Dictyostelium cells lacking the intracellular pH regulator NHE1 have defective chemotaxis. A modifier screen and reconstitution studies show expression of recombinant actin interacting protein 1 (Aip1) suppresses the Ddnhe1-phenotype. Aip1 promotes cofilin-dependent actin remodeling, which is likely a major determinant in pH-dependent chemotaxis.
Abstract
Increased intracellular pH is an evolutionarily conserved signal necessary for directed cell migration. We reported previously that in Dictyostelium cells lacking H+ efflux by a Na+-H+ exchanger (NHE; Ddnhe1−), chemotaxis is impaired and the assembly of filamentous actin (F-actin) is attenuated. We now describe a modifier screen that reveals the C-terminal fragment of actin-interacting protein 1 (Aip1) enhances the chemotaxis defect of Ddnhe1− cells but has no effect in wild-type Ax2 cells. However, expression of full-length Aip1 mostly suppresses chemotaxis defects of Ddnhe1− cells and restores F-actin assembly. Aip1 functions to promote cofilin-dependent actin remodeling, and we found that although full-length Aip1 binds cofilin and F-actin, the C-terminal fragment binds cofilin but not F-actin. Because pH-dependent cofilin activity is attenuated in mammalian cells lacking H+ efflux by NHE1, our current data suggest that full-length Aip1 facilitates F-actin assembly when cofilin activity is limited. We predict the C-terminus of Aip1 enhances defective chemotaxis of Ddnhe1− cells by sequestering the limited amount of active cofilin without promoting F-actin assembly. Our findings indicate a cooperative role of Aip1 and cofilin in pH-dependent cell migration, and they suggest defective chemotaxis in Ddnhe1− cells is determined primarily by loss of cofilin-dependent actin dynamics.
INTRODUCTION
In migrating cells, a network of a filamentous actin (F-actin) at the cell front drives membrane protrusion at the leading edge. The dynamic assembly of this actin network in response to migratory cues is tightly regulated by two key molecules; the Arp2/3 complex and cofilin (Pollard and Borisy, 2003). The Arp2/3 complex builds cross-linked actin arrays by nucleating new filaments from the sides of pre-existing filaments (Goley and Welch, 2006). In mammalian cells, cofilin plays an essential role in F-actin assembly by severing filaments to generate new free barbed (plus) ends for nucleation by the Arp2/3 complex (Mouneimne et al., 2006). Cofilin activity is stimulated by dephosphorylation, dissociation from phosphoinositides in the plasma membrane, and an increase in intracellular pH (van Rheenen et al., 2007; Frantz et al., 2008; Bernstein and Bamburg, 2010). In addition to these direct regulatory mechanisms, cofilin-dependent remodeling of F-actin is enhanced by its interaction with actin-interacting protein 1 (Aip1) (Ono, 2003; Brieher et al., 2006). However, how Aip1 functions with regulated cofilin activity in cells is unclear.
Aip1 regulates F-actin dynamics only in the presence of cofilin (Okada et al., 1999; Mohri et al., 2004, 2006; Ono et al., 2004). A member of the WD repeat family with two β-propeller domains, Aip1 forms a tertiary structure with cofilin and F-actin (Voegtli et al., 2003) and lowers the critical concentration of cofilin necessary for F-actin remodeling (Brieher et al., 2006). Aip1 also is reported to cap barbed ends of cofilin-severed filaments (Okada et al., 2002; Balcer et al., 2003), although this function is controversial (Clark et al., 2006; Okada et al., 2006). Loss-of-function studies suggest that Aip1 is required for correct remodeling of F-actin. In Saccharomyces cerevisiae, an Aip1-null mutant is a synthetic lethal with mutant cofilin alleles and has thickened actin cables caused by cofilin mislocalization (Rodal et al., 1999). Depletion of UNC-78, an Aip1 homologue in Caenorhabditis elegans, results in disorganization of F-actin in the body wall muscle (Ono, 2001). Knockdown of Aip1 in mammalian cells leads to aberrant cytokinesis and inhibits directed migration (Li et al., 2007; Kato et al., 2008). Also in the motile amoebae Dictyostelium discoideum, Aip1-null cells have impaired actin-dependent processes, including phagocytosis, cytokinesis, and motility (Konzok et al., 1999).
A rapid assembly of F-actin at the leading edge is necessary for chemotaxis of amoeboid cells such as leukocytes and Dictyostelium (Parent, 2004). Dictyostelium is an important model for studying chemotactic migration because the mechanics and regulation of F-actin dynamics are similar to those in migrating mammalian cells (Sasaki and Firtel, 2006). In response to the chemoattractant cAMP, Dictyostelium cells adopt a polarized, elongated morphology with an F-actin network enriched at the leading edge. Evidence in Dictyostelium (Van Duijn and Inouye, 1991; Patel and Barber, 2005) and mammalian (Denker and Barber, 2002; Paradiso et al., 2004; Stock and Schwab, 2006) cells indicates that an increase in intracellular pH (pHi) is necessary for directed migration and for de novo assembly of F-actin at the cell front. Dictyostelium cells null for a Na+-H+ exchanger (Ddnhe1−) that regulates dynamic changes in pHi lack efficient chemotaxis and have decreased abundance of F-actin in response to cAMP.
To further understand how DdNHE1 regulates chemotaxis we used an overexpression library to screen for modifiers of the Ddnhe1− phenotype. One clone that enhanced Ddnhe1− chemotaxis and lacked de novo F-actin assembly in response to cAMP contained a C-terminal fragment of DdAip1. However, expression of full-length wild-type but not inactive DdAip1 in Ddnhe1− cells suppressed the defective chemotaxis phenotype and restored F-actin abundance. Because cofilin-dependent F-actin remodeling in migrating mammalian cells requires increased NHE1 activity and pHi (Frantz et al., 2008), our findings suggest a cooperative role of Aip1 and cofilin in pH-dependent cell migration.
MATERIALS AND METHODS
Strain, Cell Culture, and Development
Wild-type Ax2 and Ddnhe1− cells (Patel and Barber, 2005) were cultured in axenic HL5 medium supplemented with 100 U/ml penicillin and 100 μg/ml streptomycin. Plasmid constructs were introduced into Ax2 and Ddnhe1− cells by electroporation (Knecht and Pang, 1995), and cells expressing DdAip1 constructs were selected in HL5 medium containing 10 μg/ml G418. Cells expressing Lifeact-monomeric red fluorescent protein (mRFP) were selected with 20 μg/ml hygromycin. For submerged development, exponentially growing cells were washed twice with PB buffer (20 mM K2HPO4/KH2PO4, pH 6.8) and were allowed to develop at a density of 1 × 106 cells/cm2 in PB buffer. Time-lapse images were acquired at 15-min intervals for 20 h by using an Axiovert S-100 microscope (Carl Zeiss, Jena, Germany).
Library Screening
An overexpression cDNA library (provided by Douglas Robinson, Johns Hopkins University, Baltimore, MD) described previously (Robinson and Spudich, 2000) was amplified and electroporated into Ddnhe1− cells. pREP, a helper plasmid containing an open reading frame (ORF) was cotransformed because these cells have no ORF required for the replication of Ddp2-based pLD1A15SN used for library construction (Robinson and Spudich, 2000). Transformed cells were selected by resistance to 10 μg/ml G418 for 3–4 d, plated onto PB buffer agar plates with a suspension of heat-killed bacteria, and plaque size was scored. Plasmids were recovered from cells using the DNA Mini-prep kit (QIAGEN, Valencia, CA), and cDNA in the plasmids was amplified by polymerase chain reaction (PCR) by using primers LD1SM, 5′-AAAAGTCGACCCACGCGTCC-3′ and LD13, 5′-CGCGTTTATTTATTTAGCGGCCGCCC-3′ and then subcloned into pCR2.1-TOPO vector (Invitrogen, Carlsbad, CA). Purification from Escherichia coli DH5α and DNA sequencing revealed a cDNA fragment of DdAip1 in one clone. The developmental morphology of selected clones was observed under submerged conditions and expression of developmental markers at 0 and 6 h after development was determined by reverse transcriptase-PCR using primers for cAR1 (forward, 5′-GTAATTACATATGGTAG-3′ and reverse, 5′-GAAATTAATGGTAAAC-3′) and for Gα2 (forward, 5′-GTGTCAAACAGGCAATG-3′ and reverse, 5′-GTCATAACACGAGTATG-3′). TalA expression, an internal control, was determined using primers GGATCCATGGATTACAAAAAAAAACATAGACC (forward) and GACGTCGTATGGGTAAAATGATGTCATACC (reverse).
Plasmid Construction
The plasmid containing full-length of DdAip1 tagged with green fluorescent protein (GFP) (DdAip1-FL; a generous gift from Annette Muller-Taubenberger, Ludwig Maximilians University, Munich, Germany) was used for expression in Dictyostelium and as a template for PCR to generate mutant DdAip1. The expression vector for DdAip1-Δ382 was constructed by PCR amplification using primers AIP1-S, 5′-GGATCCATGGATGATAGTGTTA-3′ and Aip1-R2, 5′-GGATCCTTAATTTGATACATACCA-3′ and ligation into a BamHI site of pTX-FLAG vector. To express an F-actin reporter in cells, the Lifeact-mRFP sequence (Riedl et al., 2008) was amplified by PCR using primers Lifeact-F, 5′-AAAAGATCTAAAAAATGGGTGTCGCAGATTTG-3′ and Lifeact-R2, 5′-TTTCTCGAGTTAAGCGCCTGTGCTATG-3′ and cloned into BglII and XhoI sites of EXP5(+) vector containing a hygromycin-resistant cassette. For cloning of rat Aip1, cDNA was synthesized using total RNA from MTLn3 rat adenocarcinoma cells and was used as a template for PCR. Aip1-FL was amplified by PCR using primers RnAip1-F, 5′-GGATCCATGCCGTACGAGATCAA-3′ and RnAip1-R, 5′-CTCGAGTTCAGTAGGTFATTGT-3′ and Aip1-Δ382 was amplified using primers RnAip1-TF, ATGGACGACACAGTGCGGTATACT-3′ and RnAip1-R. For production of recombinant proteins, PCR products were cloned into BamHI and XhoI sites of pGEX6P-2 vector (GE healthcare, Little Chalfont, Buckinghamshire, United Kingdom). DdAip1 with alanine substitutions of E125, E167, F181, and F193 (DdAip1-4X) was generated using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) using these forward primers (Daip1E125A-S, 5′-GTTGGTGATGGTAAAGCTAGATTTGGTGCAGCC-3′; Daip1E167A-S, 5′-GCCGCCACTGGTAGTGCTGATTTTGCAGTC-3′; Daip1F181A-S, 5′-GGTCCACCATTCAAAGCTCAAAAGAATATTGCC-3′; and Daip1F193A-S, 5′-GGTGATTTCACTCGTGCTGTAAATTGTGTTAG-3′).
Chemotaxis Assay
To induce chemotaxis competence, cells were suspended at a density of 1 × 107 cells/ml in PB buffer and pulsed with 30 nM cAMP every 6 min for 5 h (Mendoza and Firtel, 2006). For chemotaxis assays, aliquots of 5-h developed cells were harvested and transferred to 30-mm dishes. cAMP (100 μM) was delivered by a micropipette, and cell movement was recorded at 6-s intervals for 30 min by using an Axiovert S-100 microscope (Carl Zeiss). Cell movement was tracked using ImageJ software (National Institutes of Health, Bethesda, MD), with a manual tracking plug-in. Chemotaxis parameters were calculated by Prism software (GraphPad Software, San Diego, CA), and statistical significance was determined by unpaired two-tailed t test.
Actin Polymerization Assay
Abundance of F-actin was determined using two methods. To test clones from the library screen we used Coomassie staining of Triton X-100–insoluble fractions as described previously (Patel and Barber, 2005). To test cells expressing recombinant Aip1 we measured fluorescence of lysates prepared from cells labeled with rhodamine phalloidin as described previously (Chen et al., 2003). Cells pulsed with cAMP for 5 h were resuspended at a density of 3 × 107 cells/ml in PB buffer and incubated with 5 mM caffeine for 20 min to inhibit endogenous cAMP production. After being exposed to a uniform concentration of 2 μM exogenous cAMP, 3 × 106 cells were removed at the indicated times, fixed with 3.7% formaldehyde, and incubated in buffer containing 10 mM PIPES, 0.2% Triton X-100, 20 mM K2HPO4/KH2PO4, 5 mM EGTA, 2 mM MgCl2, and 0.4 U of rhodamine-phalloidin (Invitrogen) for 1 h. A Triton X-100–insoluble fraction was obtained by centrifugation and rhodamine-phalloidin bound to F-actin was eluted in methanol overnight. The fluorescence intensity of rhodamine-phalloidin was measured using a SpectraMax M5 plate reader (GE Healthcare).
F-actin Localization
F-actin localization was determined using two approaches. For live cell imaging, cells expressing Lifeact-mRFP were developed by pulsing with cAMP for 5 h and then loaded onto Dunn chemotaxis chambers (Hawksley Technology, Lancing, United Kingdom) in the presence of a cAMP gradient (0–5 μM). Fluorescence images were acquired every 5 s for 10 min with a 60× numerical aperture 1.20 objective lens (Plan Apo; Nikon, Tokyo, Japan) on a TE2000 inverted microscope (Nikon) equipped with a spinning-disk unit (CSU10; Yokogawa Electronics, Tokyo, Japan), 488-nm and 561-nm lasers, and a CoolSNAP HQ2 camera (Photometrics, Tucson, AZ). We also used phalloidin labeling of fixed cells chemotaxing toward cAMP released from a pipette, as described for chemotaxis assays. For analysis of F-actin localization, fluorescence intensity of rhodamine-phalloidin was measured along the cell perimeter using ImageJ. Fluorescence intensity in arbitrary units in 20 sectors around the cell starting from the cell rear was determined.
In Vitro Aip1 and Cofilin Binding
Recombinant wild-type and mutant rat Aip1 expressed as a glutathione-s-transferase-tagged fusion in E. coli BL21 (DE3) was purified using glutathione-Sepharose 4B (GE Healthcare) according to manufacturer's instructions. Recombinant wild-type cofilin was expressed and purified as described previously (Frantz et al., 2008). For binding assays, 20 μM cofilin was incubated for 1 h with 0.2 μM GST or GST-Aip1 bound to glutathione-Sepharose beads in incubation buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM MgCl2, and 0.1% NP40). After washing with incubation buffer, unbound cofilin was removed and beads-bound cofilin was boiled in the SDS-sample buffer and resolved by SDS-polyacrylamide gel electrophoresis (PAGE).
Actin Cosedimentation Assay
Rabbit muscle actin (Cytoskeleton, Denver, CO) was polymerized in 50 mM KCl, 2 mM MgCl2, and 1 mM ATP, and 10 μg was incubated with 0.5 μM GST and GST fusion proteins for 30 min at 24°C. F-actin was pelleted by centrifugation at 100,000 × g for 20 min, and proteins in supernatant and pellet fractions were resolved by 12% SDS-PAGE.
Immunoblotting
Exponentially growing cells were lysed by sonicating in lysis buffer (1× phosphate-buffered saline, protease inhibitors) and proteins in total cell lysates were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes. Membranes were probed with antibodies for GFP (JL-8, 1:1000; Clontech, Mountain View, CA) and β-actin (C4, 1:5000; Millipore Bioscience Research Reagents, Temecula, CA), and bound antibody was detected by enhanced chemiluminescence (Pierce Chemical, Rockford, IL).
RESULTS
A C-Terminal Fragment of DdAip1 Enhances Impaired Chemotaxis of Ddnhe1− Cells
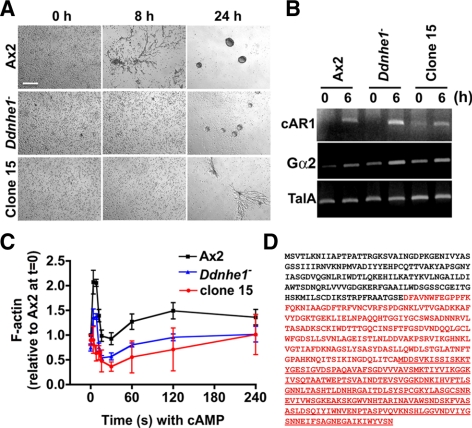
To determine how DdNHE1 regulates chemotaxis of Dictyostelium cells, we developed an easily scored assay to test for phenotypic changes. Although expression of DdNHE1 is very low in vegetative cells, we found that Ddnhe1− cells produced substantially smaller plaques than wild-type Ax2 cells on a heat-killed bacterial lawn. We screened a cDNA overexpression library previously designed to identify suppressors of cytokinesis-defective mutants (Robinson and Spudich, 2000) and scored for plaque formation. Although no clones were identified with a suppressed phenotype, one clone (clone 15) had an enhanced phenotype in which the plaques were smaller than those produced by DdNhe1− cells. We analyzed the development of clone 15 by determining stream formation, which is an index of efficient chemotaxis. When starved under nonnutrient phosphate buffer, Ax2 cells formed streams at 8 h and aggregates at 24 h (Figure 1A). Ddnhe1− cells had delayed stream formation. Although aggregates were seen at 24 h, they were smaller than those with Ax2 cells. Development of clone 15 was more delayed than Ddnhe1− cells (Figure 1A), with stream formation but no aggregates at 24 h. Development and stream formation require induced expression of several aggregation genes, including the G protein-coupled cAMP receptor cAR1 and the G protein subunit Gα2. Expression of these genes at 6 h of development was similar in the three strains, indicating that starvation-induced gene expression was unaffected (Figure 1B).
Figure 1.
Modifier screen reveals the C-terminal fragment of Aip1 enhances defects in development and F-actin assembly in Ddnhe1− cells. (A) Developmental morphologies under nonnutrient buffer at the indicated times. Clone 15 cells formed aggregates more slowly than parental Ddnhe1− cells. Bar, 100 μm. (B) Expression of early developmental genes cAR1 and Gα2 determined by RT-PCR was similar in clone 15 and Ax2 cells. TalA expression was used as a loading control. (D) Time-dependent increase in total F-actin in response to cAMP was abolished in clone 15 cells. Data are expressed as the means ± SEM of three independent experiments. (D) Amino acid sequence of DdAip1. The sequence in red denotes a truncated DdAip1 sequence recovered from clone 15. The underlined sequence denotes DdAip1-Δ382 predicted from a methionine start site.
In response to chemotactic cues, Dictyostelium cells have a well-characterized biphasic increase in F-actin, including a rapid and transient first peak at 4–8 s and a slower and more prolonged second peak at 30–180 s (Condeelis et al., 1988; Chen et al., 2003). We showed previously that in Ddnhe1− cells the first peak is reduced by 50% and the second peak is largely absent (Patel and Barber, 2005). Clone 15 had markedly less de novo increases in F-actin. Most notably, there was no first peak, which further indicated the Ddnhe1− phenotype was enhanced (Figure 1C).
Sequencing of the recovered plasmid from clone 15 revealed that it contained a C-terminal fragment of a gene encoding DdAip1 (Figure 1D). Because the recovered cDNA did not include the full-length DdAip1 sequence but contained a methionine residue at position 382, we predicted it encoded a DdAip1-Δ382 fragment (Figure 1D). This finding was revealing with regard to the role of Aip1 in promoting cofilin-dependent actin dynamics (Ono, 2003), and our previous data showing that NHE1 activity and increased pHi are necessary for cofilin-mediated F-actin remodeling in migrating mammalian fibroblasts (Frantz et al., 2008).
Chemotaxis Defect of Ddnhe1− Cells Is Enhanced by DdAip1-Δ382 but Suppressed by Full-Length DdAip1
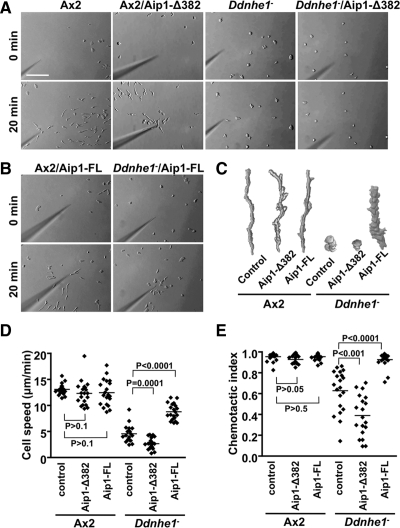
To confirm that DdAip1-Δ382 enhances the phenotype of Ddnhe1− cells, DdAip1-Δ382 tagged with a FLAG epitope was transformed into Ax2 (Ax2/DdAip1-Δ382) and Ddnhe1− (Ddnhe1−/DdAip1-Δ382) cells. We showed previously that the expression of Gα2 and cAR1 in Ddnhe1− cells is similar between 3 and 6 h (Patel and Barber, 2005), and hence determined chemotaxis toward a point source of cAMP in cells starved for 5 h. Ax2 cells rapidly adopted a polarized morphology with leading edge membrane protrusions oriented toward cAMP (Figure 2, A and C, and Supplemental Video 1). They migrated efficiently with a speed of 13.09 ± 0.23 μm/min (Figure 2D) and a chemotactic index of 0.95 ± 0.010 (Figure 2E). Ddnhe1− cells failed to adopt a polarized morphology, and although they did extend membrane protrusions, these were not directed toward the micropipette (Figure 2, A and C, and Supplemental Video 2). In addition, Ddnhe1− cells moved randomly and more slowly with a speed of 4.54 ± 0.36 μm/min (Figure 2D) and a chemotactic index of 0.63 ± 0.045 (Figure 2E). Ddnhe1−/DdAip1-Δ382 cells remained rounded without visible membrane protrusions and showed limited movement (Figure 2, A and C, and Supplemental Video 4). Their speed of 2.67 ± 0.24 μm/min (Figure 2D) and chemotactic index of 0.39 ± 0.045 (Figure 2E) were significantly less than those of Ddnhe1− cells (p = 0.0001, n = 20), suggesting a more defective chemotaxis. In contrast, expression of DdAip1-Δ382 in Ax2 cells (Ax2/DdAip1-Δ382) had no effect on chemotaxis, including polarity (Figure 2, A and C, and Supplemental Video 3), speed (Figure 2D), and chemotactic index (Figure 2E). Immunoblotting confirmed comparable expression of DdAip1-Δ382 in Ax2 and Ddnhe1− cells (Supplemental Figure 2A). These findings and our screening data suggest that DdAip1-Δ382 enhances the impaired chemotaxis phenotype of Ddnhe1− cells.
Figure 2.
Impaired chemotaxis of Ddnhe1− cells is enhanced by DdAip1-Δ382 but suppressed by DdAip1-FL. Chemotactic movement toward a micropipette tip filled with cAMP was recorded for 30 min. (A) Images of Ax2 and Ddnhe1− cells without and with heterologous expression of DdAip1-Δ382. (B) Images of Ax2 and Ddnhe1− cells expressing DdAip1-FL. (C) Morphology and tracking of the indicated cell types determined by drawing and overlapping images of a single cell from frames taken at 1-min intervals. (D) Speed of the indicated cell types, expressed as total distance moved divided by total moving time. The speed of Ddnhe1− cells is significantly decreased by DdAip1-Δ382 but increased by DdAip1-FL. (E) chemotactic index of the indicated cells types, determined as net distance moved divided by total moving distance during the time period. The chemotactic index of Ddnhe1− cells is significantly decreased by DdAip1-Δ382 but increased by DdAip1-FL. Each dot in the scatter plots represents an individual cell in time-lapse images, and data are representative of three independent experiments.
Having confirmed that DdAip1-Δ382 enhanced Ddnhe1− chemotaxis, we next determined the effect of expressing full-length DdAip1 (DdAip1-FL) in Ax2 and Ddnhe1− cells (Ax2/DdAip1-FL and Ddnhe1−/DdAip1-FL, respectively). In contrast to DdAip1-Δ382, impaired chemotaxis of Ddnhe1− cells was mostly restored by expression of DdAip1-FL. Ddnhe1−/DdAip1-FL cells adopted a polarized morphology, although they were less elongated than Ax2 cells (Figure 2, B and C, and Supplemental Video 6). Their speed (8.79 ± 0.35 μm/min) and chemotactic index (0.93 ± 0.016) were 67 and 98%, respectively, of Ax2 cells. (Figure 2, D and E). For Ax2/DdAip1-FL cells, migration speed (Figure 2D; p > 0.5, n = 20; Supplemental Video 5) and chemotactic index (Figure 2E; p > 0.05, n = 15) were not significantly different from Ax2 cells. Comparable expression of DdAip1-FL in Ax2 and Ddnhe1− cells was confirmed by immunoblotting (Supplemental Figure 2B). These data indicate that in contrast to DdAip1-Δ382, full length of DdAip1 suppresses impaired chemotaxis of Ddnhe1− cells.
Because our library screen did not identify DdAip1-FL as a modifier of the Ddnhe1− phenotype, we tested plaque formation on heat-killed bacteria by cells transformed with DdAip1-FL and DdAip1-Δ382. After 2 d, Ax2 cells formed large plaques with fruiting bodies, whereas Ddnhe1− cells formed smaller plaques and did not aggregate until 2 d (Supplemental Figure 1). As expected, Ddnhe1−/DdAip1-Δ382 cells formed smaller plaques than Ddnhe1− cells, whereas Ddnhe1−/DdAip1-FL cells formed larger plaques with some aggregates compared with Ddnhe1− cells (Supplemental Figure 1). These data indicate that expression of DdAip1-FL partially rescues defective plaque formation by Ddnhe1− cells. Because the library we screened contains inserts with an average size of 1.1–1.3 kb (Robinson and Spudich, 2000), we suspect that the 1.8-kb DdAip1-FL might not be expressed or is expressed in low abundance.
DdAip1-FL Restores F-actin Assembly but Not Polarity of Ddnhe1− Cells
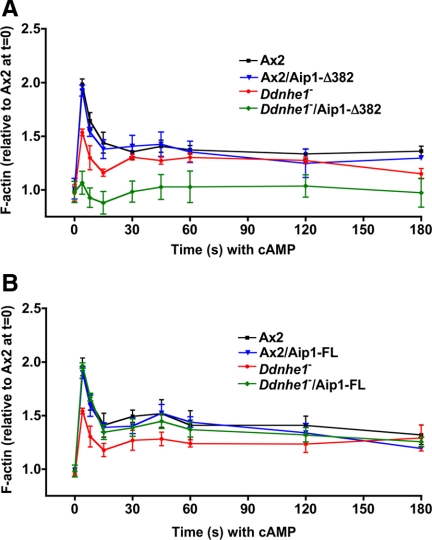
Because clone 15 had markedly attenuated F-actin with cAMP, we measured the kinetics of F-actin in response to uniform cAMP. Ax2 and Ax2/DdAip1-Δ382 cells had a similar amount of F-actin in the first and second phases (Figure 3A). However, as with clone 15, Ddnhe1−/DdAip1-Δ382 cells showed a markedly attenuated peak of F-actin in both first and second phases (Figures 1B and 3A). In contrast to Ddnhe1−/DdAip1-Δ382 cells, Ddnhe1−/DdAip1-FL cells showed restored F-actin abundance in both first and second phases to levels seen with Ax2 cells (Figure 3B). The kinetics and abundance of F-actin in Ax2/DdAip1-FL cells were similar to Ax2 cells (Figure 3B). Of importance, all strains had a similar abundance of F-actin in the absence of cAMP (0 time), which confirms our previous findings in Dictyostelium (Patel and Barber, 2005) and mammalian (Frantz et al., 2008) cells that NHE1 activity is necessary for de novo assembly of F-actin in response to chemoattractants but not for steady-state amounts of F-actin.
Figure 3.
Stimulated F-actin assembly in Ddnhe1− cells is attenuated by DdAip1-Δ382 but rescued by DdAip1-FL. Total F-actin was determined by fluorescence of rhodamine-phalloidin in Triton X-100–insoluble fractions of cells at the indicated times (seconds) after addition of cAMP and expressed relative to F-actin of Ax2 at time 0 in the absence of cAMP. (A) Expression of DdAip1-Δ382 markedly attenuated the first and second peaks of F-actin in Ddnhe1− cells but had no effect in Ax2 cells. (B) Expression of DdAip1-FL in Ddnhe1− cells restores abundance of F-actin in the first peak to that in Ax2 cells.
Although DdAip1-FL restored chemotaxis and F-actin assembly in Ddnhe1− cells, it only partially rescued polarity. Morphological polarity was scored by roundness, which is an index for lack of elongated shape and front-back asymmetry of Dictyostelium cells (van Es et al., 2001). A higher percentage of roundness indicates less polarized cells. Starved Ax2 cells were mostly elongated, had a roundness index of 47 ± 2.3% and extended pseudopods at the front toward the point source of cAMP (Figure 4, A and B). In contrast, Ddnhe1− cells had a higher roundness index of 71 ± 1.4%, and they extended smaller pseudopods that were not restricted to the cell front. The morphology of Ax2/DdAip1-Δ382 and Ax2/DdAip1-FL was similar to that of Ax2 cells. Ddnhe1−/DdAip1-Δ382 cells showed increased roundness of 78 ± 2.1% compared with Ddnhe1− cells (Figure 4B). Ddnhe1−/DdAip1-FL cells were more polarized than Ddnhe1− cells with a significantly decreased roundness index of 59 ± 3.3% compared with Ddnhe1− cells. However, Ddnhe1−/DdAip1-FL cells were still less polarized than Ax2 (47 ± 2.3%) or Ax2/DdAip1-FL cells (45 ± 2.8%) (Figure 4, A and B). We also determined F-actin localization in migrating cells as another index of cell polarity. Cells transformed with the F-actin reporter Lifeact (Riedl et al., 2008) were starved for 5 h, transferred to Dunn chemotaxis chambers, and Lifeact-mRFP fluorescence was imaged in cells migrating across a cAMP gradient. Fluorescence, an index of F-actin, was restricted to pseudopods at the leading edge of migrating Ax2 cells but was localized in multiple protrusions around the cortex of Ddnhe1− cells (Figure 4C). In Ddnhe1−/DdAip1-FL cells, fluorescence was seen at the leading edge, but also at lateral edges (Figure 4C). Similar findings were obtained using phalloidin-rhodamine labeling of F-actin (Supplemental Figure 3). Together, these data suggest that although expression of DdAip1-FL mostly restores the chemotaxis index of Ddnhe1− cells, it only partially restores their leading-edge localization of F-actin and their polarity.
Figure 4.
DdAip1-FL does not restore impaired polarity of Ddnhe1− cells. (A) Morphology of cells migrating toward cAMP. Arrows indicate the direction of cell migration. Bar, 10 μm. (B) Cell roundness, calculated as 100 × 4π × cell area/(cell perimeter)2 indicated that Ddnhe1− cells were more elongated with expression of DdAip1-FL. Data are expressed as the means ± SEM from 10 cells in three independent time-lapse sequences. (C) Dynamics of F-actin in chemotaxing cells expressing Lifeact-mRFP was determined by live cell fluorescence imaging. Arrows indicate direction of increasing cAMP gradient in the Dunn chemotaxis chamber. Lifeact-mRFP fluorescence is restricted to leading edge pseudopods of Ax2 cells but is seen in numerous protrusions not localized to the direction of the cAMP gradient in Ddnhe1− cells. In Ddnhe1−/DdAip1-FL cells, fluorescence is localized at the front and at lateral edges of migrating cells. Bar, 10 μm.
Aip1-FL but Not Aip1-Δ382 Binds F-actin
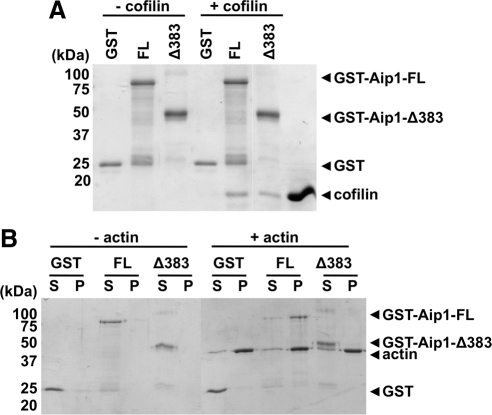
The established function of Aip1 is to enhance cofilin severing or disassembly of actin filaments (Ono, 2003). We showed previously that F-actin severing and the generation of new free barbed ends by cofilin are attenuated in migrating fibroblasts lacking NHE1 activity and having low pHi (Frantz et al., 2008). Hence, our current data suggest that expression of DdAip1-FL may restore F-actin assembly in Ddnhe1− cells by enhancing reduced cofilin activity at low pHi. However, to understand why the Ddnhe1− chemotaxis phenotype is suppressed by DdAip1-FL but enhanced by DdAip1-Δ382 we asked whether these two proteins differed in their ability to bind cofilin and F-actin. Mutagenesis studies indicate that Aip1 residues important for binding to cofilin and F-actin are located in both of N- and C-terminal propellers (Clark et al., 2006; Okada et al., 2006; Clark and Amberg, 2007). Because Dictyostelium has six cofilin isoforms and it is unknown which isoforms binds DdAip1, we used recombinant rat cofilin and Aip1 to test binding. We found that cofilin bound to GST-tagged Aip1-FL and to Aip1-Δ383 but not to GST alone (Figure 5A). These data are significant because they show that the C terminus of Aip1 is sufficient to bind cofilin in the absence of the N terminus. We tested whether binding is regulated by changes in pH but found that binding was pH-independent between pH 6.8–7.8 (data not shown). Using a sedimentation assay, we found that Aip1-FL but not Aip1-Δ383 copelleted with F-actin (Figure 5B), which indicates that the N-terminal propeller is required for binding to F-actin. These data suggest that Aip1 suppression of the Ddnhe1− chemotaxis phenotype probably requires its binding to F-actin.
Figure 5.
C-Terminal fragment of Aip1 binds cofilin but not F-actin. (A) Binding of GST-Aip1 full length (FL) and C-terminal fragment (Δ383) and cofilin (20 μM) in the absence of actin was determined by GST-pull-down assay. Cofilin binds both Aip1-FL and -Δ383, but not GST alone. (B) Binding of Aip1 to F-actin in the absence of cofilin was determined by using an actin cosedimentation assay. Polymerized actin (10 μM) was incubated with 0.5 μM Aip1-FL and -Δ383 for 1 h. After centrifugation, the supernatant (S) and the pellet (P) fractions were separated and resolved by SDS-PAGE. Aip1-FL but not -Δ383 binds to actin filaments.
Inactive DdAip1 Does Not Restore Impaired Chemotaxis or F-actin Assembly of Ddnhe1− Cells
A mutant “inactive” C. elegans Aip1 (UNC-78) containing alanine substitution of four residues (E126, D168, F182, and F192) in the N-terminal β-propeller is unable to enhance cofilin-dependent depolymerization and to suppress an unc-78-null phenotype (Mohri et al., 2006). Because these four residues are conserved in DdAip1 (E125, E167, F181, and F193), we determined effects of a similar mutant (DdAip1-4X) tagged at the N terminus with GFP to further understand the difference between DdAip1-FL and DdAip1-Δ382 on chemotaxis of Ddnhe1− cells. Immunoblotting for GFP indicated similar expression of DdAip1-4X in Ax2 and Ddnhe1− cells (Supplemental Figure 2B). Like the homologous C. elegans mutant, DdAip1-4X bound to cofilin and F-actin (Supplemental Figure 4), suggesting that protein structure is retained. Under submerged condition, starved Ax2 cells streamed and formed tight aggregates at 20 h (Figure 6A and Supplemental Video 7). Expression of DdAip1-FL or 4X did not change aggregation timing of Ax2 cells (data not shown). Streaming and aggregation were delayed in Ddnhe1− cells but restored with expression of DdAip1-FL (Figure 6A and Supplemental Videos 8 and 9). However, expression of DdAip1-4X had no effect on streaming and aggregation of Ddnhe1− cells (Figure 6A and Supplemental Video 10), indicating the phenotype was not suppressed or enhanced. In addition, chemotaxis (Figure 6B) and F-actin kinetics (Figure 6C) of Ddnhe1− cells were similar in the absence and presence of DdAip1-4X. These data suggest that the ability of Aip1-FL to restore efficient chemotaxis in Ddnhe1− cells requires its activity in enhancing cofilin function.
Figure 6.
Inactive DdAip1 does not restore impaired chemotaxis or F-actin assembly of Ddnhe1− cells. (A) Analysis of developmental morphology indicated that Ddnhe1−/DdAip1-FL cells form aggregates at 20 h like Ax2 cells, but aggregate formation is not different in Ddnhe1−/DdAip1-4X cells and Ddnhe1− cells. (B) DdAip1-4X cells did not rescue chemotaxis defect of Ddnhe1− cells. The outline of seven cells was drawn and overlapped from the images taken at 1-min intervals. The red dot indicates the position of a micropipette containing cAMP. (C) Time-dependent F-actin amount in response to cAMP showed that expression of DdAip1-FL but not DdAip1-4X rescued attenuated F-actin assembly of Ddnhe1− cells.
DISCUSSION
Aip1 is recognized as a cofactor for cofilin and enhances cofilin-dependent F-actin dynamics during endocytosis, cytokinesis, and cell movement. Aip1 facilitates severing and depolymerization of actin filaments only when they are decorated with cofilin (Ono et al., 2004), it lowers the amount of cofilin necessary for F-actin disassembly (Brieher et al., 2006), and it caps free barbed ends of cofilin-severed filaments (Okada et al., 2002; Balcer et al., 2003). Genetic evidence also indicates a functional interaction between Aip1 and cofilin. In yeasts, loss of aip1 is a synthetic lethal with cofilin mutants and cells have mislocalized cofilin (Rodal et al., 1999). In C. elegans, deletion of unc-78 enhances a motility defect of unc-60B, a cofilin homologue, and induces mislocalization of cofilin to actin aggregates (Ono, 2001). In addition, depletion of active cofilin blocks association of Aip1 with actin in Xenopus cells (Tsuji et al., 2009), and expression of active cofilin restores cytokinesis and migration that are impaired by knockdown of Aip1 in mammalian cells (Kato et al., 2008). In addition to Aip1, cofilin activity is also enhanced by increased pH (Bernstein and Bamburg, 2004). In motile mammalian fibroblasts, increased severing activity of cofilin for the assembly of new actin filaments requires increased H+ efflux by the plasma membrane Na+-H+ exchanger NHE1 (Frantz et al., 2008). We now show that in Dictyostelium cells lacking Ddnhe1 impaired F-actin assembly and defective chemotaxis are restored by expression of full-length Aip1.
Our findings on a genetic interaction between DdAip1 and DdNHE1 suggest that defective chemotaxis in Ddnhe1− cells may be determined primarily by loss of cofilin-dependent actin dynamics. A recently recognized function of Aip1 is to lower the critical concentration of active cofilin necessary for F-actin remodeling (Brieher et al., 2006). Hence, we predict that heterologously expressed Aip1-FL increases cofilin activity that is likely low in Ddnhe1− cells because pHi is markedly less than in Ax2 cells (Patel and Barber, 2005). In support of this prediction, expression of Aip1-FL suppresses chemotaxis defects and restores attenuated F-actin assembly of Ddnhe1− cells but has no effect in wild-type Ax2 cells. These findings further suggest that the role of DdAip1 in chemotaxis is a separate but compensatory pathway for cofilin regulation, such as seen with phosphatidylinositol 3-kinase and TORC2 for AKT regulation (Kamimura et al., 2008). Although most studies on Aip1 function describe enhancement of cofilin-mediated F-actin disassembly in vitro, Aip1-FL restores F-actin assembly in Ddnhe1− cells, which is consistent with a critical role of cofilin-dependent filament severing in generating new free barbed ends for de novo F-actin assembly (Mouneimne et al., 2006). Okreglak and Drubin (2010) recently showed that the annealing of actin oligomers formed by Aip1 and cofilin is a physiologically relevant pathway for F-actin assembly, suggesting a role of Aip1 and cofilin in actin filament assembly in cells. Biphasic F-actin assembly in response to cAMP is attenuated in Ddnhe1− cells (Patel and Barber, 2005), and we found that the first phase is more dramatically rescued by Aip1-FL. Similarly, biphasic formation of free barbed ends in response to growth factors (Mouneimne et al., 2006) is attenuated in mammalian NHE1-deficient cells and only the first phase is restored by expression of a mutant pH-insensitive cofilin (Frantz et al., 2008). Our findings suggest that the first phase of actin assembly in response to cAMP may be cofilin dependent and that Aip1-FL restores attenuated cofilin-dependent actin dynamics in Ddnhe1− cells. Because the Dictyostelium genome includes six cofilins, it is difficult to know which isoform is specifically enhanced by Aip1 in cells, although actin disassembly by Dictyostelium cofilin 1 is enhanced by Aip1 (Aizawa et al., 1999).
Cofilin dependence for suppression of the Ddnhe1− phenotype by Aip1-FL is also supported by our findings that Aip1-4X has no effect on impaired chemotaxis and F-actin assembly in Ddnhe1− cells. In C. elegans, a 4X mutant of UNC-78 analogous to Aip1-4X lacks cofilin-dependent actin disassembly and fails to rescue the phenotype of unc-78-null mutants (Mohri et al., 2006). Although Aip1 has both severing and capping activity, UNC-78-4X abolishes severing activity but not capping (Mohri et al., 2006). Hence, rescue of the Ddnhe1− phenotype by Aip1-FL but not Aip1-4X suggests the importance of severing but not capping activity in the absence of DdNHE1. We found that expression of the C-terminal fragment Aip1-Δ382 markedly enhances defects in chemotaxis and F-actin assembly of Ddnhe1− cells but has no effect on Ax2 cells. Thus, we suspect that Aip1-Δ382 acts as a partial dominant negative, with effects seen when cofilin activity is limited in Ddnhe1− cells but not in Ax2 cells. We initially predicted this dominant negative effect might reflect increased F-actin capping in cells with limited filament severing activity of cofilin because of decreased pHi. However, although Aip1-Δ382 retains binding to cofilin it is unable to bind F-actin. A more likely interpretation of why Aip1-Δ382 enhances defects in chemotaxis and F-actin assembly of Ddnhe1− cells is that Aip1-Δ382 may sequester the limited amount of active cofilin in cells with low pHi or prevent free cofilin from binding to F-actin. However, Aip1-4X has little effect in Ddnhe1− cells, although it has no activity but still binds cofilin. We speculate that in contrast to Aip1-Δ382, the 4X mutant preferentially complexes with F-actin-bound cofilin, consistent with previous findings that the 4X mutation does not disrupt cofilin binding to F-actin but inhibits filament severing by cofilin (Mohri et al., 2006). Because the mechanism whereby Aip1 promotes cofilin activity is still unknown, it is difficult to speculate on the functional significance of these differences between Aip1-Δ382 and Aip1-4X.
Although Aip1-FL restores attenuated F-actin kinetics in Ddnhe1− cells, it does not completely rescue cell polarity. During chemotaxis, Ddnhe1− cells expressing Aip1-FL are less elongated than Ax2 cells and actin-rich pseudopods are not restricted to the leading edge, which probably contributes to their slower speed compared with Ax2 cells. In response to chemoattractant, the first phase of actin polymerization in Dictyostelium cells is predicted to be important for establishing morphological asymmetry and the second phase drives membrane protrusion (Chen et al., 2003). However, the kinetics and magnitudes of both F-actin phases are similar in Ax2 cells and in Ddnhe1− cells expressing Aip1-FL, but expression of Aip1-FL does not limit F-actin assembly to the leading edge. Local activation of cofilin at the leading edge of migrating fibroblast cells is necessary to maintain cell polarity during directional migration (Dawe et al., 2003), which may explain why Ddnhe1− cells are more elongated with Aip1-FL and more rounded with Aip1-Δ382. In motile mammalian fibroblasts, H+ efflux by NHE1 is necessary for directional polarity (Denker and Barber, 2002) and forms a bistable positive feedback loop with the low molecular weight GTPase Cdc42 (Frantz et al., 2007). Despite the critical role of Cdc42 in cell polarity of mammalian cells, its Dictyostelium orthologue has not been identified and how H+ efflux determines polarity in Dictyostelium cells is unresolved. Because cofilin localization in protrusions is retained in Ddaip1-null cells (Konzok et al., 1999), our data suggest that Aip1 does not function to restrict F-actin assembly at the cell front.
Increased pHi is an evolutionarily conserved signal necessary for directed cell migration. The ability of Aip1-FL to nearly restore impaired chemotaxis in Ddnhe1− cells highlights the critical importance of pH-dependent cofilin activity that is likely not limited to Dictyostelium cells. Cofilin-dependent F-actin remodeling and regulatory pathways promote migration of invasive cancer cells (Wang et al., 2007), and Aip1 also enhances directed migration of mammalian cells (Li et al., 2007; Kato et al., 2008). Moreover, increased pHi is a hallmark of most cancer cells (Cardone et al., 2005; Harguindey et al., 2005). Hence, our findings on NHE1 and Aip1 in amoeboid cell migration reveal new insights in understanding the mechanism of cofilin-dependent invasive migration of cancer cells.
Supplementary Material
ACKNOWLEDGMENTS
We thank Doug Robinson for providing the overexpression library and Annette Muüller-Taubenberger for the Aip1-GFP construct. We thank Torsten Wittmann for technical assistance and members of the Barber and Wittmann laboratories for helpful discussions. This work was supported by National Institutes of Health grant GM-58642 (to D.L.B.) and was conducted in a facility constructed with support from Research Facilities Improvement Program grant C06 RR16490 from the National Institutes of Health National Center for Research Resources.
Abbreviations used:
- Aip1
actin-interacting protein 1
- Dd
Dictyostelium discoideum
- NHE1
Na+-H+ exchanger isoform 1.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-12-1058) on July 28, 2010.
REFERENCES
- Aizawa H., Katadae M., Maruya M., Sameshima M., Murakami-Murofushi K., Yahara I. Hyperosmotic stress-induced reorganization of actin bundles in Dictyostelium cells over-expressing cofilin. Genes Cells. 1999;4:311–324. doi: 10.1046/j.1365-2443.1999.00262.x. [DOI] [PubMed] [Google Scholar]
- Balcer H. I., Goodman A. L., Rodal A. A., Smith E., Kugler J., Heuser J. E., Goode B. L. Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr. Biol. 2003;13:2159–2169. doi: 10.1016/j.cub.2003.11.051. [DOI] [PubMed] [Google Scholar]
- Bernstein B. W., Bamburg J. R. A proposed mechanism for cell polarization with no external cues. Cell Motil. Cytoskeleton. 2004;58:96–103. doi: 10.1002/cm.20001. [DOI] [PubMed] [Google Scholar]
- Bernstein B. W., Bamburg J. R. ADF/cofilin: a functional node in cell biology. Trends Cell Biol. 2010;20:187–195. doi: 10.1016/j.tcb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieher W. M., Kueh H. Y., Ballif B. A., Mitchison T. J. Rapid actin monomer-insensitive depolymerization of Listeria actin comet tails by cofilin, coronin, and Aip1. J. Cell Biol. 2006;175:315–324. doi: 10.1083/jcb.200603149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone R. A., Casavola V., Reshkin S. J. The role of disturbed pH dynamics and the Na+/H+ exchanger in metastasis. Nat. Rev. Cancer. 2005;5:786–795. doi: 10.1038/nrc1713. [DOI] [PubMed] [Google Scholar]
- Chen L., Janetopoulos C., Huang Y. E., Iijima M., Borleis J., Devreotes P. N. Two phases of actin polymerization display different dependencies on PI(3,4,5)P3 accumulation and have unique roles during chemotaxis. Mol. Biol. Cell. 2003;14:5028–5037. doi: 10.1091/mbc.E03-05-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M. G., Amberg D. C. Biochemical and genetic analyses provide insight into the structural and mechanistic properties of actin filament disassembly by the Aip1p cofilin complex in Saccharomyces cerevisiae. Genetics. 2007;176:1527–1539. doi: 10.1534/genetics.107.072066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M. G., Teply J., Haarer B. K., Viggiano S. C., Sept D., Amberg D. C. A genetic dissection of Aip1p's interactions leads to a model for Aip1p-cofilin cooperative activities. Mol. Biol. Cell. 2006;17:1971–1984. doi: 10.1091/mbc.E05-10-0956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condeelis J., Hall A., Bresnick A., Warren V., Hock R., Bennett H., Ogihara S. Actin polymerization and pseudopod extension during amoeboid chemotaxis. Cell Motil. Cytoskeleton. 1988;10:77–90. doi: 10.1002/cm.970100113. [DOI] [PubMed] [Google Scholar]
- Dawe H. R., Minamide L. S., Bamburg J. R., Cramer L. P. ADF/cofilin controls cell polarity during fibroblast migration. Curr. Biol. 2003;13:252–257. doi: 10.1016/s0960-9822(03)00040-x. [DOI] [PubMed] [Google Scholar]
- Denker S. P., Barber D. L. Cell migration requires both ion translocation and cytoskeletal anchoring by the Na-H exchanger NHE1. J. Cell Biol. 2002;159:1087–1096. doi: 10.1083/jcb.200208050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz C., Barreiro G., Dominguez L., Chen X., Eddy R., Condeelis J., Kelly M. J., Jacobson M. P., Barber D. L. Cofilin is a pH sensor for actin free barbed end formation: role of phosphoinositide binding. J. Cell Biol. 2008;183:865–879. doi: 10.1083/jcb.200804161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz C., Karydis A., Nalbant P., Hahn K. M., Barber D. L. Positive feedback between Cdc42 activity and H+ efflux by the Na-H exchanger NHE1 for polarity of migrating cells. J. Cell Biol. 2007;179:403–410. doi: 10.1083/jcb.200704169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley E. D., Welch M. D. The Arp2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- Harguindey S., Orive G., Luis Pedraz J., Paradiso A., Reshkin S. J. The role of pH dynamics and the Na+/H+ antiporter in the etiopathogenesis and treatment of cancer. Two faces of the same coin–one single nature. Biochim. Biophys. Acta. 2005;1756:1–24. doi: 10.1016/j.bbcan.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Kamimura Y., Xiong Y., Iglesias P. A., Hoeller O., Bolourani P., Devreotes P. N. PIP3-independent activation of TorC2 and PKB at the cell's leading edge mediates chemotaxis. Curr. Biol. 2008;18:1034–1043. doi: 10.1016/j.cub.2008.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A., Kurita S., Hayashi A., Kaji N., Ohashi K., Mizuno K. Critical roles of actin-interacting protein 1 in cytokinesis and chemotactic migration of mammalian cells. Biochem. J. 2008;414:261–270. doi: 10.1042/BJ20071655. [DOI] [PubMed] [Google Scholar]
- Knecht D., Pang K. M. Electroporation of Dictyostelium discoideum. Methods Mol. Biol. 1995;47:321–330. doi: 10.1385/0-89603-310-4:321. [DOI] [PubMed] [Google Scholar]
- Konzok A., Weber I., Simmeth E., Hacker U., Maniak M., Muller-Taubenberger A. DAip1, a Dictyostelium homologue of the yeast actin-interacting protein 1, is involved in endocytosis, cytokinesis, and motility. J. Cell Biol. 1999;146:453–464. doi: 10.1083/jcb.146.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Brieher W. M., Scimone M. L., Kang S. J., Zhu H., Yin H., von Andrian U. H., Mitchison T., Yuan J. Caspase-11 regulates cell migration by promoting Aip1-Cofilin-mediated actin depolymerization. Nat. Cell Biol. 2007;9:276–286. doi: 10.1038/ncb1541. [DOI] [PubMed] [Google Scholar]
- Mendoza M. C., Firtel R. A. Assaying chemotaxis of Dictyostelium cells. Methods Mol. Biol. 2006;346:393–405. doi: 10.1385/1-59745-144-4:393. [DOI] [PubMed] [Google Scholar]
- Mohri K., Ono K., Yu R., Yamashiro S., Ono S. Enhancement of actin-depolymerizing factor/cofilin-dependent actin disassembly by actin-interacting protein 1 is required for organized actin filament assembly in the Caenorhabditis elegans body wall muscle. Mol. Biol. Cell. 2006;17:2190–2199. doi: 10.1091/mbc.E05-11-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohri K., Vorobiev S., Fedorov A. A., Almo S. C., Ono S. Identification of functional residues on Caenorhabditis elegans actin-interacting protein 1 (UNC-78) for disassembly of actin depolymerizing factor/cofilin-bound actin filaments. J. Biol. Chem. 2004;279:31697–31707. doi: 10.1074/jbc.M403351200. [DOI] [PubMed] [Google Scholar]
- Mouneimne G., DesMarais V., Sidani M., Scemes E., Wang W., Song X., Eddy R., Condeelis J. Spatial and temporal control of cofilin activity is required for directional sensing during chemotaxis. Curr. Biol. 2006;16:2193–2205. doi: 10.1016/j.cub.2006.09.016. [DOI] [PubMed] [Google Scholar]
- Okada K., Blanchoin L., Abe H., Chen H., Pollard T. D., Bamburg J. R. Xenopus actin-interacting protein 1 (XAip1) enhances cofilin fragmentation of filaments by capping filament ends. J. Biol. Chem. 2002;277:43011–43016. doi: 10.1074/jbc.M203111200. [DOI] [PubMed] [Google Scholar]
- Okada K., Obinata T., Abe H. XAIP 1, a Xenopus homologue of yeast actin interacting protein 1 (AIP1), which induces disassembly of actin filaments cooperatively with ADF/cofilin family proteins. J. Cell Sci. 1999;112:1553–1565. doi: 10.1242/jcs.112.10.1553. [DOI] [PubMed] [Google Scholar]
- Okada K., Ravi H., Smith E. M., Goode B. L. Aip1 and cofilin promote rapid turnover of yeast actin patches and cables: a coordinated mechanism for severing and capping filaments. Mol. Biol. Cell. 2006;17:2855–2868. doi: 10.1091/mbc.E06-02-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okreglak V., Drubin D. G. Loss of Aip1 reveals a role in maintaining the actin monomer pool and an in vivo oligomer assembly pathway. J. Cell Biol. 188:769–777. doi: 10.1083/jcb.200909176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S. The Caenorhabditis elegans unc-78 gene encodes a homologue of actin-interacting protein 1 required for organized assembly of muscle actin filaments. J. Cell Biol. 2001;152:1313–1319. doi: 10.1083/jcb.152.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S. Regulation of actin filament dynamics by actin depolymerizing factor/cofilin and actin-interacting protein 1, new blades for twisted filaments. Biochemistry. 2003;42:13363–13370. doi: 10.1021/bi034600x. [DOI] [PubMed] [Google Scholar]
- Ono S., Mohri K., Ono K. Microscopic evidence that actin-interacting protein 1 actively disassembles actin-depolymerizing factor/Cofilin-bound actin filaments. J. Biol. Chem. 2004;279:14207–14212. doi: 10.1074/jbc.M313418200. [DOI] [PubMed] [Google Scholar]
- Paradiso A., Cardone R. A., Bellizzi A., Bagorda A., Guerra L., Tommasino M., Casavola V., Reshkin S. J. The Na+-H+ exchanger-1 induces cytoskeletal changes involving reciprocal RhoA and Rac1 signaling, resulting in motility and invasion in MDA-MB-435 cells. Breast Cancer Res. 2004;6:R616–628. doi: 10.1186/bcr922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent C. A. Making all the right moves: chemotaxis in neutrophils and Dictyostelium. Curr. Opin. Cell Biol. 2004;16:4–13. doi: 10.1016/j.ceb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Patel H., Barber D. L. A developmentally regulated Na-H exchanger in Dictyostelium discoideum is necessary for cell polarity during chemotaxis. J. Cell Biol. 2005;169:321–329. doi: 10.1083/jcb.200412145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T. D., Borisy G. G. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Riedl J., et al. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D. N., Spudich J. A. Dynacortin, a genetic link between equatorial contractility and global shape control discovered by library complementation of a Dictyostelium discoideum cytokinesis mutant. J. Cell Biol. 2000;150:823–838. doi: 10.1083/jcb.150.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal A. A., Tetreault J. W., Lappalainen P., Drubin D. G., Amberg D. C. Aip1p interacts with cofilin to disassemble actin filaments. J. Cell Biol. 1999;145:1251–1264. doi: 10.1083/jcb.145.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A. T., Firtel R. A. Regulation of chemotaxis by the orchestrated activation of Ras, PI3K, and TOR. Eur. J. Cell Biol. 2006;85:873–895. doi: 10.1016/j.ejcb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Stock C., Schwab A. Role of the Na/H exchanger NHE1 in cell migration. Acta Physiol. 2006;187:149–157. doi: 10.1111/j.1748-1716.2006.01543.x. [DOI] [PubMed] [Google Scholar]
- Tsuji T., Miyoshi T., Higashida C., Narumiya S., Watanabe N. An order of magnitude faster AIP1-associated actin disruption than nucleation by the Arp2/3 complex in lamellipodia. PLoS One. 2009;4:e4921. doi: 10.1371/journal.pone.0004921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duijn B., Inouye K. Regulation of movement speed by intracellular pH during Dictyostelium discoideum chemotaxis. Proc. Natl. Acad. Sci. USA. 1991;88:4951–4955. doi: 10.1073/pnas.88.11.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es S., Wessels D., Soll D. R., Borleis J., Devreotes P. N. Tortoise, a novel mitochondrial protein, is required for directional responses of Dictyostelium in chemotactic gradients. J. Cell Biol. 2001;152:621–632. doi: 10.1083/jcb.152.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rheenen J., Song X., van Roosmalen W., Cammer M., Chen X., Desmarais V., Yip S. C., Backer J. M., Eddy R. J., Condeelis J. S. EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J. Cell Biol. 2007;179:1247–1259. doi: 10.1083/jcb.200706206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegtli W. C., Madrona A. Y., Wilson D. K. The structure of Aip1p, a WD repeat protein that regulates cofilin-mediated actin depolymerization. J. Biol. Chem. 2003;278:34373–34379. doi: 10.1074/jbc.M302773200. [DOI] [PubMed] [Google Scholar]
- Wang W., Eddy R., Condeelis J. The cofilin pathway in breast cancer invasion and metastasis. Nat. Rev. Cancer. 2007;7:429–440. doi: 10.1038/nrc2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.