT GGG TAG; V237L, GGA CAC TAA AGA TCT GGA TGC CCT GCT GAA G; V269L, CCT GCA GGC CCT GCT GGA GGA AAA TAT TAT TTC C; I426L, GGA GCG TGA GAG CCT CCT GAA GCT GCT GGA TGG. All constructs have been confirmed by enzyme digestion and DNA sequencing with additional information available upon request.
Yeast two-hybrid assay
Yeast Y190 cells were co-transformed with the GAL4 DBD fusion plasmids (pAS-hRARα, pAS-hRARγ, pGBT-hRARα, pAS-hTRα, pAS-hERα, pGBT-VDR or pGBT-PXR) and the GAL4 AD fusion plasmids (pACT2 vector or pACT-hADA3), together with 4 µg of single-strand salmon sperm DNA according to manufacturer’s instructions (Clontech, Palo Alto, CA, USA). Yeast transformants were spread onto synthetic complete plates lacking tryptophan and leucine (SC-Trp-Leu) and incubated for 3 days at 30°C. Colonies were picked and grown in selection media for additional 24 h at 30°C. Aliquots of 100 µl from each sample were taken and added to fresh selection media. For each sample, one aliquot received cognate ligand (atRA, E2 or T3) and the other aliquot received vehicle control (DMSO). After 12 h, yeast cells were harvested and resuspended in Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, pH 7.0) and permeated with 0.05% SDS. An aliquot of 100 µl Z buffer containing 4 mg/ml ONPG (o-nitrophenyl β-d-galactopyronoside) was added and incubated at 30°C until the appearance of yellowish color. The reactions were stopped by adding 100 µl of 1 M Na2CO3 and the incubation time was recorded. The samples were centrifuged and the OD420 values of the supernatants were measured. The β-galactosidase unit was calculated according to the OD420 value, cell numbers and incubation time.
GST pull-down assay
GST and GST-hADA3 fusion proteins were expressed in Escherichia coli BL-21 cells and purified by glutathione agarose beads (GE Healthcare Biosciences, Uppsala, Sweden). Human RARα, ERα and AR were in vitro translated and radiolabeled with [35S]-methionine using the TNT Quick Coupled Transcription/Translation System (Promega, Madison, WI, USA). For the GST pull-down experiment, 5 µg of bead-conjugated fusion protein was incubated with 4 µl of in vitro translated 35S-labeled protein with moderate shaking at 4°C overnight as previously described (28). Bound proteins were washed three times with fresh binding buffer containing 0.1% NP40, and beads were collected and subjected to SDS–PAGE electrophoresis. Gels were then stained with Coomassie Blue, dried and detected by autoradiography.
Cell culture and transient transfection reporter assay
HEK293 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. One day prior to transfection, cells were seeded in 12-well plates at 50 000 cells per well in phenol red free Dulbecco’s modified Eagle’s medium supplemented with 10% charcoal resin-stripped fetal bovine serum. Transfection was performed using standard calcium/phosphate method as described (29). After transfection, cells were washed with phosphate-buffered saline and re-fed with fresh medium containing vehicle alone or vehicle plus 50 nM atRA and incubated for an additional 48 h. Cells were then harvested and analyzed for luciferase and β-galactosidase activities.
RNA interference
The effect of hADA3 short hairpin RNA (shRNA) on RARα transcriptional activity was analyzed in HEK293 cells. The hADA3, PIASy and a control shRNAs were expressed from the pLL3.7 lentiviral vector (30). The sequence for the hADA3 shRNA is 5′-GAT GAG GCT GAG CAT TAC A-3′ starting at nucleotide 543/amino acid 181 of hADA3 (NM_006354). The sequence for PIASy shRNA is 5′-GCT CTA CGG AAA GTA CTT A-3′ starting at nucleotide 330/amino acid 110 of the human PIASy (BC029874). The control shRNA sequence is 5′-GGT CCG GCT CCC CCA AAT G-3′, which has no homologous sequences in the vertebrate transcriptome. After 24 h of transfection with the shRNA expression plasmid, the HEK293 cells containing transiently transfected hRARα and the luciferase reporter gene were treated with 50 nM atRA for 12 h and analyzed for luciferase activity.
Generation of ADA3 stable knockdown cells
Recombinant lentiviruses were produced by co-transfecting HEK 293T cells with the lentivirus expression plasmid and lentiviral packaging constructs pLP1, pLP2 and the pLP/VSV-G plasmids using Lipofectamine 2000 (Invitrogen). Infectious lentiviruses were harvested at 48 h post-transfection from the culture supernatants, centrifuged to eliminate cell debris and then filtered through 0.22-μm filters. For transducing lentiviral constructs, 50% confluent MCF7 cells were fed with virus containing medium for 2 days to generate 40% infected MCF7 cells by visualizing GFP expression. The top 20% of GFP positive cells were subsequently collected using fluorescence-activated cell sorting analysis to retrieve the highest GFP expressed cells. The lentivirus-infected MCF-7 cells with siCTRL and siADA3 were grown in serum free DMEM for overnight and treated with or without 2 µM atRA for 3 h. Total RNA was extracted using Trizol reagents (Invitrogen), and 2 µg RNA was reverse transcribed using Superscript II reverse transcriptase (Invitrogen) with random oligo(dT) primers. RT products were amplified by PCR using the following primer pairs: for the RARβ2 coding sequence (421 bp), forward, 5′-GGA ACG CAT TCG GAA GGC TT-3′ and reverse, 5′-AGC ACT TCT GGA GTC GAC AG-3′; for the hADA3 coding sequence, forward, 5′-TGT GCC GCA TAC TAA GTC C-3′ and reverse, 5′-GCT TCA GGA TGC TCT CAC GCT-3′; for the control GAPDH coding sequence (212 bp), forward, 5′-GTG GAT ATT GTT GCC ATC A-3′ and reverse, 5′-GAC TCC ACG ACG TAC TCA-3′. PCR products were then separated by electrophoresis on 1.5% agarose gel and visualized by ethidium bromide staining.
Chromatin immunoprecipitation assay
HEK293 cells were transiently transfected with FLAG-tagged hRARα and HA-tagged hADA3 according to the manufacturer’s instruction (Lipofectamin 2000, Invitrogen). After treated with 2 µM atRA for 3 h, cells were incubated with 1% formaldehyde in culture media for 10 min at 37°C. The formaldehyde cross-linked cells were washed twice with ice-cold PBS and lysed in SDS buffer [1% SDS, 10 mM EDTA, 50 mM Tris–HCl, pH 8.1 and protease inhibitor cocktail (Roche)]. Nuclei were collected, resuspended in lysis buffer and sonicated 10 times with 15-s pulses on ice. Supernatant were then diluted in 10-fold Dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1, 16.7 mM NaCl) and pre-cleared with 80 µl of salmon sperm DNA-protein G-agarose (Upstate) for 2 h at 4°C. The sheared DNA mixture was subjected to overnight incubation with 1 µg of anti-FLAG antibody (F3165, Sigma), anti-HA antibody (1815015, Roche), anti-RNA polymerase II antibody (05-623B, Upstate) or an equal amount of normal mouse immunoglobulin G (Santa Cruz Biotechnology) at 4°C. Immunoprecipitation was performed using protein G agarose for 1 h at 4°C. The protein/DNA cross-links were reversed by heating at 65°C for 6 h, and then the DNA were extracted using QIAquick PCR purification kit (Qiagen). For PCR reaction, 3 µl from a 40 µl DNA preparation was used for amplifications. The DNA product was analyzed using a primer set against RARβ2 promoter, 5′-AAG CTC TGT GAG AAT CCT G-3′ and 5′-GGA TCC TAC CCC GAC GGT G-3′ (GenBank accession number AF275948). The 228 bp RARβ2-promoter were visualized on a 1.5% agarose gel.
Molecular modeling
The initial model for RARα/hADA3 complex was constructed based on the crystal structure of the RARβ/RXRα complex with an LxxLL peptide from the TRAP220 co-activator (PDB: IXDK) (31). Due to the high sequence identity shared between RARα and RARβ (∼88%), the distinct residues on RARβ were directly mutated to the respective residues on hRARα and the coordinates of the NR box 4 peptide of hADA3 were built from those of the TRAP220 peptide. The resulting models were subjected to energy minimization for 2500 steps of steepest descent geometry optimization followed by 2500 steps of conjugate gradient optimization with RARα LBD held fixed, except for the residues making contact with the ADA3 peptide. The energy minimized structural model of the RARα/ADA3 complex was then utilized as the initial structure for Molecular Dynamics (MD) simulation. The model was first solvated by explicit water molecules, followed by 5000 steps energy minimization. The temperature of the system was heated gradually from 100 to 300 K for 200 ps and kept constant throughout the later simulation. Furthermore, a 500 ps equilibration step was conducted at 300 K and 1 atmosphere constant pressure to adjust the solvent density. Finally, another 2 ns MD simulation was run for data collection with snapshots obtained every 2 ps.
RESULTS
Human ADA3 enhances the transcriptional activity of hRARα in mammalian cells
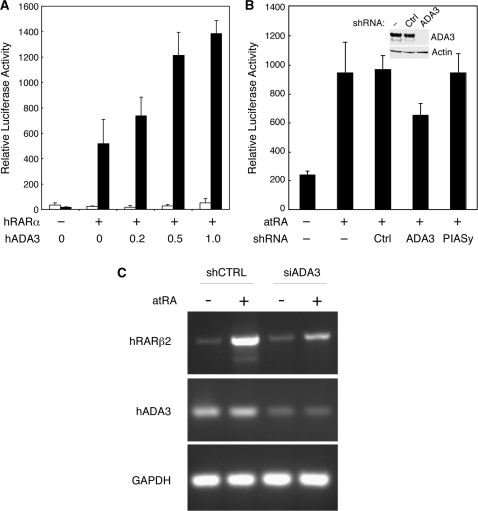
To investigate the role of hADA3 in RAR signaling, we measured the effects of hADA3 overexpression and silencing on hRARα-mediated transcriptional activation, using a transient transfection reporter assay in the human embryonic kidney HEK293 cells. As shown in Figure 1A, ectopically expressed hADA3 enhanced the activation of RARα-driven transcription in the presence of atRA in an ADA3 concentration-dependent manner, suggesting that hADA3 may function as a co-activator for RARα. To determine whether endogenous hADA3 contributes to the RARα-mediated transcriptional activation, we knocked down the endogenous ADA3 in HEK293 cells by shRNA using a lentivirus vector (Figure 1B, inset). While the control shRNAs had no effect, silencing of hADA3 significantly reduced RARα-mediated transactivation (Figure 1B). Therefore, both the endogenous and ectopically expressed hADA3 are involved in enhancing RARα-mediated transcriptional activation. Additionally, we examined the effect of ADA3 knockdown on the RARα downstream gene regulation. ADA3 knockdown cells were created using lentivirus-based siRNA system in MCF7 cells. As shown in Figure 1C, the expression of RARβ2, upon its induction by atRA, was much lower in the ADA3 knockdown cells as compared with control cells. This result suggests that hADA3 acts as an RARα co-activator regulating its target gene expression.
Figure 1.
The human ADA3 enhances transcriptional activity of RARα in mammalian cells. (A) Ectopic overexpression of human ADA3 enhances transcriptional activation mediated by hRARα. HEK293 cells were cotransfected with pCMX-hRARα, pDR5-tk-Luc reporter and indicated amounts (in µg) of pCMX-hADA3. Cells were incubated in the absence (white bars) or presence (black bars) of 50 nM atRA. The relative luciferase activities were determined from three independent samples after normalization to the internal β-galactosidase control. (B) Endogenous human ADA3 contributes to RARα-mediated transcriptional activation. HEK293 cells were cotransfected with pCMX-hRARα and pDR5-tk-Luc reporter plus pLL3.7 shRNA expression vector without an insert (−), or with the control (Ctrl), hADA3 or PIASy targeting shRNA. The relative luciferase activities were determined from three independent samples after normalization to the internal β-galactosidase control. The small inset in the graph is a western blot showing the reduction of ADA3 by the pLL3.7-hADA3 shRNA, which has no effect on the expression of endogenous β-actin. (C) Expression of RARβ2 is impaired in ADA3 knockdown cells. Total RNA was extracted from each of the control (siCTRL) or siADA3-infected stable clones without or with atRA treatment and the RNA was subjected to RT-PCR using primers specific for RARβ2 or ADA3 coding sequence. GAPDH was used as an internal control.
ADA3 interacts directly with liganded RARα
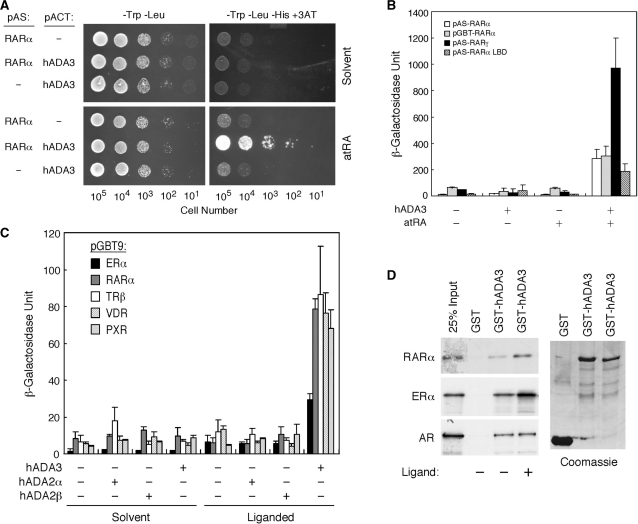
Next, we tested if hADA3 interacts directly with RARα. Yeast Y190 cells expressing both the GAL4 DBD-hRARα and GAL4 AD-hADA3 fusions proliferated well on a -Trp-Leu-His+3AT (3-aminotriazole) plate in the presence of atRA (Figure 2A). In contrast, cells without the hRARα or hADA3 fusions were unable to grow under the same conditions regardless of atRA. As controls, all transformants proliferated equally well on -Trp-Leu plates (left panels). This ADA3/RARα interaction was confirmed by using different GAL4 DBD-RAR constructs (Figure 2B), suggesting that hADA3 interacts specifically with RARs in a ligand-dependent manner.
Figure 2.
ADA3 binds to RARα in a ligand-dependent manner. (A) Yeast two-hybrid assay showing ligand-dependent RARα-ADA3 interaction. Full-length hRARα was expressed as GAL4 DBD fusion from pAS1 vector, while full-length hADA3 as GAL4 AD fusion from pACT2 vector. Yeast Y190 cells harboring the indicated plasmids were spotted onto SC-Trp-Leu or SC-Trp-Leu-His+3AT (20 mM) plates, in the absence (solvent) or presence of atRA (100 nM). The numbers of cells spotted are shown at the bottom. (B) Interaction of hADA3 with various RAR plasmids in yeast two-hybrid assay. Yeast transformants containing indicated combinations of the specified RAR plasmid and pACT-hADA3 were grown on SC-Trp-Leu media supplemented without (−) or with (+) 1 µM of atRA. (C) Ligand-dependent interactions of ADA3 with various NRs in yeast two-hybrid assay. Y190 cells were transformed with the GAL4 DBD fusion of various indicated NRs together with the indicated GAL4 AD fusion of hADA3, hADA2α or hADA2β. Transformants were grown in SC-Trp-Leu selection media in the presence or absence of cognate ligand (10 nM E2 for ERα, 10 nM atRA for RARα, 10 nM T3 for TRβ, 10 nM vitamin D3 for VDR and 10 µM rifampicin for PXR). (D) Interactions of hADA3 with NRs in vitro by GST pull-down assay. Purified GST proteins were incubated with [35S]-labeled hRARα, hERα or hAR without or with ligands (1 µM atRA for hRARα, 100 nM E2 for hERα and 100 nM DHT for hAR).
We next examined ADA family proteins, including hADA3, hADA2α and hADA2β, for their interaction potentials with various NRs. Interestingly, only hADA3 was able to interact with these NRs in the presence of cognate ligands, while hADA2α/β show little interaction regardless of ligands (Figure 2C). These results suggest hADA3 subunit as a potential mediator in the recruitment of PCAF/ADA complex to ligand-activated NRs. The interactions between hADA3 and NRs were further demonstrated in vitro using GST pull-down assays (Figure 2D), where hRARα and hERα showed specific ligand-enhanced interactions with hADA3 while hAR exhibited a ligand-independent interaction. These results suggest that hADA3 interacts directly with several NRs in a hormone-dependent manner in vivo and in vitro.
ADA3 binds to the co-activator pocket of RARα
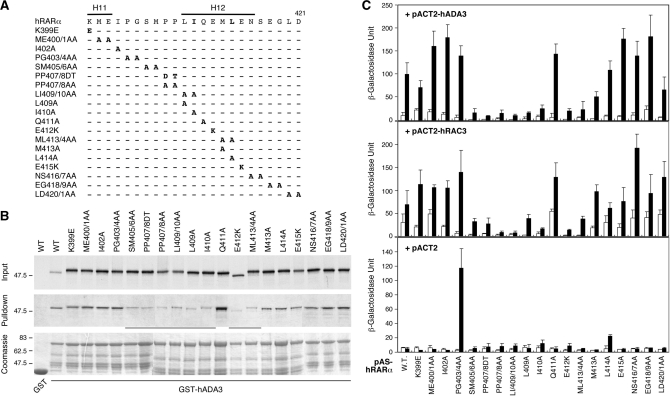
The H12 helix at the carboxyl-terminal end of NR plays a critical role in the formation of a ligand-induced co-activator-binding pocket (17). To determine if the co-activator pocket is responsible for ADA3 binding, we systematically mutated each amino acid spanning the H12 helix of hRARα (Figure 3A). These RARα mutants were labeled with [35S]-methionine and tested by GST pull-down assay for interaction with GST-hADA3 in the presence of atRA in vitro. As shown in Figure 3B, SM405/6AA, PP407/8DT, LI409/10AA, L409A, I410A, E412K and ML413/4AA mutations all interrupted RARα interaction with hADA3, suggesting that H12 helix is essential for ADA3 binding. In contrast, the Q411A mutant within the H12 helix did not affect the interaction, suggesting that the side chain of Q411 is not critical for ADA3 interaction.
Figure 3.
ADA3 interacts with the co-activator pocket of RARα. (A) Summary of site-directed mutants surrounding hRARα H12 helix. Substituted amino acids for individual mutations are as indicated at the mutated residues. (B) A GST pull-down assay showing interactions of GST-hADA3 with [35S]-hRARα and its mutants. Approximately 20% of the labeled proteins used in the pull-down reaction are shown in the input panel. The pull-down panel shows the amount of labeled RARα that interacted with GST-hADA3 or GST alone (first lane) in the presence of 1 µM of atRA. (C) Comparison of the interactions of RARα H12 mutants between hADA3 and RAC3 in yeast two-hybrid assay. Yeast Y190 cells were co-transformed with each indicated pAS-hRARα construct shown at bottom together with pACT-hADA3, pACT-RAC3 or pACT2 empty vector. The transformed cells were grown in SC-Trp-Leu media containing either solvent (open bars) or 100 nM of atRA (solid bars) for 36 h. The β-galactosidase units reflect interactions between various hRARα constructs with empty vector, RAC3 or ADA3, respectively.
To compare the RARα interaction profiles between ADA3 and p160 co-activator, yeast two-hybrid assays were carried out to determine the interaction of RARα H12 mutants with ADA3 and RAC3 (Figure 3C). We found that the ligand-induced interaction patterns of hRARα and hADA3 in yeast cells were consistent with that of the GST pull-down assay. Moreover, the hRARα interacting profiles with hADA3 and RAC3 were similar (Figure 3C). As a control, except for the PG403/4AA mutant, none of these GAL4 DBD-fused hRARα mutants displayed significant background activation, (Figure 3C, +pACT2 panel). The H12 requirement and similar interacting profile to RAC3 suggest that hADA3 interacts with the co-activator-binding pocket of hRARα.
ADA3 interaction mutants of RARα display impaired transcriptional activity
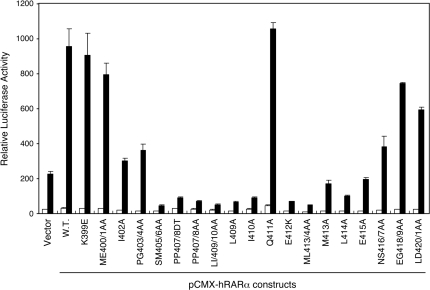
To strengthen the idea that ADA3 is a transcriptional co-activator for RARα through a mechanism involving direct protein–protein contact, we assessed the transcriptional potentials of the RARα H12 mutants by transient transfection reporter assay. Since endogenous ADA3 may function as a co-activator for RARα, this experiment was conducted in the absence of ADA3 overexpression. As shown in Figure 4, the RARα H12 mutants that are defective in ADA3 interaction also displayed impaired transactivation function. These data suggest that ADA3 interaction may contribute to RARα transcriptional activity. Interestingly, most of these ADA3 interaction mutants also display dominant-negative effects as they suppressed the reporter gene expression below control level, possibly due to sequestration of endogenous cofactors. Although the contribution of other transcriptional co-activators, such as the p160 proteins, cannot be undermined, these results support the idea of ADA3 as a co-activator for RARα.
Figure 4.
ADA3 interaction deficiency mutants of RARα display impaired transcriptional function in mammalian cells. The transcriptional activities of the hRARα H12 mutants shown in Figure 3A were determined by transient transfection reporter assay in HEK293 cells. Each of the indicated pCMX-hRARα constructs was cotransfected with the pDR5-tk-Luc reporter and the β-galactosidase control plasmid. Transfected cells were treated with either solvent (white bars) or 50 nM atRA (black bars). The relative luciferase activities were determined from three independent samples after normalization to the internal β-galactosidase control.
Putative LxxLL motifs of ADA3 are involved in interaction with RARα
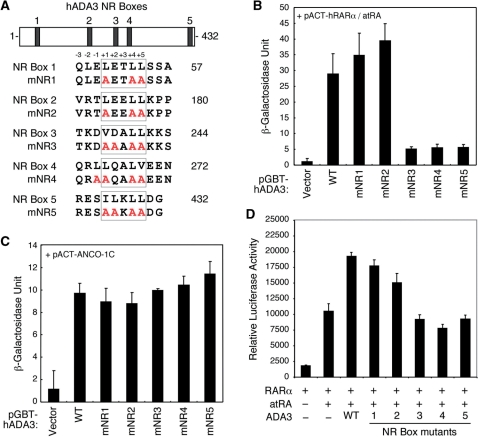
Several NR co-activators, including the p160 and DRIP220, utilize highly conserved LxxLL motifs to interact with the co-activator-binding pocket of the receptor. Previously, two putative LxxLL motifs were identified in the mouse ADA3, although they were excluded from mediating interaction with NRs (25). As shown in Figure 5A, we have located five putative LxxLL consensus sequences in the hADA3 sequence. We tested whether these putative LxxLL motifs are involved in interaction with RARα. Site-directed mutations of conserved hydrophobic amino acids within each motif were created and tested for interaction with liganded RARα in a yeast two-hybrid assay. Intriguingly, alterations of NR box 3 (mNR3), 4 (mNR4) and 5 (mNR5) showed diminished interaction with RARα in the presence of atRA (Figure 5B). In contrast, alterations in NR boxes 1 (mNR1) and 2 (mNR2) had little effect. As a control (Figure 5C), none of these NR box mutations of ADA3 affected its interaction with ANCO-1 (32). These results suggest that hADA3 may utilize specific LxxLL motifs for interaction with the co-activator pocket of RARα. Additionally, the effect of NR box mutation on the hRARα co-activator function of ADA3 was investigated by transient transfection reporter gene assay. Consistently, NR boxes 3, 4 and 5 mutants failed to enhance transactivation of hRARα (Figure 5D). These results suggest that the three NR boxes of hADA3 are involved in interaction and coactivation with hRARα.
Figure 5.
Three NR boxes in ADA3 are required for RARα interaction. (A) Amino acid alignment of the five putative hADA3 NR boxes and their corresponding mutants. Relative positions of individual NR boxes are marked in the hADA3 schematic structure on top. The amino acid positions relative to the LxxLL core motif are shown on top of the peptide sequence. (B) Yeast two-hybrid interactions of pACT-hRARα with pGBT-hADA3 wild-type or NR box mutants. Vector indicates blank pGBT9 vector. Cells were treated with 50 nM atRA for 24 h before β-galactosidase assay. Mutations of NR boxes 1 and 2 had no effect on RARα interaction, while mutations of NR boxes 3, 4 and 5 abolished the interaction. (C) Yeast two-hybrid interactions of pACT-ANCO-1C with pGBT-hADA3 wild-type and NR box mutants. None of the ADA3 NR box mutations affected the interaction between hADA3 with ANCO-1. (D) ADA3 NR boxes 3, 4 and 5 mutations abolished the coactivation function of hADA3 on hRARα. HEK293 cells were cotransfected with pCMX-hRARα along with indicated pCMX-hADA3 constructs. Transfected cells were treated with 100 nM atRA or solvent as control. Relative luciferase activity was determined from three independent experiments after normalization with co-transfected β-galactosidase internal control.
‘Gain-of-function’ mutants and association of ADA3 with RAR target genes
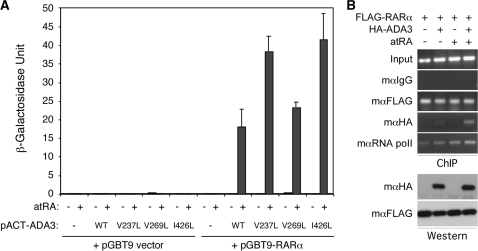
The three putative NR boxes of ADA3 that are involved in interaction with RARα are atypical LxxLL motifs that may not be optimal for binding NRs. Therefore, we mutated the NR box 3 sequence from VDALL to LDALL (creating the V237L mutant), the box 4 LQALV to LAQLL (creating the V269L mutant) and the box 5 ILKLL to LLKLL (creating the I426L mutant). These potential ‘gain-of-function’ mutations were tested for interaction with hRARα in yeast two-hybrid assay. In this experiment, we found that all three mutations enhanced the interaction of ADA3 with liganded hRARα, although the effect of NR box 4 mutation was less significant (Figure 6A). Together with the ‘loss-of-function’ mutants, these ‘gain-of-function’ mutants strongly indicate that ADA3 indeed interacts with hRARα via a mechanism involving NR boxes.
Figure 6.
(A) ‘Gain-of-function’ mutations of ADA3 NR boxes enhance interaction with RARα. We mutated the ADA3 NR Box 3 VDALL to LDALL (V237L), Box 4 LQALV to LQALL (V269L) and Box 5 ILKLL to LLKLL (I426L). The mutations were generated on the pACT-ADA3 construct and tested for interaction with pGBT9-RARα in yeast two-hybrid assay. The pGBT9, pACT2 (−) vectors and the pACT-ADA3 wild-type (WT) were used as controls. The transformed colonies were grown in SC-Trp-Leu media containing either solvent (open bars) or 100 nM of atRA (solid bars) for 36 h and β-galactosidase activities were determined from three independent cultures. (B) ADA3 associates with RARβ2 promoter in a hormone-dependent manner. Transiently transfected HEK293 cells were serum-starved and then stimulated with or without 2 µM atRA for 3 h. The cells were subjected to in vivo ChIP assay. The FLAG, HA and RNA polymerase II antibodies were used in the immunoprecipitation. Mouse IgG was used as a negative control, whereas input chromatin was used as loading control for the PCR reactions.
Furthermore, to determine if hADA3 could be recruited to RAR target genes, chromatin immunoprecipitation (ChIP) assay was performed. We transfected FLAG-tagged hRARα and HA-tagged hADA3 into both HEK293 and HeLa cells and conducted ChIP assays using monoclonal anti-FLAG and -HA antibodies on the nature hRARβ2 target promoter. In this experiment, the hRARβ2 promoter region that contains RAR responsive element was immunoprecipitated as expected with the anti-FLAG antibody, but not with the control IgG (Figure 6B). The same hRARβ2 promoter region was also immunoprecipitated with anti-HA antibody, but this occurred only in the presence of HA-hADA3 co-transfection and atRA treatment. Similar results were obtained in both HEK293 and HeLa cells. These data strongly suggest that ADA3 indeed can associate with RAR target gene promoter in vivo in a hormone-dependent manner.
A structural model of the ADA3/RARα complex
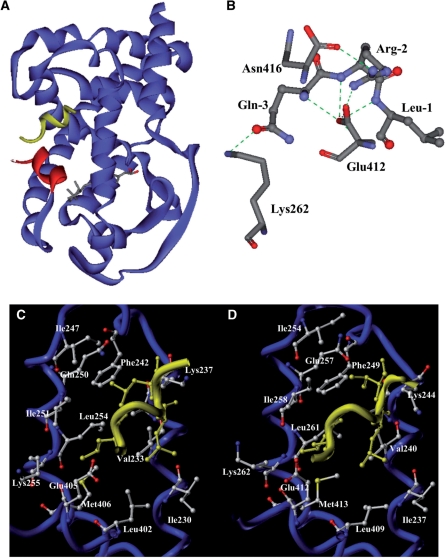
To corroborate on the findings that the LxxLL motifs of ADA3 and the co-activator pocket of RARα are involved in their interaction, we employed a molecular modeling technique to deduce the most favorable structural model of the hADA3 NR box 4 peptide complexed with the hRARα LBD. An existing crystal structure of the hRARβ/TRAP220 complex (31) was used as a molecular template for building the hRARα/ADA3 NR box 4 peptide structural model. The ADA3 NR box 4 (QRLLQALVEE) was chosen because it is essential for interaction with RARα and bears the highest similarity (70%) to the TRAP220 NR box 2 (RHKILHRLLQEGS) used in the crystal study (31). The resultant model was further refined using MD and energy minimization. The sequence of the NR box 4 was modeled as a short two-turn α-helix. The average structure derived from a 2 ns MD simulation indicates that the ADA3 peptide mimicked the observed binding mode of the LxxLL motif to other NR LBDs, and interacted with the RARα LBD through a hydrophobic groove formed by helices 3, 4, 5 and 12 (Figure 7A). The proper position of the peptide was locked by two conserved charge clamp residues, Lys244 and Glu412 positioned at opposite ends of the cleft via their electrostatic interactions with backbone atoms on the ends of the motif helix. A complex network of hydrogen bonds is formed between the amino-terminal residues of the peptide and the surrounding residues on the receptor (Figure 6B). In addition to the classical charge clamp interaction between Glu412 and the amide nitrogen atom of Leu-1, two additional electrostatic interactions are formed: one between Lys262 and Gln-3 and another between Asn416 and Arg-2. These two electrostatic interactions may explain the observed differential influence of mutated NR boxes and their interactions with hRARα (Figure 5). This hydrogen bond network can only form in NR boxes 3, 4 and 5, which augments the van der Waals contacts at the core hydrophobic interface and helps stabilize the protein–peptide complex. However, these interactions are missing from NR boxes 1 and 2 due to the presence of hydrophobic residues at positions -2 and -3, respectively. Compared with the binding of the LxxLL motif from TRAP220 to hRARβ, the helix formed by ADA3 NR box 4 is shifted toward H12 by ∼1.7Å (Figure 6C and D). Side chains of several residues undergo conformational changes upon binding of the ADA3 peptide. The carboxyl group of Glu412 is rotated ∼180 degrees around the CB–CA bond in order to accommodate the shift of the ADA3 peptide toward H12. In this binding mode, residues of Leu+1, Leu+4 and Val+5 of the ADA3 peptide comprise the core hydrophobic interface with the corresponding hydrophobic groove of the RARα co-activator pocket. Their predicted interacting residues on the receptor are listed in Table 1.
Figure 7.
A structural model of the ADA3 NR box 4/RARα LBD complex. (A) An overview of the RARα-ADA3 NR box 4 complex. The RARα LBD is shown as blue ribbons except helix 12 is colored in red. The peptide from ADA3 NR box 4 is shown in yellow. (B) Hydrogen-bond interaction map around the N-terminal of the ADA3 NR box 4 peptide. The green dashed lines represent hydrogen bonds. The three N-terminal residues from ADA3 peptide Leu-1, Arg-2 and Gln-3, along with the Lys262, Glu412 and Asn416 residues from RARα, are shown as indicated. (C, D) Structural comparison between hRARβ/TRAP220 and hRARα/ADA3 complexes. Helices 3, 4, 5 and 12 are displayed as blue coils and co-activator motifs are colored in yellow. Residues on the receptor interacting with peptides are depicted in stick representation and colored by atom-type (carbon: grey; nitrogen: blue; oxygen: red). The residues from peptides are colored in yellow and, for clarity, only residues making significant interactions with the LBD are shown. (C) Structure of the RARβ/TRAP220 NR box 2 complex. (D) Structure of the RARα/ADA3 NR box 4 complex.
Table 1.
The hRARα residues predicted to make contacts with the core hydrophobic residues of hADA3 NR box 4
| Helix | Residues |
|---|---|
| Helix 3 | Ile237, Val240, Ala 243, Lys244 |
| Helix 4 | Phe249 |
| Helix 5 | Ile254, Gln257, Ile258, Leu261, Lys262 |
| Helix 12 | Leu409, Glu412, Met413 |
The hRARα residues are listed according to their relative locations within indicated helices of hRARα LBD
DISCUSSION
In this study, we have determined the role of human ADA3 in RARα-mediated transcriptional activation and investigated the underlying molecular mechanisms. We show that hADA3 interacts directly with hRARα and enhances its transactivation function. The interaction between ADA3 and RARα is ligand-dependent. RARα interacts specifically with ADA3 but not other ADA subunits, while ADA3 may interact with other NRs. Several crucial amino acid residues on the transactivation helix (H12) of RARα co-activator pocket are identified as essential for the interaction with ADA3, as well as for the transactivation function of RARα. Furthermore, three putative LxxLL NR boxes of ADA3 are found to be critical for interaction with liganded RARα, and ADA3 is capable of associating with RAR target gene promoter in a hormone-dependent manner. We propose a structural model of the ADA3 NR box 4/RARα LBD complex to illustrate the mode of interactions. These data suggest that hADA3 is involved in RARα-mediated transactivation, likely through a mechanism of direct physical interactions between the NR boxes of ADA3 and the co-activator pocket of liganded RARα.
The involvement of ADA3 in NR signaling was first demonstrated in Saccharomyces cerevisiae when yeast ADA3 was isolated as an RXRα-interacting protein (24). The yeast ADA adaptor complex is also important for transcriptional activation by GR (33,34) and TR (35), where the yeast ADA2 seems to play a more important role. The yeast ADA3 is thought to mediate transactivation by RXRα and ERα in yeast cells through direct physical interaction and recruitment of the SAGA complex (24). In that study, yeast ADA3 was shown to interact in a ligand-dependent manner with RXRα, TRα and ERα, but not with RARα. Surprisingly, the same group reported that mouse ADA3 does not interact with NRs, even though it contains potential LxxLL motifs (25). Inconsistent data were reported regarding the roles of human and mouse ADA3 in terms of their interactions and function with human ERα/β and RXRα and hRARα in mammalian cells (22,26). Interestingly, yeast ADA3 was implicated in mediating RARα transactivation through interaction with the p160 co-activator RAC3/AIB1 and recruitment of GCN5 co-activator complex (27). Our current results demonstrate a strong ligand-dependent direct interaction between hADA3 and RARα in vivo and in vitro. We show that human ADA3, but not ADA2α or β, is capable of interaction with several NRs including ERα, TRβ, VDR, PXR and AR. This is consistent in part with prior reports supporting a direct role of hADA3 in ER signaling (22,26). However, it is noted that ADA2, instead of ADA3, was previously implicated in TRβ signaling (35), although our current data suggest that TRβ interacts with hADA3 but not ADA2α or 2β (Figure 2C). To our surprise, ADA3 seems to bind to RAR subtypes with different affinity. In particular, RARγ shows a 3-fold better interaction with ADA3 as compared to RARα (Figure 2B). We reason that the ω loop in the RARγ, which is absent in RARα and RARβ, might facilitate the conformational change of the RAR/ADA3 interaction. Since RARα and RARβ have been shown to be associated with the development of acute promyelocytic leukemia and squamous cell cancers, whereas RARγ is associated with retinoid effects on mucocutaneous tissues and bone, our results suggest that ADA3 may be involved in multiple physiological effects mediated by different subtypes of RARs. While this study focuses on dissecting the molecular interaction between ADA3 and RARα, the attribution of RAR target gene expression to different co-activators has been an important, yet challenging topic, especially due to the existence of overlapping co-activators, and the fact that cell is able to compensate the loss of a particular co-activator. This is exemplified in our ADA3 knockdown experiment where RAR reporter gene expression was affected but not abolished (Figure 1B).
To corroborate on the role of human ADA3 in RARα signaling, we have investigated the molecular basis of their interactions. Several co-activators, including the p160 proteins, are known to utilize a simple LxxLL NR box for interaction with a structurally conserved co-activator pocket located on the surface of a receptor LBD (12,29,36,37). The co-activator pocket is formed by helices 3, 4, 5 and 12 in response to ligand-induced conformational change, with helix 12 serving as a dynamic lip that closes up upon ligand binding (28,38). Our data demonstrate that the co-activator pocket of RARα is indeed required for ADA3 binding. In addition, we have identified three potential LxxLL motifs in hADA3, whose disruption clearly abolished the interaction between ADA3 and RARα (Figure 5). Importantly, ‘gain-of-function’ mutations of these NR boxes that optimize the NR-binding potentials of the LxxLL motif enhanced interaction with RARα (Figure 6A). The fact that the optimization of NR box 4 consensus sequence has the least effect on enhancing interaction with RARα is consistent with the idea that this motif is nearly optimal in interaction with RARα as demonstrated by molecular modeling (Figure 7). Whether these NR boxes are essential for interactions with other NRs, and the relative importance and the stoichiometry of these motifs, remain to be determined. The reason that a single NR box mutation in ADA3 abrogated interaction with RAR may be multifaceted. One plausible explanation is that all three NR boxes are involved in the interaction, while each of them is required for the formation of a stable complex in vivo. Thus, mutation in a single NR box would result in the destabilization of the ADA3/RAR complex.
Given that ADA3 is a key subunit of PCAF co-activator complexes, it is reasonable to speculate that ADA3 may act as a molecular bridge to mediate the recruitment of co-activator complexes to ligand-activated receptors. Our data show that not only the transcriptional activity of RARα was enhanced by overexpression of ADA3, silencing of endogenous ADA3 also diminished reporter gene expression (Figure 1). Therefore, we suggest that ADA3 may indeed function as a transcriptional co-activator for RARα. We further speculate that ADA3 may play a role in bringing its associated-co-activator complexes to other NRs. Nonetheless, there are likely receptor and receptor-subtype selectivity for ADA3, because its interaction with AR was independent of ligand and a better interaction with RARγ subtype was observed. Despite the positive epigenetic regulation of gene transcription, ADA3 also directly binds and acetylates P53, reducing Mdm2-mediated ubiquitination of P53 and subsequently stabilizing p53 expression (39). In this regard, ADA3 may also function as an RAR acetyltransferase and affect its protein turnover rate. Various possibilities warrant detailed study and will be addressed in future work.
The co-activator binding characteristics of RARα between ADA3 and RAC3 were analyzed and compared using a series of systematic helix 12 mutations in RARα (Figure 3). Intriguingly, the interacting pattern is highly related to RARα transactivation capability (Figure 4), suggesting that RARα activity is controlled by these cofactors. Additionally, the lack of ADA3 or RAC3 binding seems to correlate with a dominant negative function of RARα, possibly due to sequestration of endogenous cofactors such as RXR. This observation is consistent with a prior finding where disrupting TRβ and SRC1 interaction resulted in a dominant negative regulation in TRβ (40). Similarly, the transcriptional regulation of Hepatocyte Nuclear Factor-4α is highly dependent on its physical interaction with PGC-1 and SRC3 (41). Thus, we hypothesize that ADA3 utilizes a similar binding pocket as p160 co-activators in regulating RAR signaling. While ADA3 and p160 co-activators share the same binding property to RARα, it is conceivable that these two co-activators may compete with each other if present at the same time. It will be of special interest to know whether there is specific physiological condition(s) that governs a potential binding switch from p160 to ADA3, or vice versa.
With the help of molecular modeling, we have created a structural model of the hADA3 NR box 4 peptide/hRARα LBD complex (Figure 7). This thermodynamically stable complex is reminiscent of the TRAP220 NR box 2/hRARβ LBD complex that was previously solved by X-ray crystallography (31). Similar to other co-activator complex, the primary interaction comes from the three core hydrophobic residues of the LxxLL motif, whose side chains interpolate deeply into the co-activator pocket. Many hydrophobic contacts are formed with residues from RARα LBD helices 3, 4, 5 and 12 (Table 1). Nearly all of these residues are conserved amongst different NRs and required for binding with other co-activators (28,29,36,38,42,43). In agreement with this model, our data show that the three predicted contacting residues in H12, Leu409, Glu412 and Met413 are all required for ADA3 interaction (Figure 3). Similarly, Val240 in helix 3, Phe249 in helix 4 and Leu261 in helix 5 are also important for ADA3 binding (data not shown). It is interesting to note that additional residues within and around H12 are also important for ADA3 binding. Based on this model, a complex hydrogen bond network is predicted between the N-terminal of the NR box 4 peptide and the surrounding residues on the receptor (Figure 7B). Importantly, two additional electrostatic interactions between Arg-2 and Asn416 and Gln-3 and Lys262 may explain the differential affinity of the five LxxLL sequences for their interaction with RARα. The Leu-2 and Val-3 residues on NR box 1 and 2, respectively, may prevent formation of hydrogen bond, thus this hydrogen bond network may only occur with NR boxes 3, 4 and 5.
Taken together, we have provided strong evidence that support an important role of hADA3 as a transcriptional co-activator for RARα in mammalian cells. These data suggest a mechanism through which the mammalian ADA3 may direct the action of multi-functional co-activator complexes toward modulating NR signaling.
FUNDING
The University Professor fund from UMDNJ; the National Institutes of Health (DK52888 to J.D.C.); Department of Defense (DOD W81XWH-08-1-0143 to J.D.C.); USEPA-funded Environmental Bioinformatics and Computational Toxicology Center (ebCTC) under the STAR Grant (GAD R 832721-010 to W.J.W.). Funding for open access charge: DOD and GAD grants.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Shih-Chieh Tsai, Percy Luk Yeung and Kai-Hsiung Chang for discussion during the course of this study, and also Michael Chisamore for critical reading and comments on the article. We also thank Xiaofang Yang for the initial work on creating the RARα H12 mutants and their transactivation data.
REFERENCES
- 1.Altucci L, Leibowitz MD, Ogilvie KM, de Lera AR, Gronemeyer H. RAR and RXR modulation in cancer and metabolic disease. Nat. Rev. Drug Discov. 2007;6:793–810. doi: 10.1038/nrd2397. [DOI] [PubMed] [Google Scholar]
- 2.Olefsky JM. Nuclear receptor minireview series. J. Biol. Chem. 2001;276:36863–36864. doi: 10.1074/jbc.R100047200. [DOI] [PubMed] [Google Scholar]
- 3.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 5.Leo C, Li H, Chen JD. Differential mechanisms of nuclear receptor regulation by receptor-associated coactivator 3. J. Biol. Chem. 2000;275:5976–5982. doi: 10.1074/jbc.275.8.5976. [DOI] [PubMed] [Google Scholar]
- 6.Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 7.Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 8.Lonard DM, O'Malley BW. Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol. Cell. 2007;27:691–700. doi: 10.1016/j.molcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Onate SA, Tsai SY, Tsai MJ, O'Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 10.Hong H, Kohli K, Trivedi A, Johnson DL, Stallcup MR. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc. Natl Acad. Sci. USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong H, Kohli K, Garabedian MJ, Stallcup MR. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol. Cell. Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leo C, Chen JD. The SRC family of nuclear receptor coactivators. Gene. 2000;245:1–11. doi: 10.1016/s0378-1119(00)00024-x. [DOI] [PubMed] [Google Scholar]
- 13.McKenna NJ, Lanz RB, O'Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 14.Xu L, Glass CK, Rosenfeld MG. Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 15.Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 16.Hu X, Li Y, Lazar MA. Determinants of CoRNR-dependent repression complex assembly on nuclear hormone receptors. Mol. Cell. Biol. 2001;21:1747–1758. doi: 10.1128/MCB.21.5.1747-1758.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, Rosenfeld MG, Willson TM, Glass CK, Milburn MV. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 18.Ogryzko VV, Kotani T, Zhang X, Schiltz RL, Howard T, Yang XJ, Howard BH, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 19.Eberharter A, Sterner DE, Schieltz D, Hassan A, Yates J.R., III, Berger SL, Workman JL. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:6621–6631. doi: 10.1128/mcb.19.10.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang T, Kobayashi T, Takimoto R, Denes AE, Snyder EL, el-Deiry WS, Brachmann RK. hADA3 is required for p53 activity. EMBO J. 2001;20:6404–6413. doi: 10.1093/emboj/20.22.6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar A, Zhao Y, Meng G, Zeng M, Srinivasan S, Delmolino LM, Gao Q, Dimri G, Weber GF, Wazer DE, et al. Human papillomavirus oncoprotein E6 inactivates the transcriptional coactivator human ADA3. Mol. Cell. Biol. 2002;22:5801–5812. doi: 10.1128/MCB.22.16.5801-5812.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng G, Zhao Y, Nag A, Zeng M, Dimri G, Gao Q, Wazer DE, Kumar R, Band H, Band V. Human ADA3 binds to estrogen receptor (ER) and functions as a coactivator for ER-mediated transactivation. J. Biol. Chem. 2004;279:54230–54240. doi: 10.1074/jbc.M404482200. [DOI] [PubMed] [Google Scholar]
- 23.Li CW, Dinh GK, Zhang A, Chen JD. Ankyrin repeats cofactors interact with ADA3 and modulate its coactivator function. Biochem. J. 2008;413:349–357. doi: 10.1042/BJ20071484. [DOI] [PubMed] [Google Scholar]
- 24.vom Baur E, Harbers M, Um SJ, Benecke A, Chambon P, Losson R. The yeast Ada complex mediates the ligand-dependent activation function AF-2 of retinoid X and estrogen receptors. Genes Dev. 1998;12:1278–1289. doi: 10.1101/gad.12.9.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benecke A, Gaudon C, Garnier JM, vom Baur E, Chambon P, Losson R. ADA3-containing complexes associate with estrogen receptor alpha. Nucleic Acids Res. 2002;30:2508–2514. doi: 10.1093/nar/30.11.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng M, Kumar A, Meng G, Gao Q, Dimri G, Wazer D, Band H, Band V. Human papilloma virus 16 E6 oncoprotein inhibits retinoic X receptor-mediated transactivation by targeting human ADA3 coactivator. J. Biol. Chem. 2002;277:45611–45618. doi: 10.1074/jbc.M208447200. [DOI] [PubMed] [Google Scholar]
- 27.Brown K, Chen Y, Underhill TM, Mymryk JS, Torchia J. The coactivator p/CIP/SRC-3 facilitates retinoic acid receptor signaling via recruitment of GCN5. J. Biol. Chem. 2003;278:39402–39412. doi: 10.1074/jbc.M307832200. [DOI] [PubMed] [Google Scholar]
- 28.Ghosh JC, Yang X, Zhang A, Lambert MH, Li H, Xu HE, Chen JD. Interactions that determine the assembly of a retinoid X receptor/corepressor complex. Proc. Natl Acad. Sci. USA. 2002;99:5842–5847. doi: 10.1073/pnas.092043399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leo C, Yang X, Liu J, Li H, Chen JD. Role of retinoid receptor coactivator pockets in cofactor recruitment and transcriptional regulation. J. Biol. Chem. 2001;276:23127–23134. doi: 10.1074/jbc.M100462200. [DOI] [PubMed] [Google Scholar]
- 30.Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Zhang M, Ihrig MM, McManus MT, et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 31.Pogenberg V, Guichou JF, Vivat-Hannah V, Kammerer S, Perez E, Germain P, de Lera AR, Gronemeyer H, Royer CA, Bourguet W. Characterization of the interaction between retinoic acid receptor/retinoid X receptor (RAR/RXR) heterodimers and transcriptional coactivators through structural and fluorescence anisotropy studies. J. Biol. Chem. 2005;280:1625–1633. doi: 10.1074/jbc.M409302200. [DOI] [PubMed] [Google Scholar]
- 32.Li CW, Dinh GK, Zhang A, Chen JD. Ankyrin repeats-containing cofactors interact with ADA3 and modulate its co-activator function. Biochem. J. 2008;413:349–357. doi: 10.1042/BJ20071484. [DOI] [PubMed] [Google Scholar]
- 33.Henriksson A, Almlof T, Ford J, McEwan IJ, Gustafsson JA, Wright AP. Role of the Ada adaptor complex in gene activation by the glucocorticoid receptor. Mol. Cell. Biol. 1997;17:3065–3073. doi: 10.1128/mcb.17.6.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wallberg AE, Neely KE, Gustafsson JA, Workman JL, Wright AP, Grant PA. Histone acetyltransferase complexes can mediate transcriptional activation by the major glucocorticoid receptor activation domain. Mol. Cell. Biol. 1999;19:5952–5959. doi: 10.1128/mcb.19.9.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anafi M, Yang YF, Barlev NA, Govindan MV, Berger SL, Butt TR, Walfish PG. GCN5 and ADA adaptor proteins regulate triiodothyronine/GRIP1 and SRC-1 coactivator-dependent gene activation by the human thyroid hormone receptor. Mol. Endocrinol. 2000;14:718–732. doi: 10.1210/mend.14.5.0457. [DOI] [PubMed] [Google Scholar]
- 36.Darimont BD, Wagner RL, Apriletti JW, Stallcup MR, Kushner PJ, Baxter JD, Fletterick RJ, Yamamoto KR. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McInerney EM, Rose DW, Flynn SE, Westin S, Mullen TM, Krones A, Inostroza J, Torchia J, Nolte RT, Assa-Munt N, et al. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 39.Nag A, Germaniuk-Kurowska A, Dimri M, Sassack MA, Gurumurthy CB, Gao Q, Dimri G, Band H, Band V. An essential role of human Ada3 in p53 acetylation. J. Biol. Chem. 2007;282:8812–8820. doi: 10.1074/jbc.M610443200. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, Takeshita A, Misiti S, Chin WW, Yen PM. Lack of coactivator interaction can be a mechanism for dominant negative activity by mutant thyroid hormone receptors. Endocrinology. 1998;139:4197–4204. doi: 10.1210/endo.139.10.6218. [DOI] [PubMed] [Google Scholar]
- 41.Iordanidou P, Aggelidou E, Demetriades C, Hadzopoulou-Cladaras M. Distinct amino acid residues may be involved in coactivator and ligand interactions in hepatocyte nuclear factor-4alpha. J. Biol. Chem. 2005;280:21810–21819. doi: 10.1074/jbc.M501221200. [DOI] [PubMed] [Google Scholar]
- 42.Feng W, Ribeiro RC, Wagner RL, Nguyen H, Apriletti JW, Fletterick RJ, Baxter JD, Kushner PJ, West BL. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science. 1998;280:1747–1749. doi: 10.1126/science.280.5370.1747. [DOI] [PubMed] [Google Scholar]
- 43.Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, Rosenfeld MG, Willson TM, Glass CK, Milburn MV. Ligand binding and co-activator assembly of the peroxisome proliferator- activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]