Abstract
Fibrin polymerizes via noncovalent and dynamic association of thrombin-exposed “knobs” with complementary “holes.” Synthetic knob peptides have received significant interest as a means for understanding fibrin assembly mechanisms and inhibiting fibrin polymerization. Nevertheless, the inability to crystallize short peptides significantly limits our understanding of knob peptide structural features that regulate dynamic knob:hole interactions. In this study, we used molecular simulations to generate the first predicted structure(s) of synthetic knobs in solution before fibrin hole engagement. Combining surface plasmon resonance (SPR), we explored the role of structural and electrostatic properties of knob “A” mimics in regulating knob:hole binding kinetics. SPR results showed that association rates were most profoundly affected by the presence of both additional prolines as well as charged residues in the sixth to seventh positions. Importantly, analyzing the structural dynamics of the peptides through simulation indicated that the 3Arg side chain orientation and peptide backbone stability each contribute significantly to functional binding. These findings provide insights into early fibrin protofibril assembly dynamics as well as establishing essential design parameters for high-affinity knob mimics that more efficiently compete for hole occupancy, parameters realized here through a novel knob mimic displaying a 10-fold higher association rate than current mimics.
Introduction
The activation and polymerization of the blood-circulating protein fibrinogen, a 340-kDa glycoprotein with 6 polypeptide chains (AαBβγ)2, is the primary homeostatic mechanism preventing excessive blood loss after vascular injury. This process is initiated by the activated serine protease thrombin, which specifically cleaves 4 N-terminal arginyl-glycine motifs on the 2 adjacent Aα and Bβ chains of fibrinogen, releasing 2 sets of fibrinopeptides A and B (FpA and FpB) and exposing cryptic fibrin polymerization knobs “A” and “B,” respectively.1–5 The newly exposed fibrin knobs noncovalently interact with complementary “holes”' within the 2 distal C-terminal regions of the γ and β chains (complementary holes “a” and “b,” respectively) to initiate fibrin protofibril assembly. Understanding the fundamentals of this dynamic and noncovalent knob:hole interaction will lead to both a more thorough understanding of fibrin assembly mechanisms and the establishment of design criteria for superior anticoagulants with high polymerization hole affinity to inhibit fibrin assembly.
Evidence for fibrin knob:hole interactions was first conclusively shown when fibrin polymerization was inhibited by synthetic knob A tripeptides (Gly-Pro-Arg) competing for fibrin holes.6,7 Characterization of the equilibrium binding affinities of both knob A and B peptide variants to fibrinogen showed that the knob A peptides (ie, GPRV and GPRP) have higher affinities to fibrinogen than knob B peptides (ie, GHRP and AHRP) under calcium-free conditions.6–8 In the presence of calcium, the binding affinity knob B mimic GHRP significantly increases to near GPRP; however, GHRP is readily displaced by the knob A mimic GPRP, suggesting that knob A interactions are stronger than knob B.6 Further evaluation of knob B peptide variants (ie, GHRPY, AHRPY, and MHRPY) showed the promiscuity of hole b versus hole a because the nonglycyl knob B peptides engaged hole b, but not hole a; only N-terminal glycyl peptides bind to hole a.9,10 Recent studies elegantly conducted with fragments from fibrinogen mutants and laser tweezers–based force spectroscopy further characterized A:a, A:b, and B:b interactions with native fibrin fragments.11,12 Consistent with the knob peptide studies, knob A interactions seem to dominate the knob:hole interactions because the A:a interaction displays a 6- to 8-fold higher rupture force than A:b or B:b interactions.11,12 Although such steady-state, equilibrium studies have laid the foundation for understanding knob:hole interactions, investigating these binding events under dynamic conditions will provide critical information about the residence time of the noncovalent knob:hole interaction, a key determinant in fibrin assembly initiation and polymerization. In addition, understanding the fundamental structural cues within strong binding fibrin knob A mimics that drive the initial docking events and potentially stabilize the interaction (eg, enhance knob residence time within holes) will further establish design criteria required to develop superior anticoagulants that compete for hole occupancy.
Examination of the crystal structure of fibrinogen/fibrin hole regions (D fragment) with associated knob A peptides clearly established electrostatic interactions and hydrogen bonding between engaged knob peptides and holes a and b.13–15 Crystal structures of D fragment generated with either GPRP or GPRVVE knob A peptides indicate that the 1Gly and 3Arg residues engage the same residues on the γ chain with minimal structural differences between the GPRP and GPRVVE.13,14 This observation leads one to question why GPRP displays a 4-fold greater affinity (KD) for D fragment than for GPRV.6,8 Laudano and Doolittle6 speculated that the higher affinity of GPRP was due to the 4-Pro residue potentially stabilizing the backbone of the GPRP, thereby reducing the degrees of freedom and the number of potential conformations. However, the structural properties of the knob peptides in aqueous environments before hole engagement have not been explicitly examined largely because of the inability to crystallize small peptides for structural x-ray studies. Such knowledge is critical for rationally designing knob peptides, as well as fully synthetic analogs, with superior anticoagulant properties than is currently available (ie, GPRP). Molecular modeling and molecular dynamics (MD) simulations are an emerging approach that explores the conformational landscape of short peptides enabling one to assess molecular structural differences that influence functional binding parameters.16,17
In this study, we investigated fibrin knob peptide:hole interactions with both experimental and theoretical modeling approaches to elucidate factors that influence the kinetic binding dynamics (ka and kd) of knob A peptide variants to fibrin holes. We focused on (1) kinetic modeling of the binding interaction and (2) structural characterization of the peptides in solution. Previous binding affinity studies have contributed significantly to the current understanding of knob:hole interactions; however, as described earlier, these seminal experiments were performed under equilibrium conditions in which the details of critical dynamic interactions are overlooked. Using surface plasmon resonance (SPR), we evaluated the kinetic binding interactions of fibrin knob peptides with fibrinogen/fibrin holes and investigated appropriate kinetic binding models to describe the knob:hole interaction. On the basis of past literature, we examined a set of knob A peptide variants of 7 to 8 residues in length to evaluate 2 properties hypothesized to influence binding kinetics (Table 1). The first property included sequences with backbone “stabilizing” residue configurations such as a single Pro18,19 or Pro-Pro.20 Second, we chose residues that would alter the charge distribution across the chain similar to those observed on the native knob A chain (ie, arginine and glutamic acid). Subsequent molecular modeling and dynamic simulations of each peptide facilitated structural comparisons of the peptide conformations in solution. In this report, we correlate molecular/structural properties of the knob peptide residues with functional kinetic binding parameters to gain a better understanding of knob characteristics that contribute to knob:hole interactions and identified potential criteria for the rational design of enhanced knob variants. Illustrating this point, we identified and report here a novel knob peptide mimic with a unique element (Pro-Phe-Pro) that enhances the association rate to polymerization holes nearly 1 order of magnitude.
Table 1.
Experimental knob A peptides and corresponding properties
| Peptide sequence | Property | Net charge |
|---|---|---|
| GPRVVERC | Mimics native sequence through seventh residue | +1 |
| GPRVVAAC | Mimics native sequence minus additional charged residues | +1 |
| GPRPAAC | Stabilized backbone | +1 |
| GPRPFPAC | Stabilized backbone | +1 |
| GPRPPERC | Stabilized backbone and additional charged residues | +1 |
| GPSPAAC | Negative control; known dysfibinrogen mutant | 0 |
Methods
Fibrin knob A peptides
Peptide sequences included GPRVVAAC, GPRVVERC, GPRPAAC, GPRPPERC, GPRPFPAC, and GPSPAAC (GenScript Inc; Table 1). The peptide sequences were designed with a carboxyl-terminal cysteine residue to permit sulfhydryl-targeted reactions for future conjugation chemistries.
Preparation of fibrinogen D fragment
Human fibrinogen (Enzyme Research Laboratories) at 2 mg/mL was digested with 0.1 U/mL human plasmin (Enzyme Research Laboratories) in HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) + CaCl2 buffer (150mM NaCl, 5mM CaCl2, 25mM HEPES; pH 7.4) overnight at room temperature. D fragment was isolated as previously described, with slight modifications.21 Briefly, the plasmin-digested fibrinogen and GPRPAA beads were incubated for 30 minutes, with occasional agitation. The unbound proteins and protein fragments were removed with excessive washing with HEPES + CaCl2 buffer. D fragment was eluted with 1M sodium bromide and 50mM sodium acetate (pH 5.3). Eluted samples were pooled together and exchanged back into HEPES + CaCl2 buffer with a centrifugal filter (molecular weight cutoff, 10 000 Da). D fragment was verified by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and stored at −80°C until use.
Binding kinetics with SPR
The Biacore 2000 (Biacore Lifesciences, GE Healthcare) was used investigate kinetic binding constants (ka and kd) of knob A peptide variants for fibrinogen D fragment. Briefly, D fragment was covalently immobilized to gold-coated SPR sensor chips via self-assembled monolayer surface chemistry to generate a nonfouling surface with a controlled density of reactive carboxylic acid groups. Mixed self-assembled monolayers were generated on gold-coated chips as described previously22 by incubating with a 1-mM mixture of tri(ethylene glycol)–terminated alkanethiols (HS-(CH2)11–(OCH2CH2)3–OH; ProChimia) and carboxylic acid–terminated alkanethiols (HS-(CH2)11–(OCH2CH2)6–OCH2COOH) for 4 hours. On loading the senor chip into the Biacore 2000, the carboxylic acid–terminated alkanethiol in all 4 flow cells was activated by flowing 200mM 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (Sigma-Aldrich) and 50mM N-hydroxysuccinimide (Sigma-Aldrich; 5 μL/minute for 10 minutes). Immediately after activation, D fragment was immobilized in 3 flow cells (5 μL/minute for 10 minutes) to achieve 1800 to 2000 resonance units (1 resonance unit ∼ 1 pg/mm2). Unreacted N-hydroxysuccinimide groups were quenched in all 4 flow cells (3 sample cells and 1 reference cell) with 20mM ethanolamine (10 μL/minute for 10 minutes). On stabilization of the baseline signal, kinetic binding experiments were run in duplicate with the peptide variants as the flow analytes. Five various concentrations for each peptide (0.94μM to 150μM) were flowed at 25 μL/minute for 4 minutes immediately followed by a 10-minute dissociation phase. Between each run, the surface was regenerated with 1M sodium bromide and 50mM sodium acetate (pH 6.0). SPR experiments were performed 3 times with varying peptide injection order to rule out binding trends associated with injection sequence. Peptide solutions were incubated with tris(2-carboxyethyl)phosphine immobilized on agarose beads (Thermo Fisher Scientific Inc) to ensure reduction of any disulfide bonds between C-terminal cysteines. Mass spectrometry analysis (Fast Atom Bombardment) of peptide solutions showed that the peptides did not dimerize over the course of the SPR experiment (supplemental Figure 4, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
SPR analysis and evaluation
SPR sensorgrams were analyzed with the aid of Scrubber 2 and ClampXP software (Center for Biomolecular Interactions Analysis, University of Utah).23–25 Before analysis, all sensorgrams were inspected for abnormalities (ie, baseline drift, air spikes, or irregular deviations) and excluded. Reference cell responses were subtracted from corresponding active response curves. Double-referenced curves were acquired by further subtracting the reference cell blank buffer injections from each reference-subtracted response curve.26 All double-referenced curves were normalized by the molecular weight of each peptide and multiplied by 1000 to account for minor variations in response because of molecular weight. The resulting curves were then analyzed and fitted to the kinetic models. Kinetic modeling and simulations were performed with ClampXP software with the Langmuir 1:1 model or the heterogeneous ligand model; globally fitted parameters were determined for each kinetic dataset per peptide. Equilibrium binding constants were calculated from fitted kinetic constants. Goodness of fit for each model was determined by evaluating the residual plots and residual sum of squares.26
MD simulations
Classical MD simulations were performed with 5 knob A peptides, GPRVVAAC, GPRVVERC, GPRPAAC, GPRPPERC, and GPRPFPAC. Because the crystal structure of each of these peptides within the fibrin hole has not been determined experimentally, the initial peptide structures were rendered in Swiss-PDB Viewer (Swiss Institute of Bioinformatics27) with the backbone torsion angles of the first 3 residues constrained to an “active” peptide conformation obtained from previously published D fragment crystal structures (PDB code: 2HPC and 2FFD14). Before MD simulations, the structure of each peptide was minimized with 10 iterations of steepest descent (500 steps) energy minimization in vacuo. Each peptide was placed in the center of a water box (Visual Molecular Dynamics software28) supplemented with Na+ and Cl− ions to achieve electric neutrality, mimicking experimental conditions (∼340 mOsmol/L). The models were initially minimized for 1000 steps with the backbone atoms fixed, followed by 1000 steps of minimization with harmonic restraints on the α carbon atoms. After energy minimization, each system was heated to 310K over a period of 20 picoseconds with harmonic restraints on the α carbons. Next, with the restraints still active, each system was equilibrated at constant temperature (310°K) and pressure (1 atm) for 100 picoseconds. The restraints were then removed, and the equilibration was continued for 200 picoseconds. The production runs were carried out for 10 nanoseconds under constant temperature and pressure conditions, ie the NPT ensemble. Temperature was maintained at 310K, pressure at 1 atm. Short-range nonbonded interactions were cut off at a distance of 1.2 nm (12 Å) with a switching function between 1.0 and 1.2 nm (10 and 12 Å). The particle mesh Ewald method was used to compute electrostatics.29 All bonds involving hydrogen atoms were constrained with the use of the SHAKE algorithm,30 which allowed for an integration time step of 2 femtoseconds. All simulations were performed with NAMD Version 2.6 (Theoretical and Computational Biophysics Group at University of Illinois at Urbana-Champaign31) with the use of the CHARMM22 force field parameter set.32
MD simulation analysis
Clustering analysis.
A hierarchical cluster analysis was performed on the trajectory data from each peptide MD simulation. A trajectory for clustering was obtained by taking every 100th frame (100-picosecond interval) from a 10-nanosecond production run. Next, we calculated the root mean squared deviation (RMSD) between every frame in the trajectory to generate a dissimilarity matrix. RMSD was calculated on the basis of the backbone atoms after optimal superposition. The agnes function in the cluster package supplied with the R statistical software package (R Project) was used to construct a hierarchy of clusterings from the dissimilarity matrix. The clusters were visualized in VMD with the use of the Cluster plugin. On the basis of the resulting dendrograms generated from the cluster analysis (supplemental Figure 3), representative trajectory conformations from the 2 most populated cluster groupings at the third level were used to compare both conformational and electrostatic properties between each peptide.
Structure/conformation and electrostatic properties comparisons.
For structural/conformational comparison, representative conformations from the top 2 populated clusters were superimposed onto either GPRP or GPRV peptides in an active conformation within hole a as obtained from previous published D fragment crystal structures (PDB code: 2HPC and 2FFD,14 respectively); GPRPxxx peptides were compared with active GPRP and GPRVxxx peptides were compared with active GPRV. After least-squares superpositioning the first 3 residues of each conformation along the backbone, the RMSD of the first 3 residues was calculated with the active GPRP or GPRV as the reference; both the backbone and total atom RMSDs were calculated. For electrostatic comparisons, electrostatic potential surface maps for representative conformations from the top populated cluster group for each peptide were generated with Adaptive Poisson-Boltzmann Solver with the use of the CHARMM22 force field parameter set.33
Results
Kinetic binding models
To investigate the dynamic binding profile between the fibrinogen/fibrin holes and knob peptide variants, we used SPR. Binding interactions were evaluated by flowing the knob peptides over an immobilized surface of D fragment (Figure 1). By immobilizing D fragment as opposed to full-length fibrinogen, we simplified the kinetic binding model to a heterogeneous 2-site ligand model (ie, 1 hole a and hole b per ligand) as opposed to a 4-site model (ie, 2 of each hole a and hole b per ligand). We modeled the data with the use of both a Langmuir 1:1 model and a heterogeneous ligand model to compare previously established binding affinities to a more dynamic 2-site model. However, the complexity of the heterogeneous model fitted parameters for sites 1 and 2 (ie, maximal binding response, ka, and kd) limits direct designation or assignment of holes a or b to site 1 or 2. An additional mass transport limited model was tested as well (data not shown), but it did not fit the experimental data for any of the peptides. All peptide SPR data were fit except for the negative control peptide, GPSPAAC, in which minimal binding response was observed.
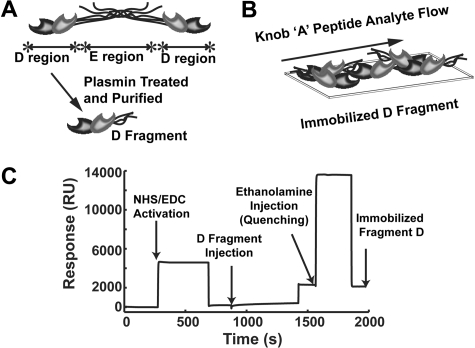
Figure 1.
SPR experimental protocol. (A) Schematic representation of fibrinogen and the 2 major regions, E and D. Plasmin treated fibrinogen, and purification of the fragments generates D fragment. (B) Surface plasmon resonance (SPR) experimental set-up with D fragment immobilized to an SPR chip acting as the ligand and the knob A peptides flow across the surface as the analyte. (C) Representative SPR sensorgram for D fragment immobilization where the carboxyl-terminated self-assembled monolayers were activated by 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC)/N-hydroxysuccinimide (NHS), enabling amine-targeted immobilization of D fragment. Ethanolamine quenched any unreacted carboxyl groups and rid the surface of nonspecifically bound D fragment.
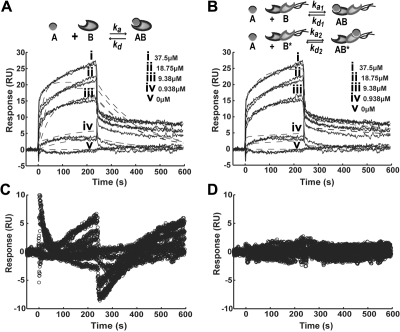
In comparing model simulation results, the heterogeneous ligand model fit the experimental binding data far better than the Langmuir 1:1 model. Here, we present response, simulation curves, and residual plots for a single set of experimental data (GPRPFPAC; Figure 2); plots for all 5 peptides are provided in supplemental Figures 1 and 2. Looking specifically at the residual sum of squares, the range for the 1:1 Langmuir model (1.497-2.197; Table 2) was higher than the heterogeneous ligand model (0.9437-1.474; Table 3), suggesting that the fitted heterogeneous ligand model deviated less from the experimental data. In addition, graphically plotting the residuals over time showed that the residuals for 1:1 Langmuir model followed a systematic trend (Figure 2C), indicative of fitting an inappropriate model to the experimental data.34 In contrast, the residuals for the heterogeneous model were lower and more randomly distributed (Figure 2D), indicating that this model adequately describes the binding response curves.23,34 On the basis of these analyses and observations, further comparisons of binding parameters were performed with the results from the heterogeneous ligand model.
Figure 2.
Kinetic model comparison. Experimental sensorgram of GPRPFPAC fitted with (A) Langmuir 1:1 model or (B) heterogeneous ligand model. Corresponding residuals plots for (C) Langmuir model or (D) heterogeneous ligand model. Solid lines indicate experimental SPR response curves; dashed lines, fitted model curves.
Table 2.
Langmuir 1:1 model, fitted parameters
| Parameter | GPRPAAC | GPRPFPAC | GPRPPERC | GPRVVERC | GPRVVAAC |
|---|---|---|---|---|---|
| Bmax (RU) | 21.47 ± 0.19* | 17.74 ± 0.08 | 19.20 ± 0.21 | 16.27 ± 0.08 | 18.88 ± 0.18 |
| ka (M−1s−1) × 10−3 | 1.14 ± 0.03 | 12.5 ± 0.26 | 1.29 ± 0.03 | 0.28 ± 0.004 | 0.09 ± 0.001 |
| kd (s−1) × 103 | 11.71 ± 0.17 | 7.27 ± 0.08 | 9.92 ± 0.13 | 23.43 ± 0.23 | 10.73 ± 0.04 |
| RSS | 2.197 | 2.092 | 1.885 | 1.521 | 1.497 |
Bmax indicates maximal binding capacity of D fragment; RU, resonance unit; ka, association rate; kd, dissociation rate; RSS, residual sum of squares.
Mean ± SD (all such values).
Table 3.
Heterogeneous ligand model, fitted parameters
| Parameter | GPRPAAC | GPRPFPAC | GPRPPERC | GPRVVERC | GPRVVAAC |
|---|---|---|---|---|---|
| Bmax (RU) | 19.38 ± 0.23* | 17.59 ± 0.13 | 14.58 ± 0.23 | 25.61 ± 0.28 | 8.36 ± 0.24 |
| ka1 (M−1s−1) × 10−3 | 2.84 ± 0.14 | 21.72 ± 0.73 | 3.22 ± 0.12 | 1.07 ± 0.03 | 0.62 ± 0.04 |
| kd1 (s−1) × 103 | 12.83 ± 0.49 | 81.10 ± 2.23 | 58.73 ± 1.86 | 57.67 ± 2.24 | 30.08 ± 1.51 |
| KD1 (μM)† | 4.53 | 3.73 | 18.23 | 53.89 | 48.51 |
| B*max (RU) | 5.91 ± 0.08 | 11.34 ± 0.08 | 5.92 ± 0.13 | 10.33 ± 0.23 | 13.52 ± 0.32 |
| ka2 (M−1s−1) × 10−3 | 1.05 ± 0.09 | 1.81 ± 0.04 | 1.01 ± 0.03 | 0.26 ± 0.01 | 0.04 ± 0.001 |
| kd2 (s−1) × 103 | 8.95 ± 1.05 | 1.96 ± 0.04 | 2.96 ± 0.09 | 4.07 ± 0.06 | 1.00 ± 0.07 |
| KD2 (μM)† | 8.52 | 1.08 | 2.93 | 15.71 | 25.00 |
| RSS | 1.474 | 0.9887 | 0.9437 | 0.9817 | 1.266 |
Bmax indicates maximal binding capacity of D fragment; RU, resonance unit; ka, association rate; kd, dissociation rate; RSS, residual sum of squares.
Mean ± SD (all such values).
Calculated KD from fitted ka and kd values in which KD = kd/ka.
Fitted binding affinity parameters
The fitted parameters (Bmax, ka, and kd) for each knob peptide variant for the heterogeneous ligand model are displayed in Table 3. In addition, the sensorgram plots in Figure 3A show the contribution each fitted parameter set has on the overall combined 2-site model. For example, for GPRPFPAC, the fit for the AB complex (site 1) encompassed a fast association rate, presumably accounting for the large initial response, whereas the slower association rate for the AB* complex (site 2) contributes to the slower response for the duration of the injection. Broadly comparing all the knob peptide variant parameters, the 4Pro peptides (ie, GPRPAAC, GPRPFPAC, and GPRPPERC) had much faster association rates (ka1 = 2.84-21.72 × 103M−1s−1; ka2 = 1.01-1.81 × 103M−1s−1) than the 4Val peptides (ie, GPRVVAAC and GPRVVERC; ka1 = 0.62-1.07 × 103M−1s−1; ka2 = 0.04-0.26 ×103M−1s−1).
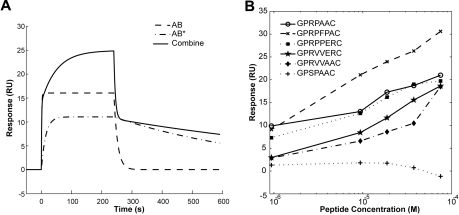
Figure 3.
Contribution of AB and AB* binding in 2-site model and Maximal binding response. (A) Simulation sensorgrams generated by the heterogeneous ligand model for GPRPFPAC. The combine response is a sum of analyte-ligand complexes AB and AB*. (B) Maximal binding response (resonance unit; RU) for the corresponding peptide concentration (molar, M). GPSPAAC, GPRPAAC, GPRPFPAC, GPRPPERC, GPRVVERC, and GPRVVAAC.
In comparing the 4Pro variants, one of the most striking differences was the nearly 10-fold increase in ka1 for GPRPFPAC (21.72 × 103M−1s−1) compared with GPRPAAC (2.84 × 103M−1s−1) and GPRPPERC (3.22 × 103M−1s−1); however, for kd1, GPRPAAC (12.83 × 10−3 s−1) displayed a 6-fold slower rate compared with GPRPFPAC (81.10 × 10−3 s−1). In contrast, for the second binding site the ka2 rate for GPRPFPAC (1.81 × 103 M−1s−1) was only moderately faster than GPRPAAC (1.05 × 103 M−1s−1) and GPRPPERC (1.01 × 103 M−1s−1), whereas the kd2 for GPRPFPAC (1.96 × 10−3 s−1) was nearly 8-fold slower than GPRPAAC (8.95 × 10−3 s−1). These simulation results indicate that GPRPFPAC has a higher affinity to the first and second binding sites and additionally dissociates more slowly from the second binding site, thus translating to longer engagement in fibrinogen holes compared with the other variants tested.
We also investigated the effect additional charged residues in the sixth and seventh position had on functional binding characteristics by comparing GPRVVAAC and GPRVVERC. For the association rates, GPRVVERC had a 2-fold increase over GPRVVAAC in ka1 (1.07 × 103M−1s−1 vs 0.62 × 103M−1s−1, respectively) and a 6-fold increase in ka2 (0.26 × 103M−1s−1 vs 0.04 × 103M−1s−1, respectively). However, the dissociation rates for GPRVVERC were 2-fold faster than GPRVVAAC for kd1 (57.67 × 10−3 s−1 vs 30.08 × 10−3 s−1, respectively) and 4-fold faster for kd2 (4.07 × 10−3 s−1 vs 1.00 × 10−3 s−1, respectively). These results collectively imply that, although the additional charged residues (ie, 6Glu and 7Arg) may enhance the affinity of the knob peptide to the binding holes, it may also result in an increased rate of dissociation.
Equilibrium dissociation constants
Using the fitted kas and kds from the kinetic models, we calculated the equilibrium dissociation constants (KDs; Table 3). These results are also represented graphically by plotting the SPR binding maximum for each variant versus injection concentration (Figure 3B). The lowest KDs were observed in the peptide variants with a 4Pro (GPRPFPAC < GPRPAAC < GPRPPERC < GPRVVERC <GPRVVAAC). In comparing the specific KDs for each site between the 4Pro variants, KD1 for GPRPFPAC (3.73μM) and GPRPAAC (4.53μM) was significantly lower than GPRPPERC (18.23μM). However, for the second site, the KD2 values for GPRPFPAC (1.08μM) and GPRPPERC (2.93μM) were significantly lower than GPRPAAC (8.52μM). This result further indicates that GPRPFPAC interacts and engages the hole domains more readily than the other variants tested, even the gold standard GPRP mimic (GPRPAAC). In comparing GPRVVAAC with GPRVVERC, the addition of charged residues, 6Glu and 7Arg, decreased KD2 from 25.00μM to 15.71μM, whereas KD1 was relatively similar.
MD simulation analysis
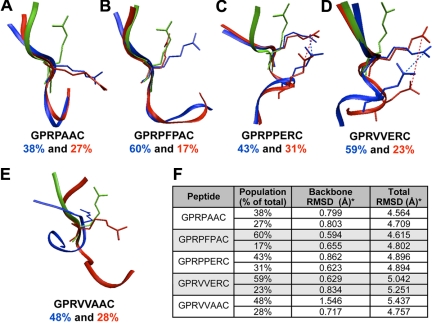
MD simulations were performed to compare conformational structures of the peptides immersed in an aqueous environment before engagement with fibrin holes. Because of the large amount of data (ie, conformations) in a 10-nanosecond MD trajectory, a hierarchical clustering method was used to select a smaller set of conformations representative of the entire MD trajectory (supplemental Figure 3). From this cluster analysis, representative conformations from the 2 most populated clusters for each peptide were used to compare structural features between experimental peptides. The representative conformations for each peptide were then superimposed on either active GPRP or GPRV peptides obtained from D fragment crystal structures (PDB code: 2HPC and 2FFD,14 respectively; Figure 4). The superposition of simulation conformations with active conformation was evaluated by calculating the RMSD for atoms (backbone and all atoms) from the first 3 residues of the simulation conformations in reference to the known active conformations. The lower the RMSD, the better the alignment to the reference point. Ranking the peptides from lowest to highest weighted average RMSD along the backbone was GPRPFPAC (0.0607 nm [0.607 Å]) less than GPRVVERC (0.0670 nm [0.670 Å]) less than GPRPPERC (0.0761 nm [0.761 Å]) less than GPRPAAC (0.0801 nm [0.801 Å]) less than GPRVVAAC (0.1241 nm [1.241 Å]). It appears that the Pro-Phe-Pro residues in GPRPFPAC help stabilize the backbone of the first 3 residues to a conformation similar to active GPRP. Similarly, although GPRVVERC and GPRPPERC were chosen to investigate the alterations in electrostatic charge, a salt bridge developed between the 3Arg and 6Glu side chains potentially stabilizing the backbone. We also calculated the RMSD of all the atoms (ie, backbone and side chain atoms) in the first 3 residues; here, the weighted average RMSD ranking from lowest to highest was GPRPFPAC (0.4656 nm [4.656 Å]) less than GPRPAAC (0.4657 nm [4.657 Å]) less than GPRPPERC (0.4895 nm [4.895 Å]) less than GPRVVERC (0.5101 nm [5.101 Å]) less than GPRVVAAC (0.5186 nm [5.186 Å]). This RMSD ranking inversely correlated with the experimental binding affinity data (ie, lower RMSD, higher binding affinity). Considering this correlation, we evaluated the orientation of the side chain groups in comparison to the active conformation, particularly the orientation of 3Arg, which is required for binding of fibrin holes. Conventional terminology for torsional side chain angle defines χ1 as the angle between Ni-Cαi-Cβi-Cγi35; the 3 common rotamer classifications are gauche− (0° to 120°), trans (120° to 240°), and gauche+ (−120° to 0°).36 In the active conformation for both GPRP and GPRV, the 3Arg χ1 angle is in a gauche+ conformation. However, in assessing the χ1 angle of the 3Arg side chain for GPRPAAC and GPRVVAAC, we noted the angle was predominantly in a trans conformation during the simulation (ie, pointing toward the carboxyl-terminus of the sequence; Figure 4A,E). In contrast, the 3Arg group in GPRPFPAC was mobile, but predominantly in the gauche+ conformation and rarely the trans conformation (Figure 4B). We speculate that the bulky side chain on 5Phe sterically hinders electrostatic interactions between 3Arg and the C-terminus as observed in GPRVVAAC and GPRPAAC. In addition, the salt bridge formation between 3Arg and 6Glu in GPRVVERC and GPRPPERC appeared to stabilize the 3Arg side chain in the gauche+ conformation (Figure 4C-D). Collectively, these observations suggest that the 3Arg rotameric classifications depended on properties of the downstream residues.
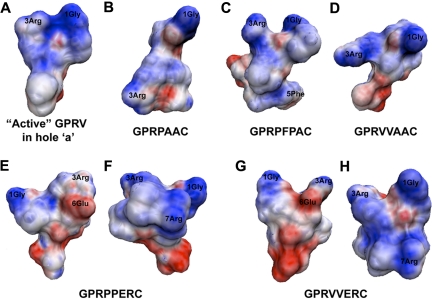
Figure 4.
Structural analysis. Representative trajectories of the top 2 (blue and red) most populated groups from hierarchical cluster analysis superimposed on the active GPRP or GPRV conformation (green); GPRPxxx peptides were compared with active GPRP and GPRVxxx peptide were compared with active GPRV. (A) GPRPAAC, (B) GPRPFPAC, (C) GPRPPERC, (D) GPRVVERC, and (E) GPRVVAAC. (F) The population percentage represents the percentage of total number trajectories in 1 conformational cluster; the top 2 populated clusters at the fourth level of the hierarchical cluster are reported. RMSD calculations for the first 3 residues were in reference to the active conformation (ie, GPRP or GPRV); both backbone and total RMSDs were calculated after optimal superposition along the backbone.
As previously mentioned, knob:hole interactions are driven by electrostatic interactions. Therefore, we generated electrostatic potential surface maps to display the charge distributions for each peptide variant (Figure 5). The active conformation of GPRV has a noticeably positively charged N-terminus generated by 1Gly and 3Arg (Figure 5A; GPRP was not shown, but it has similar structure/map properties). For the peptide variants, we noted that the electrostatic mapping was directly related to the 3Arg side chain rotamer classification, in which the gauche+ 3Arg conformations maintained the concentrated positive charge around 1Gly and 3Arg (ie, GPRPFPAC; Figure 5C). However, with 3Arg in the trans conformation (ie, GPRPAAC and GPRVVAAC), the positive charge was more broadly distributed from the N-terminus across to the C-terminus (Figure 5B,D). The addition of −ERC in the sixth through eighth residues resulted in 2 alterations (Figure 5E-H). First, although the salt bridge between 3Arg and 6Glu stabilized 3Arg in the gauche+ conformation providing a concentrated positive charge at the N-terminus, the presence of 6Glu contributed a slightly negative charge near 3Arg (Figure 5E,G). Second, the additional Arg residue in the seventh position redistributed the positive charge more broadly across the peptide chain (Figure 5F,H). Collectively, these MD simulations and subsequent analyses provided snapshots at the molecular level into potential intrachain interactions that occur in aqueous environments and may contribute to the initial binding interactions with fibrin holes.
Figure 5.
Electrostatic potential surface maps. Electrostatic potential surface maps were for the representative trajectory from most occupied cluster grouping. (A) GPRV in the active conformation, (B) GPRPAAC, (C) GPRPFPAC, (D) GPRVVAAC, (E) GPRPPERC, (F) GPRPPERC rotated 180° about the vertical axis, (G) GPRVVERC, and (H) GPRVVERC rotated 180° about the vertical axis.
Discussion
In an effort to both describe, for the first time, fibrin knob structure in solution (ie, before complimentary hole engagement) and explore potential factors that affect the initial docking/binding of fibrin knobs to holes, we modeled the binding kinetics of fibrin knob peptide:hole interactions and investigated structural properties of the peptide variants in aqueous environments. Results from this study provide significant insights into the structural dynamics of knob sequences and the role of structure in defining the dynamics of knob:hole interactions that govern fibrin assembly and fiber structure. Furthermore, these studies enabled the discovery of a novel knob mimic that displays an association rate to fibrin polymerization holes an order of magnitude higher than any previously published knob sequence.
Three decades have passed since Laudano and Doolittle7 first reported that the short tripeptide (GPR) derived from knob A binds to fibrinogen. By further extending the peptide sequence to 4 residues, Doolittle noted that the synthetic tetrapeptide GPRP (20μM) had a higher affinity to fibrinogen compared with the human knob A mimic GPRV (75μM) and that both knob A variants bound to more than 3 sites on fibrinogen.6–8 Recent x-ray crystallographic studies of D fragment in the presence of either GPRP or GPRVVE clearly verified that both knob A mimics were capable of occupying both holes a and b.14 These studies also established the remarkably high degree of structural similarity between GPRP and GPRV in the engaged position, but they were unable to elucidate why GPRP displays significantly higher affinity for fibrin holes. Previous reports have speculated that the enhanced binding affinity of GPRP over GPRV may be due to restrictions the 4Pro imparts on the peptide backbone,6 yet this has been an unverified theory until now. We addressed this critical gap by characterizing the binding kinetics of a set of knob A variants designed to specifically investigate the effect additional backbone stabilizing residues (ie, Pro and Phe) and/or electrostatic charged residues (ie, 6Arg and 7Glu mimicking the native human knob A sequence)37 have on the binding dynamics of knob:hole interactions. We were able to capture the more complex and dynamic interactions within each hole using a heterogeneous ligand model, a model that correlates with Doolittle's initial findings that A:b interactions, particularly knob A peptide:hole b interactions can and do occur.8,14 Furthermore, we discovered a novel peptide, GPRPFPAC, with the highest reported affinity to the hole domains, even surpassing the binding activity of the gold standard GPRP mimic (GPRPAAC).
Recent SPR studies have investigated the interaction between adsorbed fibrinogen and the N-terminal disulfide knot (NDSK) of differentially activated fibrin (FpA and FpB removed = desAB-NDSK; only FpA removed = desA-NDSK).38 The investigators reported KDs of 5.8μM and 3.7μM for desA-NDSK and desAB-NDSK, respectively. The slightly higher affinity of desAB-NDSK was attributed to B:b interactions because coinjection of knob B peptides with desA-NDSK or desAB-NDSK hindered only the desAB-NDSK interaction with fibrinogen.38 Even though this elegant study established the presence of B:b binding interactions, these SPR experiments were performed under equilibrium conditions (ie, low flow rates) and determined only a single equilibrium binding constant.26 Nonetheless, dynamic off-rates of desA-NDSK (8.6 × 10−4 s−1) and desAB-NDSK (1.35 × 10−3 s−1) from fibrinogen have been calculated from bond-strength measurements recorded with laser tweezers–based force spectroscopy.12 Surprisingly, these off-rates are similar to the kds measured for the knob peptides in the present study (∼ 10−3 s−1). Although there are inherent differences between the experimental parameters of SPR and laser tweezers–based force spectroscopy, the agreement of dynamic rates may provide further evidence that the knob:hole interaction is predominately mediated by the first few residues on the knob N-termini.
Performing MD simulations of the peptides in an aqueous environment lent substantial insight into potential peptide structural conformations and intrachain interactions that may influence binding properties. We acknowledge, however, the basic limitations of MD simulations: conformational sampling and the energy function. Moreover, we used theoretical models as starting structures, based on the published active conformations of GPRP and GPRV, because the structures for the peptide variants used in this study cannot be determined experimentally. Additional modeling would need to be performed to fully address these concerns. However, on the basis of our simulations, we noticed 2 main characteristics that correlated with functional binding affinities, first, the orientation of the 3Arg side chain, and second, backbone stability. For the superior binding peptide GPRPFPAC, the 3Arg side chain χ1 angle was maintained in the gauche+ rotameric conformation, and the weighted average of the backbone RMSD from the active conformation was the lowest. Meanwhile, the 3Arg group in the −ERC peptides (GPRPPERC and GPRVVERC) was stabilized in the gauche+ conformation by a salt bridge ionic interaction between 3Arg and 6Glu, thereby also stabilizing the backbone. However, the salt bridge may initially restrict 3Arg from interacting with residues on the hole domains. Surprisingly, the 3Arg side chain for GPRPAAC, a high-affinity peptide, was predominately observed in the trans conformation; a similar 3Arg rotameric conformation was observed for GPRVVAAC. Notably, the peptides used in the experimental and subsequent MD simulations were not amidated so as to facilitate future conjugation chemistries. In doing so, a negative charge was generated at the carboxyl-terminus, allowing presumably weak ionic interactions between a nonrestricted positively charged Arg side chain and the C-terminus as observed with GPRPAAC and GPRVVAAC. Despite this, the RMSD of the backbone for GPRPAAC was less than GPRVVAAC, indicating that the 4Pro residue probably stabilizes the peptide backbone. The resulting electrostatic potential was then subsequently influenced by the structural conformations of the side chains. This modeling analysis ultimately suggested that the conformation of knob peptides within an aqueous environment before contact with a hole domain contributes to the propensity of binding that occurs. This theory may translate to the N-terminal knobs on the more complex native fibrin monomer; however, significant additional experimentation would be required.
Using molecular dynamic simulations coupled with experimental SPR, we report for the first time, to our knowledge, fundamental knob structural determinants that drive knob:hole binding dynamics and provide potential knob design criteria. Exemplifying these criteria, we report a novel knob mimic with enhanced affinity. Collectively, we believe our studies provide additional insights for developing higher affinity peptides that bind and disrupt the native knob:hole interaction more effectively than previously reported knob peptides.
Supplementary Material
Acknowledgments
We thank Dr Shuming Nie for access to and assistance with the Biacore 2000. We also thank Dr Steven Harvey for the intellectual discussions and insights on molecular modeling and dynamics. The MD work was performed with the use of the computational resources of the Interactive High Performance Computing Laboratory at Georgia Institute of Technology.
This work was supported by the W. H. Coulter Foundation (GTF125000120) and the National Institutes of Health (1R21EB008463; T.H.B.) and by the NIH FIRST Post-Doctoral Fellowship (K12 GM000680; S.E.S.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.E.S, J.J.G., and T.H.B designed research, analyzed and interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thomas H. Barker, W. H. Coulter Department of Biomedical Engineering, Georgia Institute of Technology/Emory University, 313 Ferst Dr, Atlanta, GA 30332; e-mail: thomas.barker@bme.gatech.edu.
References
- 1.Bettelheim FR, Bailey K. The products of the action of thrombin on fibrinogen. Biochim Biophys Acta. 1952;9(5):578–579. doi: 10.1016/0006-3002(52)90213-8. [DOI] [PubMed] [Google Scholar]
- 2.Bailey K, Bettelheim FR, Lorand L, Middlebrook WR. Action of thrombin in the clotting of fibrinogen. Nature. 1951;167(4241):233–234. doi: 10.1038/167233a0. [DOI] [PubMed] [Google Scholar]
- 3.Lorand L, Middlebrook WR. Studies on fibrino-peptide. Biochim Biophys Acta. 1952;9(5):581–582. doi: 10.1016/0006-3002(52)90215-1. [DOI] [PubMed] [Google Scholar]
- 4.Lorand L, Middlebrook WR. The action of thrombin on fibrinogen. Biochem J. 1952;52(2):196–199. doi: 10.1042/bj0520196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laurent TC, Blomback B. On the significance of the release of 2 different peptides from fibrinogen during clotting. Acta Chemica Scand. 1958;12(9):1875–1877. [Google Scholar]
- 6.Laudano AP, Doolittle RF. Studies on synthetic peptides that bind to fibrinogen and prevent fibrin polymerization. Structural requirements, number of binding sites, and species differences. Biochemistry. 1980;19(5):1013–1019. doi: 10.1021/bi00546a028. [DOI] [PubMed] [Google Scholar]
- 7.Laudano AP, Doolittle RF. Synthetic peptide derivatives that bind to fibrinogen and prevent the polymerization of fibrin monomers. Proc Natl Acad Sci U S A. 1978;75(7):3085–3089. doi: 10.1073/pnas.75.7.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laudano AP, Doolittle RF. Influence of calcium ion on the binding of fibrin amino terminal peptides to fibrinogen. Science. 1981;212(4493):457–459. doi: 10.1126/science.7209542. [DOI] [PubMed] [Google Scholar]
- 9.Doolittle RF, Chen A, Pandi L. Differences in binding specificity for the homologous gamma- and beta-chain “holes” on fibrinogen: exclusive binding of Ala-His-Arg-Pro-amide by the beta-chain hole. Biochemistry. 2006;45(47):13962–13969. doi: 10.1021/bi061219e. [DOI] [PubMed] [Google Scholar]
- 10.Doolittle RF, Pandi L. Probing the beta-chain hole of fibrinogen with synthetic peptides that differ at their amino termini. Biochemistry. 2007;46(35):10033–10038. doi: 10.1021/bi7010916. [DOI] [PubMed] [Google Scholar]
- 11.Litvinov RI, Gorkun OV, Galanakis DK, et al. Polymerization of fibrin: direct observation and quantification of individual B:b knob-hole interactions. Blood. 2007;109(1):130–138. doi: 10.1182/blood-2006-07-033910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Litvinov RI, Gorkun OV, Owen SF, Shuman H, Weisel JW. Polymerization of fibrin: specificity, strength, and stability of knob-hole interactions studied at the single-molecule level. Blood. 2005;106(9):2944–2951. doi: 10.1182/blood-2005-05-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spraggon G, Everse SJ, Doolittle RF. Crystal structures of fragment D from human fibrinogen and its crosslinked counterpart from fibrin. Nature. 1997;389(6650):455–462. doi: 10.1038/38947. [DOI] [PubMed] [Google Scholar]
- 14.Betts L, Merenbloom BK, Lord ST. The structure of fibrinogen fragment D with the ‘A’ knob peptide GPRVVE. J Thromb Haemost. 2006;4(5):1139–1141. doi: 10.1111/j.1538-7836.2006.01902.x. [DOI] [PubMed] [Google Scholar]
- 15.Bowley SR, Merenbloom BK, Okumura N, et al. Polymerization-defective fibrinogen variant gammaD364A binds knob “A” peptide mimic. Biochemistry. 2008;47(33):8607–8613. doi: 10.1021/bi8000769. [DOI] [PubMed] [Google Scholar]
- 16.Lexa KW, Alser KA, Salisburg AM, et al. The search for low energy conformational families of small peptides: searching for active conformations of small peptides in the absence of a known receptor. Int J Quantum Chem. 2007;107(15):3001–3012. [Google Scholar]
- 17.Ho BK, Dill KA. Folding very short peptides using molecular dynamics. Plos Comput Biol. 2006;2(4):228–237. doi: 10.1371/journal.pcbi.0020027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthews BW, Nicholson H, Becktel WJ. Enhanced protein thermostability from site-directed mutations that decrease the entropy of unfolding. Proc Natl Acad Sci U S A. 1987;84(19):6663–6667. doi: 10.1073/pnas.84.19.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schimmel PR, Flory PJ. Conformational energies and configurational statistics of copolypeptides containing L-proline. J Mol Biol. 1968;34(1):105–120. doi: 10.1016/0022-2836(68)90237-4. [DOI] [PubMed] [Google Scholar]
- 20.Vanhoof G, Goossens F, Demeester I, Hendriks D, Scharpe S. Proline motifs in peptides and their biological processing. FASEB J. 1995;9(9):736–744. [PubMed] [Google Scholar]
- 21.Kostelansky MS, Betts L, Gorkun OV, Lord ST. 2.8 angstrom crystal structures of recombinant fibrinogen fragment D with and without two peptide ligands: GHRP binding to the “b” site disrupts its nearby calcium-binding site. Biochemistry. 2002;41(40):12124–12132. doi: 10.1021/bi0261894. [DOI] [PubMed] [Google Scholar]
- 22.Petrie TA, Capadona JR, Reyes CD, Garcia AJ. Integrin specificity and enhanced cellular activities associated with surfaces presenting a recombinant fibronectin fragment compared to RGD supports. Biomaterials. 2006;27(31):5459–5470. doi: 10.1016/j.biomaterials.2006.06.027. [DOI] [PubMed] [Google Scholar]
- 23.Morton TA, Myszka DG. Kinetic analysis of macromolecular interactions using surface plasmon resonance biosensors. Methods Enzymol. 1998;295:268–294. doi: 10.1016/s0076-6879(98)95044-3. [DOI] [PubMed] [Google Scholar]
- 24.Morton TA, Myszka DG, Chaiken IM. Interpreting complex binding kinetics from optical biosensors: a comparison of analysis by linearization, the integrated rate equation, and numerical integration. Anal Biochem. 1995;227(1):176–185. doi: 10.1006/abio.1995.1268. [DOI] [PubMed] [Google Scholar]
- 25.Myszka DG, Morton TA. CLAMP: a biosensor kinetic data analysis program. Trends Biochem Sci. 1998;23(4):149–150. doi: 10.1016/s0968-0004(98)01183-9. [DOI] [PubMed] [Google Scholar]
- 26.Myszka DG. Improving biosensor analysis. J Mol Recognit. 1999;12(5):279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 27.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 28.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 29.Darden T, York D, Pedersen L. Particle mesh Ewald - an N. log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98(12):10089–10092. [Google Scholar]
- 30.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical-integration of Cartesian equations of motion of a system with constraints - molecular-dynamics of N-alkanes. J Comput Phys. 1977;23(3):327–341. [Google Scholar]
- 31.Phillips JC, Braun R, Wang W, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacKerell AD, Bashford D, Bellott M, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 33.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cornish-Bowden A. Detection of errors of interpretation in experiments in enzyme kinetics. Methods. 2001;24(2):181–190. doi: 10.1006/meth.2001.1179. [DOI] [PubMed] [Google Scholar]
- 35.Chandrasekaran R, Ramachandran GN. Studies on the conformation of amino acids, XI: analysis of the observed side group conformation in proteins. Int J Protein Res. 1970;2(4):223–233. [PubMed] [Google Scholar]
- 36.Ponder JW, Richards FM. Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. J Mol Biol. 1987;193(4):775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- 37.Blomback B, Blomback M, Hessel B, Iwanaga S, Reuterby J. Primary structure of human fibrinogen and fibrin, 1: cleavage of fibrinogen with cyanogen bromide. Isolation and characterization of Nh2-terminal fragments of alpha-(a) chain. J Biol Chem. 1972;247(5):1496–1512. [PubMed] [Google Scholar]
- 38.Geer CB, Tripathy A, Schoenfisch MH, Lord ST, Gorkun OV. Role of ‘B-b’ knob-hole interactions in fibrin binding to adsorbed fibrinogen. J Thromb Haemost. 2007;5(12):2344–2351. doi: 10.1111/j.1538-7836.2007.02774.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.