Abstract
GATA-3 and c-Myb are core elements of a transcriptionally active complex essential for human Th2 cell development and maintenance. We report herein mechanistic details concerning the role of these transcription factors in human peripheral blood Th2 cell development. Silencing c-Myb in normal human naive CD4+ cells under Th2 cell-promoting conditions blocked up-regulation of GATA-3 and interleukin-4, and in effector/memory CD4+ T cells, decreased expression of GATA-3 and Th2 cytokines. In primary T cells, c-Myb allows GATA-3 to autoactivate its own expression, an event that requires the direct interaction of c-Myb and GATA-3 on their respective binding sites in promoter of GATA-3. Immunoprecipitation revealed that the c-Myb/GATA-3 complex contained Menin and mixed lineage leukemia (MLL). MLL recruitment into the c-Myb-GATA-3-Menin complex was associated with the formation Th2 memory cells. That MLL-driven epigenetic changes were mechanistically important for this transition was suggested by the fact that silencing c-Myb significantly decreased the methylation of histone H3K4 and the acetylation of histone H3K9 at the GATA-3 locus in developing Th2 and CD4+ effector/memory cells. Therefore, c-Myb, GATA-3, and Menin form a core transcription complex that regulates GATA-3 expression and, with the recruitment of MLL, Th2 cell maturation in primary human peripheral blood T cells.
Introduction
CD4+ T helper cells play a central role in immune responses. They are a heterogeneous group of cells that are further classified into 3 main subsets, Th1, Th2, and Th17, based on their cytokine production profiles and effector functions.1 Th1 cells produce interferon-γ (IFN-γ) and tumor necrosis factor and regulate cell-mediated immune responses to intracellular pathogens. Th2 cells produce interleukin-4 (IL-4), IL-5, and IL-13, which are essential for the generation of antibodies and elimination of extracellular pathogens. Th17 cells compose a recently identified Th subpopulation that appears to play an essential role in protection against certain extracellular pathogens, such as Klebsiella pneumoniae, and in the pathogenesis of autoimmunity.2 Some of the transcription factors that govern Th1/Th2/Th17 cell fate decisions have been identified; and of these, GATA-3 and T-bet appear to be the key regulators of Th2 and Th1 cell development, respectively.3,4
GATA-3 is a member of the zinc finger transcription factor family.5 In addition to its prominent role in regulating Th2 cell differentiation, it is also essential for thymic T-cell development.6,7 In peripheral blood T helper cells, IL-4–mediated activation of STAT6 induces GATA-3 mRNA expression, an early event in Th2 cell differentiation. Interestingly, ectopic expression of a small amount of GATA-3 can induce robust expression of endogenous GATA-3, independent of IL-4 signaling and STAT6 activation,8–11 suggesting the presence of an IL-4/STAT6-independent autoregulatory feedback loop. As GATA-3 levels rise, the cells commit to Th2 development and begin to acquire the Th2 phenotype, all at the expense of Th1 cell development.5 The mechanistic details of how GATA-3 expression is regulated during Th2 cell development and how it apparently primes its own expression have not been fully elucidated. Here we report that c-Myb, a transcription factor previously reported to play a direct role in the regulation of GATA-3 expression,12 plays an essential role in assembling a transcriptional complex composed of GATA-3, Menin, and c-Myb, which allows GATA-3 to autoregulate its own expression. These studies provide a more detailed, mechanisms-oriented explanation for the pivotal role of c-Myb in allowing GATA-3 to regulate the development and maintain viability of Th2 cells in peripheral blood.
Methods
Lymphocyte preparation and cell culture
Normal human peripheral blood CD4+ T lymphocytes were obtained from consenting donors via the Human Immunology Core Facility at the University of Pennsylvania School of Medicine. Naive and memory/effector cells were sorted by CD45RO or CD45RA MicroBeads (Miltenyi Biotec), or phycoerythrin anti–human CD45RA antibody (BD Biosciences). Freshly isolated cells were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum. Naive CD4+ T cells were cultured with IL-12 (10 ng/mL), IL-2 (20 ng/mL), and anti-IL-4 antibody (10 μg/mL) or with IL-4 (25-40 ng/mL), IL-2 (20 ng/mL), and anti-IL-12 antibody (5-10 μg/mL) to promote, respectively, Th1 or Th2 cell formation in 3 to 10 days. CD3/CD28 antibodies (Dynabeads CD3/CD28 T-cell expander, Invitrogen) were used for every primary T-cell stimulation. Jurkat and 293T cells were cultured in RPMI 1640 or Dulbecco modified Eagle medium supplemented with 10% fetal calf serum, respectively.
Lentivirus vector construct for c-myb RNAi
c-myb-1(tgttattgccaagcactta), c-myb-3 (ctgcctggacgaactgata), and control c-myb-1 scramble (ctttatacgtagtcataag) siRNA sequences were inserted into the H1UG lentivirus vector (a gift of Dr E. J. Brown, University of Pennsylvania), which was modified from FUGW vector.13 The mixed lentivirus constructs expressing c-myb-1 and c-myb-3 shRNA were transduced into human primary CD4+ T cells (supplemental data and supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Dual-luciferase reporter assay
Six reporter constructs were prepared by inserting the human GATA-3 promoter into pGL3 (Promega). pG3P-L: −2039 to +587 relative to the transcription start site; pG3P-S: −148 to +587 bp; and pG3P-M: −148 to +587 contained one mutated myb binding site as illustrated in Figure 2C; pG3P-GBmut1 and 2: −148 to +587 with mutated GATA-3 binding site as shown in Figure 4A. The pcDNA3 vectors containing full-length c-myb (1-5 μg) and/or full-length GATA-3 (1-5 μg) were cotransfected with phRL (0.01-0.02 μg) and appropriate promoter upstream of luciferase in pGL3 (0.5-1.0 μg) into human primary T cells using Human T cell Nucleofector kit (Lonza Walkersville) or into 293T cells using Lipofectamine 2000 (Invitrogen). Firefly and Renilla luciferase activities were measured with a Dual-Luciferase reporter assay kit (Promega) using a luminometer.
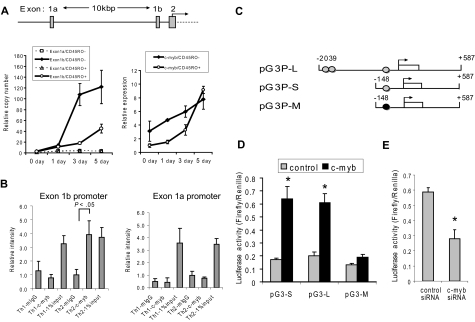
Figure 2.
c-Myb activates GATA-3 expression by binding to a canonical site within the GATA-3 exon 1b promoter in naive CD4 T cells under Th2 cell-promoting conditions. (A) Top graphic: GATA-3 promoter locus. Transcription can initiate from either exon 1a or exon 1b. The translational start site begins within exon 2. (Bottom left graph) The amount of exon 1a or exon 1b mRNA, as measured by quantitative RT-PCR, in naive (CD45RO−) or effector/memory (CD45RO+) CD4+ T cells before stimulation (0 day) and 1, 3, and 5 days after Th2 cell-promoting stimulatory conditions. The copy number was calculated for each species using a standard curve for exon 1a or exon 1b. The amount of exon 1a mRNA was adjusted to 1.0 to show the relative copy number. (Bottom right graph) The relative expression of c-myb mRNA in the same cells as the graph on the left. Data are mean ± SD of triplicate determinations. (B) Purified nucleoprotein complex was obtained from CD4+CD45RO− cells cultured under Th1- or Th2-promoting conditions and then used for ChIP assays using either anti–c-Myb (c-Myb) or mouse IgG (mIgG). The precipitated DNA fractions were analyzed by quantitative RT-PCR with primers specific to the GATA-3 upstream exon 1a (right graph) or downstream exon 1b (left graph) promoter region. *P < .05 compared with the value of murine (m) IgG. (C) Reporter constructs used for promoter analysis. Numbers represent nucleotides referenced relative to the start of transcription (arrow) from exon 1b. Gray circle represents c-Myb binding site; and black circle, mutated c-Myb binding site. (D) Dual-Luciferase reporter assays were performed in human peripheral T cells 24 hours after cotransfection with each of the pGL3–GATA-3 promoter constructs, and either a c-Myb expression construct (c-myb) or an empty plasmid (control). T cells were stimulated with CD3/CD28 antibodies, IL-2, and IL-4 for 8 hours before the cells were collected. Data are representative of 5 independent determinations. (E) The siRNA against c-Myb or control siRNA were cotransfected with the pGL3–GATA-3 promoter construct into human primary T cells. Dual-Luciferase reporter assays were performed 24 hours after the transfection. Data are representative of at least 2 independent experiments. *P < .05.
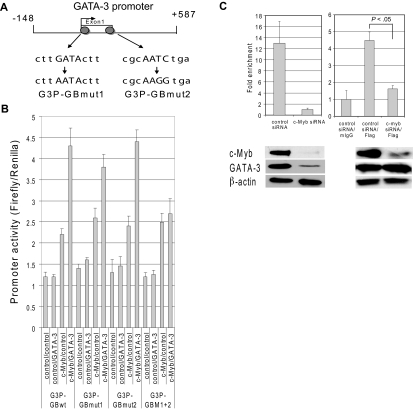
Figure 4.
Assembling the c-Myb/GATA-3 complex on the GATA-3 promoter. (A) A schematic diagram of the GATA-3 promoter region, including the minimal promoter. The region illustrated, nts −148 to +587, contains the start of transcription (arrow) from exon 1b and was used for promoter analysis experiments. Gray circles represent GATA binding sites. The sequences below the promoter scheme show the wild-type and mutated GATA binding sequences. (B) Reporter assays carried out with pG3P-S-GATA-3 wild-type minimal promoter vectors (G3P-GBwt), the reporter constructs with one mutated GATA-3 binding site (G3P-GBmut1 and G3P-GBmut2), or both mutated GATA-3 binding sites (G3P-GBmut1 + 2). The promoter vectors were cotransfected with c-Myb, GATA-3, and/or control expression vectors into to 293T cells and Dual-Luciferase activities were determined as described in the Dual Luciferase Reporter Assay method. (C) Purified nucleoprotein complex was obtained from Jurkat cells 48 hours after transfection of siRNAs with or without pcDNA GATA-3-Flag tag expression vector. ChIP assays were carried out with a GATA-3 antibody for endogenous GATA-3 or with a Flag antibody for exogenous GATA-3. The precipitated DNA fractions were analyzed by quantitative RT-PCR with primers specific to the GATA-3 downstream minimal promoter region. The bands at the bottom of each graph show c-Myb, GATA-3, and β-actin protein expression as determined by Western blot 48 hours after transfection of siRNAs with or without pcDNA GATA-3-Flag tag expression vector. Data are representative of 3 independent experiments.
Immunoprecipitation
Primary CD4+ T-cell lysates were prepared in a buffer containing 0.5% CA630, 50mM Tris (pH 7.5), 150mM NaCl, and protease inhibitor cocktails (complete mini, Roche Diagnostics), and equal volumes of the extract were incubated with 4 μg of the respective antibody with gentle rotation overnight at 4°C. Wild-type c-Myb, c-Myb-Flag tag, wild-type GATA-3, GATA-3 Flag-tag, 325-truncated c-Myb with Flag-tag, and Menin proteins were in vitro translated from their respective pcDNA expression vector using TNT Quick Coupled Transcription/Translation Systems (Promega). These proteins were incubated with the same amount of anti-Flag M2-agarose (Sigma-Aldrich) with gentle rotation for 4 hours at 4°C.
ChIP and Re-ChIP assay
Chromatin immunoprecipitation (ChIP) assays were completed using a ChIP Assay kit (Millipore) as previously described.14 CD4+ naive or effector/memory T cells were cultured under Th1 or Th2 conditions for 3 to 5 days except for histone modification ChIP assays. The cells were restimulated with IL-4 and IL-2 at 6 hours before crosslinking with formaldehyde. Immunoprecipitation was performed with anti-c-Myb (clone 1-1; Millipore), anti-GATA-3 (a mixture of MAB2605, R&D Systems; and HG3-31, Santa Cruz Biotechnology), anti p300 (clone RW128, Millipore), anti-CBP (Bethyl Laboratories), anti-MLL1 (Bethyl Laboratories), anti-Menin (Bethyl Laboratories), and anti-Flag M2 (Sigma-Aldrich) antibodies. The precipitated DNA fractions were then amplified by quantitative real-time polymerase chain reaction (RT-PCR) using standard protocols with SYBR Green and GATA-3 downstream or upstream promoter specific primers (forward: 5′-GGGTTTGGGTTGCAGTTTCCTTGT-3′; reverse: 5′-GCGACGCAACTTAAGGAGGTTCTA-3′ or forward: 5′-CGCCAGATCTGTCAGTTTCA-3′; reverse: 5′-AGGAGAAACAGCGAGGGAAT-3′, respectively). Re-ChIP assays were completed using Re-ChIP-IT, Magnetic Chromatin ReImmunoprecipitation Kit according to the instruction manual (Active Motif). In ChIP assays for histone modification, CD4+ naive and effector/memory T cells were stimulated under Th2-promoting conditions for 7 and 4 days, respectively. Antibodies against dimethylated and trimethylated H3K4 (ab6000), acetyl K9 (ab4441), and dimethyl K9 (ab1220) (Abcam) were used for the ChIP assays probing the GATA-3 locus in primary CD4+ T cells and Jurkat cells (experimental details in supplemental data).
Statistical analyses
Statistical comparisons of the data were completed using the 2-tailed Student t test. The level of significance was set at P values less than .05 in all cases.
Results
Silencing c-Myb decreases GATA-3 and Th2 cytokines gene expression in primary human Th2 cells
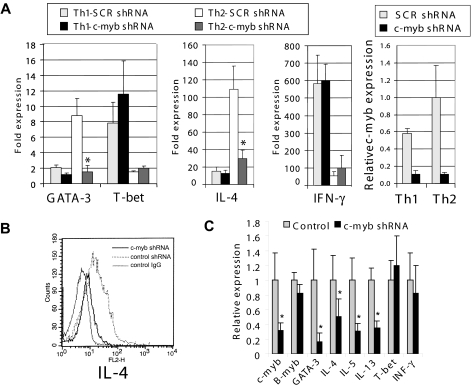
c-Myb expression is very high in immature hematopoietic cells and is down-regulated during terminal differentiation.15,16 In peripheral blood, naive and effector/memory T lymphocytes express c-Myb expression at low levels. However, after stimulation with a combination of IL-2, anti-CD3, and anti-CD28 antibodies, c-Myb expression in naive and effector/memory T cells increases dramatically. The biologic significance of this increase is still unclear. Therefore, to begin to understand the ramifications of this observation, a short hairpin RNA (shRNA) was used to silence c-Myb expression in human peripheral effector/memory CD4+ T cells after stimulation with anti-CD3/CD28 antibodies and IL-2. This strategy yielded a sequence-specific, 85% knockdown of c-Myb expression (supplemental Figure 2). Effects on candidate downstream gene targets of c-Myb were screened using a RT2 Profiler PCR Array Human Th1-Th2-Th3 (SA Biosciences). Of the 84 genes represented on the array (supplemental Table 1), the largest changes were noted in GATA-3, and in the expression of Th2 cytokine genes, a result that was confirmed by rescreening the original cell populations using quantitative RT-PCR.
After initial screening by PCR array and quantitative RT-PCR, we next determined whether c-Myb contributes to human Th2 cell development and maintenance from naive CD4+ T cells using the same c-Myb silencing strategy. Before transduction with lentiviral vectors, human primary CD4+ T cells were stimulated with anti-CD3/CD28 antibodies for 16 hours, followed by IL-2 for 5 hours. After transduction, the cells were sorted to isolate CD4+CD45RO− (naive) and CD4+CD45RO+ (effector/memory) T cells, which were then stimulated under Th1 or Th2 cell-promoting conditions. Naive human peripheral blood CD4+ T cells expressing the c-myb shRNA demonstrated 90% and 73% less up-regulation of GATA-3 and IL-4 mRNA, respectively, compared with cells expressing control shRNA, when cultured under Th2 cell-promoting conditions (Figure 1A). Flow cytometric analysis also showed that intracellular IL-4 expression was diminished in CD4+ cells stimulated under Th2-promoting conditions (Figure 1B). In contrast, silencing c-Myb did not decrease T-bet mRNA expression or IFN-γ expression in cells induced to undergo Th1 cell formation (Figure 1A).
Figure 1.
c-Myb contributes Th2 cell development and maintenance through regulation of GATA-3 expression. (A) Normal human peripheral blood CD4+ cells were transduced with lentivirus constructs expressing c-myb shRNA and GFP. Cells expressing GFP were sorted from nonexpressing cells and then further selected using MACS MicroBeads for CD4+CD45RO− cells. These were then cultured under Th1- or Th2-promoting conditions for 7 days. c-myb, GATA-3, T-bet, IL-4, and IFN-γ expression was measured by quantitative RT-PCR before and after stimulation. (B) Intracellular IL-4 levels in the cells analyzed in panel A. Before flow cytometric analysis, the cells were cultured for an additional 6 hours with phorbol myristate acetate (50 ng/mL), calcium ionomycin (500 ng/mL), and a protein transport inhibitor (BD GolgiStop). The histogram shows intracellular IL-4 in cells stimulated under Th2 cell-promoting conditions. Dotted line represents cells transfected with control shRNA; black line, cells transfected with c-myb shRNA; and gray line, control cells stained with mouse IgG-phycoerythrin (antibody control). (C) CD4+CD45RO+ (effector/memory) cells were transduced with control, or c-myb shRNA expression lentivirus and then stimulated with CD3/CD28 beads and IL-2 for 5 days. Poststimulation mRNA expression levels were measured by quantitative RT-PCR. Data are representative of at least 2 independent experiments. *P < .05.
In primary effector/memory CD4+ T cells, which includes established Th2 cells, c-Myb suppression with shRNA also decreased GATA-3 mRNA expression by approximately 85%, whereas the suppression of IL-4 expression was only approximately 50% (Figure 1C). Other Th2 cytokines, IL-5, and IL-13 decreased significantly as well, whereas T-bet and IFN-γ mRNA expression did not change. In toto, these results suggest that c-Myb not only exerts important functions during Th2 cell development but that it also plays a role in the maintenance of established Th2 cells. A more mechanisms-based understanding of these observations was then sought by carrying out the next series of experiments.
c-Myb activates GATA-3 expression by binding to a canonical site within the GATA-3 exon 1b promoter in naive CD4 T cells under Th2 cell-promoting conditions
Two different promoters are known to regulate GATA-3 expression17 (Figure 2A). Recent papers have shown that the GATA-3 exon 1a promoter is a direct target for Notch signaling and is critical for GATA-3 expression.18,19 To investigate the possibility that c-Myb might also play a direct role in regulating GATA-3 expression, we first measured the expression of mRNA transcribed from GATA-3 exon 1a and 1b, in human primary naive CD4+ cells stimulated under Th2-promoting conditions. On days 3 and 5 after stimulation, expression of GATA-3 exon 1b mRNA was dramatically increased, whereas expression of exon 1a mRNA did not increase significantly (Figure 2A). In addition, exon 1b mRNA expression remained concordant with c-myb expression (Figure 2A). These results strongly suggested that, in primary naive CD4+ T cells stimulated under Th2 promoting conditions, exon 1b promoter was used preferentially and c-Myb might therefore bind in this locus.
To test this hypothesis, we performed ChIP assays to determine whether, and where, c-Myb bound to the GATA-3 promoter in primary human CD4 cells during Th-subset development. Naive CD4+ T cells were cultured under Th1 or Th2 cell-promoting conditions for 3 to 5 days. A ChIP assay was then performed (Figure 2B), which revealed that in naive cells the exon 1b promoter was used by c-Myb in cells stimulated under Th2 cell-promoting conditions, but not under Th1 cell-promoting conditions (Figure 2B). Interestingly, the GATA-3 exon 1a promoter preferred by Notch18,19 was not occupied by c-Myb under either Th1- or Th2-promoting conditions (Figure 2B). These results corroborate the mRNA expression data that suggested that c-Myb binds the downstream GATA-3 promoter under Th2 cell-promoting conditions.
We next sought to more precisely define the c-Myb binding site within the GATA-3 (exon 1b) promoter and took advantage of previously reported investigations to accomplish this task.20,21 For example, Gregoire and Romeo analyzed a region in the human GATA-3 gene transcription initiation region from nts −8025 to +598 and showed that the −96/+598 DNA fragment is the GATA-3 minimal promoter.21 The GATA-3 promoter region that includes the minimal promoter was highly conserved in mammals, including human, mouse, rat, and dog, and revealed only one potential canonical c-Myb binding site. Additional scanning of the GATA-3 promoter locus revealed 2 closely spaced potential c-Myb binding sites around nucleotide −1837 (Figure 2C). Therefore, we carried out Dual-Luciferase reporter assays with 3 GATA-3 promoter constructs of various length and sequence (Figure 2C). pG3P-S included the minimal promoter, which contains a highly conserved c-Myb binding site (−124 relative to the transcription start site). pG3P-M had the same promoter region as pG3P-S, except that the Myb binding site was mutated. pG3P-L composed the −2039 to +587 region relative to the transcription start site, which encompassed the well-conserved Myb binding site at position −124, along with the 2 closely spaced Myb binding sites at −1837.
A c-Myb expression construct and each of the pG3P vectors were cotransfected into resting primary human T cells. After 16 hours, the cells were stimulated with CD3/CD28 antibodies, IL-2, and IL-4 for 8 hours. At 24 hours after transfection, a Dual-Luciferase reporter assay was carried out. c-Myb overexpression increased luciferase activity approximately 3.8-fold in human primary T cells cotransfected with pG3P-S, compared with when human primary T cells were cotransfected with empty vector (control; Figure 2D). In contrast, when the luciferase reporter was driven by pG3P-M, the promoter with the mutated Myb binding site, luciferase activity increased only approximately 1.5-fold in the cells compared with the control. Curiously, pG3P-L did not have any greater effect on promoter activity than when pG3-S was cotransfected with c-Myb (Figure 2D). In aggregate, these results suggest that c-Myb activates the GATA-3 promoter in human primary T cells under Th2 cell-promoting conditions by binding to the conserved Myb binding site at position −124. We additionally determined the effect of silencing c-Myb on the activity of pG3P-S GATA-3 promoter in human T cells. A Dual-Luciferase assay was carried out using the methodology just described. We found that silencing c-Myb decreased pG3P-S reporter activity by approximately 50% in the cells (Figure 2E). Because the transfection efficiency of the siRNA was only approximately 50% as well (data not shown), we considered this decrease significant.
GATA-3 and c-Myb act cooperatively to activate the GATA-3 promoter
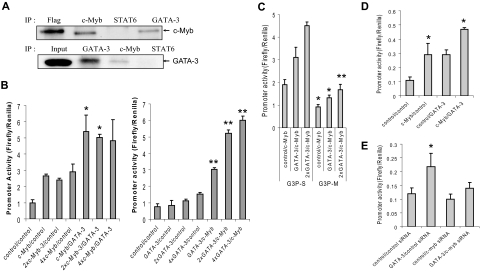
Having established that c-Myb binds the GATA-3 promoter in developing Th2 cells, we next sought to develop a more complete understanding of how such binding is regulated. To begin to address this question, we carried out immunoprecipitation experiments using material derived from primary human naive CD4+ T cells stimulated under Th2 cell-promoting conditions. Because it is known that the IL-4/STAT6 pathway induces GATA-3 expression in naive CD4+ T cells and that GATA-3 is essential for Th2 cell development,5,22 we examined whether c-Myb, STAT6, and/or GATA-3 itself interacted under Th2 cell-promoting conditions. We found that, although c-Myb and GATA-3 could be found in the same immunoprecipitated complex, STAT6 was not detected and therefore was not apparently incorporated into the c-Myb/GATA-3 complex (Figure 3A). Several reports have indicated that GATA-3 can regulate its own expression independent of IL-4 signaling and STAT6 activation.8–11 Because the results shown in Figure 3A suggested that c-Myb and GATA-3 were physically associated with each other, we wondered whether c-Myb might be playing a role in the ability of GATA-3 to autoactivate its own expression. To address this question, we first sought to determine whether c-Myb protein alone was sufficient to activate the GATA-3 promoter. We used the Dual-Luciferase reporter assay (described in the preceding paragraph) in 293T cells because these cells express neither endogenous c-Myb nor GATA-3. The results revealed that c-Myb could activate the minimal GATA-3 promoter approximately 3-fold compared with a control vector in the absence of GATA-3, whereas coexpression of GATA-3 along with c-Myb increased luciferase activity approximately 5.4-fold (Figure 3B). Dose-response studies suggested that, once a threshold amount of c-Myb was present, increasing amounts did not further activate the promoter, alone, or in combination with GATA-3. In contrast, once c-Myb was expressed in the system, increasing amounts of GATA-3 led to increasing amounts of luciferase activity, up to approximately 8-fold (Figure 3B). The results indicate that c-Myb alone can activate the GATA-3 promoter; but when combined with GATA-3 this capability is substantially enhanced.
Figure 3.
GATA-3 and c-Myb act cooperatively to activate the GATA-3 promoter. (A) Western Blots of immunoprecipitation (IP) carried out on lysates from human primary CD4+CD45RO− cells stimulated for 3 to 5 days under Th2 cell-promoting conditions. Flag indicates Flag-tagged c-Myb expressed in Jurkat cell lysate and IP with anti-Flag antibody (positive control); and Input, total GATA-3 present in cell lysate from stimulated CD4+ T cells. The blots shown are representative of 3 independent experiments. (B) Reporter assays carried out in 293T cells 48 hours after transfection with pG3P-S (wild-type Myb binding site) in the presence of c-Myb and/or GATA-3 expression constructs, or control (empty) vectors. Data in the left and right graphs are representative of 2 and 5 independent experiments, respectively. (C) Reporter assays carried out with pG3P-S or pG3P-M (mutated Myb binging site) as described in Figure 2C. (D) Reporter assays performed in human peripheral T cells with pG3P-S in the presence of c-Myb and/or GATA-3 expression constructs, or (E) siRNA against c-myb, 24 hours after transfection. (C-E) Data are representative of at least 2 independent determinations. *P < .05. **P < .01.
To further explore the contribution of c-Myb to activation of the GATA-3 promoter, we carried out a Dual-Luciferase reporter assay in 293T cells using the pG3P-M construct. Mutating the c-Myb binding site diminished the ability of GATA-3 and c-Myb to up-regulate GATA-3-driven expression from the pG3P-S reporter plasmid in 293T cells, again showing the cooperative nature of the interaction between these proteins (Figure 3C). This observation leads us to hypothesize that c-Myb plays an important role in the ability of GATA-3 to autoactivate its own promoter.
To better understand the physiologic relevance of the interaction between c-Myb and GATA-3, we performed similar reporter assays in human primary T cells, under Th2 cell-promoting conditions, using the same c-Myb and GATA-3 expression constructs. We again observed an additive effect of c-Myb and GATA-3 on reporter activity (Figure 3D). Silencing c-Myb with siRNA decreased the promoter's activity in the presence of GATA-3, but only partially, compared with primary T cells cotransfected with GATA-3 and control siRNA (Figure 3E).
Assembling the c-Myb/GATA-3 complex on the GATA-3 promoter
To understand the nature of the c-Myb/GATA-3 interaction on a more mechanistic level, we sought to identify functional GATA binding site(s) within GATA-3's promoter and the effect of c-Myb on GATA-3's ability to bind to these sites. The approximately 735 nt GATA-3 minimal promoter of pG3P-S vector contains 2 possible well-conserved GATA binding sites, CTTGATACTT and TCAGATTGCG.23 To determine the functional importance of each GATA binding site in the promoter, we mutated each individually, or both sites simultaneously (Figure 4A), and then performed Dual-Luciferase reporter assays in the absence and presence of c-Myb and/or GATA-3. Mutating the individual GATA binding sites did not change the promoter activity. However, when both GATA binding sites were mutated simultaneously, the addition of GATA-3 to c-myb did not activate luciferase activity to a greater extent than c-Myb alone (Figure 4B). Taken together, these data indicate that the 2 GATA binding sites complement each other and lead us to suggest that GATA-3 minimally requires one functional GATA binding site to activate its own promoter in the presence of c-Myb.
Having determined that GATA-3 and c-Myb act cooperatively to activate the GATA-3 promoter, we next sought to determine the nature of this interaction. In particular, we wished to know whether GATA-3 was capable of binding its own promoter in the absence of c-Myb or whether c-Myb needed to recruit GATA-3, either at the locus or in the form of a precomplex, for binding to take place. We therefore silenced c-Myb in Jurkat cells using siRNA and then performed ChIP assays with GATA-3 antibody. Because, as expected, GATA-3 expression decreased dramatically in Jurkat cells in which c-Myb was silenced, the amount of DNA that was immunoprecipitated by anti-GATA-3 decreased 90% compared with control siRNA (Figure 4C left panel). Interestingly, when we rescued GATA-3 expression in the cells in which c-Myb had been silenced with an expression vector expressing Flag-tagged GATA-3 (pcDNA3 GATA-3-Flag), GATA-3 still did not bind to its own promoter in the absence of c-Myb. Therefore, the ability of GATA-3 to bind to its own promoter is highly dependent on the expression of c-Myb.
c-Myb binds to GATA-3 through Menin
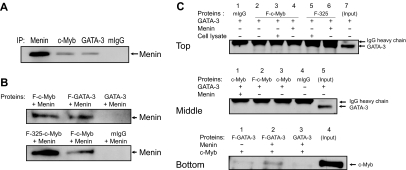
We next asked whether the c-Myb/GATA-3 interaction required a cofactor or was able to take place in the absence of a facilitating protein. To address this question, we in vitro translated c-Myb and GATA-3 individually and then mixed both proteins in vitro to determine whether they could form a stable complex. Despite numerous repetitions, we were unable to demonstrate any direct interaction between c-Myb and GATA-3 (data not shown). Accordingly, because recent work from our laboratory has shown that c-Myb interacts with mixed lineage leukemia (MLL) through the adapter protein Menin, a product of the MEN1 gene, in leukemic cells and stimulated human primary T cells,24 we explored the possibility that Menin might serve as an adaptor protein for the c-Myb/GATA-3 protein association.
We first examined whether Menin was associated with c-Myb and GATA-3 in primary CD4+ T cells stimulated to undergo Th2 cell development. In such cells, immunoprecipitation revealed that Menin was associated with both c-Myb and GATA-3 (Figure 5A). Next, we evaluated whether Menin bound to c-Myb and GATA-3 directly using in vitro synthesized proteins. When the pure proteins were allowed to interact in a cell-free solution, Menin protein was coimmunoprecipitated with Flag-tagged c-Myb, as well as Flag tagged GATA-3- fusion proteins, using an anti-Flag antibody (Figure 5B top panel). When we used a truncated c-Myb protein that lacked its negative regulatory domain (F-325; 1-325 amino acids of c-Myb),25 the interaction between c-Myb and Menin appeared stronger, suggesting that the negatively regulatory domain of the protein had some influence on the ability of c-Myb and Menin to interact (Figure 5B bottom panel). Finally, we sought to determine whether the c-Myb/GATA3 interaction could take place in the absence of Menin or whether other cellular proteins, besides Menin, might facilitate the interaction between c-Myb and GATA-3. To address these questions, additional F-c-Myb or F-325 immunoprecipitates were carried out with in vitro synthesized Menin, and GATA-3, or with a CD4+ T-cell lysate, prepared after culturing cells under Th2 cell-promoting conditions for 5 days (Figure 5C). As shown in Figure 5C (top and middle panels), where the GATA-3 band is visible just below the IgG heavy chain band in the Western blot, in the absence of Menin, neither c-Myb nor GATA-3 protein is immunoprecipitated (Figure 5C top panel, lane 2; middle panel, lane 2). Note that there are no flag-tagged proteins in lanes 1 and 3 of the middle panel, which serve as additional controls for this experiment. In Figure 5C (bottom panel), we carried out the immunoprecipitations with an anti-Flag antibody and then blotted with an anti-c-Myb antibody instead of a GATA-3 antibody. This was done to resolve any ambiguity in band identification resulting from the fact that the GATA-3 and IgG heavy chain bands run very close together. In this bottom panel, one can see that the direct c-Myb-GATA-3 interaction is weak (lane 1); but in the presence of Menin (lane 2), the interaction is considerably strengthened and therefore much more easily observed in the Western blot. In aggregate, these findings strongly suggest that c-Myb binds to GATA-3 through Menin and that the interaction is at least partially inhibited by c-Myb's negative regulatory domain.
Figure 5.
c-Myb binds to GATA-3 through the adaptor protein Menin. (A) Immunoprecipitation (IP) was performed with human primary CD4+ cells stimulated for 5 days under Th2 cell-promoting conditions. Anti-Menin, anti-c-Myb, and anti-GATA-3 antibodies and mouse IgG (mIgG) were used for the IPs. The Western blot was performed with anti-Menin antibody. The blot is representative of 2 independent experiments. (B-C) IPs were carried out with anti-Flag antibodies. Proteins used were synthesized by in vitro translation, except for the top panel of Figure 5C, where CD4+ T-cell lysate stimulated for 5 days under Th2 cell-promoting conditions was also used. The precipitated proteins were detected with Menin (B), GATA-3 (C, top and middle panels), or c-Myb antibodies (C, bottom panel). F indicates Flag-tag; and F-325, truncated c-Myb protein without its negative regulatory domain (1-325 amino acids of c-Myb). The blots are representative of at least 2 independent experiments.
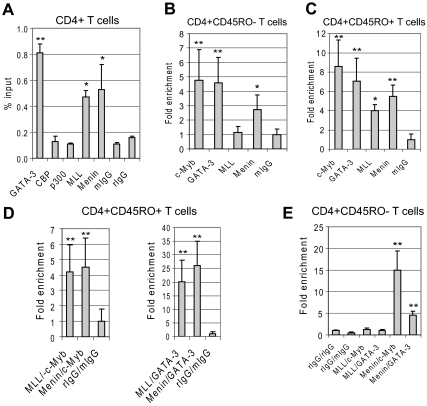
c-Myb, GATA-3 and Menin bind the GATA-3 promoter in CD4+ naive cells under Th2 cell-promoting conditions, whereas MLL binds to the same promoter region with c-Myb, GATA-3 and Menin in CD4+ effector/memory cells
Having established that the interaction between c-Myb, and GATA-3 is facilitated by Menin and knowing that Menin also facilitates the interaction between c-Myb and MLL,24 we carried out ChIP assays to determine whether c-Myb and Menin played a role in the reported recruitment of MLL to the GATA-3 promoter locus,26,27 and if so, under what circumstances. We first performed a ChIP with CD4+ T cells stimulated for 3 days with IL-2, CD3/28 antibodies, and IL-4. CBP and p300 antibodies were also added to the ChIP reaction because of reports that these proteins function as c-Myb coactivator proteins.28,29 The ChIP assays carried out with material from primary human CD4+ T cells stimulated under Th2 cell-promoting conditions revealed that GATA-3, MLL, and Menin bound to the GATA-3 promoter, but CBP and p300 did not under these experimental conditions (Figure 6A). Next, we evaluated protein binding to the GATA-3 promoter in CD4+ naive and effector/memory T cells. We found that c-Myb, GATA-3, and Menin bound to the GATA-3 promoter, but MLL did not, in naive CD4+ T cells stimulated with IL-2, IL-4, and CD3/CD28 antibodies (Figure 6B). In contrast, MLL bound to the promoter along with c-Myb, GATA-3, and Menin in effector/memory CD4+ T cells stimulated with IL-2, IL-4, and CD3/CD28 antibodies (Figure 6C). We found no evidence to support direct binding of GATA-3 to MLL by immunoprecipitation assays (data not shown). Therefore, we carried out a Re-ChIP assay to determine whether MLL, Menin, and GATA-3 associated in the same region of the GATA-3 promoter in effector/memory cells. The Re-ChIP assay with stimulated CD4+ effector/memory T cells showed that GATA-3 bound to the same locus in the GATA-3 promoter when precipitated with either MLL or Menin polyclonal antibodies (Figure 6D). c-Myb also bound in the same region. However, neither GATA-3 nor c-Myb bound the locus where MLL antibody precipitated in naive CD4+ T cells after 3-day stimulation under Th2-promoting conditions (Figure 6E).
Figure 6.
c-Myb, GATA-3, and Menin bind to GATA-3's minimal promoter in CD4+ naive cells under Th2 cell-promoting conditions, whereas MLL binds the same promoter region in a complex with c-Myb, GATA-3, and Menin in CD4+ effector/memory cells.(A) Purified nucleoprotein complex was obtained from primary human whole CD4+ cells after the stimulation with IL-4, IL-2, and antibodies against CD3/CD28 for 3 days. ChIP assays were carried out with GATA-3, CBP, p300, MLL1, and Menin antibodies and mouse IgG (mIgG) or rabbit IgG (rIgG). The precipitated DNA fractions were analyzed by quantitative RT-PCR with primers specific to the GATA-3 downstream minimal promoter region. (B) ChIP assays were performed with primary human CD4+ CD45RO− (naive) cells after stimulation with IL-4, IL-2, and antibodies against CD3/CD28. (C) ChIP assays were carried out with primary human CD4+ CD45RO+ (effector/memory) cells after a 3-day stimulation with IL-4, IL-2, and antibodies against CD3/CD28. (D-E) Re-ChIP assays were carried out with human CD4+ CD45RO+ (D) or CD4+ CD45RO− (E); cells stimulated as described in panel C. The first ChIP assays were performed with antibodies against MLL or Menin. The second ChIP (Re-ChIP) assays were done with antibodies against c-Myb or GATA-3. Controls for the Re-ChIP assay were performed with antirabbit IgG (rIgG) and antimouse IgG (mIgG) antibodies. Data are representative of at least 2 independent experiments. *P < .05. **P < .01.
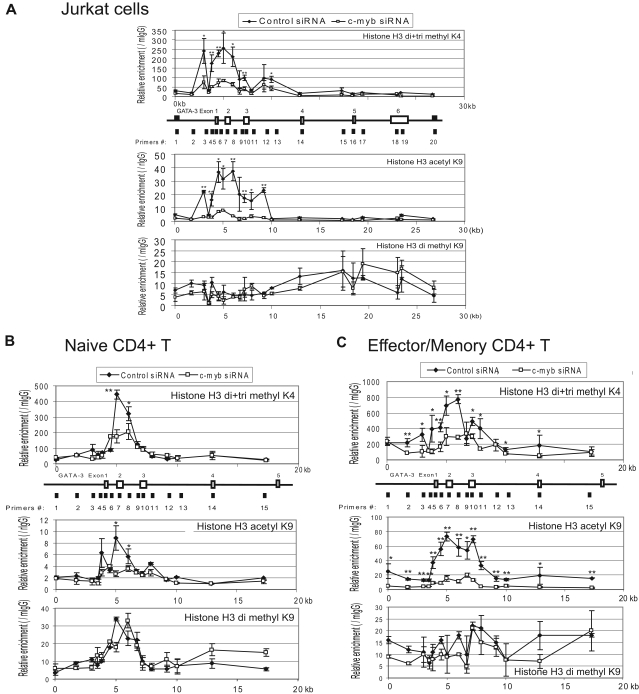
c-Myb silencing decreases the methylation of histone H3K4 and the acetylation of histone H3K9 at GATA-3 locus in Jurkat cells, and primary human CD4+ naive and effector/memory T cells when stimulated under Th2-promoting conditions
We next examined whether c-Myb could influence histone modifications of the GATA-3 gene locus. To do this, we silenced c-Myb expression in Jurkat cells, as well as in primary human CD4+ CD45RA+ T cells, and CD4+ CD45RA− T cells stimulated under Th2 cell-promoting conditions as described in “Lymphocyte preparation and cell culture” and “ChIP and Re-ChIP assay.” We then performed ChIP assays with antibodies directed to histone H3 dimethylated and trimethylated K4, histone H3 acetyl K9, histone H3 dimethylated K9, and 20 primer pairs suggested by Primer3 (http://frodo.wi.mit.edu/primer3; supplemental Table 2) for sequences predicted to amplify well by quantitative RT-PCR within the approximately 27 kb GATA-3 locus. The ChIPs revealed that, compared with control siRNA-treated cells, silencing c-Myb led to decreased levels of H3K4 dimethylation and trimethylation, as well as H3K9 acetylation in the region of the GATA-3 promoter. These changes were observed in Jurkat cells, as well as in stimulated human CD4+ T lymphocytes where a bona fide c-Myb binding site is known to exist (Figure 7). Interestingly, histone changes were most marked in stimulated effector/memory CD4+ T lymphocytes (Figure 7C). Of note, silencing c-Myb did not affect the level of histone H3K9 dimethylation. In aggregate, these results demonstrate that c-Myb makes a significant contribution to chromatin remodeling at the GATA-3 gene locus in human primary Th2 cells during their development, and as we interpret our data, in particular during their transition into memory cells.
Figure 7.
c-Myb silencing decreases the methylation of histone H3K4 and the acetylation of histone H3K9 at GATA-3 locus in Jurkat cells, and primary human CD4+ naive and effector/memory cells with the stimulation under Th2-promoting conditions. Representative result of ChIP assays carried out with a series of primer pairs covering the GATA-3 gene locus in Jurkat cells (A), primary human CD4+ CD45RA+ naive T cells (B), and CD4+CD45RA− effector/memory T cells (C) infected with lentivirus expressing either c-Myb or control (scrambled sequence) shRNA. Cells were stimulated under Th2-promoting conditions for 7 days and 4 days, respectively, before conducting the ChIP assays. The ChIP assays were carried out with antibodies directed against dimethylated and trimethylated histone H3K4 (top graph in each panel), acetyl H3K9 (middle graph in each panel), and dimethyl H3K9 (bottom graph in each panel) and analyzed by quantitative RT-PCR. The graphs show the relative specific antibody-mediated enrichment compared with control mouse or rabbit IgG. Each point represents the mean ± SD of 3 replicate determinations. In each top panel of the figure, the line drawing immediately underneath the baseline of each graph illustrates (A) approximately 27 kbp or (B-C) approximately 20 kbp of the GATA-3 gene locus. The previously validated, highly conserved c-Myb binding site is situated between primer pairs 4 and 5. Two independent experiments were performed with similar results. *P < .05. **P < .01.
Discussion
Using a variety of experimental techniques, including transient gene silencing with oligonucleotides,30 expression of dominant negative c-Myb protein,31,32 and tissue-specific loss of floxed c-Myb in Cre-producing mouse strains,33–36 it has been demonstrated unequivocally that c-Myb is important at multiple points during T-cell development.7 Maurice et al demonstrated that c-Myb regulates T helper cell lineage commitment in developing mouse thymocytes, at the same time that it appears to block development of cytotoxic T cells via regulation of GATA-3.12 However, most of these studies have been carried out on cells in the thymic developmental compartments and, in addition, have focused on murine cells. As a result, beyond correlative observations, such as those demonstrating that proliferating human T cells require c-Myb for cell cycle progression,30 there is a paucity of mechanistic information on c-Myb's functions in activated human CD4+ peripheral blood T cells. To address this knowledge gap, we carried out biochemical, immunoaffinity, and carefully controlled RNA interference studies in normal human peripheral blood lymphocytes to study the role of c-Myb in regulating peripheral blood T-cell fate generally, and Th2 cell differentiation more specifically.
Th2 cell differentiation is dependent on a regulated expression of the GATA-3 transcription factor.3 In naive T cells, GATA-3 expression is detectable but very low. On binding IL-4, a STAT6-dependent signal transduction cascade is initiated that leads to the up-regulation of GATA-3 expression.37 We think that this initial up-regulation is indeed a “priming” stimulus because thereafter GATA-3 levels increase dramatically, in what previous investigations have suggested is an IL-4/STAT6-independent event.8–11 The studies presented in this manuscript suggest that it is the formation of a highly active transcription complex composed minimally of c-Myb and GATA-3 on the GATA-3 promoter, which allows this marked up-regulation to occur (Figures 1–2). Because the binding of GATA-3 to its own promoter is required, we have further shown that GATA-3 up-regulation is properly considered autoregulatory in nature (Figures 3–4). Finally, we have also determined that, although GATA-3 binding to its own promoter is required for the enhanced expression of GATA-3 during Th2 cell development, it is not by itself sufficient to induce the levels of GATA-3 that are observed during this process (Figure 3). Rather, our data demonstrate that GATA-3 is active within a transcriptional complex that contains c-Myb (Figure 4) and Menin (Figure 5) in naive CD4+ cells undergoing Th2 cell differentiation, and with c-Myb, Menin, and MLL in CD4+ effector/memory cells (Figure 6). The importance of c-Myb in assembling this transcriptional complex, and the potential physiologic significance of the regulatory loop it enables, was demonstrated by the fact that silencing c-myb gene expression blocked the autoactivation of GATA-3 expression, and which in turn aborts Th2 cell differentiation. This observation led us to hypothesize that the relatively “high” levels of c-Myb in naive T cells (compared with effector/memory T cells) may enable a “default” pathway, which favors Th2 cell development over Th1 or Th17 cells.
That the marked up-regulation of GATA-3 noted during Th2 cell development might require the assembly of a multiprotein coactivator complex was in some respects predicted by previous observations in the literature. For example, GATA-3 and Smad3 are known to physically interact in the context of transforming growth factor-β-mediated gene expression38; and, given the nature of Smad3, this interaction might reasonably be expected to require additional stabilizing proteins for effective promoter contacts.38,39 In another example, nuclear factor-κB (NF-κB) is known to be activated by T-cell receptor engagement, and activation of important downstream target genes, such as GlNAc6ST-1, a gene that encodes a sulfotransferase required for the synthesis of 6-sulfated cell recognition glycans, is dependent on physical interaction with GATA-3.40 Das et al have also shown that in NF-κB p50-deficient mice, CD4+ T cells failed to induce GATA-3 expression under Th2 priming condition but maintained unimpaired T-bet and IFN-γ expression under Th1 differentiation conditions.41 These reports initially suggested the possibility that a c-Myb/GATA-3 coactivator complex might include Smad3 or NF-κB. However, we were unable to demonstrate the presence of either transcription factor in c-Myb/GATA-3 immunoprecipitates (data not shown) and therefore turned our attention to other candidates.
We noted with interest several reports indicating that transcriptionally active proteins more usually associated with myelopoiesis also play important roles in lymphoid cell development. For example, Yamashita et al have demonstrated a crucial role for MLL in the maintenance of GATA-3 gene expression and the formation of Th2 memory cells.26 Specifically, this group reported that MLL+/− CD4+ T cells could differentiate normally into antigen-specific effector Th1/Th2 cells in vitro, but the ability of MLL+/− memory Th2 cells to produce Th2 cytokines was reduced. The reduced expression of MLL in memory Th2 cells was also associated with decreased GATA3 expression as well as impaired maintenance of GATA3 locus histone modifications. Because MLL, a member of the Trithorax gene family, functions as a histone methyltransferase for H3K4,42 this finding suggested that MLL was affecting local gene expression by epigenetic modification of the locus. Pertinent to this report, our group has recently found that c-Myb interacts with MLL in leukemic cells through Menin.24 Menin, the product of the MEN1 gene mutated in familial multiple endocrine neoplasia type 1, is often found in MLL histone methyltransferase complexes42 and plays an important role as an adapter protein, along with LEDGF, in allowing functional MLL complexes to assemble.43 We were further intrigued by several papers that reported that Menin is able to interact with several different partners affecting T-cell development, including NF-κB, JunD, and Smad3, as well as MLL.44,45
For these reasons, and the fact that we have recently shown that Menin recruits c-Myb into the MLL complex in human leukemia cells,24 we investigated the possibility that Menin might also be involved in assembling the Myb-GATA3 transcription complex and found that this was indeed the case (Figures 5–6). Importantly, the c-Myb-Menin-MLL complexes were shown to have biologic function, as demonstrated by ChIP assays performed on primary human CD4+ effector/memory T cells after silencing c-Myb with siRNA. Specifically, these ChIPs revealed considerably diminished H3K4 methylation and H3K9 acetylation at the GATA-3 locus (Figure 7). Interestingly, when c-Myb was silenced in naive CD4+ cells after 7 days of stimulation under Th2 cell-promoting conditions, histone modifications were not nearly as marked as those observed in effector/memory T cells, especially with regard to H3K9 acetylation. These results led us to postulate that the recruitment of MLL probably occurs as effector cells develop into memory cells.
In conclusion, the results of our studies led us to hypothesize that the Myb-Menin-GATA-3 complex may play an important role in the developmental choice, and differentiation, of T-cell subsets from naive T cells. Specifically, we postulate that the IL-4/STAT6 signaling pathway initiates GATA-3 expression, leading to the development of Th2 cells. Subsequently, GATA-3 activates its own promoter by forming an autoregulatory complex with c-Myb and Menin that markedly increases GATA-3 expression. During the process of generating memory Th2 cell, the GATA-3/c-Myb complex recruits MLL. These events sustain GATA-3 expression and are permissive of cytokine production as a result of the changes in histone modifications at the GATA-3 locus (Memory Th2; illustrated in supplemental Figure 3). The details that explain how c-Myb-Menin-GATA-3 can recruit MLL to the GATA-3 locus in CD4+ effector/memory T cells at specific developmental windows are of particular interest to our group, as are the postulated epigenetic and transcriptional changes that result from this recruitment. Uncovering these details should greatly clarify how naive CD4+ T cells become memory Th2 cells. These questions are currently being addressed in our laboratory and will be the focus of future reports.
Supplementary Material
Acknowledgments
The authors thank Dr Susan Shetzline for critical reading of the manuscript and helpful editorial suggestions and Dr Daniel F. Heitjan, Department of Biostatistics and Statistics, Biostatistics Core Facility, Abramson Cancer Center, University of Pennsylvania School of Medicine, for review of the manuscript and advice on appropriate statistical analysis of the data.
This work was supported by the National Institutes of Health (RO1-CA101859; A.M.G.) and the Abramson Cancer Center at the University of Pennsylvania (State of Pennsylvania Department of Health Tobacco Settlement Grant).
Footnotes
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: Y.N. designed and performed the majority of the research and wrote the paper; A.C.B. assisted in lentivirus experiments and data analysis; S.J. aided experimental design and assisted with ChIP experiments; S.I.R. and Y.S. assisted with experimental design; M.S. assisted in performing ChIP assays; and A.M.G. designed research, supervised all the experiments completed herein, and edited the final drafts of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation of S.I.R. is Fox Chase Cancer Center, Department of Medical Oncology, Philadelphia, PA.
Correspondence: Alan M. Gewirtz, Division of Hematology/Oncology, Department of Medicine, Rm 713, BRB II/III, University of Pennsylvania School of Medicine, 421 Curie Blvd, Philadelphia, PA 19104; e-mail: gewirtz@mail.med.upenn.edu.
References
- 1.Dong C. Diversification of T-helper-cell lineages: finding the family root of IL-17-producing cells. Nat Rev Immunol. 2006;6(4):329–333. doi: 10.1038/nri1807. [DOI] [PubMed] [Google Scholar]
- 2.Bettelli E, Korn T, Oukka M, Kuchroo VK. Induction and effector functions of T(H)17 cells. Nature. 2008;453(7198):1051–1057. doi: 10.1038/nature07036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89(4):587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 4.Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295(5553):338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- 5.Zhou M, Ouyang W. The function role of GATA-3 in Th1 and Th2 differentiation. Immunol Res. 2003;28(1):25–37. doi: 10.1385/IR:28:1:25. [DOI] [PubMed] [Google Scholar]
- 6.Ho IC, Pai SY. GATA-3: not just for Th2 cells anymore. Cell Mol Immunol. 2007;4(1):15–29. [PubMed] [Google Scholar]
- 7.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8(1):9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ouyang W, Lohning M, Gao Z, et al. Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity. 2000;12(1):27–37. doi: 10.1016/s1074-7613(00)80156-9. [DOI] [PubMed] [Google Scholar]
- 9.Lee HJ, Takemoto N, Kurata H, et al. GATA-3 induces T helper cell type 2 (Th2) cytokine expression and chromatin remodeling in committed Th1 cells. J Exp Med. 2000;192(1):105–115. doi: 10.1084/jem.192.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou M, Ouyang W, Gong Q, et al. Friend of GATA-1 represses GATA-3-dependent activity in CD4+ T cells. J Exp Med. 2001;194(10):1461–1471. doi: 10.1084/jem.194.10.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ranganath S, Murphy KM. Structure and specificity of GATA proteins in Th2 development. Mol Cell Biol. 2001;21(8):2716–2725. doi: 10.1128/MCB.21.8.2716-2725.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maurice D, Hooper J, Lang G, Weston K. c-Myb regulates lineage choice in developing thymocytes via its target gene Gata3. EMBO J. 2007;26(15):3629–3640. doi: 10.1038/sj.emboj.7601801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295(5556):868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 14.Nakata Y, Shetzline S, Sakashita C, et al. c-Myb contributes to G2/M cell cycle transition in human hematopoietic cells by direct regulation of cyclin B1 expression. Mol Cell Biol. 2007;27(6):2048–2058. doi: 10.1128/MCB.01100-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westin EH, Gallo RC, Arya SK, et al. Differential expression of the amv gene in human hematopoietic cells. Proc Natl Acad Sci U S A. 1982;79(7):2194–2198. doi: 10.1073/pnas.79.7.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oh IH, Reddy EP. The myb gene family in cell growth, differentiation and apoptosis. Oncogene. 1999;18(19):3017–3033. doi: 10.1038/sj.onc.1202839. [DOI] [PubMed] [Google Scholar]
- 17.Asnagli H, Afkarian M, Murphy KM. Cutting edge: identification of an alternative GATA-3 promoter directing tissue-specific gene expression in mouse and human. J Immunol. 2002;168(9):4268–4271. doi: 10.4049/jimmunol.168.9.4268. [DOI] [PubMed] [Google Scholar]
- 18.Amsen D, Antov A, Jankovic D, et al. Direct regulation of Gata3 expression determines the T helper differentiation potential of Notch. Immunity. 2007;27(1):89–99. doi: 10.1016/j.immuni.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang TC, Yashiro-Ohtani Y, Del Bianco C, Knoblock DM, Blacklow SC, Pear WS. Notch directly regulates Gata3 expression during T helper 2 cell differentiation. Immunity. 2007;27(1):100–110. doi: 10.1016/j.immuni.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.George KM, Leonard MW, Roth ME, et al. Embryonic expression and cloning of the murine GATA-3 gene. Development. 1994;120(9):2673–2686. doi: 10.1242/dev.120.9.2673. [DOI] [PubMed] [Google Scholar]
- 21.Gregoire JM, Romeo PH. T-cell expression of the human GATA-3 gene is regulated by a non-lineage-specific silencer. J Biol Chem. 1999;274(10):6567–6578. doi: 10.1074/jbc.274.10.6567. [DOI] [PubMed] [Google Scholar]
- 22.Grogan JL, Locksley RM. T helper cell differentiation: on again, off again. Curr Opin Immunol. 2002;14(3):366–372. doi: 10.1016/s0952-7915(02)00340-0. [DOI] [PubMed] [Google Scholar]
- 23.Merika M, Orkin SH. DNA-binding specificity of GATA family transcription factors. Mol Cell Biol. 1993;13(7):3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin S, Zhao H, Yi Y, Nakata Y, Kalota A, Gewirtz AM. c-Myb binds MLL through menin in human leukemia cells and is an important driver of MLL-associated leukemogenesis. J Clin Invest. 2010;120(2):593–606. doi: 10.1172/JCI38030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanei-Ishii C, Ninomiya-Tsuji J, Tanikawa J, et al. Wnt-1 signal induces phosphorylation and degradation of c-Myb protein via TAK1, HIPK2, and NLK. Genes Dev. 2004;18(7):816–829. doi: 10.1101/gad.1170604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamashita M, Hirahara K, Shinnakasu R, et al. Crucial role of MLL for the maintenance of memory T helper type 2 cell responses. Immunity. 2006;24(5):611–622. doi: 10.1016/j.immuni.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 27.Nakayama T, Yamashita M. Initiation and maintenance of Th2 cell identity. Curr Opin Immunol. 2008;20(3):265–271. doi: 10.1016/j.coi.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 28.Dai P, Akimaru H, Tanaka Y, et al. CBP as a transcriptional coactivator of c-Myb. Genes Dev. 1996;10(5):528–540. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- 29.Oelgeschlager M, Janknecht R, Krieg J, Schreek S, Luscher B. Interaction of the coactivator CBP with Myb proteins: effects on Myb-specific transactivation and on the cooperativity with NF-M. EMBO J. 1996;15(11):2771–2780. [PMC free article] [PubMed] [Google Scholar]
- 30.Gewirtz AM, Anfossi G, Venturelli D, Valpreda S, Sims R, Calabretta B. G1/S transition in normal human T-lymphocytes requires the nuclear protein encoded by c-myb. Science. 1989;245(4914):180–183. doi: 10.1126/science.2665077. [DOI] [PubMed] [Google Scholar]
- 31.Allen RD, 3rd, Bender TP, Siu G. c-Myb is essential for early T cell development. Genes Dev. 1999;13(9):1073–1078. doi: 10.1101/gad.13.9.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pearson R, Weston K. c-Myb regulates the proliferation of immature thymocytes following beta-selection. EMBO J. 2000;19(22):6112–6120. doi: 10.1093/emboj/19.22.6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emambokus N, Vegiopoulos A, Harman B, Jenkinson E, Anderson G, Frampton J. Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. EMBO J. 2003;22(17):4478–4488. doi: 10.1093/emboj/cdg434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bender TP, Kremer CS, Kraus M, Buch T, Rajewsky K. Critical functions for c-Myb at three checkpoints during thymocyte development. Nat Immunol. 2004;5(7):721–729. doi: 10.1038/ni1085. [DOI] [PubMed] [Google Scholar]
- 35.Lieu YK, Kumar A, Pajerowski AG, Rogers TJ, Reddy EP. Requirement of c-myb in T cell development and in mature T cell function. Proc Natl Acad Sci U S A. 2004;101(41):14853–14858. doi: 10.1073/pnas.0405338101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hooper J, Maurice D, Argent-Katwala MJ, Weston K. Myb proteins regulate expression of histone variant H2A.Z during thymocyte development. Immunology. 2008;123(2):282–289. doi: 10.1111/j.1365-2567.2007.02697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 38.Blokzijl A, ten Dijke P, Ibanez CF. Physical and functional interaction between GATA-3 and Smad3 allows TGF-beta regulation of GATA target genes. Curr Biol. 2002;12(1):35–45. doi: 10.1016/s0960-9822(01)00623-6. [DOI] [PubMed] [Google Scholar]
- 39.Massague J, Chen YG. Controlling TGF-beta signaling. Genes Dev. 2000;14(6):627–644. [PubMed] [Google Scholar]
- 40.Chen GY, Sakuma K, Kannagi R. Significance of NF-kappaB/GATA axis in tumor necrosis factor-alpha-induced expression of 6-sulfated cell recognition glycans in human T-lymphocytes. J Biol Chem. 2008;283(50):34563–34570. doi: 10.1074/jbc.M804271200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Das J, Chen CH, Yang L, Cohn L, Ray P, Ray A. A critical role for NF-kappa B in GATA3 expression and TH2 differentiation in allergic airway inflammation. Nat Immunol. 2001;2(1):45–50. doi: 10.1038/83158. [DOI] [PubMed] [Google Scholar]
- 42.Yokoyama A, Wang Z, Wysocka J, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24(13):5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14(1):36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agarwal SK, Kennedy PA, Scacheri PC, et al. Menin molecular interactions: insights into normal functions and tumorigenesis. Horm Metab Res. 2005;37(6):369–374. doi: 10.1055/s-2005-870139. [DOI] [PubMed] [Google Scholar]
- 45.Balogh K, Racz K, Patocs A, Hunyady L. Menin and its interacting proteins: elucidation of menin function. Trends Endocrinol Metab. 2006;17(9):357–364. doi: 10.1016/j.tem.2006.09.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.