Phosphatidylinositol (PI) is unique in the abundance of stearic acid at the sn-1 position. This fatty acid is thought to be incorporated through fatty acid remodeling. Here, we identified a phospholipase and acyltransferases involved in the fatty acid remodeling at the sn-1 position of PI and provide a link between the sn-1 fatty acid of PI and asymmetric cell division.
Abstract
Phosphatidylinositol (PI), an important constituent of membranes, contains stearic acid as the major fatty acid at the sn-1 position. This fatty acid is thought to be incorporated into PI through fatty acid remodeling by sequential deacylation and reacylation. However, the genes responsible for the reaction are unknown, and consequently, the physiological significance of the sn-1 fatty acid remains to be elucidated. Here, we identified acl-8, -9, and -10, which are closely related to each other, and ipla-1 as strong candidates for genes involved in fatty acid remodeling at the sn-1 position of PI. In both ipla-1 mutants and acl-8 acl-9 acl-10 triple mutants of Caenorhabditis elegans, the stearic acid content of PI is reduced, and asymmetric division of stem cell-like epithelial cells is defective. The defects in asymmetric division of these mutants are suppressed by a mutation of the same genes involved in intracellular retrograde transport, suggesting that ipla-1 and acl genes act in the same pathway. IPLA-1 and ACL-10 have phospholipase A1 and acyltransferase activity, respectively, both of which recognize the sn-1 position of PI as their substrate. We propose that the sn-1 fatty acid of PI is determined by ipla-1 and acl-8, -9, -10 and crucial for asymmetric divisions.
INTRODUCTION
Phosphatidylinositol (PI) is a versatile lipid that not only serves as a structural component of cellular membranes, but also plays important roles in signal transduction through distinct phosphorylated derivatives of the inositol head group (Di Paolo and De Camilli, 2006). The pathway for de novo synthesis of PI begins with the acylation of glycerol 3-phosphate (G3P) at the sn-1 position by G3P acyltransferase to form lysophosphatidic acid (lysoPA). LysoPA is further acylated at the sn-2 position by lysoPA acyltransferase to form phosphatidic acid (PA), which serves as a general precursor for all phospholipids (Dircks and Sul, 1999; Wendel et al., 2009). PA is then converted to cytidinediphosphodiacylglycerol (CDP-DAG), which combines with inositol to form PI. The newly synthesized PI possesses a mono- or di-unsaturated fatty acid at the sn-2 position (Akino and Shimojo, 1970; Holub and Kuksis, 1971a, 1972). In contrast, three-fourths or more of membrane PI obtained from mammalian tissues are constituted by the 1-stearoyl-2-arachidonoyl (18:0/20:4) species (Holub and Kuksis, 1971b; Baker and Thompson, 1972). Considerable attention has been paid to how membrane PI retains such a high proportion of arachidonoyl molecular species at the sn-2 position. It has been widely accepted that arachidonic acid (AA) is incorporated into PI by fatty acid remodeling in which the sn-2 acyl chain is replaced with AA by the sequential actions of phospholipase A2 (PLA2) and lysoPI acyltransferase (LPIAT) after de novo synthesis of PI (Akino and Shimojo, 1970; Holub and Kuksis, 1971a, 1972; Luthra and Sheltawy, 1976). In an RNA interference (RNAi)-based genetic screen, we recently identified mboa-7/LPIAT, a member of the membrane-bound O-acyltransferases (MBOAT) family, as an acyltransferase that selectively incorporates polyunsaturated fatty acids (PUFAs), such as AA and eicosapentaenoic acid (EPA), into the sn-2 position of PI (Lee et al., 2008). In this screen, we used the model organism Caenorhabditis elegans in which 1-stearoyl-2-eicosapentaenoyl (18:0/20:5) PI is the major PI species (Supplementary Figure 1; Lee et al., 2008). In C. elegans mboa-7/LPIAT mutants, EPA attached at the sn-2 position of PI was remarkably reduced and was replaced with other fatty acids such as oleic acid (18:1), indicating that fatty acid remodeling is crucial for determining the mature PI species.
On the other hand, many lines of evidence suggest that fatty acid remodeling also occurs at the sn-1 position of PI. In experiments with rabbit alveolar macrophage microsomes, newly synthesized PI from [14C]G3P contains palmitic acid (16:0), oleic acid (18:1), and linoleic acid (18:2) at the sn-1 position, but very low levels of stearic acid (18:0), the predominant fatty acid esterified at the sn-1 position of tissue PI (Nakagawa et al., 1989). In addition, incubation of rat liver microsomes with dipalmitoyl (16:0/16:0) CDP-DAG and [3H]inositol results in the rapid synthesis of [3H]PI and [3H]lysoPI, the latter of which is subsequently reacylated with stearic acid (18:0) at the sn-1 position (Darnell et al., 1991a,b). This suggests that dipalmitoyl [3H]PI formed by the microsomes is rapidly hydrolyzed by phospholipase A1 (PLA1) to produce sn-2-acyl lysoPI, which is then reacylated with stearic acid by acyltransferase, which is present in the microsomes. In fact, a high level of stearoyl-CoA:sn-2-acyl lysoPI acyltransferase activity was detected in rat liver microsomes (Holub and Piekarski, 1979; Darnell and Saltiel, 1991). These data strongly suggest that the fatty acid composition at the sn-1 position of PI is determined by the sequential actions of PLA1 and acyltransferase. However, the genes involved in this process have not been identified.
We previously showed that ipla-1, an intracellular PLA1 family member (Inoue and Aoki, 2006), acts as a regulator of asymmetric cell divisions in C. elegans (Kanamori et al., 2008). Loss of ipla-1 causes defects in asymmetric cell-fate determination and orientation of division of stem cell-like epithelial cells, called seam cells. We also found that a mutation of tbc-3 or mon-2, both of which are thought to be involved in endosome-to-Golgi retrograde transport (Lafourcade et al., 2004; Gillingham et al., 2006), suppresses the seam cell phenotypes of ipla-1 mutants. However, the role of ipla-1 in phospholipid fatty acid metabolism has not been determined. In this work, we analyzed the phospholipid fatty acid composition of ipla-1 mutants and found that the sn-1 fatty acid composition of PI was significantly altered in the mutants. Furthermore, we identified evolutionarily conserved acyltransferases that are required for normal fatty acid composition of the sn-1 acyl moiety of PI.
MATERIALS AND METHODS
Materials
PI and lysoPI from bovine liver,lysoPC from egg yolk, dioleoyl PA, dioleoyl phosphatidylcholine (PC), dioleoyl PE, 1-palmitoyl-2-oleoyl phosphatidylserine (PS) were purchased from Avanti Polar Lipids (Alabaster, AL). Phosphatidylglycerol (PG) from egg yolk was purchased from Sigma-Aldrich (St. Louis, MO). Dipalmitoyl PI was purchased from Serdary Research Laboratories (London, ON, Canada). [1-14C]palmitoyl-CoA, [1-14C]stearoyl-CoA, [1-14C]oleoyl-CoA, and [1-14C]arachidonoyl-CoA were purchased from American Radiolabeled Chemicals (St. Louis, MO). Rhizopus delemer lipase was purchased from Seikagaku (Tokyo, Japan). PLA2 from honey bee venom was purchased from Sigma-Aldrich.
General Methods and Strains
Worm cultures, genetic crosses, and other C. elegans methods were performed according to standard protocols (Brenner, 1974) except where otherwise indicated. The following mutations and transgenes were used: ipla-1(xh13), acl-8 acl-9(tm2290), acl-10(tm1045), mon-2(xh22), tbc-3(xh23), acs-20(tm3232); acs-22(tm3236) (Kage-Nakadai et al., 2010), wIs51[scm::gfp], osIs10[scm::wrm-1::venus], xhEx3517[acl-10p::gfp; rol-6(su1006)], xhEx3518[acl-10p::acl-10::gfp; rol-6(su1006)], xhEx3523[acl-10p::acl-10::gfp, dpy-7p::ipla-1::mCherry], xhEx3526[dpy- 7p::ipla-1::mCherry], xhEx3529[acl-10p::acl-10::mCherry], tmEx1920[acs-20p:: acs-20::egfp, gcy-10p::DsRed] (Kage-Nakadai et al., 2010), xhEx3514[scm::acl-10; Pges-1::dsREDm], xhEx3521[dpy-7p::mouse LYCAT; Pges-1::dsREDm], xhIs3501[dpy-7p::acl-10; Pges-1::dsREDm]. The mutant alleles acl-8 acl-9(tm2290) and acl-10(tm1045) were isolated in this study by TMP (trimethylpsoralen)/UV method (Gengyo-Ando and Mitani, 2000). The following primers for acl-8 acl-9 deletion screen were used: 5′-CGA CTG TGC TTC TCG ACT AA-3′; 5′-TAG TGC GGA AGA GAA CTT GT-3′; 5′-TCC TCA CTT CTC GGA ACT GT-3′; and 5′-AGG CAC CTC ATA GTG GTT GC-3′, and the following primers for acl-10 deletion screen were used: 5′-TCG AGG AGG AAA CAC CTT CT-3′; 5′-CTA CTT GCA TCC TGC TCG TT-3′; 5′-CGT CCA TTA CTC GGA TGG TT-3′; and 5′-AAT GGA CTT CTC GTG GAC TT-3′. Some of the strains used in this work were obtained from Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN). All mutations and were backcrossed at least five times before further analysis.
Cloning of C. elegans acl-10 and Mouse LYCAT
acl-10 cDNA (GenBank accession number NM_073570) was amplified by PCR from a C. elegans cDNA library using the primers, 5′-CAG AAG CTA GCA TGA TGA GGA TTC CAT GTC-3′ and 5′-AAA ATG GTA CCT TAT ATA GAA GAA GAT GAT-3′, and were cloned into pPD49.78 (a gift from Dr. A. Fire, Stanford University) at the NheI and KpnI sites. Mouse LYCAT cDNA (GenBank accession number NM_001081071) was amplified from a cDNA library derived from mouse heart using the primers, 5′-CCC GGG TAC CGA ATT CAC CAT GGA GCA GAA GCT GA-3′ and 5′-GGC CAT CGA TCT CGA GTT ACT CAT TTT TCT TTG AAT-3′, and cloned into pCAGGS-MCS vector (N-terminal Myc tag) at the EcoRI and XhoI sites.
Constructs and Transgenic Worms
For each construct, more than three independent transgenic lines were analyzed. pIR1 (acl-10p::gfp), pIR2 (acl-10p::acl-10::gfp), pIR3 (dpy-7p::acl-10), pIR4 (scm::acl-10), pIR5 (dpy-7p::ipla-1::mCherry), pIR6 (acl-10p::acl-10::mCherry) and pIR7 (dpy-7p::mouse LYCAT) were prepared as follows. pIR1 (acl-10p::gfp): 0.6-kb genomic fragment immediately upstream the ATG initiation codon of acl-10 was PCR amplified using the primers 5′-AAA TGC TGC AGA ATC GGA TAA AGA AAG GTG-3′ and 5′-GGA ATG GAT CCC ATT TCA ACT TCT GGA TGT G-3′, and cloned into pPD95.67 (NLS-; a derivative of pPD95.67, a gift from A. Fire, Stanford University School of Medicine) at the PstI and BamHI sites. pPD95.67 (NLS-) was constructed by removal of nuclear localization signal (NLS) from pPD95.67. pIR2 (acl-10p::acl-10::gfp): 3.3-kb genomic fragment corresponding to 0.6 kb immediately upstream the ATG initiation codon of acl-10 and the full-length acl-10 (2.7 kb) was PCR amplified using the primers 5′-AAA TGC TGC AGA ATC GGA TAA AGA AAG GTG-3′ and 5′-CCC GGG GAT CCG CTA TAG AAG AAG ATG ATG GC-3′, and cloned into pPD95.67 (NLS-) at the PstI and BamHI sites. pIR3 (dpy-7p::acl-10): acl-10 cDNA was subcloned under a dpy-7 promoter in a pTK030 (Kanamori et al., 2008) at the SmaI and NotI sites. pIR4 (scm::acl-10): acl-10 cDNA was subcloned under a scm promoter in a pTK020 (Kanamori et al., 2008) at the SmaI and NotI sites. pIR5 (dpy-7p::ipla-1::mCherry): dpy-7p::ipla-1 was PCR amplified from pTK030 (Kanamori et al., 2008) and was cloned into unc-122p::arl-8::mCherry (pYB109; Nakae et al., 2010) at the BamHI and NotI sites. pIR6 (acl-10p::acl-10::mCherry): acl-10p::acl-10 (described above) was PCR amplified using the primers 5′-AAA TGG GAT CCA ATC GGA TAA AGA AAG GTG-3′ and 5′-GCC AAT CCC GGC GGC CGC CTA CCG GTA CCC TCC AAG GGT CCT CCT TTG GGT CCT TTG GCC AAT CCC GGG GGT CGG CTA TAG AAG AAG ATG ATG GCG-3′, and was cloned into pYB109 (Nakae et al., 2010) at the BamHI and NotI sites. pIR7 (dpy-7p::mouse LYCAT): Mouse LYCAT cDNA was PCR amplified using the primers 5′-GGG ATG GTC TCC TGG AAG GGA-3′ and 5′-AGA ATA GGG CGC GGC CGC TTA CTC ATT TTT CTT TG-3′, and cloned under a dpy-7 promoter in a pTK030 at the SmaI and NotI sites. The primers used were designed based on the codon-usage preference in C. elegans (Stenico et al., 1994).
Lipid Analysis
Total lipids of synchronized L4 stage worms were extracted by the method of Bligh and Dyer (Bligh and Dyer, 1959). The fatty acid composition of each phospholipid was determined by gas chromatography (GC) as described previously (Lee et al., 2008) with slight modifications. Briefly, phospholipids were separated from total lipids by one-dimensional TLC on silica gel 60 plates (Merck Biosciences, Darmstadt, Germany) in chloroform:methanol:acetic acid (65:25:13, vol/vol). The area of silica gel corresponding to each phospholipid (PC, PE, and PS+PI) was scraped off the plates. The PS+PI fraction was reextracted and separated by TLC in chloroform:ethanol:water:triethylamine (30:35:7:35, vol/vol), and the areas of silica gel corresponding to PS and PI were scraped off the plates. Each phospholipid fraction was then treated with dehydrated methanol:acetyl chloride (10:1) to produce the fatty acid methyl esters. The fatty acid methyl ester derivatives were further analyzed with a GC 353B gas chromatograph equipped with a flame ionization detector (GL Sciences, Tokyo, Japan). Liquid chromatography/electrospray ionization-mass spectrometry (LC/ESI-MS) analysis was performed as described previously (Ban et al., 2007) using 1,2-dipalmitoyl PI as internal standard.
PLA Assay
PLA assay using recombinant IPLA-1 was performed as described previously (Morikawa et al., 2007), except that human embryonic kidney (HEK) 293 cells were used instead of HeLa cells. Purified recombinant protein was incubated with liposomes containing each phospholipid (dioleoyl PA, dioleoyl PC, dioleoyl PE, 1-palmitoyl-2-oleoyl PS, and dipalmitoyl PI) at 37°C for 3 h. The released free fatty acid level was measured with the ACS-ACOD method (NEFA C-Test kit 279–75401, Wako Chemical, Osaka, Japan).
Acyltransferase Assay
sn-2-acyl-1-lysophospholipids (sn-2-acyl lysophospholipids) were prepared as described previously (Lee et al., 2008) using dioleoyl PC, dioleoyl PE, sn-1-palmitoyl-2-oleoyl PS, bovine liver PI, and egg yolk PG. Each sn-2-acyl lysophospholipid was immediately used for acyltransferase assay. Because sn-2-acyl lysophospholipid is known to easily isomerize to sn-1-acyl-2-lysophospholipid (sn-1-acyl lysophospholipid), it is possible that 14C-acyl donor is incorporated into sn-1-acyl lysophospholipid and sn-2-acyl lysophospholipid (Supplementary Figure 2A). Thus, an accurate measure of sn-2-acyl lysophospholipid acyltransferase activity could only be obtained by determining the position that had been acylated. The acyltransferase reaction mixtures contained 80 μM lysophospholipid, 12.5 μM [14C]acyl-CoA (55 mCi/mmol), and 50 μg of microsomal protein of C. elegans in a total volume of 0.8-ml assay buffer (0.15 M KCl, 0.25 M sucrose, 50 mM potassium phosphate buffer [pH 6.8]). After incubation at 20°C for 5 min, reactions were stopped by the addition of 2 ml of methanol. Total lipid was extracted by the method of Bligh and Dyer, and separated by one-dimensional TLC on silica gel 60 plates (Merck) in chloroform/methyl acetate/1-propanol/methanol/0.25% KCl (25/25/25/10/9, vol/vol). To check the positional specificity, the radiolabeled product was reextracted from the TLC plates and treated with bee venom PLA2 (Supplementary Figure 2). The distribution of radioactivity between the resultant sn-1-acyl lysophospholipid and free fatty acids was assessed after TLC in chloroform/methanol/acetic acid/water (50:30:8:4, vol/vol). Acyl-CoA:sn-2-acyl lysophospholipid acyltransferase activity was determined by the radioactivity of sn-1-acyl lysophospholipid (Supplementary Figure 2, B and C). Acyl-CoA:sn-1-acyl lysoPI acyltransferase assay was performed essentially as described previously (Lee et al., 2008), except that 50 μg of microsomal protein and 80 μM lysoPI from bovine liver were used for the acyltransferase assay. Acyltransferase activity of the HEK 293 cell microsomes was measured similarly, except that the incubation temperature was 37°C.
Cell Culture and Transfection
HEK 293 cells were maintained in Dulbecco's modified eagle medium (DMEM) supplemented with 10% fetal calf serum, penicillin (100 U/ml), streptomycin (100 mg/ml), and l-glutamine (2 mM). Transfection of the plasmid DNA into cells was performed using LipofectAMINE 2000 (Invitrogen, San Diego, CA) according to the manufacturer's protocol.
Microscopy and Phenotypic Analysis
Animals were mounted on a 5% agar pad on a glass slide and immobilized in 0.02 M azide. Fluorescence images were obtained using an Axio Imager M1 (Carl Zeiss MicroImaging, Japan) microscope equipped with a digital CCD camera. Confocal images were obtained using a Zeiss LSM510META confocal microscope system (Carl Zeiss MicroImaging, Thornwood, NY). The orientation of seam cell division was analyzed as previously described (Kanamori et al., 2008). Briefly, a line was drawn between the two daughter nuclei and the angle between this line and the A-P axis was calculated. The angle outside the range of ±10° was defined as abnormal.
Western Blot Analysis
Mix-stage populations of worms were collected and sonicated in SET buffer (10 mM Tris-HCl, pH 7.4, 1 mM EDTA, and 250 mM sucrose) with protease inhibitors (5 μg/ml leupeptin, 5 μg/ml pepstatin A, 5 μg/ml aprotinin, and 1 mM PMSF). After sonication, the lysates were centrifuged at 1000 × g for 10 min at 4°C. The resulting supernatants were further centrifuged at 100,000 × g for 60 min at 4°C, and the supernatant and sedimented fractions (S100 and P100, respectively) were subjected to SDS-PAGE and immunoblotting. The antibodies used in the immunoblots and their dilutions were anti-IPLA-1 polyclonal antibody (Kanamori et al., 2008), 1:20; anti-mCherry mAb (632543, Clontech), 1:1000; anti-α-tubulin mAb (DM1A, Sigma) 1:5000; and anti-green fluorescent protein (GFP) mAb (JL-8, Clontech), 1:1000.
RESULTS
ipla-1 Mutation Causes Reduced Stearic Acid Content of PI
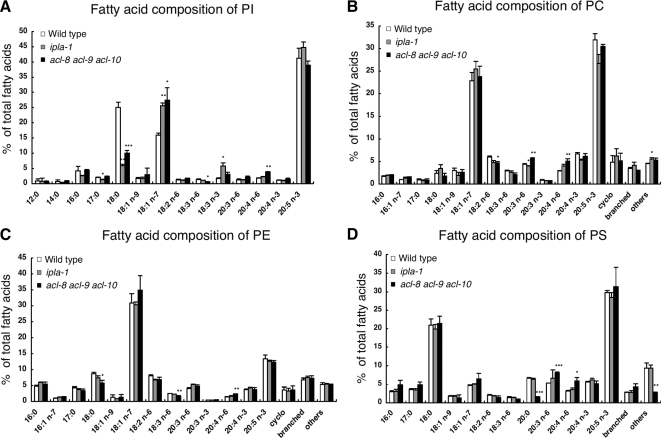
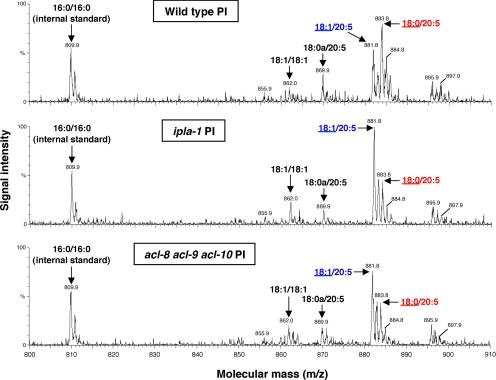
PLA1 is an enzyme that hydrolyzes fatty acids attached at the sn-1 position of phospholipids. The C. elegans genome contains one intracellular PLA1 family member named ipla-1 (Kanamori et al., 2008). To determine the role of ipla-1 in phospholipid metabolism, we first analyzed the fatty acid composition of phospholipids in wild-type and ipla-1 mutants by GC (Figure 1, A–D). GC analysis of PC, PE, PS, and PI revealed that the ipla-1 mutation significantly affected the fatty acid composition of PI, but not the fatty acid compositions of PC, PE, or PS. In ipla-1 mutants, the amount of stearic acid (18:0), which is the major fatty acid in the sn-1 position of PI, was reduced to 6.0% of total PI fatty acids compared with 25.1% in wild-type animals. Conversely, the amount of vaccenic acid (18:1n-7) in PI increased to 25.6% of total PI fatty acids compared with 16.0% in wild-type animals. The amount of EPA (20:5), the predominant fatty acid at the sn-2 position of PI in C. elegans (Supplementary Figure 1; Lee et al., 2008) was not affected. Consistent with this observation, LC/ESI-MS analysis revealed that the amount of 18:0/20:5-PI was reduced and instead, 18:1/20:5-PI was increased in ipla-1 mutants (Figure 2). These data indicate that ipla-1 is required for the incorporation of 18:0 into the sn-1 position of PI.
Figure 1.
Fatty acid composition of PI (A), PC (B), PE (C), and PS (D) from wild-type, ipla-1 mutants and acl-8 acl-9 acl-10 triple mutants. GC analysis was used to measure the individual fatty acid species. cyclo, total cyclopropane fatty acids (17: Δ, 19: Δ); branched, branched fatty acids (15:0 iso, 15:0 ante, 16:0 iso, 17:0 iso, 17:0 ante); others, unidentified fatty acids. Bars, mean ± SEM of at least three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
Figure 2.
MS analysis of PI molecular species. Negative ionization LC/ESI-MS spectra of PI molecular species of wild-type (top), ipla-1 mutants (middle), and acl-8 acl-9 acl-10 triple mutants (bottom). Assigning specific molecular species to m/z values was based on their calculated theoretical monoisotopic masses and verified by MS/MS. Lower case a refers to alkyl ether linkage.
Fatty Acid Composition of PI in acl-8 acl-9 acl-10 Triple Mutants Is Similar to That in ipla-1 Mutants
Deacylation-reacylation of phospholipids by PLA and acyltransferase is assumed to be important for acquiring the appropriate fatty acid composition of membrane phospholipids (Lands, 1958; Waku and Nakazawa, 1972; Lands, 2000). So far, several acyltransferases that incorporate fatty acids into the sn-2 position of lysophospholipids have been identified and are classified into two gene families, the 1-acylglycerol-3-phosphate O-acyltransferase (AGPAT) family and the MBOAT family (Shindou and Shimizu, 2009). We have isolated all mutants of AGPAT family members in C. elegans (acl-1-14; Supplementary Table 1) and found that the triple mutants of acl-8, -9, and -10 (Supplementary Figure 3), which are closely related to each other (Supplementary Figure 4), exhibited vulval defects similar to those of ipla-1 mutants (see below). This observation led us to examine the fatty acid composition of phospholipids in acl-8 acl-9 acl-10 triple mutants [acl-8 acl-9 (tm2290) acl-10 (tm1045)].
In acl-8 acl-9 acl-10 mutants, the amount of stearic acid (18:0) in PI was reduced (25.1% in wild-type vs. 10.0% in acl-8 acl-9 acl-10 mutants), whereas the amount of vaccenic acid (18:1n-7) in PI increased (16.0% in wild-type vs. 27.5% in acl-8 acl-9 acl-10 mutants) in a similar manner to ipla-1 mutants (Figure 1A). The amount of EPA (20:5) in PI was unaltered. LC/ESI-MS analysis showed that 18:0/20:5-PI was replaced by 18:1/20:5-PI in acl-8 acl-9 acl-10 mutants like it was in ipla-1 mutants. No drastic changes were observed in the fatty acid compositions of other phospholipids such as PC, PE, and PS (Figure 1, B–D). These data indicate that acl-8, -9, and -10 are also involved in the incorporation of 18:0 into the sn-1 position of PI.
Phenotypes of acl-8 acl-9 acl-10 Triple Mutants Are Similar to Those of ipla-1 Mutants
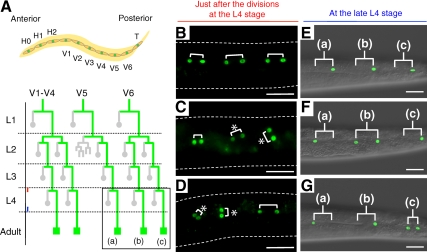
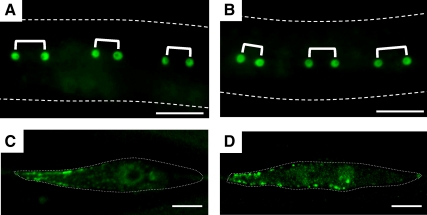
We previously demonstrated that ipla-1 mutants exhibited defects in vulval morphology (Supplementary Figure 5, A and B) and cell division of lateral epithelial cells, termed seam cells (Figure 3 and Table 1; Kanamori et al., 2008). In the course of constructing the acl-8 acl-9 acl-10 triple mutants used above, we noticed that acl-8 acl-9 acl-10 mutants, like ipla-1 mutants, showed vulval defects, including protruding and bursting vulvae (Supplementary Figure 5, A–C). To address whether acl-8 acl-9 acl-10 mutants also show abnormal cell division of seam cells, we analyzed the seam cell divisions of acl-8 acl-9 acl-10 mutants using a seam cell marker, scm::gfp. During the larval stages, seam cells divide asymmetrically in a stem cell-like manner, producing an anterior daughter cell that fuses with a major epithelial syncytium (hyp7) and loses the expression of scm::gfp, and a posterior daughter cell that assumes the seam cell fate again and continues to express scm::gfp (Figure 3A). Each seam cell division is oriented parallel to the anterior-posterior axis (A-P axis). Because orientation of seam cell division is randomized relative to the A-P axis in ipla-1 mutants (Kanamori et al., 2008), we first analyzed the orientation of seam cell division. In wild-type worms, all seam cells divided parallel to the A-P axis as judged by scm::gfp, which is expressed in the nuclei of both daughter cells just after the division (Figure 3B and Table 1). On the other hand, ipla-1 mutants exhibited aberrant orientation of seam cell divisions as reported previously (Figure 3C and Table 1; Kanamori et al., 2008). acl-8 acl-9 acl-10 triple mutations also caused misorientation of seam cell divisions similar to that observed in ipla-1 mutants (i.e., the seam cells divided obliquely, and sometimes at right angles to the A-P axis; Figure 3D and Table 1). Next, we investigated the cell lineage pattern of seam cells at the L4 stage in acl-8 acl-9 acl-10 mutants. As mentioned above, in wild-type worms, all posterior daughter cells maintained expression of scm::gfp and assumed the seam cell fate again (Figure 3E). In contrast, the seam cell division became symmetric or was reversed (the fates of daughters were the opposite of what they are in the wild type) in ipla-1 mutants (62%, n = 53; Figure 3F; Kanamori et al., 2008). Similarly, the asymmetry of the divisions was often disrupted in acl-8 acl-9 acl-10 mutants (67%, n = 25; Figure 3G). These observations indicate that acl-8 acl-9 acl-10 mutants, like ipla-1 mutants, are defective in orientation and cell-fate determination of seam cell divisions.
Figure 3.
ipla-1 mutants and acl-8 acl-9 acl-10 triple mutants are defective in the orientation and asymmetric cell-fate determination of seam cell divisions. Anterior is toward the left. (A, top) Schematic arrangement of seam cells on each side in an early L1 larva. (A, bottom) The postembryonic division pattern of V seam cells. The expression pattern of scm::gfp, which is specifically expressed in the nuclei of seam cells, is indicated by green lines. The box marks the lineages analyzed in this study: V5.pppp lineage (a), V6.papp lineage (b), and V6.pppp lineage (c). Gray circles represent anterior daughters that fuse with the epithelial syncytium, hyp7, and green squares denote seam cells. Red and blue lines indicate the developmental stages corresponding to those of B–D and E–G, respectively. (B–D) Fluorescent images of scm::gfp just after the divisions at the L4 stage. The shapes of the worms are indicated by dotted lines. Three pairs of daughter cells are shown with brackets. (B) Wild-type. All seam cells divide parallel to the A-P axis. (C and D) Representative scm::gfp images of ipla-1 mutants (C) and acl-8 acl-9 acl-10 triple mutants (D). In ipla-1 mutants and acl-8 acl-9 acl-10 triple mutants, the seam cell division is randomly oriented relative to the A-P axis (C and D; *). (E–G) Seam cells at the late L4 stage visualized by scm::gfp. Merged fluorescence and differential interference contrast images are shown. The letters (a), (b), and (c) correspond to those of A. (E) Wild-type. The posterior daughter cells adopt the seam cell fate in all the three lineages. (F and G) Representative scm::gfp images of ipla-1 mutants (F) and acl-8 acl-9 acl-10 triple mutants (G). The asymmetry of the divisions is often disrupted in ipla-1 mutants and acl-8 acl-9 acl-10 triple mutants. Scale bars, 20 μm.
Table 1.
Orientation of seam cell division
| Strain | % of cell divisions with abnormal orientation | n |
|---|---|---|
| Wild type | 0 | 110 |
| ipla-1 | 52.4 | 107 |
| acl-8 acl-9 | 0 | 104 |
| acl-10 | 43.7 | 112 |
| acl-8 acl-9 acl-10 | 40.6 | 114 |
| ipla-1; acl-8 acl-9 acl-10 | 46.7 | 107 |
| acl-8 acl-9 acl-10; tbc-3 | 5.7 | 141 |
| acl-8 acl-9 acl-10; mon-2 | 5.5 | 109 |
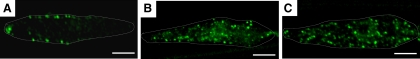
In C. elegans, the Wnt/β-catenin asymmetry pathway determines the cell fate of most asymmetric divisions (Mizumoto and Sawa, 2007b). In asymmetric division of seam cells, Wnt is expressed posterior to the mother cell before dividing, and this polarity information is converted to anterior cortical localization of β-catenin WRM-1 (Nakamura et al., 2005; Takeshita and Sawa, 2005; Mizumoto and Sawa, 2007a). The asymmetrical localization of WRM-1 in the mother cell is ultimately responsible for the asymmetric transcription in the two daughter cells after division of the mother cell. To understand the nature of the defects in the asymmetric division of acl-8 acl-9 acl-10 mutants, we examined the subcellular localization of WRM-1::GFP in mother seam cells at the L4 stage in acl-8 acl-9 acl-10 mutants. In wild-type seam cells, punctate fluorescence was clearly visible near the cell membrane in the anterior half of the cells (Figure 4A). On the other hand, the cortical localization of WRM-1::GFP was randomized in ipla-1 mutants as described previously (Figure 4B; Kanamori et al., 2008). The asymmetric cortical localization of WRM-1::GFP was also disrupted in acl-8 acl-9 acl-10 mutants, in which the localization of WRM-1::GFP was symmetric or enriched anteriorly or posteriorly in seam cells before divisions, and occasionally, we observed cytoplasmic puncta of WRM-1::GFP similar to those in ipla-1 mutants (Figure 4, B and C). These results indicate that both ipla-1 and acl-8, -9, -10 are required for the formation and/or maintenance of cortical asymmetry of WRM-1 before the divisions of the seam cells.
Figure 4.
Cortical asymmetry of β-catenin WRM-1 is disrupted in acl-8 acl-9 acl-10 triple mutants similar to that in ipla-1 mutants. Confocal images showing WRM-1::GFP localization in the V6.pppp cell of wild-type (A), ipla-1 mutants (B) and acl-8 acl-9 acl-10 triple mutants (C) before division of the cell. The anterior and dorsal sides of the cells are on the left and bottom, respectively. Scale bars, 5 μm. The shapes of the seam cells are indicated by dotted lines.
Suppressor Mutations of ipla-1 Suppress the Seam Cell Defects of acl-8 acl-9 acl-10 Triple Mutants
In a previous genetic screen, we isolated two alleles, tbc-3(xh23) and mon-2(xh22), as suppressors of the seam cell defects of ipla-1 mutants (Kanamori et al., 2008). mon-2 encodes a homologue of an ARF GEF-like protein and tbc-3 encodes a homologue of a Rab GAP, both of which have been reported to regulate endosome-to-Golgi retrograde transport (Lafourcade et al., 2004; Gillingham et al., 2006). We also found that a seam cell-specific RNAi for the retromer complex, which is also known to control intracellular retrograde transport (Seaman, 2005), suppresses the seam cell phenotypes of ipla-1 mutants, suggesting that reduction of endosome-to-Golgi retrograde transport in seam cells rescues the seam cell defects of ipla-1 mutants (Kanamori et al., 2008). In this study, we found that mutation of tbc-3 did not appreciably change the PI molecular species in ipla-1 mutants (Supplementary Figure 6), suggesting that the mutation suppresses seam cell phenotypes by abnormal membrane traffic downstream of altered PI molecular species.
To address whether the seam cell defects of acl-8 acl-9 acl-10 mutants are also mediated by retrograde transport, we crossed acl-8 acl-9 acl-10 mutants with tbc-3(xh23) or mon-2(xh22) mutants and analyzed seam cell divisions. The orientation of seam cell divisions was restored in both acl-8 acl-9 acl-10; tbc-3 and acl-8 acl-9 acl-10; mon-2 mutants. The percent of the seam cells in which the angle between the A-P axis and the direction of cell division was more than 10° was 6% in the acl-8 acl-9 acl-10; tbc-3 and acl-8 acl-9 acl-10; mon-2 mutants, whereas it was 41% in the acl-8 acl-9 acl-10 mutants (Figure 5, A and B; Table 1). The abnormal localization of WRM-1::GFP observed in the mother seam cells of acl-8 acl-9 acl-10 mutants was also rescued by a mutation of tbc-3(xh23) or mon-2(xh22) (Figure 5, C and D). These results suggest that the seam cell defects of ipla-1 mutants and acl-8 acl-9 acl-10 mutants occur through the same pathway, which is mediated by retrograde transport.
Figure 5.
Seam cell defects of acl-8 acl-9 acl-10 mutants are suppressed by a mutation of tbc-3 or mon-2. Anterior is oriented toward the left. (A and B) Randomized orientation of seam cell division is suppressed in acl-8 acl-9 acl-10; tbc-3 mutants (A) and acl-8 acl-9 acl-10; mon-2 mutants (B). Scale bars, 20 μm. (C and D) Anterior cortical localization of WRM-1::GFP is also restored in acl-8 acl-9 acl-10; tbc-3 mutants (C) and acl-8 acl-9 acl-10; mon-2 mutants (D). Scale bars, 5 μm.
Recombinant IPLA-1 Hydrolyzes a Fatty Acid Attached to PI
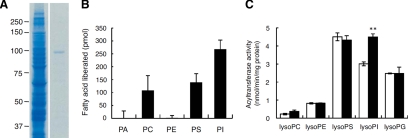
Mammals have at least three members of the intracellular PLA1 family, namely PA-PLA1, KIAA0725, and p125, all of which show significant homology to an ipla-1 gene product (IPLA-1; Higgs et al., 1998; Nakajima et al., 2002; Kanamori et al., 2008). PA-PLA1 and KIAA0725 have been reported to hydrolyze the sn-1 fatty acids attached to PI in in vitro assays (Higgs and Glomset, 1996; Morikawa et al., 2007). To determine whether C. elegans IPLA-1 also has hydrolytic activity toward PI, we prepared recombinant IPLA-1 by expressing it in HEK cells (Figure 6A, see Materials and Methods). As shown in Figure 6B, the purified recombinant IPLA-1 showed hydrolytic activity toward PI. PS and PC were also hydrolyzed by IPLA-1, but PE or PA did not serve as a substrate (Figure 6B).
Figure 6.
Enzymatic assay of IPLA-1 and ACL-10. (A) Purification of IPLA-1 expressed in HEK 293 cells was verified by SDS-PAGE followed by Coomassie Brilliant Blue staining. Left, lysates of HEK 293 cells transfected with ipla-1 cDNA in the pFLAG-CMV2 vector (Kanamori et al., 2008). Right, FLAG-IPLA-1 purified with anti-FLAG M2 affinity gel (Sigma) and eluted with buffer containing the FLAG peptide. (B) Substrate specificity of IPLA-1. The following phospholipids were used for substrates; dioleoyl PA, dioleoyl PC, dioleoyl PE, 1-palmitoyl-2-oleoyl PS, and dipalmitoyl PI. For details of the assay procedure, see Materials and Methods. (C) Substrate specificity of ACL-10. Acyltransferase activities of wild-type (□) or ACL-10–expressing transgenic worms (xhIs3501[dpy-7p::acl-10]; ■) were measured using [14C]stearoyl-CoA as an acyl donor and the indicated sn-2-acyl lysophospholipids as acyl acceptors. Bars, mean ± SEM of at least three independent experiments. **p < 0.01.
ACL-10 Possesses Acyltransferase Activity toward the sn-1 Position of PI
acl-10 single mutants showed defects in vulval morphology and seam cell divisions comparable to those of acl-8 acl-9 acl-10 triple mutants, whereas acl-8 acl-9 double mutants exhibited no abnormalities in vulval morphology or seam cell divisions (Table 1 and Supplementary Figure 5, C–E and I–K), indicating that acl-10 predominantly contributes to vulval morphology and seam cell divisions. We therefore focused on acl-10 and examined whether an acl-10 gene product (ACL-10) has LPIAT activity toward the sn-1 position of PI. The membrane fraction of ACL-10-expressing transgenic worms (xhIs3501[dpy-7p::acl-10]; see Materials and Methods) showed increased sn-2-acyl LPIAT activity with stearoyl-CoA as an acyl donor (Figure 6C). Acyltransferase activities against other sn-2-acyl lysophospholipids, such as lysoPC, lysoPE, lysoPS, and lysoPG, did not increase significantly (Figure 6C), indicating that ACL-10 prefers sn-2-acyl lysoPI as an acyl acceptor. Increased sn-2-acyl LPIAT activity was also observed when we used palmitoyl-CoA (16:0-CoA) and oleoyl-CoA (18:1n-9-CoA), but not arachidonoyl-CoA (20:4n-6-CoA), as acyl donors (data not shown). These data indicate that ACL-10 has acyltransferase activity toward the sn-1 position of PI with a preference for saturated and mono-unsaturated fatty acids. In the membrane fraction of acl-8 acl-9 acl-10 mutants, sn-2-acyl LPIAT activity was significantly reduced when we used stearoyl-CoA (18:0-CoA) as the acyl donor (Supplementary Figure 7). However, it was not reduced when we used arachidonoyl-CoA (20:4n-6-CoA), which is the preferred acyl donor of MBOA-7/LPIAT. This result indicates that ACL-8, -9, and -10 contribute to sn-2-acyl LPIAT activity with stearoyl-CoA, but not with arachidonoyl-CoA, in C. elegans. The observation that appreciable activity was still detected in the membrane fraction of acl-8 acl-9 acl-10 mutants suggests the existence of other sn-2-acyl LPIAT in the worms.
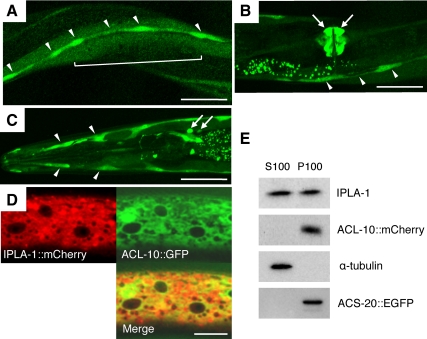
IPLA-1 and ACL-10 Are Expressed in the Endoplasmic Reticulum of Epithelial Cells
We also examined the expression pattern of acl-10 by using a transcriptional GFP fusion gene. Strong acl-10p::GFP expression was observed in seam cells throughout development (Figure 7A). Expression of GFP was also observed in other epithelial cells, such as vulval epithelial cells and the major epithelial syncytium hyp7, and in several head neurons including AIY interneurons (Figure 7, B and C). To analyze the intracellular localizations of IPLA-1 and ACL-10, we generated a transgenic strain expressing IPLA-1::mCherry and ACL-10::GFP under the control of the epidermal-specific dpy-7 promoter and acl-10 own promoter, respectively. IPLA-1::mCherry and ACL-10::GFP rescued the phenotypes of ipla-1 mutants and acl-8 acl-9 acl-10 mutants, respectively (Supplementary Figure 5, F and G), indicating that these fusion proteins are functional. As shown in Figure 7D, IPLA-1::mCherry and ACL-10::GFP were distributed in an endoplasmic reticulum (ER)-like reticular pattern throughout the cytoplasm and were partially colocalized. We also found that both IPLA-1 and ACL-10 partially colocalized with an ER marker, ACS-20::EGFP (Supplementary Figure 8; Kage-Nakadai et al., 2010). Furthermore, an immunoblot analysis revealed that both IPLA-1 and ACL-10 were present in the membrane fraction (Figure 7E). IPLA-1 was also present in the soluble fraction (Figure 7E), as was its mammalian homologue, KIAA0725 (Nakajima et al., 2002; Morikawa et al., 2009). These data indicate that ACL-10 and a portion of IPLA-1 are localized at the ER membrane in epithelial cells.
Figure 7.
Expression of ipla-1 and acl-10. (A–C) Confocal images of transgenic worms expressing acl-10p::GFP. Seam cells (A–C: arrowheads), epithelial syncytium hyp7 (A: bracket), vulval epithelium (B: arrows), and several neurons including AIYL/R (C: arrowheads) in the head region at the adult stage. Scale bars, 40 μm. (D) Confocal images of epithelial cells of a transgenic L4 worm expressing IPLA-1::mCherry and ACL-10::GFP. Bar, 20 μm. (E) Subcellular fractionation of IPLA-1 and ACL-10. Lysate of transgenic worms expressing ACL-10::mCherry and ACS-20::EGFP (acs-20;acs-22;xhEx3529[acl-10p::acl-10::mCherry]; tmEx1920[acs-20p::acs-20::egfp]) were subjected to S100 (cytosol)/P100 (membrane) fractionation and processed for immunoblotting with anti-IPLA, anti-mCherry (ACL-10), anti-α-tubulin (cytoplasmic marker), and anti-GFP (ER marker ACS-20) antibodies. acs-20 encodes a very long chain fatty acid acyl-CoA synthetase that is known to be localized in ER membranes. acs-22 is a homologous gene of acs-20. acs-20p::acs-20::egfp transgene fully rescues the phenotypes of acs-20;acs-22 double mutants (Kage-Nakadai et al., 2010).
DISCUSSION
Identification of ipla-1 and acl-8, -9, -10 as Strong Candidates for Enzymes Involved in Fatty Acid Remodeling at the sn-1 Position of PI
PI is a membrane phospholipid that has a unique fatty acid composition in that 1-stearoyl-2-arachidonoyl species is predominant in mammals (Holub and Kuksis, 1971b; Baker and Thompson, 1972). C. elegans, in which EPA is a major PUFA in membrane phospholipids and AA is a minor component, possesses 1-stearoyl-2-eicosapentaenoyl PI as the major molecular species (Lee et al., 2008). These molecular species are thought to be formed by a fatty acid remodeling reaction after the de novo synthesis of PI (Holub and Kuksis, 1971a; Nakagawa et al., 1989; Darnell et al., 1991a,b). The remodeling reaction involves the hydrolysis of a fatty acyl ester bond at the sn-1 or -2 position of the newly synthesized phospholipids and subsequent incorporation of the appropriate fatty acid into the position. We recently identified the acyltransferase, named mboa-7/LPIAT, which preferentially incorporates AA and EPA into the sn-2 position of PI (Lee et al., 2008). In the present study, we identified PLA1 (ipla-1) and acyltransferases (acl-8, -9, -10) that are involved in the incorporation of stearic acid into the sn-1 position of PI. We demonstrated that 1) stearic acid (18:0) attached at the sn-1 position of PI is replaced with vaccenic acid (18:1n-7) in both ipla-1 mutants and acl-8 acl-9 acl-10 mutants, 2) ipla-1 mutants and acl-8 acl-9 acl-10 mutants show similar phenotypes (i.e., they exhibit defects in orientation of seam cell divisions, cell-fate determination of seam cells, and cortical localization of β-catenin in mother seam cells), and 3) these phenotypes are suppressed by a same mutation (tbc-3(xh23) or mon-2(xh22)) in both ipla-1 mutants and acl-8 acl-9 acl-10 mutants. Together, these data strongly suggest that ipla-1 (PLA1) and acl-8, -9, -10 (acyltransferases) function in the same pathway. We also showed that ipla-1 mutants, acl-8 acl-9 acl-10 mutants, and ipla-1; acl-8 acl-9 acl-10 quadruple mutants have seam cell defects with similar penetrance (Table 1) and have similar fatty acid compositions of PI (Supplementary Figure 9), indicating no synergism between the ipla-1 and acl-8 acl-9 acl-10 mutations. These data further support the idea that ipla-1 and acl-8, -9, -10 function in the same pathway. In in vitro analyses, IPLA-1 was capable of hydrolyzing the fatty acyl moiety of PI, and ACL-10 transferred stearic acid to the sn-1 position of PI. From these observations, we propose that ipla-1 functions as a PLA1 and that acl-8, -9, -10 function as acyltransferases in the fatty acid remodeling of the sn-1 position of PI (Supplementary Figure 10).
Calcium-independent PLA2 (iPLA2) has been assumed to play a role in fatty acid remodeling of the sn-2 position of phospholipids (Balsinde et al., 1997). Recent knockout studies, however, revealed that lack of iPLA2β causes no significant change in the fatty acid composition of membrane phospholipids (Bao et al., 2007). Knockout mice of iPLA2γ, which hydrolyze the sn-2 ester bond of PC, show marked differences in fatty acid composition of cardiolipin (Mancuso et al., 2007), although the mechanism by which the fatty acid composition of cardiolipin is changed is unclear. As for PLA1, three members of the intracellular PLA1 family (PA-PLA1, KIAA0725, and p125) have been identified and analyzed (Inoue and Aoki, 2006; Morikawa et al., 2009), although the contribution of these PLA1s to fatty acid composition of membrane phospholipids is unknown. The present results show that ipla-1 mutants, in which PI-recognizing PLA1 is disrupted, have altered fatty acid composition of PI and seam cell defects similar to those observed in the acl-8 acl-9 acl-10 mutants, which lack acyltransferases for transferring the fatty acid at the sn-1 position of PI. To our knowledge, ipla-1 is the first identified phospholipase involved in the fatty acid remodeling at the sn-1 position of membrane phospholipids. In an in vitro analysis, IPLA-1 also showed hydrolytic activity toward PS and PC in addition to PI (Figure 6B), although the ipla-1 mutation did not appreciably affect the fatty acid composition of PS and PC in vivo (Figure 1). It is possible that IPLA-1 cleaves the sn-1 fatty acyl bond of PS and PC under our in vitro condition, but not in vivo. Alternatively, lysoPS and lysoPC produced by IPLA-1 might be reacylated back to their original states by certain acyltransferases in vivo.
According to the model shown in Supplementary Figure 10, mutations of acl-8, -9, -10 are expected to accumulate lysoPI, but not to accumulate 18:1/20:5-PI. However, in acl-8 acl-9 acl-10 mutants, the amount of 18:1/20:5-PI increased (Figures 1 and 2), and lysoPI was not detected by our measuring system (data not shown). This may be because acyltransferases other than ACL-8, -9, -10 transfer 18:1, the principal fatty acid in C. elegans (Hutzell and Krusberg, 1982), to the sn-1 position of PI in acl-8 acl-9 acl-10 mutants. It is also possible that IPLA-1 and ACL-8, -9, -10 activities are coupled so that IPLA-1 is inactive when ACL-8, -9, -10 are lost.
As mentioned above, acl-10 single mutants showed seam cell defects comparable to those of acl-8 acl-9 acl-10 triple mutants, whereas acl-8 acl-9 double mutants exhibited no abnormalities (Table 1), indicating that acl-10 predominantly contributes to seam cell divisions. In acl-10 single mutants, the amount of 18:0/20:5-PI was reduced and the amount of 18:1/20:5-PI was increased compared with the amounts in wild-type worms. However, the changes were less than those observed in acl-8 acl-9 acl-10 mutants (Supplementary Figure 11A). As shown in Figure 7A, acl-10 is strongly expressed in seam cells, but is not expressed in intestinal cells or muscle cells, which form large organs in C. elegans. We also found that acl-10 mRNA obtained from the whole C. elegans body was less than that of acl-8 or -9 as judged by quantitative real-time PCR (Supplementary Figure 11B). Therefore, the local expression of acl-10 appears to be the cause of the weak change of PI molecular species in acl-10 single mutants.
Several laboratories, including ours, have identified acyltransferases that incorporate fatty acids into the sn-2 position of lysophospholipids (Shindou and Shimizu, 2009). However, the acyltransferases transferring fatty acids at the sn-1 position of lysophospholipids are largely unknown. Very recently, another acyltransferase, Psi1p, was shown to catalyze acyl transfer to the sn-1 position of PI in yeast (Le Guedard et al., 2009). In psi1 Δ mutants, the content of stearic acid at the sn-1 position of PI is reduced, but the physiological function of Psi1p has not been elucidated. We show here that the C. elegans acyltransferase toward the sn-1 position of PI is required for asymmetric cell division, the fundamental mechanism by which multicellular organisms generate cell diversity.
Asymmetric Cell Division and Fatty Acid Remodeling of PI
The present results demonstrate that both ipla-1 mutants and acl-8 acl-9 acl-10 mutants have defects in asymmetric cell divisions of stem-cell like epithelial cells, named seam cells. In C. elegans, the Wnt/β-catenin asymmetry pathway determines the cell fate of most asymmetric divisions (Mizumoto and Sawa, 2007b; i.e., Wnt expressed posterior to the mother cell induces anterior cortical localization of β-catenin in the mother cell, leading to asymmetric cell-fate determination between the two daughter cells). Both ipla-1 mutants and acl-8 acl-9 acl-10 mutants exhibit defects in anterior cortical localization of β-catenin in mother cells and cell-fate determination of the daughter cells. We also found that ipla-1 and acl-10 act cell-autonomously to regulate asymmetric cell divisions of seam cells (Kanamori et al., 2008 and Supplementary Figure 5L, respectively). These data suggest that in ipla-1 mutants and acl-8 acl-9 acl-10 mutants, the altered fatty acid composition of PI in mother cells causes abnormal intracellular distribution of β-catenin, leading to defects in cell-fate determination of the daughter cells.
How is the fatty acid composition of PI involved in asymmetric cell divisions? It has been reported that asymmetric cortical enrichment of a phosphoinositide synthesis enzyme, ppk-1 [PI(4)P5-kinase], is required for asymmetric cell division in the one-celled C. elegans embryo (Panbianco et al., 2008). We also found that seam cell-specific knockdown of ppk-1 results in defective asymmetric division of seam cells (our unpublished data). These data suggest that the cortical asymmetry of phosphoinositides is important for asymmetric cell divisions. Furthermore, a recent study revealed that in HeLa cells, PI(3,4,5)P3 is accumulated in the midsection at the cortex, and the cortical localization of PI(3,4,5)P3 plays a crucial role in the orientation of cell divisions (Toyoshima et al., 2007). These previous reports, together with our findings, suggest that the cortical localization of phosphoinositides in the mother cell is required for correctly determining the fates of the two daughter cells and for the correct orientation of the cell divisions.
It has been assumed that cell-fate determination and orientation of the two daughter cells require the polarized distribution of cortical proteins in the mother cell (Gönczy, 2008; Siller and Doe, 2009) and that the polarized distribution of cortical proteins is achieved by membrane trafficking (Bilder, 2001; Rodriguez-Boulan et al., 2005). Meanwhile, it has been shown that differences in acyl chains in membrane phospholipids are responsible for the differential lateral distribution and accumulation of lipids in membrane microdomains (i.e., the phospholipids with long and saturated acyl chains preferentially partition into more rigid or highly ordered domains, such as lipid rafts, while those with short or unsaturated tails prefer to enter more fluid regions of the bilayer; Mukherjee and Maxfield, 2004). We speculate that in the mother seam cells of ipla-1 mutants and acl-8 acl-9 acl-10 mutants, the altered fatty acid composition of PI causes abnormal localization of phosphoinositides in the membrane bilayer, leading to the missorting of cortical proteins by retrograde membrane trafficking regulated by tbc-3 and mon-2.
acl-8, acl-9, acl-10 Subfamily Genes Are Evolutionarily Conserved from C. elegans to Mammals
In this study, we showed that acl-8 acl-9 acl-10 mutants have defects in asymmetric division of stem cell-like epithelial cells. acl-8, -9, and -10 form a subfamily of the C. elegans acl/AGPAT family and are the closest homologues of LYCAT/ALCAT1, which is conserved in various species including C. elegans, zebrafish, chicken, and human (Supplementary Figure 4). So far, mammalian LYCATs have been reported to possess acyltransferase activity toward the sn-2 position of anionic lysophospholipids including lysoPI in vitro (Cao et al., 2004, 2009; Agarwal et al., 2006; Zhao et al., 2009). We revealed that mouse LYCAT transfers stearic acid to the sn-1 position of PI as well as the sn-2 position of PI at comparable levels (Supplementary Figure 12). Furthermore, we found that expression of mouse LYCAT rescues the defects in asymmetric division in acl-8 acl-9 acl-10 mutants (Supplementary Figure 5M), indicating that mouse LYCAT is a functional homologue of acl-8, -9, and -10. Interestingly, mouse LYCAT was reported to be highly expressed in hematopoietic stem cells (Wang et al., 2007), which undergo asymmetric divisions to renew themselves and produce the various progeny cells of the distinct blood lineages (Congdon and Reya, 2008; Giebel, 2008). In addition, morpholino-mediated knockdown of zebrafish LYCAT results in a reduction of blood cells (Xiong et al., 2008). Further studies are expected to reveal an evolutionarily conserved role of the sn-1 fatty acid of PI in the asymmetric division of stem cells.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Aoki (Tohoku University) for technical advice; K. Gengyo-Ando (Saitama University) for alleles of acl-8, -9, and -10; R. Y. Tsien (HHMI, University of California, San Diego) and K. Kontani (University of Tokyo) for mCherry plasmids; Y. Iino and M. Tomioka (University of Tokyo) for technical advice and helpful discussions; and H. Fukuda and Y. Toyoda for excellent technical assistance. We also thank H. Sawa (RIKEN CDB), Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN) and A. Fire (Stanford University School of Medicine) for strains and plasmids.
Abbreviations used:
- AA
arachidonic acid
- AGPAT
1-acylglycerol-3-phosphate O-acyltransferase
- CoA
coenzyme A
- EPA
eicosapentaenoic acid
- GC
gas chromatography
- GFP
green fluorescent protein
- LC/ESI-MS
liquid chromatography/electrospray ionization-mass spectrometry
- LPIAT
lysophosphatidylinositol acyltransferase
- PA
phosphatidic acid
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PI
phosphatidylinositol
- PLA
phospholipase A
- PS
phosphatidylserine
- PUFA
polyunsaturated fatty acid
- X:Yn-Z
fatty acid chain of X carbon atoms and Y methylene-interrupted cis bonds (Z indicates the position of the terminal double bond relative to the methyl end of the molecules).
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-03-0195) on July 28, 2010.
REFERENCES
- Agarwal A. K., Barnes R. I., Garg A. Functional characterization of human 1-acylglycerol-3-phosphate acyltransferase isoform 8, cloning, tissue distribution, gene structure, and enzymatic activity. Arch. Biochem. Biophys. 2006;449:64–76. doi: 10.1016/j.abb.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Akino T., Shimojo T. On the metabolic heterogeneity of rat liver phosphatidylinositol. Biochim. Biophys. Acta. 1970;210:343–346. [PubMed] [Google Scholar]
- Baker R. R., Thompson W. Positional distribution and turnover of fatty acids in phosphatidic acid, phosphinositides, phosphatidylcholine and phosphatidylethanolamine in rat brain in vivo. Biochim. Biophys. Acta. 1972;270:489–503. doi: 10.1016/0005-2760(72)90114-2. [DOI] [PubMed] [Google Scholar]
- Balsinde J., Balboa M. A., Dennis E. A. Antisense inhibition of group VI Ca2+-independent phospholipase A2 blocks phospholipid fatty acid remodeling in murine P388D1 macrophages. J. Biol. Chem. 1997;272:29317–29321. doi: 10.1074/jbc.272.46.29317. [DOI] [PubMed] [Google Scholar]
- Ban N., Matsumura Y., Sakai H., Takanezawa Y., Sasaki M., Arai H., Inagaki N. ABCA3 as a lipid transporter in pulmonary surfactant biogenesis. J. Biol. Chem. 2007;282:9628–9634. doi: 10.1074/jbc.M611767200. [DOI] [PubMed] [Google Scholar]
- Bao S., Li Y., Lei X., Wohltmann M., Jin W., Bohrer A., Semenkovich C. F., Ramanadham S., Tabas I., Turk J. Attenuated free cholesterol loading-induced apoptosis but preserved phospholipid composition of peritoneal macrophages from mice that do not express group VIA phospholipase A2. J. Biol. Chem. 2007;282:27100–27114. doi: 10.1074/jbc.M701316200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilder D. Cell polarity: squaring the circle. Curr. Biol. 2001;11:R132–R135. doi: 10.1016/s0960-9822(01)00060-4. [DOI] [PubMed] [Google Scholar]
- Bligh E. G., Dyer W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Liu Y., Lockwood J., Burn P., Shi Y. A novel cardiolipin-remodeling pathway revealed by a gene encoding an endoplasmic reticulum-associated acyl-CoA:lysocardiolipin acyltransferase (ALCAT1) in mouse. J. Biol. Chem. 2004;279:31727–31734. doi: 10.1074/jbc.M402930200. [DOI] [PubMed] [Google Scholar]
- Cao J., Shen W., Chang Z., Shi Y. ALCAT1 is a polyglycerophospholipid acyltransferase potently regulated by adenine nucleotide and thyroid status. Am. J. Physiol. Endocrinol. Metab. 2009;296:E647–E653. doi: 10.1152/ajpendo.90761.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congdon K., Reya T. Divide and conquer: how asymmetric division shapes cell fate in the hematopoietic system. Curr. Opin. Immunol. 2008;20:302–307. doi: 10.1016/j.coi.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J. C., Osterman D. G., Saltiel A. R. Fatty acid remodelling of phosphatidylinositol under conditions of de novo synthesis in rat liver microsomes. Biochim. Biophys. Acta. 1991a;1084:279–291. doi: 10.1016/0005-2760(91)90070-x. [DOI] [PubMed] [Google Scholar]
- Darnell J. C., Osterman D. G., Saltiel A. R. Synthesis of phosphatidylinositol in rat liver microsomes is accompanied by the rapid formation of lysophosphatidylinositol. Biochim. Biophys. Acta. 1991b;1084:269–278. doi: 10.1016/0005-2760(91)90069-t. [DOI] [PubMed] [Google Scholar]
- Darnell J. C., Saltiel A. R. Coenzyme A-dependent, ATP-independent acylation of 2-acyl lysophosphatidylinositol in rat liver microsomes. Biochim. Biophys. Acta. 1991;1084:292–299. doi: 10.1016/0005-2760(91)90071-o. [DOI] [PubMed] [Google Scholar]
- Di Paolo G., De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Dircks L., Sul H. S. Acyltransferases of de novo glycerophospholipid biosynthesis. Prog. Lipid Res. 1999;38:461–479. doi: 10.1016/s0163-7827(99)00012-0. [DOI] [PubMed] [Google Scholar]
- Gengyo-Ando K., Mitani S. Characterization of mutations induced by ethyl methanesulfonate, UV, and trimethylpsoralen in the nematode Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 2000;269:64–69. doi: 10.1006/bbrc.2000.2260. [DOI] [PubMed] [Google Scholar]
- Giebel B. Cell polarity and asymmetric cell division within human hematopoietic stem and progenitor cells. Cells Tissues Organs. 2008;188:116–126. doi: 10.1159/000112842. [DOI] [PubMed] [Google Scholar]
- Gillingham A. K., Whyte J. R., Panic B., Munro S. Mon2, a relative of large Arf exchange factors, recruits Dop1 to the Golgi apparatus. J. Biol. Chem. 2006;281:2273–2280. doi: 10.1074/jbc.M510176200. [DOI] [PubMed] [Google Scholar]
- Gönczy P. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat. Rev. Mol. Cell Biol. 2008;9:355–366. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- Higgs H. N., Glomset J. A. Purification and properties of a phosphatidic acid-preferring phospholipase A1 from bovine testis. Examination of the molecular basis of its activation. J. Biol. Chem. 1996;271:10874–10883. doi: 10.1074/jbc.271.18.10874. [DOI] [PubMed] [Google Scholar]
- Higgs H. N., Han M. H., Johnson G. E., Glomset J. A. Cloning of a phosphatidic acid-preferring phospholipase A1 from bovine testis. J. Biol. Chem. 1998;273:5468–5477. doi: 10.1074/jbc.273.10.5468. [DOI] [PubMed] [Google Scholar]
- Holub B. J., Kuksis A. Differential distribution of orthophosphate-32P and glycerol-14C among molecular species of phosphatidylinositols of rat liver in vivo. J. Lipid Res. 1971a;12:699–705. [PubMed] [Google Scholar]
- Holub B. J., Kuksis A. Structural and metabolic interrelationships among glycerophosphatides of rat liver in vivo. Can. J. Biochem. 1971b;49:1347–1356. doi: 10.1139/o71-195. [DOI] [PubMed] [Google Scholar]
- Holub B. J., Kuksis A. Further evidence for the interconversion of monophosphoinositides in vivo. Lipids. 1972;7:78–80. doi: 10.1007/BF02531275. [DOI] [PubMed] [Google Scholar]
- Holub B. J., Piekarski J. The formation of phosphatidylinositol by acylation of 2-acyl-sn-glycero-3-phosphorylinositol in rat liver microsomes. Lipids. 1979;14:529–532. doi: 10.1007/BF02533526. [DOI] [PubMed] [Google Scholar]
- Hutzell P. A., Krusberg L. R. Fatty acid composition of Caenorhabditis elegans and C. briggsae. Comp. Biochem. Physiol. 1982;73B:517–520. [Google Scholar]
- Inoue A., Aoki J. Phospholipase A1, structure, distribution and function. Future Lipidol. 2006;1:623–636. [Google Scholar]
- Kage-Nakadai E., Kobuna H., Kimura M., Gengyo-Ando K., Inoue T., Arai H., Mitani S. Two very long chain fatty acid Acyl-CoA synthetase genes, acs-20 and acs-22, have roles in the cuticle surface barrier in Caenorhabditis elegans. PLoS ONE. 2010;5:e8857. doi: 10.1371/journal.pone.0008857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori T., Inoue T., Sakamoto T., Gengyo-Ando K., Tsujimoto M., Mitani S., Sawa H., Aoki J., Arai H. Beta-catenin asymmetry is regulated by PLA1 and retrograde traffic in C. elegans stem cell divisions. EMBO J. 2008;27:1647–1657. doi: 10.1038/emboj.2008.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafourcade C., Galan J. M., Gloor Y., Haguenauer-Tsapis R., Peter M. The GTPase-activating enzyme Gyp1p is required for recycling of internalized membrane material by inactivation of the Rab/Ypt GTPase Ypt1p. Mol. Cell. Biol. 2004;24:3815–3826. doi: 10.1128/MCB.24.9.3815-3826.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lands W. E. Metabolism of glycerolipides; a comparison of lecithin and triglyceride synthesis. J. Biol. Chem. 1958;231:883–888. [PubMed] [Google Scholar]
- Lands W. E. Stories about acyl chains. Biochim. Biophys. Acta. 2000;1483:1–14. doi: 10.1016/s1388-1981(99)00177-8. [DOI] [PubMed] [Google Scholar]
- Le Guedard, et al. PSI1 is responsible for the stearic acid enrichment that is characteristic of phosphatidylinositol in yeast. FEBS J. 2009;276:6412–6424. doi: 10.1111/j.1742-4658.2009.07355.x. [DOI] [PubMed] [Google Scholar]
- Lee H. C., Inoue T., Imae R., Kono N., Shirae S., Matsuda S., Gengyo-Ando K., Mitani S., Arai H. Caenorhabditis elegans mboa-7, a member of the MBOAT family, is required for selective incorporation of polyunsaturated fatty acids into phosphatidylinositol. Mol. Biol. Cell. 2008;19:1174–1184. doi: 10.1091/mbc.E07-09-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra M. G., Sheltawy A. The metabolic turnover of molecular species of phosphatidylinositol and its precursor phosphatidic acid in guinea-pig cerebral hemispheres. J. Neurochem. 1976;27:1501–1511. doi: 10.1111/j.1471-4159.1976.tb02636.x. [DOI] [PubMed] [Google Scholar]
- Mancuso D. J., et al. Genetic ablation of calcium-independent phospholipase A2γ leads to alterations in mitochondrial lipid metabolism and function resulting in a deficient mitochondrial bioenergetic phenotype. J. Biol. Chem. 2007;282:34611–34622. doi: 10.1074/jbc.M707795200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumoto K., Sawa H. Cortical beta-catenin and APC regulate asymmetric nuclear beta-catenin localization during asymmetric cell division in C. elegans. Dev. Cell. 2007a;12:287–299. doi: 10.1016/j.devcel.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Mizumoto K., Sawa H. Two betas or not two betas: regulation of asymmetric division by beta-catenin. Trends Cell Biol. 2007b;17:465–473. doi: 10.1016/j.tcb.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Morikawa R., Tsujimoto M., Arai H., Aoki J. Phospholipase A1 assays using a radiolabeled substrate and mass spectrometry. Methods Enzymol. 2007;434:1–13. doi: 10.1016/S0076-6879(07)34001-9. [DOI] [PubMed] [Google Scholar]
- Morikawa R. K., Aoki J., Kano F., Murata M., Yamamoto A., Tsujimoto M., Arai H. Intracellular phospholipase A1γ (iPLA1γ) is a novel factor involved in coat protein complex I- and Rab6-independent retrograde transport between the endoplasmic reticulum and the Golgi complex. J. Biol. Chem. 2009;284:26620–26630. doi: 10.1074/jbc.M109.038869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Maxfield F. R. Membrane domains. Annu. Rev. Cell Dev. Biol. 2004;20:839–866. doi: 10.1146/annurev.cellbio.20.010403.095451. [DOI] [PubMed] [Google Scholar]
- Nakae I., Fujino T., Kobayashi T., Sasaki A., Kikko Y., Fukuyama M., Gengyo-Ando K., Mitani S., Kontani K., Katada T. The Arf-like GTPase Arl8 mediates delivery of endocytosed macromolecules to lysosomes in C. elegans. Mol. Biol. Cell. 2010;21:2434–2442. doi: 10.1091/mbc.E09-12-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa Y., Rustow B., Rabe H., Kunze D., Waku K. The de novo synthesis of molecular species of phosphatidylinositol from endogenously labeled CDP diacylglycerol in alveolar macrophage microsomes. Arch. Biochem. Biophys. 1989;268:559–566. doi: 10.1016/0003-9861(89)90323-8. [DOI] [PubMed] [Google Scholar]
- Nakajima K., Sonoda H., Mizoguchi T., Aoki J., Arai H., Nagahama M., Tagaya M., Tani K. A novel phospholipase A1 with sequence homology to a mammalian Sec23p-interacting protein, p125. J. Biol. Chem. 2002;277:11329–11335. doi: 10.1074/jbc.M111092200. [DOI] [PubMed] [Google Scholar]
- Nakamura K., Kim S., Ishidate T., Bei Y., Pang K., Shirayama M., Trzepacz C., Brownell D. R., Mello C. C. Wnt signaling drives WRM-1/beta-catenin asymmetries in early C. elegans embryos. Genes Dev. 2005;19:1749–1754. doi: 10.1101/gad.1323705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panbianco C., Weinkove D., Zanin E., Jones D., Divecha N., Gotta M., Ahringer J. A casein kinase 1 and PAR proteins regulate asymmetry of a PIP2 synthesis enzyme for asymmetric spindle positioning. Dev. Cell. 2008;15:198–208. doi: 10.1016/j.devcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Boulan E., Kreitzer G., Müsch A. Organization of vesicular trafficking in epithelia. Nat. Rev. Mol. Cell Biol. 2005;6:233–247. doi: 10.1038/nrm1593. [DOI] [PubMed] [Google Scholar]
- Seaman M. N. Recycle your receptors with retromer. Trends Cell Biol. 2005;15:68–75. doi: 10.1016/j.tcb.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Shindou H., Shimizu T. Acyl-CoA:lysophospholipid acyltransferases. J. Biol. Chem. 2009;284:1–5. doi: 10.1074/jbc.R800046200. [DOI] [PubMed] [Google Scholar]
- Siller K., Doe C. Spindle orientation during asymmetric cell division. Nat. Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- Stenico M., Lloyd A. T., Sharp P. M. Codon usage in Caenorhabditis elegans: delineation of translational selection and mutational biases. Nucleic Acids Res. 1994;22:2437–2446. doi: 10.1093/nar/22.13.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita H., Sawa H. Asymmetric cortical and nuclear localizations of WRM-1/beta-catenin during asymmetric cell division in C. elegans. Genes Dev. 2005;19:1743–1748. doi: 10.1101/gad.1322805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima F., Matsumura S., Morimoto H., Mitsushima M., Nishida E. PtdIns(3,4,5)P3 regulates spindle orientation in adherent cells. Dev. Cell. 2007;13:796–811. doi: 10.1016/j.devcel.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Waku K., Nakazawa Y. Acyltransferase activity to 1-acyl-, 1-O-alkenyl-, and 1-O-alkyl-glycero-3-phosphorylcholine in Ehrlich ascites tumor cells. J. Biochem. 1972;72:495–497. doi: 10.1093/oxfordjournals.jbchem.a129928. [DOI] [PubMed] [Google Scholar]
- Wang C. Y., Faloon P. W., Tan Z. J., Lv Y. X., Zhang P. B., Ge Y., Deng H. K., Xiong J. W. Mouse lysocardiolipin acyltransferase controls the development of hematopoietic and endothelial lineages during in vitro embryonic stem-cell differentiation. Blood. 2007;110:3601–3609. doi: 10.1182/blood-2007-04-086827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel A. A., Lewin T. M., Coleman R. A. Glycerol-3-phosphate acyltransferases: rate limiting enzymes of triacylglycerol biosynthesis. Biochim. Biophys. Acta. 2009;1791:501–506. doi: 10.1016/j.bbalip.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J. W., Yu Q. M., Zhang J. J., Mably J. D. An acyltransferase controls the generation of hematopoietic and endothelial lineages in zebrafish. Circ. Res. 2008;102:1057–1064. doi: 10.1161/CIRCRESAHA.107.163907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Chen Y. Q., Li S., Konrad R. J., Cao G. The microsomal cardiolipin remodeling enzyme acyl-CoA lysocardiolipin acyltransferase is an acyltransferase of multiple anionic lysophospholipids. J. Lipid Res. 2009;50:945–956. doi: 10.1194/jlr.M800567-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.