Abstract
The development and emergence of the hematopoietic stem cell involves a series of tightly regulated molecular events that are not well characterized. The hematopoietically expressed homeobox (Hhex) gene, a member of the homeobox gene family, is an essential regulator of embryogenesis and hematopoietic progenitor development. To investigate the role of Hhex in hematopoiesis we adapted a murine embryonic stem (ES) cell coculture system, in which ES cells can differentiate into CD41+ and CD45+ hematopoietic progenitors in vitro. Our results show that in addition to delayed hemangioblast development, Hhex−/− ES-derived progeny accumulate as CD41+ and CD41+c-kit+ cells, or the earliest definitive hematopoietic progenitors. In addition, Hhex−/− ES-derived progeny display a significantly reduced ability to develop into mature CD45+ hematopoietic cells. The observed reduction in hematopoietic maturation was accompanied by reduced proliferation, because Hhex−/− CD41+CD45−c-kit+ hematopoietic progenitors accumulated in the G2 phase of the cell cycle. Thus, Hhex is a critical regulator of hematopoietic development and is necessary for the maturation and proliferation of the earliest definitive hematopoietic progenitors.
Introduction
The hematopoietic stem cell (HSC), which maintains blood cell production throughout adult life, resides in the bone marrow. However, these hematopoietic progenitors arise in distinct anatomical sites within the embryo before colonization of the bone marrow.1 Key transcription factors are necessary to regulate the development and the emergence of functional HSCs from the mesoderm. The homeobox family of transcription factors, which is characterized by an evolutionarily conserved 60–amino acid DNA-binding homeodomain, is known to play an important role in developmental biology.2 Specifically, hematopoietically expressed homeobox (Hhex) has been reported to be an essential regulator of embryonic development and hematopoiesis.3–7
Hhex has been found to be an important transcription factor in embryogenesis because Hhex−/− mice display embryonic lethality starting at embryonic day E10.5 to E11.5 due to impaired forebrain, liver, and thyroid development.8–10 Furthermore, Hhex is expressed in areas of the embryo that contribute to both hematopoietic and vascular development.11 Hhex expression is initially seen in the blood islands of the yolk sac at the same time Flk-1 expression is initiated.9 In the adult, Hhex gene expression has been observed in many blood cell types, including multipotent progenitors, B lymphocytes, and myeloid lineages.3,12 Yet Hhex gene expression has been shown to be down-regulated on terminal differentiation of these lineages.7,11
Preliminary evidence suggests that Hhex may regulate definitive hematopoiesis.6,7,13 Specifically, Hhex is required for hematopoietic output from the hemangioblast, because Hhex−/− embryoid bodies (EBs) show a statistically significant decrease in the ability to form definitive hematopoietic colonies.6,13 In addition, although Hhex+/+ and Hhex+/− yolk sacs display similar numbers of primitive and definitive hematopoietic progenitors, the number of colonies from Hhex−/− yolk sac was dramatically reduced.6 Hhex has also been shown to play a significant role in B-cell development with Hhex−/−;RAG1−/− chimeric mice showing decreased B220+CD19+ B-cell production.4 Overall, these results suggest Hhex expression is critical for normal hematopoietic progenitor cell function, but they do not describe how Hhex transcriptional activity regulates hematopoiesis.
Given that Hhex is a key regulator of embryogenesis, it became important to further define the role of Hhex at various stages of hematopoietic development. Our work shows that Hhex is required for the initial formation of the hemangioblast, as well as, is essential for the proper maturation and proliferation of the CD41+c-kit+ progenitor.
Methods
Cells and cell culture
Murine E14Tg2a (tTA5-4) embryonic stem (ES) cells, murine Jet ES cells, and OP9 stromal cells were maintained as previously described.4,14 Jet ES lines were maintained in Dulbecco modified Eagle medium (high glucose; Invitrogen), 10% fetal calf serum (StemCell Technologies), 10−4 M 2-mercaptoethanol, 1mM sodium pyruvate, 2mM l-glutamine, 0.1mM nonessential amino acids, 100 μg/mL penicillin-streptomycin, and 1000 U/mL leukemia inhibitory factor. The E14Tg2a tTA5-4 ES line was maintained in Glasgow minimal essential medium (Invitrogen), 10% fetal calf serum (StemCell Technologies), 10−4 M 2-mercaptoethanol, 1mM sodium pyruvate, 2mM l-glutamine, 0.1mM nonessential amino acids, and 1000U/mL leukemia inhibitory factor. OP9 stromal cells were grown in α-minimum essential medium (Invitrogen), 20% fetal bovine serum (Omega Scientific), and 2mM l-glutamine.
ES-cell differentiation
ES cells were cocultured on OP9 stromal cells as previously described, with a few modifications. Our modifications included the following: (1) the irradiation of OP9 stroma, (2) the lack of a 30-minute replating step before reseeding of day 5 coculture cells onto new OP9, and (3) the harvesting of the entire coculture rather than only the floating hematopoietic cells.14 OP9 stroma were irradiated at 80 Gy (Cs137) and plated at 7.8 × 104 cells/cm2 24 hours before seeding of ES cells. On day 0, 1 × 103 undifferentiated ES cells were plated, per cm2, on confluent, irradiated layers of OP9 stroma. On day 5 of coculture, differentiating ES cells and OP9 stroma were harvested with 0.25% Trypsin–ethylenediaminetetraacetic acid (StemCell Technologies). After trypsinization, 4.31 × 104 to 5.51 × 104 total cells/cm2, were reseeded onto a new layer of confluent, irradiated OP9 cells until the end of the coculture time period (9 maximum total days).
Fluorescence-activated cell sorting analysis
Single-cell suspensions were prepared from cocultures by treating with 0.25% Trypsin–ethylenediaminetetraacetic acid and passaged through a 21-guage blunt-end needle. One million cells were preincubated with rat anti–mouse CD16/CD32 (FCR-4G8; Invitrogen) to prevent nonspecific binding of monoclonal antibody to FcR receptors. To determine expression of cell surface markers the following reagents were used: anti–CD34-fluorescein isothiocyanate (MEC14.7; AbD Serotec), anti–CD34-phycoerythrin (PE; MEC 14.7; Invitrogen), anti–CD41-PE (MWReg30; BD Biosciences), anti–c-Kit-TriColor (2B8; Caltag), anti–c-Kit-PE-indocyanine 7 (Cy7; 2B8; BD Bioscience), anti–Flk-1-allophycocyanin (APC; Avas12a1; eBioscience), anti–CD45-APC-Cy7 (30-F11; BD Bioscience), anti–B220-PE (RA3-6B2; BD Bioscience), anti–CD19-TC (6D5; AbD Serotec), anti–CD11b-APC (M1/70.15.11.5; Miltenyi Biotec). Cells were stained for 30 minutes on ice. Flow cytometry was performed either on a modified BD FACScan or LSR flow cytometer (BD Biosciences) and analyzed with the use of FlowJo Version 8.8.6 software (TreeStar Inc).
Cell-cycle and apoptosis analyses
Cell-cycle profiles were determined with the use of Hoechst 33342 (Invitrogen). Cells were first incubated with anti–CD41-PE (MWReg30; BD Biosciences), anti–c-Kit-TriColor (2B8; Caltag), anti–CD45-PE (30-F11; BD Biosciences), or anti–CD45-fluorescein isothiocyanate (YW62.3; AbD Serotec) as in “Fluorescence-activated cell sorting analysis.” Cells were then incubated with 5 μg/mL Hoechst 33342 for 45 minutes at 37°C in α-minimum essential medium (Invitrogen). Flow cytometry was analyzed with the use of the Dean-Jett-Fox model associated with the FlowJo software.
The frequency of apoptotic cells within the CD41+ and CD45+ cell populations was determined with the use of 7 amino-actinomycin D (7-AAD; BD Biosciences). Briefly, cells were stained with either anti–CD41-PE or anti–CD45-PE as in “Fluorescence-activated cell sorting analysis,” then incubated with 0.25 μg 7-AAD for 10 minutes on ice.
Quantitative real-time polymerase chain reaction
RNA from flow-sorted CD41+CD45-c-kit+ cells was extracted with the use of the RNeasy Micro kit (QIAGEN). Concentration and purity of RNA were determined by spectrophotometric analysis. cDNA synthesis was performed with SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's directions. Semiquantitative real-time polymerase chain reaction (PCR) was performed with the use of the iQ SYBR Green Supermix (Bio-Rad Laboratories Inc) and an iCycler Thermal cycler (Bio-Rad Laboratories Inc). The PCR primers used to detect GAPDH were as follows: forward, 5′-ACTCCACTCACGGCAAATTCA-3′, and reverse, 5′-CGCTCCTGGAAGATGGTGAT-3′; MLL forward, 5′-ATCAACGATAAGCCAGGATA-3′, and reverse, 5′-TAGTTTGCATGGATACCACA-3′; Cyclin D1 forward, 5′-TGAAGGAGACCATTCCCTTG-3′, and reverse, 5′-CCACTTGAGCTTGTTCACCA-3′; Cyclin B1 forward, 5′-TCTCTCAGTGCCCTCCACAG-3′, and reverse, 5′-CAAAGCACACCCCTGGAAGA-3′.
Hematopoietic sorts and colony assays
Total cocultures were stained as in “Fluorescence-activated cell sorting analysis” with the use of anti–CD41-PE, anti–c-Kit-TriColor, and anti–CD45-APC-Cy7 antibodies. CD41+CD45−c-kit+ cell populations were sorted with the use of a BD FACSAriaII Cell Sorter. A total of 25 000 cells was plated into 35-mm dishes (5 dishes, 5000 cells/dish) in MethoCult GF M3434 (StemCell Technologies). Colonies were enumerated after 7 days. The total number of cells within each colony assay (plated at 10 000 cells/dish) was determined after 7 days.
Results
Hematopoietic initiation coincides with increased Hhex expression
The murine ES cell/OP9 coculture system is a highly reproducible way to model embryonic hematopoiesis in vitro, because it recapitulates the development of the earliest hematopoietic precursors and their differentiation into erythroid, B-cell, and myeloid lineages.14,15 This coculture system provides a variety of benefits, namely that (1) the earliest definitive hematopoietic progenitors can be generated without murine embryos and (2) the differentiation of various progenitors into more committed myeloid and lymphoid lineages can be evaluated in vitro.15,16
A clear understanding of the pattern of Hhex gene expression is required to refine a testable hypothesis that centers on the precise role Hhex transcriptional activity plays in hematopoietic development. Hhex expression was measured by quantitative real-time PCR from ES-derived progeny on days 4 to 8 of coculture. Hhex exhibited a greater than 3-fold up-regulation in gene expression from the Flk-1+ hemangioblast stage to the hematopoietic precursor cell stage (days 4/5 to 8; supplemental Figure 1A-B, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). This result suggests that Hhex may play a role in the generation of the earliest hematopoietic progenitors from the hemangioblast.
Loss of Hhex expression leads to a delay in hemangioblast development
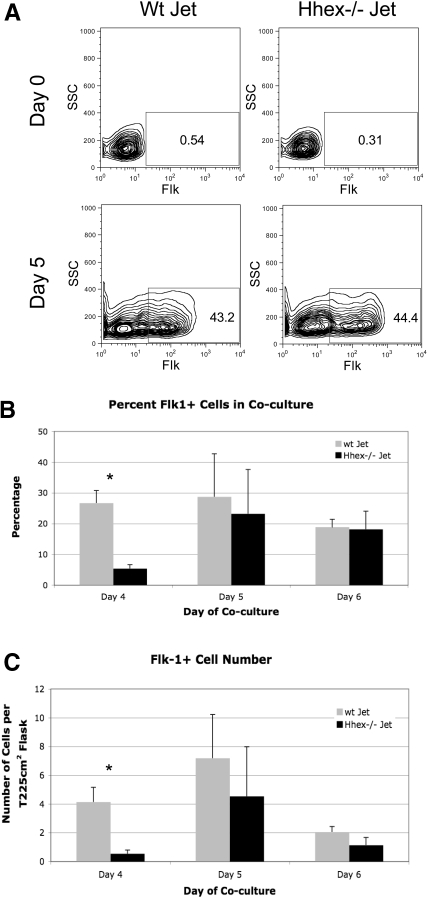
The Flk-1+ hemangioblast is defined as the common mesodermal precursor for both the hematopoietic and endothelial lineages. Given that Hhex expression is detectable at the onset of hemangioblast formation (supplemental Figure 1A-B), it became important to assess whether Hhex regulates hemangioblast development. Wild-type (wt) and Hhex−/− ES cell cocultures were harvested on days 4 through 6 of differentiation and evaluated for Flk-1 expression by flow cytometry (Figure 1A). The absence of Hhex expression significantly affects the frequency of Flk-1+ cells and the total number of Flk-1+ progenitors on day 4, compared with wt cocultures (Figure 1B-C). However, on days 5 and 6, the frequency and number of Flk-1+ progenitors are restored (Figure 1B-C). Taken together, these results indicate that loss of Hhex expression results in delayed Flk-1 hemangioblast emergence.
Figure 1.
Loss of Hhex expression affects hemangioblast development. (A) Representative flow cytometric profile of the frequency of Flk-1+ hemangioblast on days 0 and 5 of coculture from wt and Hhex−/− Jet ES cells. (B) Overall frequency of Flk-1+ cells on days 4 through 6 of coculture from wt Jet (gray) and Hhex−/− (black) Jet ES cells (n = 5). Error bars represent SD. *P ≤ .005. (C) Total number of Flk-1+ cells on days 4 through 6 of coculture. *P ≤ .005. Error bars represent SD. SSC indicates side scatter.
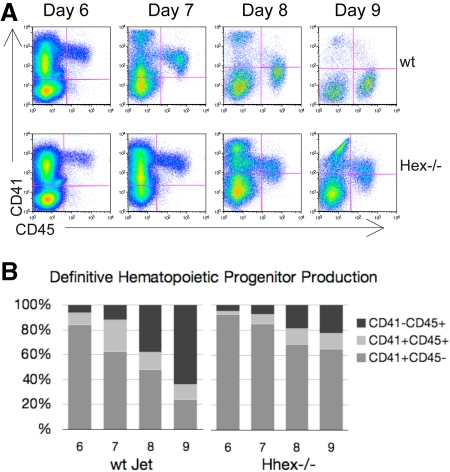
Hhex−/− progenitors persist as CD41+ definitive hematopoietic progenitors
We next wanted to determine whether loss of Hhex expression affects the development of definitive hematopoietic progenitors from the hemangioblast. In the murine embryo, CD41 expression denotes the initiation of definitive hematopoiesis.17,18 Similarly, ES cells placed in coculture begin to express CD41 after the Flk-1+ hemangioblast stage.19 As CD41+ definitive hematopoietic progenitors mature, they become CD41+CD45+, with the subsequent down-regulation of CD41. Looking at the overall pattern of hematopoietic cell development one can distinguish 3 main hematopoietic cell populations: a CD41+CD45− (early), a CD41+CD45+ (intermediate), and a CD41−CD45+ (late) progenitor population (Figure 2A). Wt hematopoietic cells show a transition from a predominantly CD41-single positive state to a CD45-single positive state from days 6 to 9 of coculture (Figure 2A-B). In contrast, a greater portion of the Hhex−/− hematopoietic cells continue to express CD41 throughout the coculture time period; such that 80% of the Hhex−/− cells still express CD41 at day 9 (Figure 2B). These data suggest a delay in progenitor maturation, associated with a loss of Hhex expression that results in the accumulation of Hhex−/− progeny as the earliest definitive hematopoietic progenitors.
Figure 2.
Hematopoietic differentiation pattern of wt and Hhex−/− Jet ES cell–derived progeny. As CD41+ definitive hematopoietic progenitors develop, they gain CD45 expression and down-regulate CD41 expression. (A) Representative flow cytometric analysis of CD41 and CD45 expression on days 6 to 9 of cocultures derived from differentiating wt and Hhex−/− Jet ES cells. (B) The proportion of each hematopoietic population from the representative experiment shown in panel A, CD41+CD45− (early progenitor) CD41+CD45+ (intermediate progenitor), and CD41−CD45+ (late progenitor), is indicated.
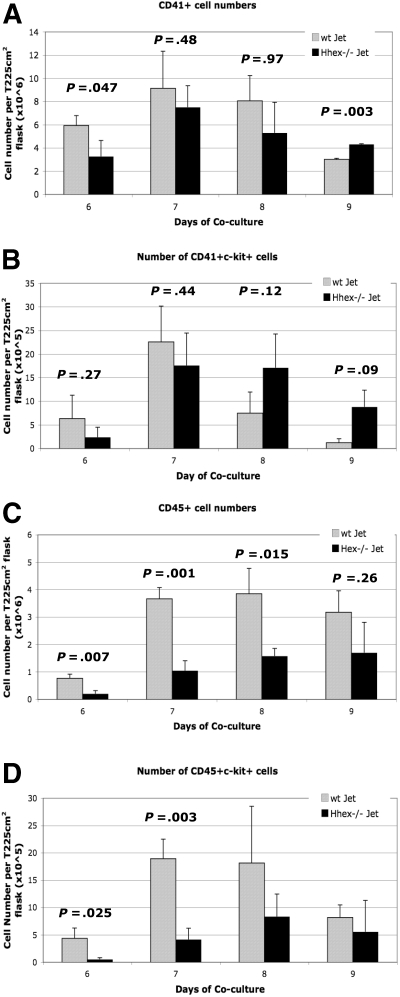
Hhex−/− ES progeny exhibit a reduced capacity to form mature CD45+ definitive progenitors
To further characterize the hematopoietic defect, the number of hematopoietic progenitors was determined for the coculture time period (supplemental Figure 2A-B). A greater number of wt cells were CD41+ on day 6 of culture compared with the Hhex−/− cells (P = .047; Figure 3A), with the trend continuing through day 8. However, by day 9, Hhex−/− progeny showed statistically higher number of CD41+ cells than wt Jet cocultures (P = .003; Figure 3A). We then determined the number of CD41+ckit+ cells, defined as the earliest definitive hematopoietic progenitors, emerging from both wt and Hhex−/− cocultures (supplemental Figure 2A-B).17 The number of wt CD41+c-kit+ cells reached maximal levels at day 7 and then declined dramatically on days 8 and 9 (Figure 3B). Interestingly, the number of Hhex−/− CD41+c-kit+ definitive hematopoietic progenitors persisted longer in coculture than those derived from wt Jet cocultures (Figure 3B). These results indicate that loss of Hhex expression also results in the significant accumulation of the earliest definitive hematopoietic progenitors.
Figure 3.
Developmental maturation of hematopoietic progenitors from wt and Hhex−/− ES cells. Hhex−/− hematopoietic cells retain the phenotype of the earliest definitive hematopoietic progenitors throughout the coculture. (A) Number of CD41+ cells found in wt Jet and Hhex−/− on days 6 to 9 of coculture (n = 5). Day 9 is significant (P < .005, 2-tailed t test). (B) Number of CD41+c-kit+ cells from wt and Hhex−/− cocultures, as determined by flow cytometry. (C) Number of CD45+ cells found in wt Jet and Hhex−/− on days 6 to 9 of coculture. The number or CD45+ cells is significantly reduced in the Hhex−/− cocultures on days 6 to 8 of coculture (2-tailed t test). (D) Number of CD45+c-kit+ cells from wt and Hhex−/− cocultures, as determined by flow cytometry. Days 6 and 7 are significant (2-tailed t test). Error bars represent SD.
As definitive hematopoietic progenitors mature they begin to express the panhematopoietic marker CD45. In coculture, all of the CD45+ hematopoietic cells develop from the CD41+ cell population.19 Coculture cells were assayed for the expression of CD45 cell surface marker (supplemental Figure 2Aiii, Biii). We observed a greater than 3-fold increase in wt CD45+ cells from day 6 to day 7, which is maintained through day 9 (Figure 3C). In contrast, the absolute number of Hhex−/− CD45+ cells is significantly reduced, compared with wt CD45+ cells, on days 6 to 8 (Figure 3C).
Further, results show that wt cells differentiate more significantly toward a more mature, adult CD45+c-kit+ progenitor phenotype than Hhex−/− cells, at days 6 and 7 (P = .025 and P = .003, respectively; Figure 3D). However, the number of wt CD45+c-kit+ cells begins to decrease, eventually reaching the number of Hhex−/− CD45+c-kit+ cells (Figure 3D). These results confirm that Hhex−/−-derived progenitors do not transition into the more mature hematopoietic progenitors at the same rate or frequency as the wt progenitors.
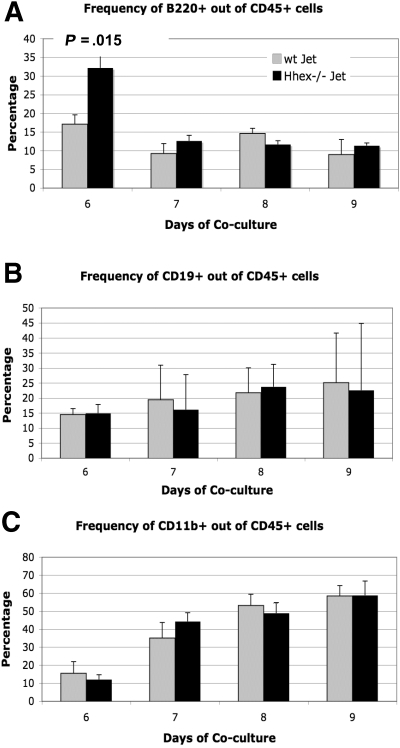
Hhex−/− ES progeny can differentiate into both myeloid and lymphoid lineages
Because loss of Hhex expression affects hematopoietic progenitor development, the next logical question is whether Hhex affects the differentiation of the hematopoietic progenitor into the various hematopoietic lineages. Previous literature suggests that loss of Hhex expression results in both myeloid and lymphoid defects.4,6,20 We examined the expression of CD19, B220 (CD45R), and CD11b within the CD45+ cell population of wt Jet and Hhex−/− cocultures to characterize the myeloid and lymphoid progeny. B220 (CD45R) staining shows a significant difference in the number of CD45+B220+ cells produced on day 6; however, this difference was not sustained over the days of coculture (Figure 4A). In general, the percentage of CD19+ and CD11b+ cells within the CD45+ population remained the same for both wt and Hhex−/− ES–derived progeny (Figure 4B-C). Hence, although loss of Hhex expression results in reduced CD45+ cell numbers, those cells that become CD45+ are still able to differentiate into more committed B and myeloid lineages at the same frequency as wt Jet CD45+ cells.
Figure 4.
Differentiation potential of wt and Hhex−/− CD45+ cells. The ability to produce lymphoid and myeloid progeny from wt and Hhex−/− CD45+ cells remains intact. (A) The frequency of CD45+B220+ B cells as determined by flow cytometry, from wt Jet and Hhex−/− cocultures on days 6 to 9. A significant change in B-cell frequency was observed on day 6 in the Hhex−/− cocultures but not on subsequent days (P < .05, 2-tailed t test). (B) The frequency of CD45+CD19+ B cells from wt Jet and Hhex−/− cocultures. (C) The frequency of CD45+CD11b+ cells from wt Jet and Hhex−/− cocultures. Error bars represent SD.
Loss of Hhex expression leads to hematopoietic colony defects
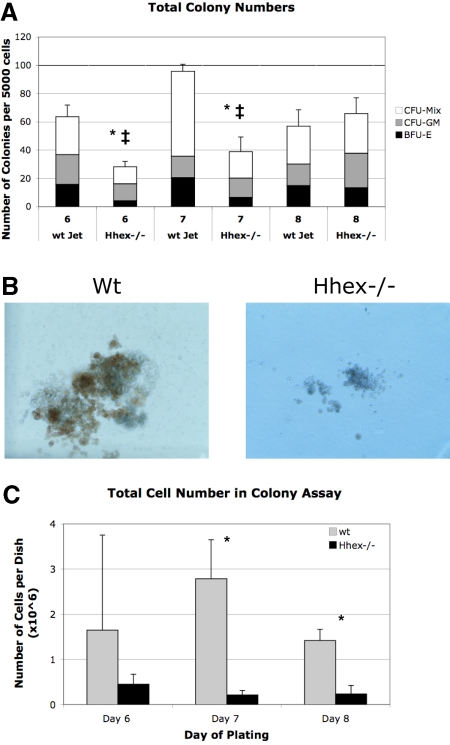
A majority of the Hhex−/− ES–derived progeny remain as CD41+CD45-c-kit+ cells, the earliest definitive hematopoietic progenitors; therefore, we reasoned that this population might be the origin of the definitive hematopoietic defect. To compare the function of these progenitors, we performed colony assay from flow sorted CD41+CD45−c-kit+ cells from days 6 to 8 cocultures. Hhex−/− CD41+CD45−c-kit+ did not show a significant change in the frequency of colony distribution (granulocyte-macrophage colony-forming unit, mixed lineage colony-forming unit, erythroid burst-forming unit) compared with wt progenitor-derived colonies (Figure 5A). However, the absolute number of colonies derived from Hhex−/− day 6 and day 7 isolated CD41+CD45−c-kit+ cells was significantly reduced (P < .001; Figure 5A asterisk). Interestingly, Hhex−/− CD41+CD45−c-kit+ show significantly increasing colony formation, over the days isolated, such that day 8 wt and Hhex−/− CD41+CD45−c-kit+ cells exhibit a similar number of total colonies. However, despite an increase in the total number of colonies, the average colony size for Hhex−/− samples on days 6, 7, and 8 was dramatically smaller, with the total number of cells found per Hhex−/− colony assay being significantly reduced (P < .05; Figure 5B-C). These results indicate that Hhex−/− hematopoietic progenitors have a reduced capacity for hematopoietic outgrowth.
Figure 5.
Colony-forming ability of wt and Hhex−/− CD41+CD45−c-kit+ cells. (A) The number of colonies obtained from day 6 to 8 sorted CD41+CD45−c-kit+ cells. Mixed lineage colony-forming unit (CFU-Mix), granulocyte-macrophage colony-forming unit (CFU-GM), and erythroid burst-forming unit (BFU-E) colonies were enumerated after 7 days in culture. Hhex−/− did not show a significant change in the percentage of each colony type (CFU-GM, CFU-Mix, BFU-E) compared with wt Jet–derived colonies. The total number of colonies derived from Hhex−/− day 6 and day 7 isolated CD41+CD45−c-kit+ cells was significantly reduced, whereas the total number of colonies from day 8 cells was relatively equivalent between the 2 samples; *P < .001. Error bars represent SD. The ‡ indicates the difference within the Hhex−/− colonies over time; P < .005. (B) Colonies derived from day 8 flow-sorted wt (left) and Hhex−/− (right) CD41+CD45−c-kit+ cells. Images were captured on Olympus IX50 inverted microscope; original magnification ×100, 10×/0.25 Ph1 objective; Olympus SC35 camera; Kodak 64T Tungsten film. (C) The total number of cells counted, from colonies obtained from day 6 to 8 sorted CD41+CD45−c-kit+ cells, after 7 days in culture. Error bars represent SD. *P < .05.
Hhex−/− hematopoietic progenitors exhibit aberrant cell-cycle profiles
The reduced ability of Hhex−/− Jet ES cells to develop into mature CD45+ cells could be due to apoptosis, cell-cycle arrest, or both. To assay for apoptotic cells, wt and Hhex−/− hematopoietic cells were stained with CD41 and CD45 and then with the DNA dye 7-AAD.21 The 7-AAD staining showed no statistically significant difference in cell viability between wt and Hhex−/− CD41+ and CD45+ cells (supplemental Figure 3A-B).
We next looked at potential cell-cycle changes associated with loss of Hhex. To assess cell-cycle profiles, the c-kit+ populations within the CD41+CD45− (early progenitor) and CD41+CD45+ (intermediate progenitor) cell populations were assayed (supplemental Figure 4A-B). Because of the low numbers of Hhex−/− CD41−CD45+ cells, it was not possible to assay cell cycle for the CD41−CD45+c-kit+ cell populations. Hoechst 33342 staining of the earliest progenitors, the CD41+CD45−c-kit+ cells, showed that the Hhex−/− progenitors appear to be abnormally accumulating in the G2 phase of the cell cycle (Table 1). Day 7 results show a significant difference in the G2 phase of the cell cycle, with a higher frequency of Hhex−/− CD41+CD45−c-kit+ cells in G2 of the cell cycle (Table 1). In addition, day 8 Hhex−/− CD41+CD45−c-kit+ on average have a higher percentage of cells in G2, in addition to exhibiting a significantly lower number of cells in S phase (Table 1).
Table 1.
Early definitive hematopoietic progenitor cell cycle
| CD41+/CD45−c-kit+ | Day 6 |
Day 7 |
Day 8 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| G1 | S | G2 | G1 | S | G2* | G1 | S† | G2 | |
| Wt | 82.9 ± 7.64 | 14.34 ± 7.44 | 2.24 ± 3.16 | 92.45 ± 10.68 | 9.76 ± 3.88 | 0.04 ± 0.06 | 79.87 ± 6.9 | 25.27 ± 3.58 | 1.72 ± 2.32 |
| Hhex−/− | 83.2 ± 5.09 | 8.71 ± 5.5 | 10.74 ± 7.3 | 93.65 ± 6. 57 | 5.81 ± 7.91 | 5.14 ± 0.41 | 79.2 ± 6.16 | 11.12 ± 5.44 | 11.51 ± 7.95 |
Cell-cycle profiles for early CD41+CD45−c-kit+ progenitors on days 6 to 8. Each cell within the table represents the average percentage within each phase of cell cycle ± the standard deviation, as determined from 3 experiments. Gating for cell cycle is shown in supplemental Figure 4. Hhex−/− CD41+CD45−c-kit+ cells are accumulating in the G2 phase of the cell cycle. Day 7 shows a significant difference in G2 phase of the cell cycle, with Hhex−/− CD41+CD45−c-kit+ cells accumulating in G2 of the cell cycle. In addition, day 8 cocultures show that wt CD41+CD45−c-kit+ have a significantly higher number of cells in S phase, but Hhex−/− CD41+CD45−c-kit+ on average still have higher number of cells in G2.
P < .005, 2-tailed t test.
P < .05, 2-tailed t test.
Similarly, the intermediate Hhex−/− CD41+CD45+c-kit+ progenitors appear to be accumulating in the G2 phase of the cell cycle. Days 7 and 8 of coculture show a significant difference in the percentage of intermediate Hhex−/− CD41+CD45+c-kit+ cells in the G2 phase (Table 2). These results suggest that G2 accumulation by the Hhex−/− definitive progenitors may account in part for the persistence of the CD41+ and CD41+c-kit+ cells in culture (Figures 2–3). Therefore, an aberrant cell-cycle status may be the underlying cause of the maturation defect and the reduced production of hematopoietic progeny.
Table 2.
Intermediate definitive hematopoietic progenitor cell cycle
| CD41+/CD45+c-kit+ | Day 7 |
Day 8 |
||||
|---|---|---|---|---|---|---|
| G1 | S | G2* | G1 | S | G2* | |
| Wt | 83.65 ± 2.05 | 17.8 ± 0.99 | 1.62 ± 2.29 | 85.97 ± 12.86 | 16.04 ± 11.19 | 0.30 ± 0.33 |
| Hhex−/− | 76.65 ± 23.83 | 15.88 ± 21.39 | 9.83 ± 0.18 | 84.8 ± 11.94 | 13.75 ± 6.58 | 3.41 ± 2.27 |
Cell-cycle profiles for intermediate CD41+CD45+c-kit+ hematopoietic progenitors. Each cell within the table represents the average percentage within each phase of cell cycle ± the standard deviation, as determined from 3 experiments. Gating for cell cycle is shown in supplemental Figure 4. Hhex−/− CD41+CD45+c-kit+ cells appear to be accumulating in G2. Days 7 and 8 of coculture show a significant difference in G2 phase of the cell cycle, with Hhex−/− CD41+CD45+c-kit+ cells accumulating the G2 phase of the cell cycle, compared with wt CD41+CD45+c-kit+ progenitors.
P < .05, 2-tailed t test.
Loss of Hhex expression leads to down-regulated Cyclin D1 expression
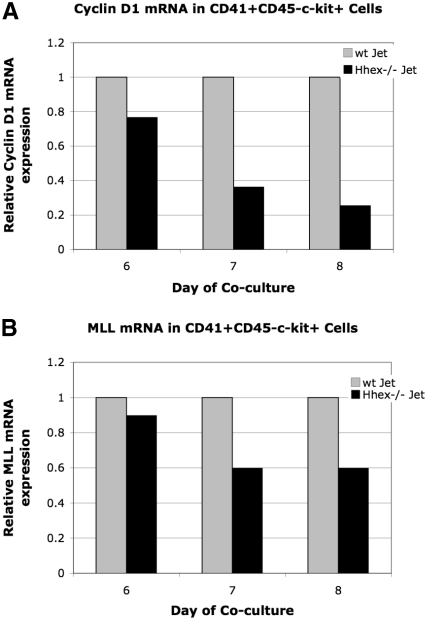
To further confirm Hhex−/− progenitor accumulation in the G2 phase of the cell cycle we decided to assess Cyclin D1 expression. Cyclin D1 is necessary for transition through G1 of the cell cycle; however, expression initiates in the G2 phase.22,23 Therefore, because early CD41+CD45−c-kit+ and intermediate CD41+CD45+c-kit+ cells accumulate at G2 (Tables 1–2), Cyclin D1 expression could be down-regulated in the Hhex−/− progenitors. Cyclin D1 expression was found to be down-regulated 1.3-, 2.7-, and 3.9-fold in days 6 through 8 CD41+CD45−c-kit+ Hhex−/− cells, respectively (Figure 6A). It is interesting to note that Cyclin B1 expression, which regulates G2/M, is the same in days 6 through 8 wt and Hhex−/− CD41+CD45−c-kit+ cells in 4 independent experiments (data not shown). These result confirm that loss of Hhex expression results in aberrant cell-cycle kinetics and indicates that Hhex is potentially directly regulating the G1 phase of the cell cycle in the hematopoietic progenitor.
Figure 6.
Relative expression of cyclin D1 and Mll mRNA. The level of Mll and cyclin D1 mRNA expression was determined by real-time PCR after normalization for GAPDH. Graphs display representative experiment. (A) Relative levels of cyclin D1 mRNA in wt and Hhex−/− CD41+CD45−c-kit+ cells. Cyclin D1 is down-regulated 1.3-, 2.7-, and 3.9-fold in CD41+CD45−c-kit+ cells at day 6 through 8, respectively. (A) Relative levels of Mll mRNA in wt and Hhex−/− CD41+CD45−c-kit+ cells. Mll was down-regulated 1.1 at day 6, and 1.6 at days 7 and 8 of coculture.
Loss of Hhex expression leads to down-regulated Mll gene expression
The generation of the earliest hematopoietic progenitors requires the proper function of various transcription factors, epigenetic modifiers, and cell-cycle regulators.24 One gene known to regulate the maturation of the definitive hematopoietic progenitor is the mixed lineage leukemia Mll gene.25 Mll encodes a DNA-binding protein, which methylates H3 lysine 4, and is frequently involved in various chromosomal translocations associated with acute myeloid leukemia.26 Interestingly, the Hhex−/−-associated definitive hematopoietic defect mirrors that reported with loss of Mll expression.6,8,13,25,27–29 Specifically, although Mll−/− EBs exhibit normal CD41+c-kit+ cell production, hematopoietic colony formation from Mll−/− CD41+c-kit+ cells is significantly reduced.25 In addition, cell-cycle analysis of Mll−/− progenitors shows aberrant cell-cycle profiles, with adult Mll−/− HSCs exhibiting increased cycling.28,29 Overall, these results suggest that Mll regulates hematopoietic progenitor development in part through cell-cycle regulation.28,29
The regulation of various Hox family members by Mll, along with the analogous Mll−/− and Hhex−/− hematopoietic defects, suggests a potential connection between Mll and Hhex.26,30 Therefore, we examine whether loss of Hhex expression leads to altered Mll or Mll-target gene expression in sorted days 6 to 8 CD41+CD45−c-kit+ cells. Mll expression, in a representative experiment, was found to be down-regulated 1.1-fold (day 6) and 1.6-fold (days 7 and 8) in the Hhex−/− CD41+CD45−c-kit+ cells (Figure 6B). Moreover, 5 independent experiments showed Mll expression was reduced 1.4- to 2-fold in day 7 Hhex−/− CD41+CD45−c-kit+ cells (data not shown). Note that the Hhex−/− progenitors begin to accumulate in G2, when expression of Mll is also decreased. These results indicate that a Hhex-regulated signaling pathway is potentially driving Mll expression within the definitive hematopoietic progenitor.
Mll is also known to regulate the expression of various Hox family members; therefore, we wanted to know whether Hox gene expression was also being altered in Hhex−/− progenitors.30 Expression of Mll-target genes, Hoxa7, Hoxa9, and Hoxb4, was assayed by real-time PCR; however, expression levels were too low for proper quantification (data not shown).
Rescue of hematopoietic defect
Hhex knockout experiments showed that Hhex is essential for progenitor maturation; therefore, we assessed whether inducible Hhex expression could rescue the definitive hematopoietic defect. Because Hhex functions in the CD41+c-kit+ cell, Hhex expression was induced before this cell population emergence. Although the exact window of CD41+c-kit+ cell generation is not known, initial cocultures show the presence of CD41+ cells at day 4 (data not shown). Therefore, we decided to induce Hhex expression, using a tetracycline (TET)–inducible system, at day 3 or day 5 of coculture. Hematopoietic rescue then was determined by changes in CD45+ cell number (supplemental Figure 5). At day 6, the number of CD45+ cells was the same for Hhex−/− control (+TET) and Hhex re-expression (−TET, day 3 and day 5) cocultures. However, induction of Hhex expression at day 3 (−TET, day 3) resulted in an increase in CD45+ cell numbers on days 7 and 8, compared with Hhex−/− control cocultures (supplemental Figure 5). Similarly, CD45+ cell number increased, relative to the control, when Hhex expression was introduced at day 5 (−TET, day 5). Note that there is delay in CD45+ cell rescue when Hhex expression is reintroduced at day 5, as opposed to day 3 (supplemental Figure 5). In addition, preliminary experiments show that reintroduction of Hhex expression (−TET, day 3) lead to a 1.6- to 2.2-fold, at day 7 or 8, up-regulation in Mll expression, over control (+TET) cocultures (data not shown). Overall, these results suggest that Hhex re-expression is able to at least partially rescue the Hhex−/− hematopoietic defect.
Discussion
Hematopoiesis begins at E7 to E8.25 within the extraembryonic yolk sac.17 After a transient wave of primitive hematopoiesis, definitive hematopoietic progenitors appear in the murine yolk sac, placenta, and aorta-gonad-mesonephros. These highly proliferative hematopoietic progenitors then migrate to the fetal liver, the site of functional HSC maturation and hematopoietic lineage expansion and differentiation. Each stage of hematopoietic development requires a coordinated effort from various transcriptional activators, repressors, and epigenetic modifiers.24 Here, our data show that Hhex carries out a gene program that is required at 2 distinct hematopoietic developmental stages, at the hemangioblast stage and for the developmental maturation of definitive hematopoietic progenitor.
To determine whether Hhex regulates the generation of hemangioblasts, cocultures were stained with Flk-1. We observed the percentage of Flk-1+ cells and the number of Flk-1+ cells was significantly different at day 4 of coculture but returned to wt levels by day 6 of coculture (Figure 1B-C). Therefore, our data suggest that Hhex plays a role in the development of the hemangioblast from mesodermal precursors. Although most literature shows that Hhex expression begins at the onset of hemangioblast development, it is critical to mention that there is conflicting data about the role of Hhex in hemangioblast development.6,13 Analysis by Kubo et al13 of day 4 Hhex+/+, Hhex+/−, Hhex−/− EBs showed no difference in the number of Flk-1+ cells; however, day 4 Hhex−/− EBs showed an increase in the number of Blast colony-forming cells (BL-CFCs) after 3 days of differentiation. Conversely, Guo et al6 did not observe a significant difference in the number of BL-CFCs formed from day 3 Hhex+/+, Hhex+/−, Hhex−/− EBs. The differences in the role of Hhex in hemangioblast formation, among all 3 works, may be due in part to the different time points analyzed.13 The increase in BL-CFC numbers observed by Kubo et al13 may be reflected in our increase in Flk-1+ cells at day 6 of coculture.
During definitive hematopoietic development, Hhex is expressed in various progenitor populations and is down-regulated on terminal differentiation.3,7,9,12 In the course of normal ES differentiation, CD41 cells develop from Flk1+ hemangioblasts.19 As these progenitors further develop, they begin to down-regulate CD41 expression and express CD45 (Figure 2). However, Hhex−/− hematopoietic cells accumulate as CD41+ and CD41+c-kit+ definitive hematopoietic progenitors, while exhibiting reduced CD45+ cell formation (Figures 2–3). Although loss of Hhex expression resulted in reduced CD45+ production, the proportion of lymphoid or myeloid progeny within the CD45+ cell population remains unchanged, suggesting that Hhex does not regulate lineage differentiation (Figure 4). The persistence of a CD41+ cell population, in the absence of increased apoptosis, suggests that Hhex regulates the maturation of the earliest definitive hematopoietic progenitors into more mature progenitors. Thus, this study identifies a key transcriptional regulator of hematopoietic progenitor maturation, which is essential for proper formation of the HSC.
Although our data suggest that Hhex is not required for proper hematopoietic differentiation, previous literature has shown that Hhex plays a significant role in B-cell development.4 Bogue et al4 show a decrease in B220+CD19+ B-cell production in the spleen and bone marrow of Hhex−/−RAG1−/− mice. In addition, Hhex−/−RAG1−/− showed a significant increase in B220−CD19+ cells, which was recently identified as the progenitor for B-1 cell population.4,31 Whereas our data show a significant increase in the percentage in Hhex−/−B220+ cells at day 6, an overall difference in B-cell potential is not observed over time (Figure 4). The difference between Bogue et al4 and our results may be due in part to the more detailed characterization of various B-cell populations by Bogue et al,4 as well as different environmental factors. In general, these reports suggest that Hhex is important for various stages of hematopoietic development.
The increased presence of the Hhex−/− definitive hematopoietic progenitors appears to be the result of aberrant cell-cycle profiles. Cell-cycle analysis of Hhex−/−-derived CD41+CD45−c-kit+ and CD41+CD45+c-kit+ cells shows that these progenitors accumulate in the G2 phase of the cell cycle (Tables 1–2). Fetal progenitor cells are highly proliferative, whereas adult HSCs are mostly quiescent. Proper cell-cycle status by fetal and adult HSCs is essential for HSC function, because knockout and knockdown of many cell-cycle regulators has been associated with various hematopoietic defects.32 Specifically, our data show that aberrant cell cycling by the Hhex−/− progenitors results in reduced colony formation and reduced colony size (Figure 5). Although little is known about cell-cycle regulation of the developing fetal progenitor, Bmi-1, Gfi-1, and p21 have been described as playing a role in fetal HSC development and adult HSC self-renewal and quiescence.33–36 The commonality of various cell-cycle regulators in fetal and adult hematopoiesis suggest Hhex may also play an additional role in regulating quiescence in the adult HSC. Further studies are required to determine whether Hhex plays a role in adult HSC function.
Although the transcriptional network responsible for the development of fetal hematopoietic progenitors into functional adult HSCs still needs to be determined, Mll is already known to regulate the maturation of the earliest definitive hematopoietic progenitors.25,37 Hhex−/− and Mll−/− hematopoietic cells display similar and unique hematopoietic defects in vitro and in vivo. Colony assays from Mll−/− and Hhex−/− yolk sac cells display reduced colony numbers and size, compared with wt yolk sac cells.6,27 In addition, although Mll−/− EBs display normal CD41+c-kit+ formation, hematopoietic colony development from Mll−/− CD41+c-kit+ cells was significantly reduced.25 Correspondingly, we observed that Hhex−/− CD41+CD45−c-kit+ exhibit reduced colony number and size (Figure 5). Initial experiments show that Mll potentially plays a role in the Hhex hematopoietic defect, because Mll expression was found to be down-regulated in Hhex−/− CD41+CD45−c-kit+ cells (Figure 6). Considering most of the literature indicates that HHEX functions as a transcriptional repressor, it is unlikely that HHEX directly regulates the expression of Mll.11,38,39 Instead, Hhex may regulate another transcriptional repressor, which controls Mll expression. These results show that Hhex regulates cell-cycle progression and maturation by the earliest definitive hematopoietic progenitors and begins to place Hhex within a transcriptional network. Future work is necessary to determine whether Mll is a direct target of Hhex-mediated transcription.
Previous literature has linked Mll to the cell-cycle machinery.28,29 MLL exhibits a biphasic expression pattern, with the timing of MLL expression being required for proper G1/S transition and M phase progression.40 Evidence from other systems suggest that Mll plays a role in G2/M progression, because HeLa cells transfected with an MLL1-antisense-oligoenucletide exhibit an increased percentage of cells in G2/M.41 Analogously, knockdown of Mll5 in HeLa cells resulted in G2/M arrest due to increased p21 expression.42 CD41+CD45−c-kit+ and CD41+CD45+c-kit+ show a statistically significant accumulation of cells in the G2 phase of the cell cycle, and these observed results suggest that G2 accumulation by Hhex−/− progenitors may in part be due to reduced Mll expression (Table 1; Figure 6). Although additional experiments are needed to more clearly define the Hhex/Mll connection, these results expand our understanding of the regulatory network required for the proper proliferation by fetal hematopoietic progenitors.
To date SCL, c-Myb, Runx1, Cdx4, Gata-2, and Mll have all been described as being required for the proper development of the earliest hematopoietic lineages.43–48 SCL/tal1 is involved in the specification of both the primitive and definitive hematopoietic lineages from the hemangioblast, whereas Runx1 is required for the emergence of all definitive hematopoietic cells.43,47 Because Hhex appears to play a role in 2 distinct developmental stages, one would hypothesize that Hhex is downstream of both SCL and Runx1 transcriptional activity. Incidentally, recent literature suggests that Scl binds to the Hhex enhancer region, suggesting a role for Hhex transcriptional regulation by Scl.49 Disruption in Mll, c-Myb, Gata-2, and Cdx4 gene expression lead to defects in definitive hematopoietic progenitor cell production and function. Additional experiments are necessary to further define the relationship between Hhex and those genes involved in fetal progenitor cell development.
Hematopoietic stem and progenitor cells go through various changes in gene expression, function, and self-renewal potential throughout developmental life. HSCs transition, shortly after birth, from a highly proliferative fetal state to a mainly quiescent adult state.33 Here, our data identify Hhex as being a unique regulator of hematopoietic development, in that it regulates 2 key different hematopoietic steps. First, Hhex is required for the initial specification of the hemangioblast from mesodermal precursors, and, second, Hhex is required for the maturation and proliferation of early definitive hematopoietic progenitors. Without proper regulation of these initial developmental steps adult stem cell function productivity is compromised, as evidence by reduced colony formation by Hhex−/− progenitors.
Recently, Hhex has been implicated in acute myeloid leukemia, with the discovery of a chromosomal translocation composed of the N-terminal region of the nucleoporin 98 to the HHEX homeodomain.50 On the basis of our findings aberrant cell-cycle progression potentially could contribute to Hhex-mediated leukemogenesis. A clear understanding of Hhex transcriptional targets in both fetal and adult HSCs could provide insights into the self-renewal program affected by the Hhex chromosomal translocation, which is relevant for providing novel targets for therapies.
In summary, our data clearly indicate that Hhex is a critical and unique regulator of hematopoietic progenitor cell maturation, mediated by its effects on proliferation. This furthers our understanding of the intrinsic molecular cues directing the development of the HSC and may provide the basis for novel therapies in a subset of leukemias.
Supplementary Material
Acknowledgments
We thank E. Montecino-Rodriguez and H. Mikkola for their technical expertise and advice.
This work was supported by a fellowship from the National Heart, Lung, and Blood Institute (F31HL087714; H.P.). The University of California, Los Angeles (UCLA) Flow Cytometry Core Facility is supported by the NIH (CA-16 042 and AI-28 697), the Jonsson Cancer Center, the UCLA AIDS Institute, and the David Geffen School of Medicine at UCLA.
The content of this work is solely the responsibility of the author and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health (NIH).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: H.P. performed research and wrote the paper; M.R.L. performed research; C.W.B. contributed new vital reagents; and J.C.G. analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Judith C. Gasson, UCLA Jonsson Comprehensive Cancer Center, PO Box 951781, 8-684 Factor Bldg, Los Angeles, CA 90095-1781; e-mail: jgasson@mednet.ucla.edu.
References
- 1.Dzierzak E, Speck NA. Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat Immunol. 2008;9(2):129–136. doi: 10.1038/ni1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawrence HJ, Largman C. Homeobox genes in normal hematopoiesis and leukemia. Blood. 1992;80(10):2445–2453. [PubMed] [Google Scholar]
- 3.Bedford FK, Ashworth A, Enver T, Wiedemann LM. HEX: a novel homeobox gene expressed during haematopoiesis and conserved between mouse and human. Nucleic Acids Res. 1993;21(5):1245–1249. doi: 10.1093/nar/21.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogue CW, Zhang PX, McGrath J, Jacobs HC, Fuleihan RL. Impaired B cell development and function in mice with a targeted disruption of the homeobox gene Hex. Proc Natl Acad Sci U S A. 2003;100(2):556–561. doi: 10.1073/pnas.0236979100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brickman JM, Jones CM, Clements M, Smith JC, Beddington RS. Hex is a transcriptional repressor that contributes to anterior identity and suppresses Spemann organiser function. Development. 2000;127(11):2303–2315. doi: 10.1242/dev.127.11.2303. [DOI] [PubMed] [Google Scholar]
- 6.Guo Y, Chan R, Ramsey H, et al. The homeoprotein Hex is required for hemangioblast differentiation. Blood. 2003;102(7):2428–2435. doi: 10.1182/blood-2003-02-0634. [DOI] [PubMed] [Google Scholar]
- 7.Jayaraman PS, Frampton J, Goodwin G. The homeodomain protein PRH influences the differentiation of haematopoietic cells. Leuk Res. 2000;24(12):1023–1031. doi: 10.1016/s0145-2126(00)00072-2. [DOI] [PubMed] [Google Scholar]
- 8.Hallaq H, Pinter E, Enciso J, et al. A null mutation of Hhex results in abnormal cardiac development, defective vasculogenesis and elevated Vegfa levels. Development. 2004;131(20):5197–5209. doi: 10.1242/dev.01393. [DOI] [PubMed] [Google Scholar]
- 9.Thomas PQ, Brown A, Beddington RS. Hex: a homeobox gene revealing peri-implantation asymmetry in the mouse embryo and an early transient marker of endothelial cell precursors. Development. 1998;125(1):85–94. doi: 10.1242/dev.125.1.85. [DOI] [PubMed] [Google Scholar]
- 10.Martinez Barbera JP, Clements M, Thomas P, et al. The homeobox gene Hex is required in definitive endodermal tissues for normal forebrain, liver and thyroid formation. Development. 2000;127(11):2433–2445. doi: 10.1242/dev.127.11.2433. [DOI] [PubMed] [Google Scholar]
- 11.Soufi A, Jayaraman PS. PRH/Hex: an oligomeric transcription factor and multifunctional regulator of cell fate. Biochem J. 2008;412(3):399–413. doi: 10.1042/BJ20080035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh B, Jacobs HC, Wiedemann LM, et al. Genomic structure, cDNA mapping, and chromosomal localization of the mouse homeobox gene, Hex. Mamm Genome. 1999;10(10):1023–1025. doi: 10.1007/s003359901152. [DOI] [PubMed] [Google Scholar]
- 13.Kubo A, Chen V, Kennedy M, Zahradka E, Daley GQ, Keller G. The homeobox gene HEX regulates proliferation and differentiation of hemangioblasts and endothelial cells during ES cell differentiation. Blood. 2005;105(12):4590–4597. doi: 10.1182/blood-2004-10-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turksen K. Embryonic Stem Cells: Methods and Protocols. Totowa, NJ: Humana Press; 2002. [Google Scholar]
- 15.Nakano T, Kodama H, Honjo T. Generation of lymphohematopoietic cells from embryonic stem cells in culture. Science. 1994;265(5175):1098–1101. doi: 10.1126/science.8066449. [DOI] [PubMed] [Google Scholar]
- 16.Nakano T, Era T, Takahashi T, Kodama H, Honjo T. Development of erythroid cells from mouse embryonic stem cells in culture: potential use for erythroid transcription factor study. Leukemia. 1997;11(Suppl 3):496–500. [PubMed] [Google Scholar]
- 17.Mikkola HK, Fujiwara Y, Schlaeger TM, Traver D, Orkin SH. Expression of CD41 marks the initiation of definitive hematopoiesis in the mouse embryo. Blood. 2003;101(2):508–516. doi: 10.1182/blood-2002-06-1699. [DOI] [PubMed] [Google Scholar]
- 18.Gekas C, Dieterlen-Lievre F, Orkin SH, Mikkola HK. The placenta is a niche for hematopoietic stem cells. Dev Cell. 2005;8(3):365–375. doi: 10.1016/j.devcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 19.Hashimoto K, Fujimoto T, Shimoda Y, Huang X, Sakamoto H, Ogawa M. Distinct hemogenic potential of endothelial cells and CD41+ cells in mouse embryos. Dev Growth Differ. 2007;49(4):287–300. doi: 10.1111/j.1440-169X.2007.00925.x. [DOI] [PubMed] [Google Scholar]
- 20.Keng VW, Yagi H, Ikawa M, et al. Homeobox gene Hex is essential for onset of mouse embryonic liver development and differentiation of the monocyte lineage. Biochem Biophys Res Commun. 2000;276(3):1155–1161. doi: 10.1006/bbrc.2000.3548. [DOI] [PubMed] [Google Scholar]
- 21.Schmid I, Uittenbogaart CH, Keld B, Giorgi JV. A rapid method for measuring apoptosis and dual-color immunofluorescence by single laser flow cytometry. J Immunol Methods. 1994;170(2):145–157. doi: 10.1016/0022-1759(94)90390-5. [DOI] [PubMed] [Google Scholar]
- 22.Stacey DW. Cyclin D1 serves as a cell cycle regulatory switch in actively proliferating cells. Curr Opin Cell Biol. 2003;15(2):158–163. doi: 10.1016/s0955-0674(03)00008-5. [DOI] [PubMed] [Google Scholar]
- 23.Guo Y, Harwalkar J, Stacey DW, Hitomi M. Destabilization of cyclin D1 message plays a critical role in cell cycle exit upon mitogen withdrawal. Oncogene. 2005;24(6):1032–1042. doi: 10.1038/sj.onc.1208299. [DOI] [PubMed] [Google Scholar]
- 24.Teitell MA, Mikkola HK. Transcriptional activators, repressors, and epigenetic modifiers controlling hematopoietic stem cell development. Pediatr Res. 2006;59(4 Pt 2):33R–39R. doi: 10.1203/01.pdr.0000205155.26315.c7. [DOI] [PubMed] [Google Scholar]
- 25.Ernst P, Mabon M, Davidson AJ, Zon LI, Korsmeyer SJ. An Mll-dependent Hox program drives hematopoietic progenitor expansion. Curr Biol. 2004;14(22):2063–2069. doi: 10.1016/j.cub.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 26.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7(11):823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 27.Hess JL, Yu BD, Li B, Hanson R, Korsmeyer SJ. Defects in yolk sac hematopoiesis in Mll-null embryos. Blood. 1997;90(5):1799–1806. [PubMed] [Google Scholar]
- 28.Jude CD, Climer L, Xu D, Artinger E, Fisher JK, Ernst P. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1(3):324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McMahon KA, Hiew SY, Hadjur S, et al. Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell. 2007;1(3):338–345. doi: 10.1016/j.stem.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 30.Ernst P, Wang J, Korsmeyer SJ. The role of MLL in hematopoiesis and leukemia. Curr Opin Hematol. 2002;9(4):282–287. doi: 10.1097/00062752-200207000-00004. [DOI] [PubMed] [Google Scholar]
- 31.Montecino-Rodriguez E, Leathers H, Dorshkind K. Identification of a B-1 B cell-specified progenitor. Nat Immunol. 2006;7(3):293–301. doi: 10.1038/ni1301. [DOI] [PubMed] [Google Scholar]
- 32.Steinman RA. Cell cycle regulators and hematopoiesis. Oncogene. 2002;21(21):3403–3413. doi: 10.1038/sj.onc.1205325. [DOI] [PubMed] [Google Scholar]
- 33.Kim I, Saunders TL, Morrison SJ. Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell. 2007;130(3):470–483. doi: 10.1016/j.cell.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng T, Rodrigues N, Shen H, et al. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science. 2000;287(5459):1804–1808. doi: 10.1126/science.287.5459.1804. [DOI] [PubMed] [Google Scholar]
- 35.Park IK, Qian D, Kiel M, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423(6937):302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 36.Zeng H, Yucel R, Kosan C, Klein-Hitpass L, Moroy T. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J. 2004;23(20):4116–4125. doi: 10.1038/sj.emboj.7600419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu BD, Hanson RD, Hess JL, Horning SE, Korsmeyer SJ. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc Natl Acad Sci U S A. 1998;95(18):10632–10636. doi: 10.1073/pnas.95.18.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guiral M, Bess K, Goodwin G, Jayaraman PS. PRH represses transcription in hematopoietic cells by at least two independent mechanisms. J Biol Chem. 2001;276(4):2961–2970. doi: 10.1074/jbc.M004948200. [DOI] [PubMed] [Google Scholar]
- 39.Swingler TE, Bess KL, Yao J, Stifani S, Jayaraman PS. The proline-rich homeodomain protein recruits members of the Groucho/Transducin-like enhancer of split protein family to co-repress transcription in hematopoietic cells. J Biol Chem. 2004;279(33):34938–34947. doi: 10.1074/jbc.M404488200. [DOI] [PubMed] [Google Scholar]
- 40.Liu H, Cheng EH, Hsieh JJ. Bimodal degradation of MLL by SCFSkp2 and APCCdc20 assures cell cycle execution: a critical regulatory circuit lost in leukemogenic MLL fusions. Genes Dev. 2007;21(19):2385–2398. doi: 10.1101/gad.1574507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mishra BP, Ansari KI, Mandal SS. Dynamic association of MLL1, H3K4 trimethylation with chromatin and Hox gene expression during the cell cycle. FEBS J. 2009;276(6):1629–1640. doi: 10.1111/j.1742-4658.2009.06895.x. [DOI] [PubMed] [Google Scholar]
- 42.Cheng F, Liu J, Zhou SH, Wang XN, Chew JF, Deng LW. RNA interference against mixed lineage leukemia 5 resulted in cell cycle arrest. Int J Biochem Cell Biol. 2008;40(11):2472–2481. doi: 10.1016/j.biocel.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 43.Shivdasani RA, Mayer EL, Orkin SH. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature. 1995;373(6513):432–434. doi: 10.1038/373432a0. [DOI] [PubMed] [Google Scholar]
- 44.Mucenski ML, McLain K, Kier AB, et al. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65(4):677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- 45.Tsai FY, Keller G, Kuo FC, et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371(6494):221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 46.Robertson SM, Kennedy M, Shannon JM, Keller G. A transitional stage in the commitment of mesoderm to hematopoiesis requiring the transcription factor SCL/tal-1. Development. 2000;127(11):2447–2459. doi: 10.1242/dev.127.11.2447. [DOI] [PubMed] [Google Scholar]
- 47.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84(2):321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 48.McKinney-Freeman SL, Lengerke C, Jang IH, et al. Modulation of murine embryonic stem cell-derived CD41+c-kit+ hematopoietic progenitors by ectopic expression of Cdx genes. Blood. 2008;111(10):4944–4953. doi: 10.1182/blood-2007-11-124644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson NK, Miranda-Saavedra D, Kinston S, et al. The transcriptional program controlled by the stem cell leukemia gene Scl/Tal1 during early embryonic hematopoietic development. Blood. 2009;113(22):5456–5465. doi: 10.1182/blood-2009-01-200048. [DOI] [PubMed] [Google Scholar]
- 50.Jankovic D, Gorello P, Liu T, et al. Leukemogenic mechanisms and targets of a NUP98/HHEX fusion in acute myeloid leukemia. Blood. 2008;111(12):5672–5682. doi: 10.1182/blood-2007-09-108175. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.