Abstract
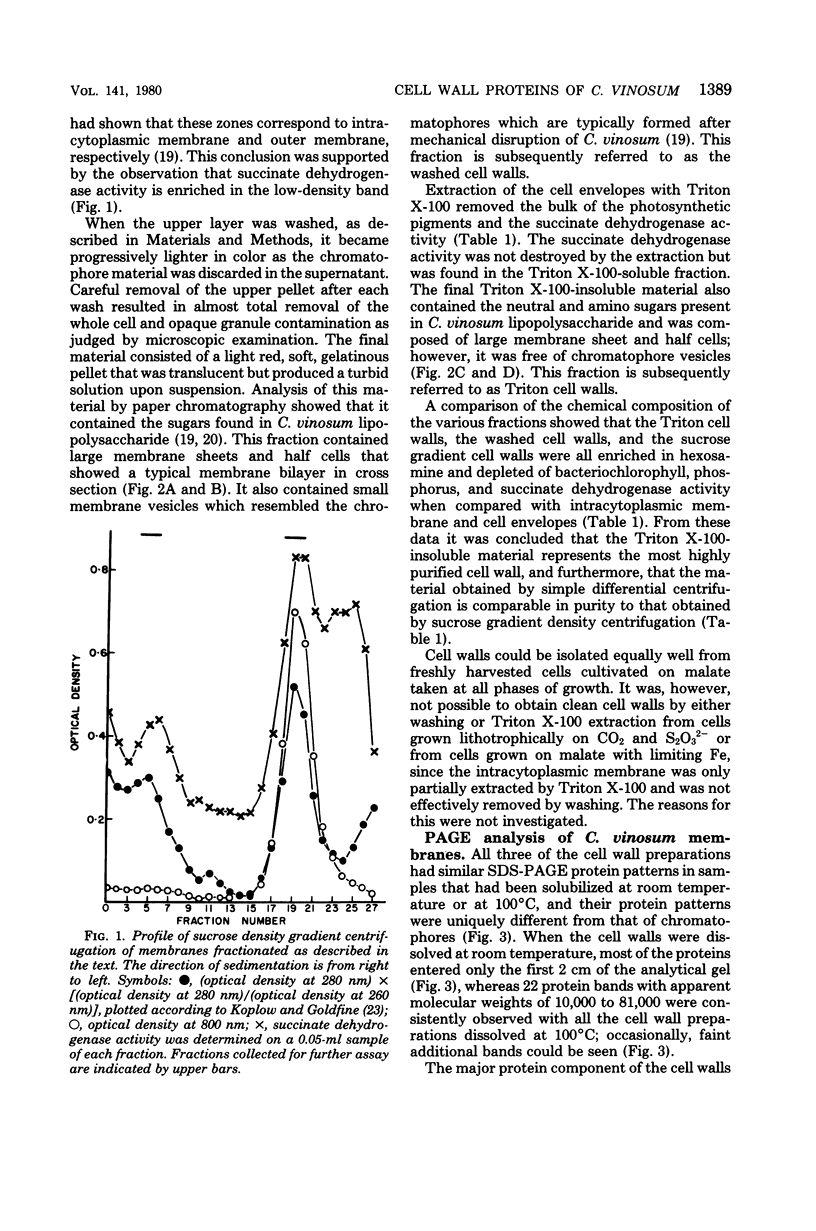
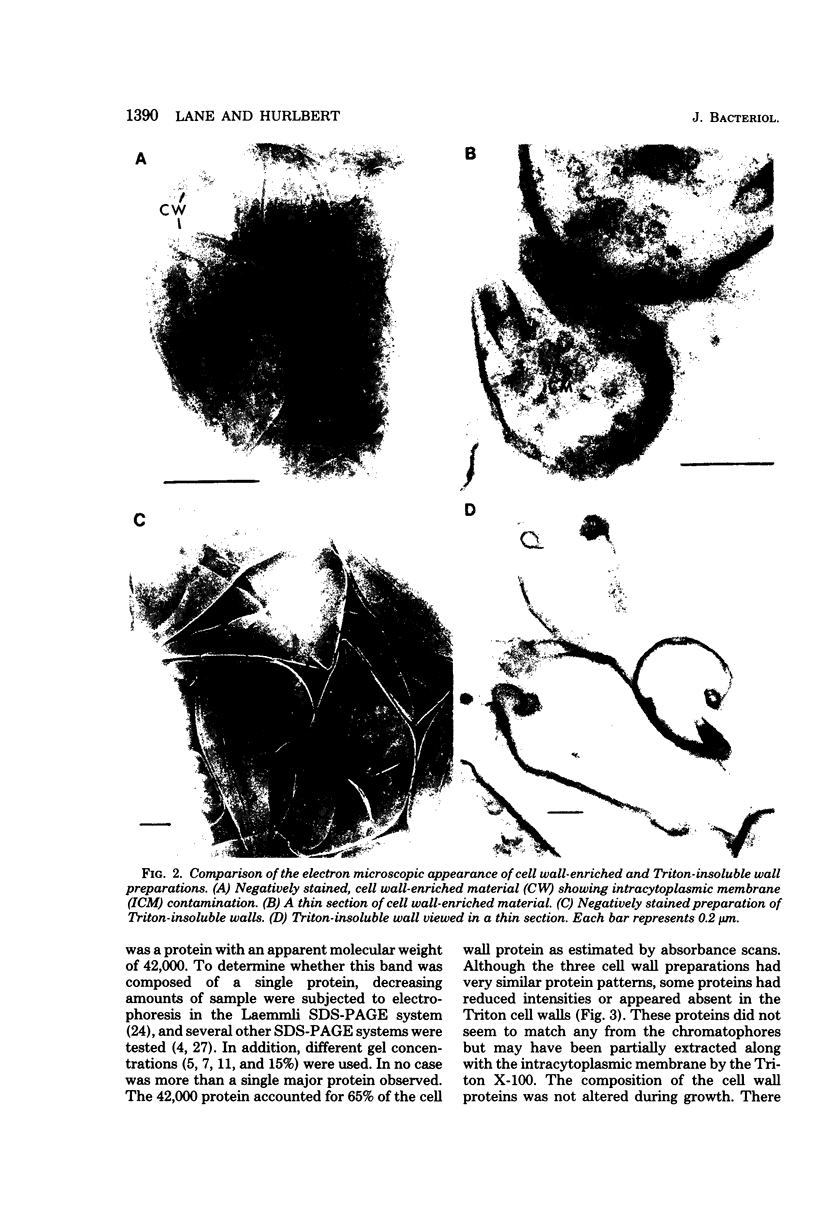
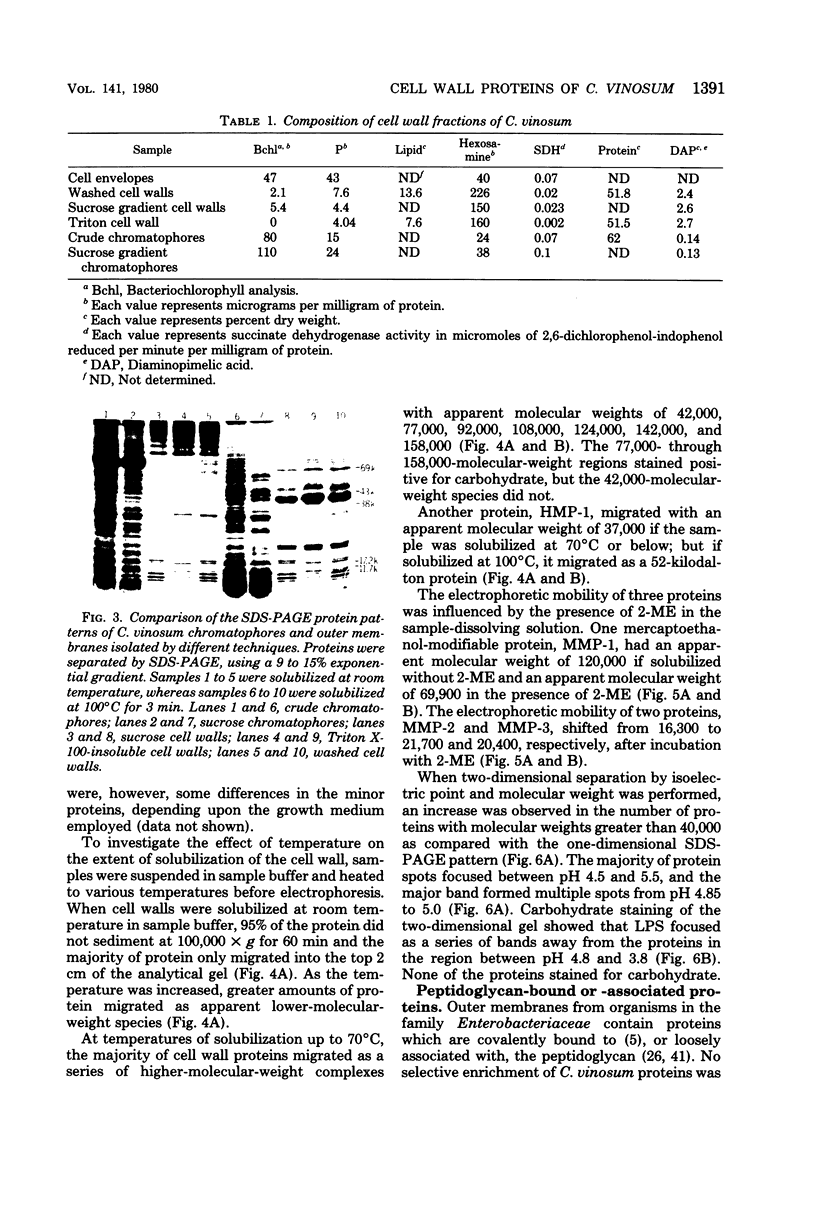
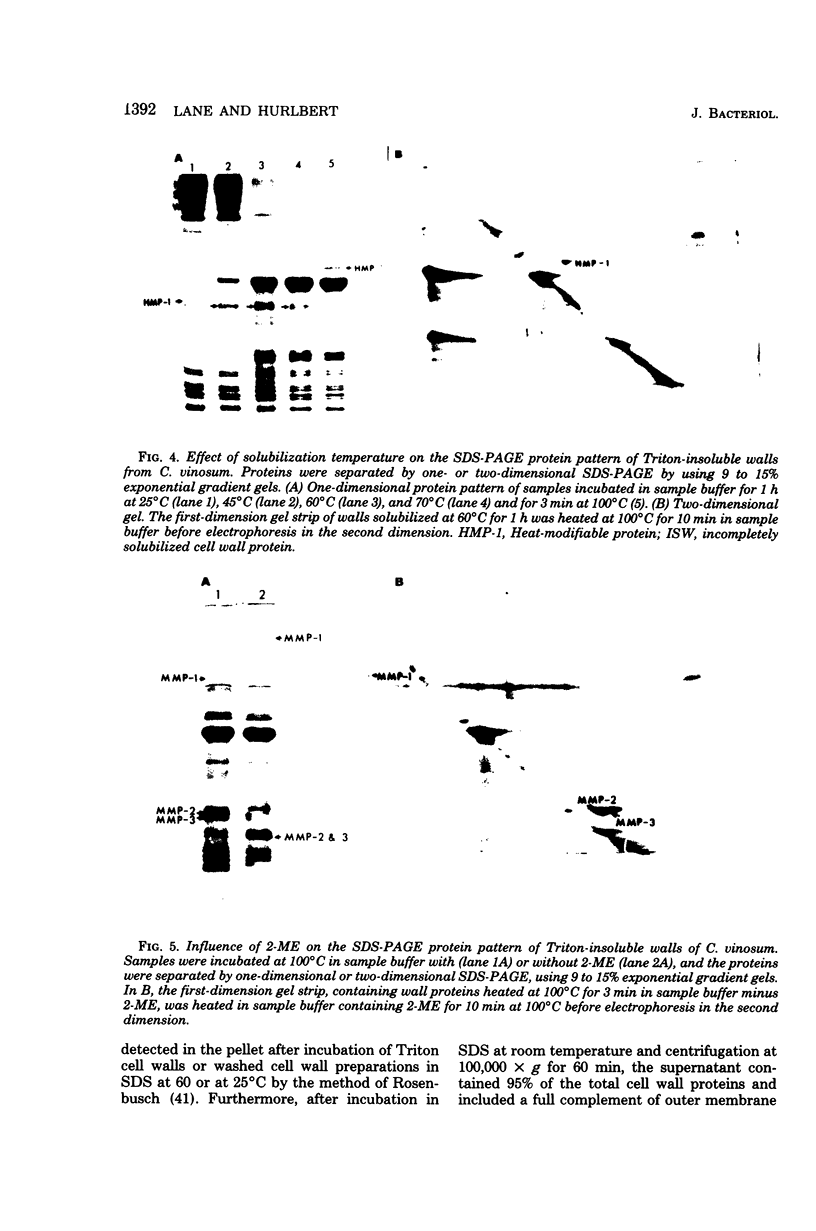
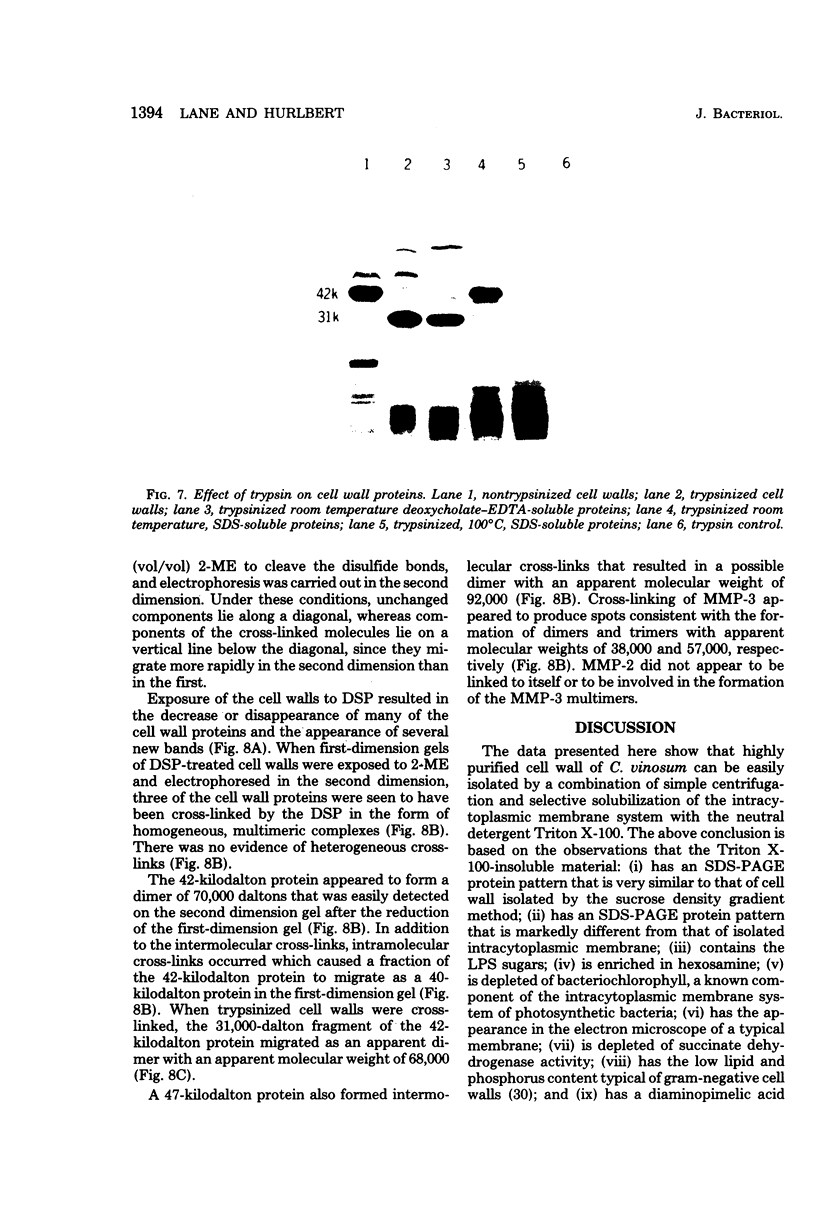
Highly purified cell walls of Chromatium vinosum were isolated by differential centrifugation, with or without Triton X-100 extraction. The isolated material had a protein composition similar to that of cell walls obtained by sucrose density gradient centrifugation. Twenty-two proteins were reproducibly detected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. A 42-kilodalton protein was shown to account for 65% of the total cell wall protein. The majority of cell wall proteins were solubilized in sodium dodecyl sulfate at room temperature; however, they existed as high-molecular-weight complexes unless heated to 45 degrees C or above. The cell wall contained one heat-modifiable protein which migrated with an apparent molecular weight of 37,400 when solubilized at 70 degrees C or below, but which migrated with an apparent molecular weight of 52,500 if solubilized at 100 degrees C. The electrophoretic mobility of three proteins was modified by 2-mercaptoethanol. The majority of C. vinosum cell wall proteins had isoelectric points between pH 4.5 and 5.5, and the 42-kilodalton protein focused at pH 4.9. No proteins were detected which were analogous to the lipoprotein or peptidoglycan-associated proteins of the Enterobacteriaceae. Nearest-neighbor analysis with a reducible, cross-linking reagent indicated that three proteins, including the 42-kilodalton protein, associated with themselves. Most of the cell wall proteins were partially accessible to proteases in both intact cells and isolated cell walls. Protease treatment of the whole cell or isolated cell wall digested approximately an 11,000-molecular-weight portion of the 42-kilodalton protein.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ames G. F., Nikaido K. Two-dimensional gel electrophoresis of membrane proteins. Biochemistry. 1976 Feb 10;15(3):616–623. doi: 10.1021/bi00648a026. [DOI] [PubMed] [Google Scholar]
- Bayer M. E. Adsorption of bacteriophages to adhesions between wall and membrane of Escherichia coli. J Virol. 1968 Apr;2(4):346–356. doi: 10.1128/jvi.2.4.346-356.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragg P. D., Hou C. Organization of proteins in the native and reformed outer membrane of Escherichia coli. Biochim Biophys Acta. 1972 Aug 9;274(2):478–488. doi: 10.1016/0005-2736(72)90193-9. [DOI] [PubMed] [Google Scholar]
- Braun V., Rehn K. Chemical characterization, spatial distribution and function of a lipoprotein (murein-lipoprotein) of the E. coli cell wall. The specific effect of trypsin on the membrane structure. Eur J Biochem. 1969 Oct;10(3):426–438. doi: 10.1111/j.1432-1033.1969.tb00707.x. [DOI] [PubMed] [Google Scholar]
- Braun V., Rehn K., Wolff H. Supramolecular structure of the rigid layer of the cell wall of Salmonella, Serratia, Proteus, and Pseudomonas fluorescens. Number of lipoprotein molecules in a membrane layer. Biochemistry. 1970 Dec 22;9(26):5041–5049. doi: 10.1021/bi00828a001. [DOI] [PubMed] [Google Scholar]
- Collins M. L., Niederman R. A. Membranes of Rhodospirillum rubrum: isolation and physicochemical properties of membranes from aerobically grown cells. J Bacteriol. 1976 Jun;126(3):1316–1325. doi: 10.1128/jb.126.3.1316-1325.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton J. W., Ingram J. M., Cheng K. J. Structure and function of the cell envelope of gram-negative bacteria. Bacteriol Rev. 1974 Mar;38(1):87–110. doi: 10.1128/br.38.1.87-110.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiRienzo J. M., Nakamura K., Inouye M. The outer membrane proteins of Gram-negative bacteria: biosynthesis, assembly, and functions. Annu Rev Biochem. 1978;47:481–532. doi: 10.1146/annurev.bi.47.070178.002405. [DOI] [PubMed] [Google Scholar]
- Dunker A. K., Rueckert R. R. Observations on molecular weight determinations on polyacrylamide gel. J Biol Chem. 1969 Sep 25;244(18):5074–5080. [PubMed] [Google Scholar]
- Gatt R., Berman E. R. A rapid procedure for the estimation of amino sugars on a micro scale. Anal Biochem. 1966 Apr;15(1):167–171. doi: 10.1016/0003-2697(66)90262-4. [DOI] [PubMed] [Google Scholar]
- Gruss P., Gmeiner J., Martin H. H. Amino-acid composition of the covalent rigid-layer lipoprotein in cell walls of Proteus mirabilis. Eur J Biochem. 1975 Sep 15;57(2):411–414. doi: 10.1111/j.1432-1033.1975.tb02315.x. [DOI] [PubMed] [Google Scholar]
- Hancock R. E., Nikaido H. Outer membranes of gram-negative bacteria. XIX. Isolation from Pseudomonas aeruginosa PAO1 and use in reconstitution and definition of the permeability barrier. J Bacteriol. 1978 Oct;136(1):381–390. doi: 10.1128/jb.136.1.381-390.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin M., Rottem S., Razin S. The outer membrane of Proteus mirabilis. I. Isolation and characterization of the outer and cytoplasmic membrane fractions. Biochim Biophys Acta. 1975 Feb 14;375(3):381–394. doi: 10.1016/0005-2736(75)90354-5. [DOI] [PubMed] [Google Scholar]
- Hirashima A., Wu H. C., Venkateswaran P. S., Inouye M. Two forms of a structural lipoprotein in the envelope of Escherichia coli. Further characterization of the free form. J Biol Chem. 1973 Aug 25;248(16):5654–5659. [PubMed] [Google Scholar]
- Hui K. M., Hurlbert R. E. Modifiable chromatophore proteins in photosynthetic bacteria. J Bacteriol. 1979 Apr;138(1):207–217. doi: 10.1128/jb.138.1.207-217.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlbert R. E. Isolation and characterization of Chromatium vinosum membranes. Arch Microbiol. 1974;101(2):169–186. doi: 10.1007/BF00455937. [DOI] [PubMed] [Google Scholar]
- Hurlbert R. E., Weckesser J., Mayer H., Fromme I. Isolation and characterization of the lipopolysaccharide of Chromatium vinosum. Eur J Biochem. 1976 Sep 15;68(2):365–371. doi: 10.1111/j.1432-1033.1976.tb10823.x. [DOI] [PubMed] [Google Scholar]
- Johnston K. H., Gotschlich E. C. Isolation and characterization of the outer membrane of Neisseria gonorrhoeae. J Bacteriol. 1974 Jul;119(1):250–257. doi: 10.1128/jb.119.1.250-257.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koplow J., Goldfine H. Alterations in the outer membrane of the cell envelope of heptose-deficient mutants of Escherichia coli. J Bacteriol. 1974 Feb;117(2):527–543. doi: 10.1128/jb.117.2.527-543.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUFT J. H. Improvements in epoxy resin embedding methods. J Biophys Biochem Cytol. 1961 Feb;9:409–414. doi: 10.1083/jcb.9.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lugtenberg B., Bronstein H., van Selm N., Peters R. Peptidoglycan-associated outer membrane proteins in gammegatine bacteria. Biochim Biophys Acta. 1977 Mar 17;465(3):571–578. doi: 10.1016/0005-2736(77)90274-7. [DOI] [PubMed] [Google Scholar]
- Lugtenberg B., Meijers J., Peters R., van der Hoek P., van Alphen L. Electrophoretic resolution of the "major outer membrane protein" of Escherichia coli K12 into four bands. FEBS Lett. 1975 Oct 15;58(1):254–258. doi: 10.1016/0014-5793(75)80272-9. [DOI] [PubMed] [Google Scholar]
- Markwell J. P., Lascelles J. Membrane-bound, pyridine nucleotide-independent L-lactate dehydrogenase of Rhodopseudomonas sphaeroides. J Bacteriol. 1978 Feb;133(2):593–600. doi: 10.1128/jb.133.2.593-600.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura T., Mizushima S. Separation by density gradient centrifugation of two types of membranes from spheroplast membrane of Escherichia coli K12. Biochim Biophys Acta. 1968 Jan 3;150(1):159–161. doi: 10.1016/0005-2736(68)90020-5. [DOI] [PubMed] [Google Scholar]
- Mizuno T., Kageyama M. Separation and characterization of the outer membrane of Pseudomonas aeruginosa. J Biochem. 1978 Jul;84(1):179–191. doi: 10.1093/oxfordjournals.jbchem.a132106. [DOI] [PubMed] [Google Scholar]
- Nakae T. Identification of the outer membrane protein of E. coli that produces transmembrane channels in reconstituted vesicle membranes. Biochem Biophys Res Commun. 1976 Aug 9;71(3):877–884. doi: 10.1016/0006-291x(76)90913-x. [DOI] [PubMed] [Google Scholar]
- Nakae T. Outer membrane of Salmonella. Isolation of protein complex that produces transmembrane channels. J Biol Chem. 1976 Apr 10;251(7):2176–2178. [PubMed] [Google Scholar]
- Nakamura K., Mizushima S. Effects of heating in dodecyl sulfate solution on the conformation and electrophoretic mobility of isolated major outer membrane proteins from Escherichia coli K-12. J Biochem. 1976 Dec;80(6):1411–1422. doi: 10.1093/oxfordjournals.jbchem.a131414. [DOI] [PubMed] [Google Scholar]
- Neville D. M., Jr, Glossmann H. Molecular weight determination of membrane protein and glycoprotein subunits by discontinuous gel electrophoresis in dodecyl sulfate. Methods Enzymol. 1974;32:92–102. doi: 10.1016/0076-6879(74)32012-5. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975 May 25;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Osborn M. J., Gander J. E., Parisi E., Carson J. Mechanism of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membrane. J Biol Chem. 1972 Jun 25;247(12):3962–3972. [PubMed] [Google Scholar]
- REYNOLDS E. S. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963 Apr;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reithmeier R. A., Bragg P. D. Cross-linking of the proteins in the outer membrane of Escherichia coli. Biochim Biophys Acta. 1977 Apr 18;466(2):245–256. doi: 10.1016/0005-2736(77)90222-x. [DOI] [PubMed] [Google Scholar]
- Reithmeier R. A., Bragg P. D. Molecular characterization of a heat-modifiable protein from the outer membrane of Escherichia coli. Arch Biochem Biophys. 1977 Jan 30;178(2):527–534. doi: 10.1016/0003-9861(77)90223-5. [DOI] [PubMed] [Google Scholar]
- Rosenbusch J. P. Characterization of the major envelope protein from Escherichia coli. Regular arrangement on the peptidoglycan and unusual dodecyl sulfate binding. J Biol Chem. 1974 Dec 25;249(24):8019–8029. [PubMed] [Google Scholar]
- Russell R. R., Johnson K. G., McDonald I. J. Envelope proteins in Neisseria. Can J Microbiol. 1975 Oct;21(10):1519–1534. doi: 10.1139/m75-224. [DOI] [PubMed] [Google Scholar]
- Russell R. R. Two-dimensional SDS-polyacrylamide gel electrophoresis of heat-modifiable outer-membrane proteins. Can J Microbiol. 1976 Jan;22(1):83–91. doi: 10.1139/m76-011. [DOI] [PubMed] [Google Scholar]
- SABATINI D. D., BENSCH K., BARRNETT R. J. Cytochemistry and electron microscopy. The preservation of cellular ultrastructure and enzymatic activity by aldehyde fixation. J Cell Biol. 1963 Apr;17:19–58. doi: 10.1083/jcb.17.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaitman C. A. Solubilization of the cytoplasmic membrane of Escherichia coli by Triton X-100. J Bacteriol. 1971 Oct;108(1):545–552. doi: 10.1128/jb.108.1.545-552.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. K., Winkler H. H. Separation of inner and outer membranes of Rickettsia prowazeki and characterization of their polypeptide compositions. J Bacteriol. 1979 Feb;137(2):963–971. doi: 10.1128/jb.137.2.963-971.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K., Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969 Aug 25;244(16):4406–4412. [PubMed] [Google Scholar]