Tom40 forms the channel of the mitochondrial preprotein translocase. This beta-barrel protein assembles with alpha-helical proteins, however little is known about the mechanism of assembly. Becker et al identified a new intermediate in Tom40 assembly and show that small alpha-helical Tom proteins associate with Tom40 directly at the SAM complex.
Abstract
The preprotein translocase of the outer mitochondrial membrane (TOM) consists of a central β-barrel channel, Tom40, and six proteins with α-helical transmembrane segments. The precursor of Tom40 is imported from the cytosol by a pre-existing TOM complex and inserted into the outer membrane by the sorting and assembly machinery (SAM). Tom40 then assembles with α-helical Tom proteins to the mature TOM complex. The outer membrane protein Mim1 promotes membrane insertion of several α-helical Tom proteins but also affects the biogenesis of Tom40 by an unknown mechanism. We have identified a novel intermediate in the assembly pathway of Tom40, revealing a two-stage interaction of the precursor with the SAM complex. The second SAM stage represents assembly of Tom5 with the precursor of Tom40. Mim1-deficient mitochondria accumulate Tom40 at the first SAM stage like Tom5-deficient mitochondria. Tom5 promotes formation of the second SAM stage and thus suppresses the Tom40 assembly defect of mim1Δ mitochondria. We conclude that the assembly of newly imported Tom40 is directly initiated at the SAM complex by its association with Tom5. The involvement of Mim1 in Tom40 biogenesis can be largely attributed to its role in import of Tom5.
INTRODUCTION
The translocase of the outer membrane (TOM complex) forms the main entry gate for nuclear-encoded mitochondrial precursor proteins (Koehler, 2004; Dolezal et al., 2006; Neupert and Herrmann, 2007; Chacinska et al., 2009; Endo and Yamano, 2009). The TOM complex consists of seven different proteins. The β-barrel of the central component Tom40 forms the protein-conducting channel (Hill et al., 1998; Suzuki et al., 2004; Becker et al., 2005). The other subunits of the TOM complex are embedded into the membrane via single transmembrane α-helices. Three receptor proteins, Tom20, Tom22, and Tom70, and three small Tom proteins, Tom5, Tom6, and Tom7, associate with Tom40 to form the mature TOM complex (Ahting et al., 1999; Stan et al., 2000; Meisinger et al., 2001). Tom20 and Tom70 initially recognize mitochondrial precursor proteins and deliver them to the central receptor Tom22 (Kiebler et al., 1993; van Wilpe et al., 1999; Yano et al., 2000; Yamano et al., 2008). The small Tom proteins are involved in regulating the stability of the TOM complex and may also participate in the transfer of precursor proteins (Alconada et al., 1995; Hönlinger et al., 1996; Dietmeier et al., 1997; Esaki et al., 2004; Schmitt et al., 2005; Sherman et al., 2005; Kato and Mihara, 2008).
All TOM subunits themselves are nuclear encoded, synthesized as precursors on cytosolic ribosomes, and imported into mitochondria (Becker et al., 2009; Walther and Rapaport, 2009; Endo and Yamano 2010). The precursor of the β-barrel protein Tom40 is initially transported across the outer membrane via the TOM complex (Model et al., 2001; Wiedemann et al., 2003). With the help of intermembrane space chaperones the precursor is transferred to the sorting and assembly machinery (SAM complex) (Hoppins and Nargang, 2004; Wiedemann et al., 2004). The SAM complex contains three main components: Sam50 (also termed Omp85/Tob55), Sam37, and Sam35 (Tob38/Tom38) (Kozjak et al., 2003; Paschen et al., 2003; Wiedemann et al., 2003; Gentle et al., 2004; Ishikawa et al., 2004; Milenkovic et al., 2004; Waizenegger et al., 2004). Sam50 has been conserved from bacteria to humans; its bacterial homolog, BamA (Omp85), forms the core of the β-barrel assembly machinery in the outer membrane of Gram-negative bacteria (Schleiff and Soll, 2005; Dolezal et al., 2006; Ruiz et al., 2006; Bos et al., 2007; Kutik et al., 2009; Walther et al., 2009; Hagan et al., 2010; Stroud et al., 2010). A β-sorting signal in the last (carboxy-terminal) β-strand of Tom40 directs the precursor to the SAM complex and initiates its membrane insertion via Sam50-Sam35 (Kutik et al., 2008). Sam37 is involved in release of the precursor protein from the SAM complex (Chan and Lithgow, 2008; Dukanovic et al., 2009). The Tom40 precursor then sequentially assembles with the α-helical Tom subunits to form the mature multi-subunit TOM complex. A fourth SAM subunit, the mitochondrial distribution and morphology protein Mdm10, has been found in association with two protein complexes, the SAM complex and the MDM complex that is involved in the formation of mitochondria-endoplasmic reticulum junctions (Boldogh et al., 2003; Meisinger et al., 2004, 2007; Kornmann et al., 2009; Wideman et al., 2010). SAM-bound Mdm10 promotes late steps of Tom40 assembly and favors association of Tom40 with Tom22 (Meisinger et al., 2004, 2006; Thornton et al., 2010; Yamano et al., 2010).
A further outer membrane protein, the mitochondrial import protein Mim1, transiently interacts with a fraction of SAM complexes (Becker et al., 2008). Mim1 promotes membrane insertion of the majority of α-helical Tom proteins. (i) The precursors of the three small Tom proteins are membrane inserted with the help of Mim1 and subsequently depend on SAM functions for assembly into the TOM complex (Stojanovski et al., 2007a; Becker et al., 2008; Thornton et al., 2010). (ii) The precursors of the receptors Tom20 and Tom70 are inserted into the outer membrane with the help of Mim1, but independently of SAM, and then assemble with a TOM core complex consisting of Tom40, Tom22, and the small Tom proteins (Meisinger et al., 2004; Milenkovic et al., 2004; Stojanovski et al., 2007a; Becker et al., 2008; Hulett et al., 2008; Popov-Celeketic et al., 2008). (iii) Additionally, Mim1-deficient mitochondria are also impaired in Tom40 assembly. Tom40 assembly stages following the SAM complex were inhibited in the mutant mitochondria (Ishikawa et al., 2004; Waizenegger et al., 2005; Becker et al., 2008). Although the involvement of Mim1 in Tom40 assembly was reported before its role in insertion of α-helical proteins was found, it is unknown how Mim1 affects the biogenesis of the β-barrel precursor.
Each of the small Tom proteins was found to be involved in the assembly pathway of Tom40. Tom5 and Tom6 were reported to associate with the Tom40 precursor after its release from the SAM core complex, forming an assembly intermediate that associates with further Tom subunits (Model et al., 2001; Wiedemann et al., 2003). Dukanovic et al. (2009) showed that Tom6 genetically and functionally interacts with Sam37; however, they did not observe a direct interaction between Tom6 and Sam37. The currently available results indicate that Tom6 stabilizes Tom40 precursor molecules and promotes their association with Tom22 (Alconada et al., 1995; Dekker et al., 1998; Dembowski et al., 2001; Dukanovic et al., 2009). In contrast, Tom7 was found to delay Tom40 assembly by promoting a release of Mdm10 from the SAM complex (Model et al., 2001; Meisinger et al., 2006). The exact molecular mechanisms underlying the function of small Tom proteins in Tom40 assembly have not been clarified.
For this study, we have addressed the role of small Tom proteins in the interaction of Tom40 with the SAM complex. We dissected the interaction of the Tom40 precursor with SAM into two stages. In a first stage, Tom40 binds to the SAM core complex. Tom5 is critical for formation of the second stage by assembling with newly synthesized Tom40 directly at the SAM complex. Interestingly, mitochondria lacking Mim1 accumulate the Tom40 precursor at the first SAM stage (i.e., before the assembly with Tom5). The Tom40 assembly defect of mim1Δ mitochondria was indeed suppressed by Tom5. Our results show that small Tom proteins assemble with newly synthesized Tom40 already at the SAM complex and explain the influence of Mim1 on Tom40 assembly by its role in the import of small Tom proteins.
MATERIALS AND METHODS
Yeast Strains
The S. cerevisiae mutant strains tom5Δ, tom6Δ, tom7Δ, sam37Δ, mdm10Δ, mim1Δ, and the corresponding wild-type strains have been described before (Alconada et al., 1995; Hönlinger et al., 1996; Dietmeier et al., 1997; Wiedemann et al., 2003; Meisinger et al., 2004; Becker et al., 2008). The viability of the double deletions sam37Δ tom5Δ, sam37Δ tom6Δ, and sam37Δ tom7Δ was assessed by a plasmid shuffling approach. The open reading frame of SAM37 was cloned into the vector pYep352 under the control of a MET25 promoter and a CYC1 terminator. A yeast strain with a disruption of the open reading frame of SAM37 by the LEU2-Marker was transformed with pYep352 encoding SAM37. Clones were selected using the URA3 marker of the plasmid. After this, a kanamycin resistance cassette was introduced in the open reading frames of TOM5, TOM6, or TOM7. Finally, the plasmid encoding SAM37 was eliminated by growth on plates containing 5-fluoroorotic acid (5-FOA).
For generation of overexpression mutants, the open reading frames of TOM5, TOM6, TOM7, MIM1, or SAM37 were introduced in a pYep352 vector. Subsequently, the yeast strains tom5Δ and sam37Δ were transformed and clones were selected by the URA3 marker of the plasmid.
Protein Import into Mitochondria
Mitochondria were isolated by differential centrifugation according to standard procedures (Stojanovski et al., 2007b). The protein concentrations were adjusted to 10 mg/ml in SEM buffer (250 mM sucrose, 1 mM EDTA, 10 mM MOPS/KOH [pH 7.2]). Mitochondria aliquots were shock-frozen in liquid nitrogen and stored at −80°C. For in vitro import assays, 35S-labeled proteins were synthesized using pGEM4Z-based constructs as template for coupled in vitro transcription and translation (Promega, Madison, WI). In a standard import assay, 50 μg mitochondria (protein amount) were incubated with the radiolabeled proteins in the presence of 2 mM NADH, 2 mM ATP, 5 mM methionine, 5 mM creatine phosphate, and 100 μg/ml creatine kinase in import buffer (3% [wt/vol] BSA, 250 mM sucrose, 80 mM KCl, 5 mM MgCl2, 10 mM MOPS/KOH [pH 7.2], 2 mM KH2PO4). Transfer on ice stopped the import reaction. Mitochondria were reisolated, washed with SEM buffer, and lysed with 1% (wt/vol) digitonin in lysis buffer (20 mM Tris/HCl [pH 7.4], 0.1 mM EDTA, 50 mM NaCl, 10% [vol/vol] glycerol) at a protein concentration of 1 mg/ml for 15 min on ice. After a clarifying spin, mitochondrial lysates were loaded onto a blue native gel as described (Dekker et al., 1998; Ryan et al., 2001; Stojanovski et al., 2007b). Protein complexes were visualized by digital autoradiography. For a two-step import assay, chemical amounts of the proteins were generated in vitro using a wheat germ based translation system (5 Prime, Hamburg, Germany). These proteins were imported into isolated mitochondria for 40 min at 25°C. Subsequently, mitochondria were reisolated and washed with SEM buffer. The import of radiolabeled precursors into these mitochondria was performed according to the conditions described above.
Antibody Shift Assays
For antibody shift assays, radiolabeled proteins were imported into isolated mitochondria. After washing, mitochondria were incubated in SEM buffer with the indicated antisera for 30 min on ice. Mitochondria were reisolated and lysed with 1% digitonin, and protein complexes were analyzed by blue native electrophoresis. For the antibody-shift of imported Flag-tagged proteins, chemical amounts of these proteins were imported into isolated mitochondria. After reisolation and washing with SEM buffer, radiolabeled proteins were imported into the mitochondria. Mitochondria were isolated and washed with SEM buffer and lysed with 1% digitonin in lysis buffer. Anti-Flag antibodies (Sigma, St. Louis, MO) or water were added to the reaction and incubated for 30 min on ice. Finally, insoluble material was removed by centrifugation and protein complexes were analyzed by blue native electrophoresis and digital autoradiography.
Miscellaneous
Western blotting transfer onto PVDF membranes and immunodecoration were performed according to standard procedures. Enhanced chemiluminescence (GE Healthcare, Piscataway, NJ) was used for the detection of the antibodies.
RESULTS
Genetic Interaction of TOM5 and SAM37
Yeast cells lacking SAM37 show a growth defect at elevated temperature (Gratzer et al., 1995; Dukanovic et al., 2009). Dukanovic et al. (2009) reported that expression of TOM6, but neither TOM5 nor TOM7, from a high copy number plasmid (pRS426) suppressed the growth defect of sam37Δ cells. When we expressed the small TOM genes via the high copy number plasmid pYEP352 (with a MET25 promoter), we unexpectedly found that overexpression of TOM5 suppressed the growth defect of sam37Δ cells as efficiently as TOM6 did (Figure 1A). Expression of TOM7 from the plasmid did not suppress sam37Δ cells (Figure 1A) in agreement with Dukanovic et al. (2009).
Figure 1.
Genetic interaction of SAM37 with genes of small Tom proteins. (A) The S. cerevisiae sam37Δ strain was transformed with the pYEP352 vector containing the indicated open reading frames. Growth of the strains was analyzed on selective medium containing glucose as energy source. (B) S. cerevisiae strains were plated on 5-FOA containing medium with glucose as energy source. 5-FOA containing medium selects for the loss of the URA3-containing pYEP352 plasmids. WT, wild-type.
To test for a synthetic interaction of the genes for TOM5 and SAM37, we generated a yeast double deletion mutant, sam37Δ tom5Δ. When SAM37 was expressed from a URA3-containing plasmid in the double mutant, the cells were viable. By transfer of the cells to 5-FOA containing plates, the plasmid was eliminated and the sam37Δ tom5Δ cells ceased growth (Figure 1B). Similarly, yeast cells with a chromosomal deletion of the essential gene TOM40 ceased growth when the plasmid encoding the wild-type copy of TOM40 was lost (Figure 1B). We conclude that deletion of TOM5 and SAM37 leads to synthetic lethality. For comparison, we also generated the double mutants sam37Δ tom6Δ and sam37Δ tom7Δ in our strain background and observed a strong synthetic growth defect of sam37Δ tom6Δ cells, whereas sam37Δ tom7Δ cells grew like the sam37Δ single deletion mutant (Figure 1B) in agreement with Dukanovic et al. (2009). These results demonstrate that not only TOM6 but also TOM5 shows a strong genetic interaction with SAM37.
Tom5 Suppresses the Defect of sam37Δ Mitochondria in Tom40 Assembly
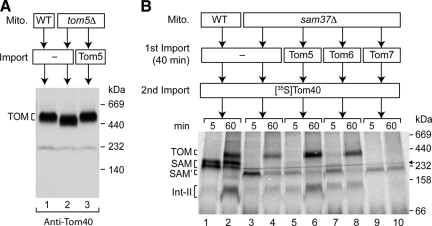
To explore the molecular mechanisms behind these phenotypes we wanted to obtain biochemical evidence for a relation of Tom5 to Sam37 (i.e., we asked whether Tom5 was able to suppress protein sorting defects in sam37Δ mitochondria). A detailed functional analysis of outer membrane protein biogenesis can be performed by an in organello system using purified mitochondria and precursor proteins that are synthesized and radiolabeled in reticulocyte lysate in the presence of [35S]methionine (Ryan et al., 2001; Stojanovski et al., 2007b). However, reticulocyte lysates typically synthesize proteins in small radiochemical amounts that are not sufficient to resolve mutant defects on a biochemical level. The expression of mitochondrial membrane proteins in E. coli cells produces large chemical amounts but usually leads to aggregation in inclusion bodies. Only few mitochondrial membrane proteins have been successfully extracted from inclusion bodies in a denatured but transport-competent form and imported into mitochondria. We thus established a system to efficiently synthesize mitochondrial proteins in vitro using a wheat germ–based translation system. To test whether Tom5 synthesized by this system was imported into mitochondria and assembled into the TOM complex, we used mitochondria that were isolated from a yeast strain lacking TOM5. When mitochondria are lysed with the nonionic detergent digitonin and separated by blue native electrophoresis, the TOM complex migrates at ∼450 kDa (Dekker et al., 1998; Meisinger et al., 2001). In tom5Δ mitochondria, the TOM complex migrates slightly faster due to the lack of this small subunit (Figure 2A, compare lane 2 to lane 1) (Dekker et al., 1998). On import of chemical amounts of Tom5, the mobility of the TOM complex was fully shifted to wild-type conditions (Figure 2A, lane 3), demonstrating that Tom5 synthesized in the wheat germ system was import-competent and assembled into the TOM complex.
Figure 2.
Chemical amounts of Tom5 and Tom6 promote Tom40 assembly in the absence of Sam37. (A) Wheat germ–synthesized Tom5 was imported into isolated tom5Δ yeast mitochondria where indicated. The mitochondria were reisolated, lysed with digitonin, and subjected to blue native electrophoresis and Western Blotting. The TOM complex was detected by a Tom40 specific antibody. (B) 35S-labeled precursor of Tom40 was imported into sam37Δ mitochondria that had been preincubated with chemical amounts of the indicated proteins. The assembly of Tom40 was analyzed by blue native electrophoresis and digital autoradiography. SAM′, small form of the SAM complex in sam37Δ mitochondria; arrow, unspecific band.
Mitochondria were isolated from sam37Δ cells and incubated with Tom5, Tom6, or Tom7 synthesized in the wheat germ system. On reisolation of the mitochondria, radiolabeled precursor of Tom40 was added in a second import reaction and its assembly was analyzed by blue native electrophoresis and autoradiography. In wild-type mitochondria, the precursor of Tom40 has been shown to form two assembly intermediates before being integrated into the mature TOM complex (Figure 2B, lanes 1 and 2) (Model et al., 2001; Wiedemann et al., 2003). The first intermediate of ∼250 kDa represents the interaction of the precursor with the SAM complex (Kozjak et al., 2003; Paschen et al., 2003; Wiedemann et al., 2003; Ishikawa et al., 2004; Milenkovic et al., 2004; Waizenegger et al., 2004) and is followed by a second, smaller intermediate, termed Int-II, that contains Tom40 and Tom5 (Wiedemann et al., 2003). In sam37Δ mitochondria, the formation of SAM, Int-II, and mature TOM complex is impaired, and the precursor of Tom40 is found in association with a smaller SAM complex that lacks Sam37 (Figure 2B, lanes 3 and 4) (Wiedemann et al., 2003; Waizenegger et al., 2004; Chan and Lithgow, 2008; Dukanovic et al., 2009). When chemical amounts of Tom5 were imported into sam37Δ mitochondria before the import of [35S]Tom40, the assembly of Tom40 into the mature TOM complex was efficiently restored (Figure 2B, lane 6). Import of Tom6 supported a partial rescue of Tom40 assembly in sam37Δ mitochondria, whereas Tom7 did not support the assembly of Tom40 (Figure 2B, lanes 7–10). Taken together, the genetic and biochemical results indicate that overexpression of Tom5 can restore the assembly of Tom40 into the TOM complex in mitochondria lacking Sam37.
Two-Stage Interaction of Tom40 with the SAM Complex Involves Tom5
It has been shown that Tom5 interacts with the precursor of Tom40 in the assembly intermediate II and that mitochondria lacking Tom5 accumulate the Tom40 precursor at the SAM complex (Model et al., 2001; Wiedemann et al., 2003). We used high-resolution blue native gels and observed that the SAM intermediate of [35S]Tom40 consisted of two distinct bands that have not been characterized so far. We termed these bands SAM-Ia and SAM-Ib, respectively (Figure 3A, lanes 1–3). In tom5Δ mitochondria, SAM-Ia was generated, whereas the formation of SAM-Ib and all following assembly stages (Int-II and mature TOM complex) was strongly inhibited (Figure 3A, lanes 4–6). To exclude indirect effects of the tom5Δ mutant, we asked whether the lack of Tom5 was responsible for the defect in Tom40 assembly and performed a two-step import experiment. We first imported chemical amounts of Tom5 into tom5Δ mitochondria and then [35S]Tom40. The addition of chemical amounts of Tom5 completely restored the formation of SAM-Ib, intermediate II and of the mature TOM complex (Figure 3B, lanes 4–6).
Figure 3.

Tom5 is involved in the two-stage interaction of the Tom40 precursor with the SAM complex. (A) 35S-labeled Tom40 was imported into wild-type (WT) and tom5Δ yeast mitochondria for the indicated periods. Mitochondria were reisolated, lysed with digitonin, and analyzed by blue native electrophoresis and autoradiography. (B) tom5Δ mitochondria were either mock treated or incubated with wheat germ–synthesized Tom5. Subsequently, the assembly of 35S-labeled Tom40 was analyzed by blue native electrophoresis and autoradiography.
To test whether Tom5 functioned after accumulation of the Tom40 precursor at the SAM-Ia stage, we first imported [35S]Tom40 in a short-term import reaction and reisolated the mitochondria. In a subsequent chase incubation, Tom40 was assembled into the TOM complex in wild-type mitochondria (Figure 4A, lane 2) but remained largely at the SAM-Ia stage in tom5Δ mitochondria (Figure 4A, lanes 3 and 4). When additional chemical amounts of Tom5 were imported in the second incubation, the Tom40 precursor was chased to the SAM-Ib stage and finally to the mature TOM complex (Figure 4A, lanes 5 and 6).
Figure 4.
Tom5 is a component of the SAM-Ib intermediate of Tom40 assembly. (A) 35S-labeled Tom40 was imported for 10 min into wild-type (WT) and tom5Δ mitochondria. Subsequently, mitochondria were reisolated and a chase reaction was performed in the absence or presence of chemical amounts of Tom5 for the indicated periods. The assembly of 35S-labeled Tom40 was analyzed by blue native electrophoresis and autoradiography. (B) Chemical amounts of Tom5Flag were imported into wild-type mitochondria. After reisolation of the mitochondria, 35S-labeled Tom40 was imported for 5 min, the mitochondria were lysed, and anti-Flag antibodies were added. The assembly of Tom40 and antibody shifts were analyzed by blue native electrophoresis and autoradiography. (C) 35S-labeled Tom40 was imported into wild-type mitochondria. Subsequently, antibody shift assays with the indicated antibodies were performed.
These results show that formation of the SAM-Ib intermediate of Tom40 requires Tom5. We then asked whether Tom5 was only required for generation of the intermediate or whether Tom5 was part of the SAM-Ib intermediate itself. To experimentally address the second possibility, chemical amounts of Flag-tagged Tom5 were imported into mitochondria. 35S-labeled Tom40 was imported into these mitochondria for a short period to allow binding of the precursor to the SAM complex. Pre-import of Tom5Flag accelerated the formation of the SAM-Ib intermediate (Figure 4B, lane 2). The mitochondria were lysed with digitonin and incubated with anti-Flag antibodies. Separation by blue native electrophoresis revealed that the mobility of accumulated [35S]Tom40 was shifted to larger molecular masses by anti-Flag (Figure 4B, lane 3), demonstrating that the Tom40 precursor was associated with Tom5Flag. To test whether endogenous Tom5 was present in the SAM-Ib intermediate, we performed a short-term import of [35S]Tom40 into wild-type mitochondria without importing additional amounts of Tom5. Antibodies directed against Tom5 were added to the mitochondria. Blue native electrophoresis showed that anti-Tom5 affected SAM-Ib and intermediate II but not SAM-Ia (Figure 4C, lane 2). The addition of control antibodies raised against the outer membrane protein Fis1 did not affect the [35S]Tom40 intermediates (Figure 4C, lane 3). We conclude that Tom5 is part of the SAM-Ib intermediate.
Taken together, the precursor of Tom40 interacts with the SAM complex in two stages, SAM-Ia and SAM-Ib. Tom5 associates with the Tom40 precursor at the SAM complex, thus forming intermediate SAM-Ib.
Tom6 Partially Substitutes for Tom5 in Maturation of Tom40
We asked whether the two other small Tom proteins also affected the formation of Tom40 intermediates at the SAM complex. Mitochondria lacking Tom6 efficiently generated the SAM-Ia intermediate but were partially impaired in the formation of SAM-Ib and the assembly of Tom40 into the TOM complex (Figure 5A). In contrast, mitochondria lacking Tom7 efficiently generated both SAM intermediates (Figure 5B) (integration of [35S]Tom40 into the mature TOM complex occurred with higher efficiency in tom7Δ mitochondria than in wild-type mitochondria as previously reported [Meisinger et al., 2006]). Tom7 functions in an antagonistic manner to Mdm10 that promotes assembly of Tom40 into the TOM complex (Meisinger et al., 2004, 2006; Yamano et al., 2010). We thus also analyzed mdm10Δ mitochondria and observed that the SAM-Ib intermediate of [35S]Tom40 was generated (Figure 5C) ([35S]Tom40 accumulated in assembly intermediate II as reported [Meisinger et al., 2004, 2006]). Thus, of the three mutant mitochondria, tom6Δ, tom7Δ, and mdm10Δ, only tom6Δ mitochondria were (partially) impaired in formation of the new SAM-Ib intermediate.
Figure 5.
Analysis of SAM-Ib formation in mitochondria lacking Tom6, Tom7, or Mdm10. 35S-labeled Tom40 was imported for the indicated periods into mitochondria isolated from (A) tom6Δ, (B) tom7Δ, or (C) mdm10Δ yeast strains. The assembly of Tom40 was analyzed by blue native electrophoresis and autoradiography.
To address whether Tom6 may play a role in formation of the SAM-Ib intermediate, we imported chemical amounts of Tom6 into isolated mitochondria before importing the [35S]Tom40 precursor. The import of Tom6 indeed promoted the formation of the SAM-Ib stage (Figure 6A, lanes 5 and 6). Chemical amounts of Flag-tagged Tom6 similarly accelerated the formation of SAM-Ib (Figure 6B, lane 2). Anti-Flag antibodies shifted part of the SAM-Ib intermediate on blue native gels (Figure 6B, lane 3), indicating that Tom6 associated with the precursor of Tom40 at the SAM-Ib stage. We thus asked whether Tom6 could replace the function of Tom5 in Tom40 maturation. We imported chemical amounts of small Tom proteins into tom5Δ mitochondria and analyzed the assembly stages of [35S]Tom40 (Figure 6C). Chemical amounts of Tom6 promoted the assembly of Tom40 into the TOM complex, although with considerably lower efficiency than Tom5 did (Figure 6C, compare lane 6 to lane 8). Moreover, Tom6 supported the formation of SAM-Ib in tom5Δ mitochondria only with low efficiency (Figure 6C, lane 7), in contrast to the efficient promotion of SAM-Ib formation in wild-type mitochondria (Figure 6, A and B), indicating that the presence of Tom5 is required for an efficient formation of SAM-Ib. Chemical amounts of Tom7 had no stimulatory effect on Tom40 assembly (Figure 6C, lanes 9 and 10). In agreement with these biochemical results, overexpression of Tom6, but not Tom7, partially rescued growth of tom5Δ cells at elevated temperature (Figure 6D).
Figure 6.
Tom6 partially substitutes for Tom5 in the assembly of Tom40. (A) Chemical amounts of Tom5 or Tom6 were imported into wild-type (WT) mitochondria. After reisolation of the mitochondria, 35S-labeled Tom40 was imported, and the mitochondria were lysed and analyzed by blue native electrophoresis and autoradiography. (B) Chemical amounts of Tom6Flag were imported into wild-type mitochondria. 35S-labeled Tom40 was imported into the reisolated mitochondria, and anti-Flag antibodies were added to the lysed mitochondria. The assembly of Tom40 was analyzed by blue native electrophoresis. (C) 35S-labeled Tom40 was imported into wild-type and tom5Δ mitochondria that had been preincubated with chemical amounts of the indicated small Tom proteins. (D) Overexpression of the indicated proteins was performed in a tom5Δ yeast strain. The growth of the indicated strains was analyzed on selective medium containing glycerol and ethanol as energy source.
Taken together, these results show that Tom6 can partially substitute for Tom5 in Tom40 assembly, yet Tom5 is the main factor required for generation of the SAM-Ib intermediate. Tom5 and Tom6 can associate with the precursor of Tom40 at the SAM-Ib stage.
Tom5 Suppresses the Defect of mim1Δ Mitochondria in Tom40 Assembly
Using the new assembly intermediate of Tom40 and mim1Δ mitochondria, we made a surprising observation. Mitochondria lacking Mim1 were not only impaired in the formation of Tom40 assembly after the SAM complex (Ishikawa et al., 2004; Waizenegger et al., 2005; Becker et al., 2008) but also in the generation of the SAM-Ib intermediate (Figure 7, lanes 3 and 4). [35S]Tom40 mainly accumulated in the SAM-Ia stage, and only little SAM-Ib was generated. Strikingly, chemical amounts of Tom5 imported into mim1Δ mitochondria efficiently restored the assembly of Tom40 (Figure 7, lanes 5 and 6). Chemical amounts of Tom6 moderately stimulated the generation of SAM-Ib, whereas Tom7 did not promote Tom40 assembly at all (Figure 7, lanes 7–10). We conclude that Tom5 can suppress the Tom40 assembly defect of mim1Δ mitochondria.
Figure 7.
Chemical amounts of Tom5 compensate for the loss of Mim1 in the assembly of Tom40. 35S-labeled Tom40 was imported into mim1Δ mitochondria that had been preincubated with chemical amounts of the indicated small Tom proteins. The import of Tom40 was analyzed by blue native electrophoresis and autoradiography.
DISCUSSION
We have identified a novel intermediate in the biogenesis of the protein import channel of the mitochondrial outer membrane. On its assembly pathway, the precursor of the β-barrel protein Tom40 has to associate with several α-helical Tom proteins, first the small Tom proteins and then the Tom receptors (Model et al., 2001; Wiedemann et al., 2003; Meisinger et al., 2004; Becker et al., 2008; Hulett et al., 2008; Popov-Celeketic et al., 2008; Dukanovic et al., 2009; Thornton et al., 2010). To date it has been assumed that the α-helical Tom proteins associate with the Tom40 precursor after its release from the SAM complex. We report that Tom5 plays a critical role in Tom40 assembly already at the SAM complex. The precursor of Tom40 stably interacts with SAM in the absence of Tom5, however further progression of assembly strongly depends on Tom5. By high-resolution blue native electrophoresis we could dissect two assembly stages of Tom40 at the SAM complex, the first one, SAM-Ia, representing the Tom5-independent binding of Tom40 to SAM, whereas the second stage, SAM-Ib, represents the assembly of Tom5 with Tom40 at the SAM complex.
The mitochondrial outer membrane contains only three proteins that are essential for cell viability under all growth conditions tested: Tom40 and the two core components Sam50 and Sam35 of the SAM complex (Baker et al., 1990; Kozjak et al., 2003; Paschen et al., 2003; Gentle et al., 2004; Ishikawa et al., 2004; Milenkovic et al., 2004; Waizenegger et al., 2004). Yeast cells lacking Tom5 are inviable at elevated temperature yet are viable at lower temperature. We asked whether and which component may be able to substitute for Tom5. We observed that overexpression of Tom6, but not Tom7, partially suppressed the growth defect of tom5Δ yeast cells. Because the small Tom proteins are subunits of the mature TOM complex, it may be argued that this genetic relation is only caused by their function in the TOM complex. However, a genetic interaction of TOM5 as well as TOM6 with SAM is indicated by two lines of evidence: the growth defect of sam37Δ cells is suppressed by overexpression of Tom5, as well as by overexpression of Tom6; and a double deletion of TOM5 and SAM37 as well as a double deletion of TOM6 and SAM37 cause strong synthetic growth defects, whereas no genetic interaction between TOM7 and SAM37 was observed (Dukanovic et al., 2009; this study). With the in organello assembly assay, we found that tom6Δ mitochondria were partially impaired in formation of the SAM-Ib stage, resembling a milder form of the defect of tom5Δ mitochondria, suggesting that Tom6 may indeed be involved in an early stage of Tom40 biogenesis at the SAM complex.
To directly define the functions of individual small Tom proteins, we established a combined genetic-biochemical approach. Mitochondrial protein sorting defects of yeast deletion mutants were complemented by synthesizing small Tom proteins in chemical amounts and importing them into isolated mitochondria. With this approach we could show that Tom5 as well as Tom6 promoted the assembly pathway of Tom40 at the SAM complex, whereas Tom7 had no stimulatory effect. By antibody shift analysis, we demonstrated that Tom5 and Tom6 associated with the precursor of Tom40 at the SAM-Ib stage, thus providing a direct biochemical explanation for the genetic connections. Tom5 plays the major role in Tom40 maturation at the SAM complex for the following reasons. First, the conversion of SAM-Ia to SAM-Ib is more severely inhibited in tom5Δ mitochondria than in tom6Δ mitochondria. Second, chemical amounts of Tom6 strongly stimulate the formation of SAM-Ib when Tom5 is present in wild-type amounts, but only partially rescue Tom40 assembly when Tom5 is absent.
The new assembly intermediate provided the possibility to address the role of Mim1 in Tom40 assembly. We found that mitochondria lacking Mim1 displayed a similar defect in assembly of the Tom40 precursor as mitochondria lacking Tom5 (i.e., accumulation of the precursor at the first stage of SAM interaction). Because Mim1 is required for the efficient import and membrane insertion of small Tom proteins (Becker et al., 2008; Thornton et al., 2010), we speculated that the role in import of small Tom proteins may explain the requirement of Mim1 for the assembly pathway of Tom40. Indeed, import of chemical amounts of Tom5 fully suppressed the Tom40 assembly defect of mim1Δ mitochondria, whereas Tom6 partially suppressed the defect—in line with the role of these small Tom proteins in Tom40 maturation at the SAM complex. We conclude that Mim1 is not directly needed for the assembly stages of Tom40 but functions via the import of small Tom proteins.
After the Mim1-dependent membrane insertion, the precursors of small Tom proteins depend on SAM functions for assembly into the TOM complex (Stojanovski et al., 2007a; Becker et al., 2008; Thornton et al., 2010). In case of Tom6, an interaction of the radiolabeled precursor with the SAM complex was observed. The Tom6 precursor associated with a module of pre-existing Tom40-Tom5 at the SAM complex; however, due to the transient nature of the intermediate the Tom6-SAM interaction was observed in only low abundance (Thornton et al., 2010) and thus it remained open whether this assembly at the SAM complex was only a special pathway for the precursor of Tom6 or whether it represented a main mechanism for the biogenesis of further Tom proteins. We show here that the two stages of Tom40 interaction with the SAM complex represent main import stages in the biogenesis pathway of this essential β-barrel precursor and conclude that the assembly of Tom40 with small Tom proteins represents an important function of the SAM complex.
ACKNOWLEDGMENTS
We thank Dr. Natalia Gebert for discussion. This work was supported by the Baden-Württemberg Stiftung, Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 746, Trinationales Graduiertenkolleg GRK 1478, Excellence Initiative of the German Federal & State Governments (EXC 294 BIOSS; GSC-4 Spemann Graduate School), Bundesministerium für Bildung und Forschung, Landesforschungspreis Baden-Württemberg, Gottfried Wilhelm Leibniz Program, and the Fonds der Chemischen Industrie.
Abbreviations used:
- 5-FOA
5-Fluoroorotic acid
- Mdm
mitochondrial distribution and morphology
- Mim1
mitochondrial import protein 1
- SAM
sorting and assembly machinery
- TOM
translocase of outer membrane.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-06-0518) on July 28, 2010.
REFERENCES
- Ahting U., Thun C., Hegerl R., Typke D., Nargang F. E., Neupert W., Nussberger S. The TOM core complex: the general protein import pore of the outer membrane of mitochondria. J. Cell Biol. 1999;29:959–968. doi: 10.1083/jcb.147.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alconada A., Kübrich M., Moczko M., Hönlinger A., Pfanner N. The mitochondrial receptor complex: the small subunit Mom8b/Isp6 supports association of receptors with the general insertion pore and transfer of preproteins. Mol. Cell Biol. 1995;15:6196–6205. doi: 10.1128/mcb.15.11.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker K. P., Schaniel A., Vestweber D., Schatz G. A yeast mitochondrial outer membrane protein essential for protein import and cell viability. Nature. 1990;348:605–609. doi: 10.1038/348605a0. [DOI] [PubMed] [Google Scholar]
- Becker L., Bannwarth M., Meisinger C., Hill K., Model K., Krimmer T., Casadio R., Truscott K. N., Schulz G. E., Pfanner N., Wagner R. Preprotein translocase of the outer mitochondrial membrane: reconstituted Tom40 forms a characteristic TOM pore. J. Mol. Biol. 2005;353:1011–1020. doi: 10.1016/j.jmb.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Becker T., Pfannschmidt S., Guiard B., Stojanovski D., Milenkovic D., Kutik S., Pfanner N., Meisinger C., Wiedemann N. Biogenesis of the mitochondrial TOM complex: Mim1 promotes insertion and assembly of signal-anchored receptors. J. Biol. Chem. 2008;283:120–127. doi: 10.1074/jbc.M706997200. [DOI] [PubMed] [Google Scholar]
- Becker T., Gebert M., Pfanner N., van der Laan M. Biogenesis of mitochondrial membrane proteins. Curr. Op. Cell Biol. 2009;21:484–493. doi: 10.1016/j.ceb.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Boldogh I. R., Nowakowski D. W., Yang H. C., Chung H., Karmon S., Royes P., Pon L. A. A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery. Mol. Biol. Cell. 2003;14:4618–4627. doi: 10.1091/mbc.E03-04-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos M. P., Robert V., Tommassen J. Biogenesis of the gram-negative bacterial outer membrane. Annu. Rev. Microbiol. 2007;61:191–214. doi: 10.1146/annurev.micro.61.080706.093245. [DOI] [PubMed] [Google Scholar]
- Chacinska A., Koehler C. M., Milenkovic D., Lithgow T., Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan N. C., Lithgow T. The peripheral membrane subunits of the SAM complex function codependently in mitochondrial outer membrane biogenesis. Mol. Biol. Cell. 2008;19:126–136. doi: 10.1091/mbc.E07-08-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker P. J., Ryan M. T., Brix J., Müller H., Hönlinger A., Pfanner N. Preprotein translocase of the outer mitochondrial membrane: molecular dissection and assembly of the general import pore. Mol. Cell Biol. 1998;18:6515–6524. doi: 10.1128/mcb.18.11.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembowski M., Künkele K. P., Nargang F. E., Neupert W., Rapaport D. Assembly of Tom6 and Tom7 into the TOM core complex of Neurospora crassa. J. Biol. Chem. 2001;276:17679–17685. doi: 10.1074/jbc.M009653200. [DOI] [PubMed] [Google Scholar]
- Dietmeier K., Hönlinger A., Bömer U., Dekker P. J., Eckerskorn C., Lottspeich F., Kübrich M., Pfanner N. Tom5 functionally links mitochondrial preprotein receptors to the general import pore. Nature. 1997;388:195–200. doi: 10.1038/40663. [DOI] [PubMed] [Google Scholar]
- Dolezal P., Likic V., Tachezy J., Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313:314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- Dukanovic J., Dimmer K. S., Bonnefoy N., Kumpe K., Rapaport D. Genetic and functional interactions between the mitochondrial outer membrane proteins Tom6 and Sam37. Mol. Cell. Biol. 2009;29:5975–5988. doi: 10.1128/MCB.00069-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T., Yamano K. Multiple pathways for mitochondrial protein traffic. Biol. Chem. 2009;390:723–730. doi: 10.1515/BC.2009.087. [DOI] [PubMed] [Google Scholar]
- Endo T., Yamano K. Transport of proteins across or into the mitochondrial outer membrane. Biochim. Biophys. Acta. 2010;1803:706–714. doi: 10.1016/j.bbamcr.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Esaki M., Shimizu H., Ono T., Yamamoto H., Kanamori T., Nishikawa S., Endo T. Mitochondrial protein import: requirement of presequence elements and Tom components for precursor binding to the TOM complex. J. Biol. Chem. 2004;279:19464–19470. doi: 10.1074/jbc.M404591200. [DOI] [PubMed] [Google Scholar]
- Gentle I., Gabriel K., Beech P., Waller R., Lithgow T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol. 2004;164:19–24. doi: 10.1083/jcb.200310092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratzer S., Lithgow T., Bauer R. E., Lamping E., Paltauf F., Kohlwein S. D., Haucke V., Junne T., Schatz G., Horst M. Mas37p, a novel receptor subunit for protein import into mitochondria. J. Cell Biol. 1995;129:25–34. doi: 10.1083/jcb.129.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan C. L., Kim S., Kahne D. Reconstitution of outer membrane protein assembly from purified components. Science. 2010;328:890–892. doi: 10.1126/science.1188919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K., Model K., Ryan M. T., Dietmeier K., Martin F., Wagner R., Pfanner N. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature. 1998;395:516–521. doi: 10.1038/26780. [DOI] [PubMed] [Google Scholar]
- Hönlinger A., Bömer U., Alconada A., Eckerskorn C., Lottspeich F., Dietmeier K., Pfanner N. Tom7 modulates the dynamics of the mitochondrial outer membrane translocase and plays a pathway-related role in protein import. EMBO J. 1996;15:2125–2137. [PMC free article] [PubMed] [Google Scholar]
- Hoppins S. C., Nargang F. E. The Tim8-Tim13 complex of Neurospora crassa functions in the assembly of proteins into both mitochondrial membranes. J. Biol. Chem. 2004;279:12396–12405. doi: 10.1074/jbc.M313037200. [DOI] [PubMed] [Google Scholar]
- Hulett J. M., Lueder F., Chan D. C., Perry A. J., Wolynec P., Likic V. A., Gooley P. R., Lithgow T. The transmembrane segment of Tom20 is recognized by Mim1 for docking to the mitochondrial TOM complex. J. Mol. Biol. 2008;376:694–704. doi: 10.1016/j.jmb.2007.12.021. [DOI] [PubMed] [Google Scholar]
- Ishikawa D., Yamamoto H., Tamura Y., Moritoh K., Endo T. Two novel proteins in the mitochondrial outer membrane mediate β-barrel protein assembly. J. Cell Biol. 2004;166:621–627. doi: 10.1083/jcb.200405138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Mihara K. Identification of Tom5 and Tom6 in the preprotein translocase complex of human mitochondrial outer membrane. Biochem. Biophys. Res. Commun. 2008;369:958–963. doi: 10.1016/j.bbrc.2008.02.150. [DOI] [PubMed] [Google Scholar]
- Kiebler M., Keil P., Schneider H., van der Klei I. J., Pfanner N., Neupert W. The mitochondrial receptor complex: a central role of MOM22 in mediating preprotein transfer from receptors to the general insertion pore. Cell. 1993;74:483–492. doi: 10.1016/0092-8674(93)80050-o. [DOI] [PubMed] [Google Scholar]
- Koehler C. M. New developments in mitochondrial assembly. Annu. Rev. Cell Dev. Biol. 2004;20:309–335. doi: 10.1146/annurev.cellbio.20.010403.105057. [DOI] [PubMed] [Google Scholar]
- Kornmann B., Currie E., Collins S. R., Schuldiner M., Nunnari J., Weissman J. S., Walter P. An ER-mitochondrial tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozjak V., Wiedemann N., Milenkovic D., Lohaus C., Meyer H. E., Guiard B., Meisinger C., Pfanner N. An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J. Biol. Chem. 2003;278:48520–48523. doi: 10.1074/jbc.C300442200. [DOI] [PubMed] [Google Scholar]
- Kutik S., Stojanovski D., Becker L., Becker T., Meinecke M., Krüger V., Prinz C., Meisinger C., Guiard B., Wagner R., Pfanner N., Wiedemann N. Dissecting membrane insertion of mitochondrial β-barrel proteins. Cell. 2008;132:1011–1024. doi: 10.1016/j.cell.2008.01.028. [DOI] [PubMed] [Google Scholar]
- Kutik S., Stroud D. A., Wiedemann N., Pfanner N. Evolution of mitochondrial protein biogenesis. Biochim. Biophys. Acta. 2009;1790:409–415. doi: 10.1016/j.bbagen.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Meisinger C., Ryan M. T., Hill K., Model K., Lim J. H., Sickmann A., Müller H., Meyer H. E., Wagner R., Pfanner N. Protein import channel of the outer mitochondrial membrane: a highly stable Tom40-Tom22 core structure differently interacts with preproteins, small Toms, and import receptors. Mol. Cell Biol. 2001;21:2337–2348. doi: 10.1128/MCB.21.7.2337-2348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisinger C., Rissler M., Chacinska A., Szklarz L. K., Milenkovic D., Kozjak V., Schönfisch B., Lohaus C., Meyer H. E., Yaffe M. P., Guiard B., Wiedemann N., Pfanner N. The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev. Cell. 2004;7:61–71. doi: 10.1016/j.devcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Meisinger C., Wiedemann N., Rissler M., Strub A., Milenkovic D., Schönfisch B., Müller H., Kozjak V., Pfanner N. Mitochondrial protein sorting: differentiation of β-barrel assembly by Tom7-mediated segregation of Mdm10. J. Biol. Chem. 2006;281:22819–22826. doi: 10.1074/jbc.M602679200. [DOI] [PubMed] [Google Scholar]
- Meisinger C., Pfannschmidt S, Rissler M., Milenkovic D., Becker T., Stojanovski D., Youngman M. J., Jensen R. E., Chacinska A., Guiard B., Pfanner N., Wiedemann N. The morphology proteins Mdm12/Mmm1 function in the major β-barrel assembly pathway of mitochondria. EMBO J. 2007;26:2229–2239. doi: 10.1038/sj.emboj.7601673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenkovic D., Kozjak V., Wiedemann N., Lohaus C., Meyer H. E., Guiard B., Pfanner N., Meisinger C. Sam35 of the mitochondrial protein sorting and assembly machinery is a peripheral outer membrane protein essential for cell viability. J. Biol. Chem. 2004;279:22781–22785. doi: 10.1074/jbc.C400120200. [DOI] [PubMed] [Google Scholar]
- Model K., Meisinger C., Prinz T., Wiedemann N., Truscott K. N., Pfanner N., Ryan M. T. Multistep assembly of the protein import channel of the mitochondrial outer membrane. Nat. Struct. Biol. 2001;8:361–370. doi: 10.1038/86253. [DOI] [PubMed] [Google Scholar]
- Neupert W., Herrmann J. M. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Paschen S. A., Waizenegger T., Stan T., Preuss M., Cyrklaff M., Hell K., Rapaport D., Neupert W. Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature. 2003;426:862–866. doi: 10.1038/nature02208. [DOI] [PubMed] [Google Scholar]
- Popov-Celeketic J., Waizenegger T., Rapaport D. Mim1 functions in an oligomeric form to facilitate the integration of Tom20 into the mitochondrial outer membrane. J. Mol. Biol. 2008;376:671–680. doi: 10.1016/j.jmb.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Ruiz N., Kahne D., Silhavy T. Advances in understanding bacterial outer-membrane biogenesis. Nat. Rev. Microbiol. 2006;4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- Ryan M. T., Voos W., Pfanner N. Assaying protein import into mitochondria. Meth. Cell Biol. 2001;65:189–215. doi: 10.1016/s0091-679x(01)65012-x. [DOI] [PubMed] [Google Scholar]
- Schleiff E., Soll J. Membrane protein insertion: mixing eukaryotic and prokaryotic concepts. EMBO Rep. 2005;6:1023–1027. doi: 10.1038/sj.embor.7400563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt S., Ahting U., Eichacker L., Granvogel B., Go N. E., Nargang F. E., Neupert W., Nussberger S. Role of Tom5 in maintaining the structural stability of the TOM complex of mitochondria. J. Biol. Chem. 2005;280:14499–14506. doi: 10.1074/jbc.M413667200. [DOI] [PubMed] [Google Scholar]
- Sherman E. L., Go N. E., Nargang F. E. Functions of the small proteins in the TOM complex of Neurospora crassa. Mol. Biol. Cell. 2005;16:4172–4182. doi: 10.1091/mbc.E05-03-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stan T., Ahting U., Dembowski M., Künkele K. P., Nussberger S., Neupert W., Rapaport D. Recognition of preproteins by the isolated TOM complex of mitochondria. EMBO J. 2000;19:4895–4902. doi: 10.1093/emboj/19.18.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski D., Guiard B., Kozjak-Pavlovic V., Pfanner N., Meisinger C. Alternative function for the mitochondrial SAM complex in biogenesis of α-helical TOM proteins. J. Cell Biol. 2007a;179:881–893. doi: 10.1083/jcb.200706043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski D., Pfanner N., Wiedemann N. Import of proteins into mitochondria. Meth. Cell Biol. 2007b;80:783–806. doi: 10.1016/S0091-679X(06)80036-1. [DOI] [PubMed] [Google Scholar]
- Stroud D. A., Meisinger C., Pfanner N., Wiedemann N. Biochemistry: assembling the outer membrane. Science. 2010;328:831–832. doi: 10.1126/science.1190507. [DOI] [PubMed] [Google Scholar]
- Suzuki H., Kadowaki T., Maeda M., Sasaki H., Nabekura J., Sakaguchi M., Mihara K. Membrane-embedded C-terminal segment of rat mitochondrial Tom40 constitutes protein-conducting pore with enriched β-structure. J. Biol. Chem. 2004;279:50619–50629. doi: 10.1074/jbc.M408604200. [DOI] [PubMed] [Google Scholar]
- Thornton N., Stroud D. A., Milenkovic D., Guiard B., Pfanner N., Becker T. Two modular forms of the mitochondrial sorting and assembly machinery are involved in biogenesis of α-helical outer membrane proteins. J. Mol. Biol. 2010;396:540–549. doi: 10.1016/j.jmb.2009.12.026. [DOI] [PubMed] [Google Scholar]
- van Wilpe S., Ryan M. T., Hill K., Maarse A. C., Meisinger C., Brix J., Dekker P. J., Moczko M., Wagner R., Meijer M., Guiard B., Hönlinger A., Pfanner N. Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature. 1999;401:485–489. doi: 10.1038/46802. [DOI] [PubMed] [Google Scholar]
- Waizenegger T., Habib S. J., Lech M., Mokranjac D., Paschen S. A., Hell K., Neupert W., Rapaport D. Tob38, a novel essential component in the biogenesis of β-barrel proteins of mitochondria. EMBO Rep. 2004;5:704–709. doi: 10.1038/sj.embor.7400183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waizenegger T., Schmitt S., Zivkovic J., Neupert W., Rapaport D. Mim1, a protein required for the assembly of the TOM complex of mitochondria. EMBO Rep. 2005;6:57–62. doi: 10.1038/sj.embor.7400318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther D. M., Rapaport D. Biogenesis of mitochondrial outer membrane proteins. Biochim. Biophys. Acta. 2009;1793:42–51. doi: 10.1016/j.bbamcr.2008.04.013. [DOI] [PubMed] [Google Scholar]
- Walther D. M., Papic D., Bos M. P., Tommassen J., Rapaport D. Signals in bacterial β-barrel proteins are functional in eukaryotic cells for targeting to and assembly in mitochondria. Proc. Natl. Acad. Sci. USA. 2009;106:2531–2536. doi: 10.1073/pnas.0807830106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N., Kozjak V., Chacinska A., Schönfisch B., Rospert S., Ryan M. T., Pfanner N., Meisinger C. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature. 2003;424:565–571. doi: 10.1038/nature01753. [DOI] [PubMed] [Google Scholar]
- Wiedemann N., Truscott K. N., Pfannschmidt S., Guiard B., Meisinger C., Pfanner N. Biogenesis of the protein import channel Tom40 of the mitochondrial outer membrane: intermembrane space components are involved in an early stage of the assembly pathway. J. Biol. Chem. 2004;279:18188–18194. doi: 10.1074/jbc.M400050200. [DOI] [PubMed] [Google Scholar]
- Wideman J. G., Go N. E., Klein A., Redmond E., Lackey S. W., Tao T., Kalbacher H., Rapaport D., Neupert W., Nargang F. E. Roles of the Mdm10, Tom7, Mdm12 and Mmm1 proteins in the assembly of mitochondrial outer membrane proteins in Neurospora crassa. Mol. Biol. Cell. 2010;21:1725–1736. doi: 10.1091/mbc.E09-10-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K., Yatsukawa Y., Esaki M., Hobbs A. E., Jensen R. E., Endo T. Tom20 and Tom22 share the common signal recognition pathway in mitochondrial protein import. J. Biol. Chem. 2008;283:3799–3807. doi: 10.1074/jbc.M708339200. [DOI] [PubMed] [Google Scholar]
- Yamano K., Tanaka-Yamano S., Endo T. Mdm10 as a dynamic constituent of the TOB/SAM complex directs coordinated assembly of Tom40. EMBO Rep. 2010;11:187–193. doi: 10.1038/embor.2009.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M., Hoogenraad N., Terada K., Mori M. Identification and functional analysis of human Tom22 for protein import into mitochondria. Mol. Cell Biol. 2000;20:7205–7213. doi: 10.1128/mcb.20.19.7205-7213.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]