A polybasic motif in the metabolic regulator lipin1 is both a membrane anchor and a nuclear localization sequence required for lipin1 function in phospholipid metabolism and adipogenesis.
Abstract
Lipins are phosphatidic acid phosphatases with a pivotal role in regulation of triglyceride and glycerophospholipid metabolism. Lipin1 is also an amplifier of PGC-1α, a nuclear coactivator of PPAR-α responsive gene transcription. Lipins do not contain recognized membrane-association domains, but interaction of these enzymes with cellular membranes is necessary for access to their phospholipid substrate. We identified a role for a conserved polybasic amino acid motif in an N-terminal domain previously implicated as a determinant of nuclear localization in selective binding of lipin1β to phosphatidic acid, using blot overlay assays and model bilayer membranes. Studies using lipin1β polybasic motif variants establish that this region is also critical for nuclear import and raise the possibility that nuclear/cytoplasmic shuttling of lipin1β is regulated by PA. We used pharmacological agents and lipin1β polybasic motif mutants to explore the role of PA-mediated membrane association and nuclear localization on lipin1β function in phospholipid metabolism and adipogenic differentiation. We identify a role for the lipin1 polybasic motif as both a lipid binding motif and a primary nuclear localization sequence. These two functions are necessary for full expression of the biological activity of the protein in intracellular lipid metabolism and transcriptional control of adipogenesis.
INTRODUCTION
Phosphatidic acid (PA) is an intracellular lipid signal (Stace and Ktistakis, 2006) and a metabolic precursor for the synthesis of glycero- and glycerophospho- lipids (Carman and Han, 2009). Lipins are Mg2+-dependent “type 1” phosphatidic acid phosphatases that dephosphorylate PA to generate diacylglycerol (DG), which can then be further acylated to form triglyceride (TG) (Reue, 2009). Studies with lipin1–deficient mice identify this lipin isoform as a key regulator of triglyceride and phospholipid metabolism linked to adiposity, TG storage, and lipoprotein synthesis (Peterfy et al., 2001). Lipin 1 overexpression is associated with increased adiposity and insulin resistance (Phan and Reue, 2005). The single lipin homolog in budding yeast (SMP2/PAH1) is a master regulator of phospholipid synthesis that controls the balance between its PA substrate and DG product, which are key intermediates in the synthesis of the major classes of glycerophospholipids in both yeast and mammalian cells (Han et al., 2006; O'Hara et al., 2006). In yeast, PAH1p has an additional indirect function as a transcriptional regulator through control of PA-dependent anchoring of the OPI1p transcriptional repressor to cytoplasmic membranes, which antagonizes its nuclear import and function to suppress expression of genes encoding proteins involved in phospholipid synthesis (Carman and Henry, 2007). Mammalian lipin1 is an amplifier of PPAR-α–responsive gene expression through a direct interaction with the PGC-1α coactivator, suggesting that the protein functions both within the nucleus and at intracellular membrane sites (Finck et al., 2006). Indeed, lipin1 was originally identified as a nuclear protein (Peterfy et al., 2001), although more recent work indicates that distribution of the protein between the nucleus and cytoplasm is heterogeneous (Peterfy et al., 2005; Bou Khalil et al., 2009; Peterfy et al., 2009). While the catalytic activity of lipin1 appears dispensable for association with PGC1-α and amplification of PPAR-α responsive gene transcription (Finck et al., 2006), the precise contributions of the lipid phosphatase activity and transcriptional coactivator function to the integrated role of lipin1 in metabolic regulation are presently largely undefined. Lipin1, its mammalian homologues Lipin 2 and Lipin 3, and PAH1p do not contain recognized membrane-association domains and can be isolated as stable soluble proteins in the absence of detergents. These enzymes readily dephosphorylate substrates presented as components of mixed phospholipid and detergent micelles, suggesting that they act preferentially on aggregated substrates (Han et al., 2006). Recruitment of lipins to cellular membranes is therefore likely to be critical and necessary for access to their phospholipid substrate. Evidence has been presented that association of lipin1 with intracellular membranes, most likely the endoplasmic reticulum, is regulated hormonally (Bou Khalil et al., 2009). The purpose of this study was to investigate mechanisms responsible for association of lipin1 with membranes to determine their relationship to those controlling movement of the protein between the nucleus and cytosol and evaluate the functional consequences of mutations that disrupt these processes.
MATERIALS AND METHODS
Mouse Husbandry
BALB/cByJ+/fld mice were a generous gift from Brian Finck (Washington University, St. Louis, MO) and bred to produce lipin-deficient (fld/fld) and wild-type (+/+ and +/fld) mice. All procedures conformed to the recommendations of Guide for the Care and Use of Laboratory Animals (Department of Health, Education, and Welfare publication number NIH 78-23, 1996) and were approved by the Institutional Animal Care and Use Committee.
cDNA Constructs
Constructs used in this study were made from a murine lipin1β cDNA with an N-terminal hemagglutinin (HA) epitope tag provided by Dr. Thurl Harris (University of Virginia, Charlottesville, VA). After insertion of this cDNA into the pENTR “Gateway” entry vector (Invitrogen, Carlsbad, CA) site-directed mutagenesis was performed using the QuikChange protocol (Stratagene, La Jolla, CA). The lipin 1 polybasic motif was fused to enhanced green fluorescent protein (GFP) using PCR and also inserted into pENTR. Plasmid, lentivirus, and baculovirus vectors were constructed by recombination of these entry vectors with appropriate destination vectors, in some cases appending additional epitope or fusion tags. Recombinant baculoviruses or lentiviruses were generated using reagent systems from Invitrogen (Carlsbad, CA).
Cell Culture and Transfections
HEK293, HepG2, and mouse embryo fibroblast (MEFs) cells were maintained in DMEM supplemented with 10% fetal bovine serum and 2 mM L-glutamine cultured in a humidified 37°C incubator with 5% CO2. Plasmid transfections were performed using lipofectamine 2000 (Invitrogen, Carlsbad, CA), according to manufacturer's instructions. MEFs were infected with recombinant lentiviruses at an estimated multiplicity of infection of 10. For radiochemical measurements of lipid metabolism 24 h after transfection, cells were incubated with DMEM (serum-free) and 1 μCi/ml [3H]palmitate acid (Amersham, Oakville, ON, Canada) or 1 μg/ml palmitate acid (Sigma-Aldrich Corp., St. Louis, MO), complexed with fatty acid-free BSA.
Lipid Analysis by Tandem Mass Spectrometry
Lipids were extracted from HepG2 cells using acidified organic solvents with the addition of lipid class-specific internal standards containing a C17 fatty acid (Avanti Polar Lipids). The lipid containing lower phases evaporated to dryness under N2 and reconstituted in 4:1 MeOH CHCl3. Molecular species of TG, DG, PA, and phosphatidylcholine (PC) were quantitated by selective reaction monitoring mode HPLC- electrospray ionization (ESI) tandem mass spectrometry using an AB Sciex (Foster City, CA) 4000 Q-Trap hybrid linear ion trap triple-quadrupole mass spectrometer equipped with a Turbo V electrospray ion source. A minimum of 16 abundant molecular species of each lipid class were detected by monitoring species specific precursor product ion pairs. DG and TG species (as ammoniated adducts) and PC species were analyzed in positive mode, while PA species were analyzed in negative mode. Recovery was determined by reference to the internal standards and quantitation accomplished by reference to calibration curves constructed using a set of synthetic standards for each lipid class obtained from Avanti Polar Lipids (Alabaster, AL) that were independently quantitated by accurate mass measurements or phosphorous determination (Pamuklar et al., 2009; Su et al., 2009). Lipid levels were normalized to measurements of total phospholipid phosphorous determined after wet digestion in percholoric acid.
TLC
Radiolabeled lipids were separated by TLC on silica gel plates developed in toluene/ether/ethanol/NH4OH (50/30/20/0.2, vol/vol/vol/vol). Lipids were identified by reference to authentic standards that were visualized by iodine staining. Material was scraped from the plates and transferred to a scintillation vial for quantitation by liquid scintillation counting.
Western Blotting
Cells were lysed in 50 mM Tris-HCl (pH 7.4), 100 μM 4-(2-aminoethyl) benzenesulfonyl fluoride, a protease inhibitor cocktail (Pierce, Rockford, IL), 4 mM MgCl2, 200 mM NaCl, 1% Triton-X 100 (Buffer A), and homogenized by sonication on ice. Equivalent volumes of the supernatants and the pellets were mixed with sample buffer, heated to 95°C for 5 min, and resolved by SDS-PAGE followed by Western blotting. Western blot was conducted using anti-HA antibody (Abcam, Cambridge, MA) or an anti–β-actin antibody (Sigma-Aldrich Corp., St. Louis, MO) and visualized using fluorescently conjugated secondary antibodies using a Licor Odyssey system (LI-COR Biosciences, Lincoln, NE).
Isolation, Culture, and Adipocyte Differentiation of MEFs
Cultures MEFs were established from E13.5 mouse embryos. Embryos were dissected from pregnant BALB/cByJ+/fld mice. The yolk sacs, heads, and internal organs were removed and used for genotyping by RT-PCR. The remaining embryo carcasses were dissected for preparation of MEFs, which were used for experiments within 6 passages. Adipocyte differentiation was induced by treatment of cells with a differentiation cocktail containing 0.25 μM dexamethasone, 1 μg/ml insulin, 0.5 mM 3-isobutyl-L-methylxanthine, and 0.1 mg/ml troglitazone (Sigma-Aldrich Corp., St. Louis, MO). Cells were incubated with this differentiation cocktail for 2 d, which was subsequently replaced with complete medium containing 2 μg/ml insulin for 4 d. Adipocyte differentiation was monitored by oil red-O staining, measurements of TG accumulation, and Western blotting and/or RNA analysis of differentiation markers. Cells were infected with lentivirus vectors for expression of GFP or the indicated lipin 1 variants before induction of differentiation.
RNA Analysis
RNA was extracted from MEFs using Trizol (Invitrogen, Carlsbad, CA) and levels of mRNAs for a series of adipocyte markers determined using real-time PCR with primer sets obtained from Applied Biosystems (Carlsbad, CA).
Oil Red-O Staining of MEFs
Cultures of differentiated MEFs were washed in PBS and then fixed in 4% paraformaldehyde (Fisher Scientific, Fair Lawn, NJ) for 30 min at room temperature. A stock solution of 0.5% oil red-O (Sigma-Aldrich Corp., St. Louis, MO) in isopropanol (wt/vol) was diluted 60:40 in water and added to fixed cells for 1 h at room temperature. Cells were washed in water before examination by light microscopy.
Protein Purification and Determination of PA Phosphatase Activity
His6-tagged lipin proteins were expressed by infection of monolayer cultures of sf9 insect cells with recombinant baculoviruses at a multiplicity of infection of ∼10. Cells were disrupted by resuspension in Buffer A without detergents, particulate material sedimented by centrifugation, and proteins purified from the soluble fraction by affinity chromatography using proBond Nickel-Chelating Resin (Invitrogen, Carlsbad, CA). PA phosphatase activity was determined using mixed micelles of PA (100 μM) and Triton X-100 (3.2 mM) (Donkor et al., 2007). [32P] phosphatidic acid was prepared by phosphorylation of 1,2-dioleoyl-sn-glycerol using [32P] ATP and E. coli diacylglycerol kinase. Reactions contained 50 mM Tris-HCl (pH 7.5), 1.0 mM MgCl2, 2 mM Triton X-100, 0.2 mM [32P]phosphatidic acid (10,000 cpm/nmol). PA Phosphatase activity was determined by quantitation of released [32P]PO42−.
Lipid Blot Assay
Equal quantities of lipids were immobilized by spotting on a nylon membrane. The membrane was blocked with 3% fatty acid free bovine serum/TBST (50 mM Tris/HCl, pH 7.5, 150 mM NaCl and 0.1% vol/vol Tween-20) for 1 h at room temperature. Purified lipin1 proteins were diluted in this blocking solution at ∼0.2 μg/ml. The membrane was incubated with the proteins overnight at 4°C on a rocking platform. After washing six times for 5min with TBST, bound proteins were then identified by incubating with a 1:4000 dilution of rabbit polyclonal to HA tag (Abcam, Cambridge, MA, USA) in blocking reagent for 1h. The membrane was then incubated with goat anti-rabbit secondary antibody (LI-COR, Biosciences, Lincoln, Nebraska, USA) diluted 1:5000 into blocking reagent for 1 h followed by six times washing for 5 min with TBST. Bound proteins were visualized using a Licor Odyssey system (LI-COR Biosciences, Lincoln, NE).
Preparation of Sucrose-Loaded Liposomes
Phosphatidylinositol 4,5-bisphosphate (PI4,5P2) was purified from bovine brain lipid extracts as described (Du et al., 2002). Other lipids were obtained from Avanti Polar Lipids (Alabaster, AL). Sucrose-loaded liposomes were prepared using minor adaptations of previously described methods (Buser and McLaughlin, 1998). Lipids were combined from stock solutions in CHCl3 and dried by rotary evaporation. Dried lipid films were hydrated in sucrose buffer containing 176 mM sucrose, 1 mM MOPS, pH 7.0 by rapid vortexing to produce multilamellar vesicles. Large unilamellar liposomes were generated by five freeze-thawing cycles and 15 extrusion steps through a double 0.1 μm nylon membrane (Avanti Polar Lipids, AL, USA) using a compressed N2 driven extruder (Northern Lipids BC, Canada) at 25°C. A trace amount of 3H-PC was included for determination of recovery through the procedure and calculation of the lipid concentration in the final preparation.
Sucrose-Loaded Liposome Binding Assay
Binding of lipin proteins to sucrose loaded liposomes was conducted by minor modifications of a previously reported procedure (Sciorra et al., 1999; Sun et al., 2004). In brief, sucrose-loaded liposomes of various lipid compositions were prepared and resuspended in buffer containing 100 mM KCl and 1 mM MOPS in microcentrifuge tubes. Proteins were added and incubated with the vesicles at room temperature for 15 min. Binding of proteins to liposomes was monitored by measuring PA phosphatase activity remaining in the supernatant after sedimentation of the liposomes by ultracentrifugation at 100,000g for 1 h and by detecting bound proteins in the liposome pellets by Western blotting.
Indirect Immunofluorescence
Cells were grown on tissue culture chamber slides (Falcon, Franklin Lakes, NJ) and transected with plasmids for expression of lipin 1 proteins. Two days after transfection, cells were fixed with 4% formaldehyde for 30 min and permeabilized for 20 min with 0.1% Triton X-100. Cells were blocked with PBS containing 1 mg/ml BSA for 1h before incubation with primary antibodies for detection of epitope tagged recombinant proteins or endogenous markers for 1h. After further washing cells were then incubated with appropriate secondary antibodies for 1 h in the dark followed by washing with PBS. Cells were stained with 300 nM 4′,6-diamidino-2-phenylindole for 5 min, followed by two washes in PBS. Coverslips were then mounted onto glass slides using ProLong Gold antifade reagent (Invitrogen, Carlsbad, CA, USA). The cells were visualized by fluorescence microscopy (Nikon Eclipse TE2000) or Confocal microscopy (Leica TCS SP5). For evaluation of the subcellular distribution of proteins between the cytosol and nucleus we examined up to 10 independent fields of cells and scored at least 100 cells for each measurement.
Association of Lipin1β with Importin-α
GST importin-α was expressed using a plasmid generously provided by Christian Faul (University of Miami, Miami, FL) and purified using glutathione sepharose 4B CL. Lipin1β-expressing cells were lysed in buffer containing 50 mM Tris-HCl, pH 7.5, 1% Triton X-100, 150 mM NaCl, 0.5% sodium deoxycholate, 1 mM NaF and 100 mmol PMSF. Lysates were incubated with glutathione sepharose immobilized GST importin-α at 4°C overnight. The resin was collected by centrifugation and washed five times in 1 ml of lysis buffer. Bound proteins were analyzed by SDS-PAGE and immunoblotting.
Statistical Analyses
Variables were compared using one-way analysis of variance. All data are presented as the means ± SD of at least three separate determinations. Statistically significant differences are identified with P values <0.05 (*), <0.005 (**), and <0.0005 (***).
RESULTS
Role of the Polybasic Motif in Nuclear Localization of Lipin1β
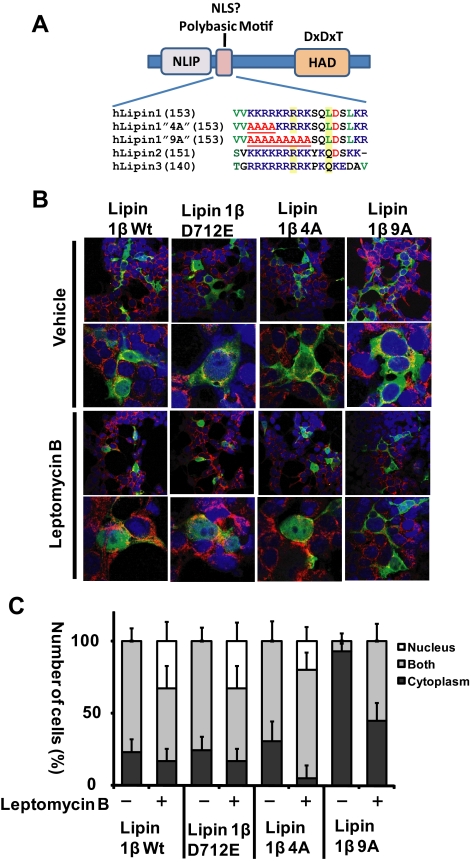
The transcriptional coactivator function of lipin1β necessarily requires localization in the nucleus. Mammalian lipins contain a conserved sequence of basic amino acid residues (a “polybasic motif”) that has been speculated to function as a nuclear localization sequence, and evidence that nuclear localization of lipin 1 is attenuated when certain of these residues were mutated has been presented (Peterfy et al., 2005; Bou Khalil et al., 2009; Peterfy et al., 2009). To examine the requirement for integrity of this polybasic motif for nuclear localization we generated variants of lipin1β in which residues within this sequence were systematically substituted with alanine. Lipin1β-4A contains alanine substitutions of the N-terminal basic residues in this motif, while Lipin1β-9A contains alanine substitutions of all contiguous basic residues within the motif (Figure 1A). We compared the subcellular localization of these variants to that of wild-type lipin1β and a catalytically inactive mutant lipin1β D712E (Figure 1, B and C). When expressed in cultured mammalian cells, we observed that the subcellular distribution of wild-type lipin1β between the cytosol and the nucleus was heterogeneous. In most cells, lipin1β partially colocalized with the endoplasmic reticulum marker calnexin, and to a lesser extent, the nuclear marker DAPI. No cells exhibited exclusive nuclear localization of wild-type lipin1β. When cells expressing wild-type lipin1β were treated with the nuclear export inhibitor leptomycin B for short times, we observed marked accumulation of lipin1β in the nucleus with a significant number of cells now exhibiting exclusively nuclear localized lipin1β. This result implies that nuclear localization of lipin1β is dynamic and that steady state levels of lipin1β in the nucleus reflect a balance between import and export. Although the ability of leptomycin B to promote nuclear localization of lipin1β was unaltered by mutation of catalytic domain sequences (lipin1β D712E) or the first four polybasic motif residues, (lipin1β 4A), exclusive nuclear localization of the lipin1β 9A mutant was not observed in any cells after treatment with leptomycin B. These results suggest that the lipin1β polybasic motif functions as a nuclear localization sequence and that last five basic residues within the motif have a dominant function in this process. In the absence of leptomycin B treatment the number of cells exhibiting partial nuclear localization of lipin1β 9A was markedly reduced in comparison to wt Lipin1β. However, leptomycin B treatment did produce a modest increase in the fraction of cells in which the lipin 1β 9A mutant exhibited partial localization to the nucleus, suggesting that when the polybasic motif is mutated the protein may contain additional weaker determinants of nuclear import that remain to be determined. Two regions of basic amino acid residues within the C terminus of lipin 1β (residues 436-444 and 598-605) are candidates for this function (Figure 1, B and C).
Figure 1.
The Lipin1β polybasic motif is required for nuclear localization. (A) Sequence alignment showing the conserved polybasic motif in mammalian lipins, and the polybasic motif mutants “9A” “4A” in lipin1β. In the 4A and 9A mutants, the polybasic motif was disrupted by substitution of conserved basic residues with four or nine alanines, respectively. In the “D712E” mutant the catalytic motif aspartic acid residue was substituted with glutamic acid. NLIP is the conserved lipin N-terminal homology domain, and HAD is the lipin catalytic domain with homology to halo acid dehalogenases. (B) HEK293 cells were transfected with vectors for expression of the indicated lipin1β variants. 24 h after transfection, cells were treated with vehicle or 40 nM leptomycin B for 4 h. Subcellular localization of HA-tagged lipin1β (green) was examined by confocal microscopy to compare the localization with the nuclear marker DAPI (blue) and the endoplasmic reticulum (ER) marker calnexin (red). (C) Percentage of the number of transfected cells examined in which lipin1β was localized in cytoplasm, nucleus, or both cytoplasm and nucleus (mean ± SD of at least 100 cells counted in 10 distinct fields).
The Lipin1β Polybasic Motif Sequence Is Sufficient for Nuclear Import
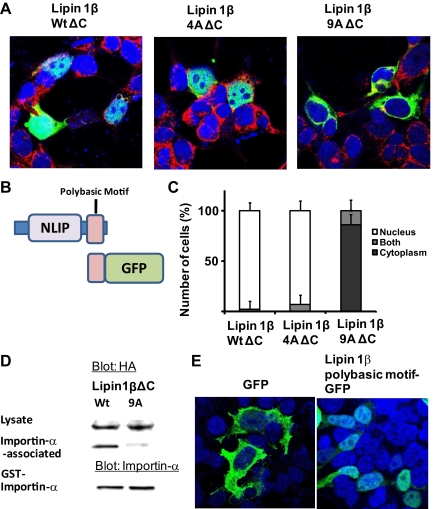
To further explore the nuclear import function of the lipin 1 polybasic motif we generated lipin1 truncation mutants in which the C terminus of the protein from residue 204 onwards that includes the haloacid dehalogenase homology (HAD) catalytic domain was deleted, leaving the N-terminal lipin homology domain (NLIP) and the polybasic motif (lipin1β ΔC). We examined their subcellular localization (Figure 2, A–C). The deletion mutant containing a wild-type polybasic motif sequence was strongly localized to the nucleus in the absence of leptomycin B treatment. The localization pattern was preserved in a lipin1β truncation mutant containing the 4A polybasic motif mutation, as observed with the full-length protein in leptomycin B-treated cells. A lipin1β truncation mutant harboring the 9A polybasic motif mutation was markedly excluded from the nucleus, with no cells exhibiting exclusive nuclear localization of this protein and only a small number of cells exhibiting partial localization to both the nucleus and cytosol, which may reflect cells with compromised nuclear integrity, for example dividing or apoptotic cells. Nuclear import requires nuclear localization sequence-mediated interaction of cargo proteins with importins (Lange et al., 2007). Consistent with its marked nuclear localization, we found that lipin1βΔC interacted strongly with a GST Importin-α fusion protein. This interaction was disrupted by the 9A mutation of the polybasic motif (Figure 2D). Finally we found that fusion of the 9-aa lipin polybasic motif sequence to the N terminus of GFP directed the fusion protein to the nucleus (Figure 2E). These results indicate that the lipin1β polybasic motif can function as an autonomous nuclear localization sequence and suggest that sequences specifying nuclear export or other interactions that are antagonistic to the import process reside in the C-terminal region of the protein. Computational analysis of the lipin1 sequence using a previously defined algorithm (la Cour et al., 2004) identifies a candidate leucine-rich nuclear export sequence centered on leucine 674 of murine lipin1β that is conserved in lipin 1α and could be a determinant of nuclear export. Unlike the truncated variants, we were unable to demonstrate robust polybasic motif-dependent interaction of full-length lipin1β with GST importin-α, suggesting that the C terminus of the protein participates in intermolecular interactions or perhaps associations with other proteins such as 14-3-3 (Peterfy et al., 2009) that impact on the nuclear localization sequence function of the polybasic motif. Further work will be needed to investigate these possibilities.
Figure 2.
Deletion of the lipin1β C terminus promotes nuclear localization. (A) Subcellular localization of truncated lipin1β (lipin1βΔC) with wt 4A and 9A polybasic motif sequences (green) compared with the nuclear marker DAPI (blue) and endoplasmic reticulum marker calnexin (red). (B) Structural organization of lipin1β C-terminal truncated mutants and lipin polybasic motif GFP fusion. (C) Percentage of the number of transfected cells examined in which lipin1β was localized in cytoplasm, nucleus, or both cytoplasm and nucleus (mean ± SD of at least 100 cells counted in 10 distinct fields). (D) Association of lipin1βΔC with a wt or “9A” polybasic motif sequence with importin-α was determined as described in the Materials and Methods. (E) Subcellular localization of GFP or a lipin 1β polybasic motif GFP fusion protein (green) and the nuclear marker DAPI (blue).
Lipin1β Polybasic Motif Mutants Are Catalytically Active but Exhibit Impaired Binding to Phosphatidic Acid
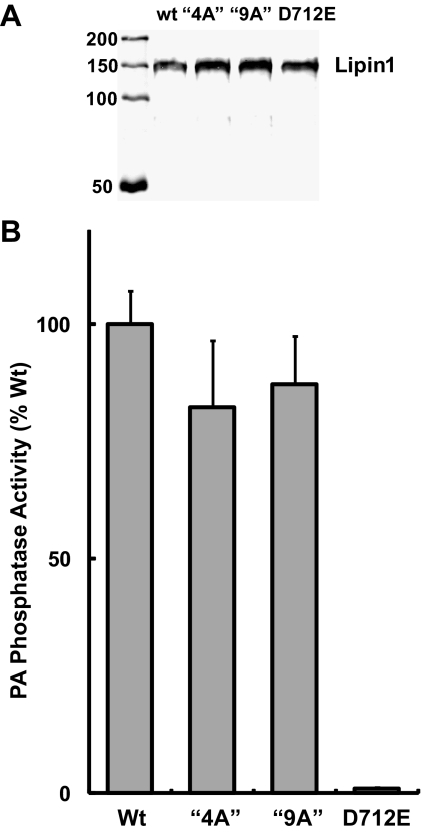
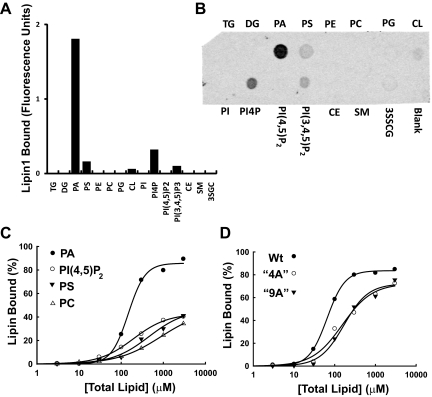
Lipin1 variants with altered nuclear or cytoplasmic subcellular localization would be of great value for probing lipin function in transcriptional regulation and lipid metabolism. We were therefore interested in evaluating catalytic activity of these lipin1β polybasic motif variants, which was accomplished using purified proteins generated in the baculovirus expression system. When assayed using mixed micelles of PA and Triton X-100 at saturating substrate concentrations, purified wild-type lipin1β and the lipin1β 4A and 9A mutants exhibited very similar specific activities while the lipin1β D712E mutant had no measurable activity as expected (Figure 3, A and B). To explore the lipid binding properties of lipin1β, we conducted experiments using membrane immobilized lipids and sucrose loaded phospholipid vesicles. We found that lipin1β exhibited a marked binding preference for PA over a series of other lipids in a blot overlay assay (Figure 4, A and B). This selectivity was preserved in measurements made using sucrose loaded bilayer liposomes (Figure 4, C and D). In this latter system, inclusion of low molar fractions of PA, but not other acidic lipids including phosphoinositides resulted in an ∼10-fold increase in affinity of lipin1β for PC vesicles. This binding does not likely involve the interaction of substrate with the catalytic site of the enzyme because catalytically inactive lipin1β D712E (in which, by analogy to related haloacid dehalogenases, coordination of a catalytic Mg2+ ion that binds the phosphate group of the substrate in the active site is impaired) also showed binding specificity for PA in both the blot overlay and liposome assays (not shown). We also examined binding of the lipin1β 4A and 9A polybasic motif mutants to sucrose loaded phospholipid vesicles. In contrast to the wild-type enzyme, the binding enhancement observed with low molar fractions of PA was abolished in both of these polybasic motif mutants (Figure 4, C and D). These results indicate that lipin1β contains structural determinant(s) that specify binding to acidic lipids, with a pronounced selectivity for PA, that is distinct from the catalytic motif and identify an important contribution of the polybasic motif to this process. Although the 4A and 9A mutants exhibited a similar decrease in binding affinity to vesciles containing 10 mol% PA, more extensive quantitative measurements of these interactions will be needed to determine precise affinities of lipin1 for PA and the overall contribution of residues within the polybasic motif to this process.
Figure 3.
Lipin 1 polybasic motif mutants are catalytically active. (A) Western blot of purified lipin1β wt and the indicated mutants expressed in insect cells using baculovirus vectors and detected by using an anti-HA antibody. (B) Specific PA phosphatase activity of wt lipin1β and mutants were determined by using [32P]-labeled PA substrate, and activity was monitored by measuring the release of water soluble [32P]-PO42−. Data are means ± SD of triplicate determinations normalized to activity of wt lipin1β.
Figure 4.
Selective binding of lipin 1 to PA in bilayer liposomes is attenuated by mutation of the polybasic motif. (A, B). The indicated lipids were immobilized on a nylon membrane (Triacylglycerol [TG]; Diacylglycerol [DG]; Phosphatidic Acid [PA]; Phosphatidylserine [PS]; Phosphatidylethanolamine [PE]; Phosphatidylglycerol [PG]; Cardiolipin [CL]; Phosphatidylinositol [PI']; Phosphatidylinositol 4-phosphate [PI4P]; Phosphatidylinositol (4,5)-bisphosphate: [PI(4,5)P2]; Phosphatidylinositol (3,4,5)-trisphosphate [PI(3,4,5P3]; Cholesterol ester [CE]; Sphingomyelin [SM]; 3-sulfogalactosylceramide [3SGC]. Binding of recombinant purified lipin1β was monitored using an HA-tag selective primary antibody and fluorescently conjugated secondary antibody with quantitation using the Li-Cor Odyssey imaging system. (C) Binding of lipin1β to sucrose loaded phosphatidylcholine (PC) vesicles containing 10 mol % of the indicated phospholipids was determined by measuring PA phosphatase activity remaining in the supernatant after sedimentation of the vesicles by ultracentrifugation. (D) Comparison of the binding of wild-type lipin1β and the 4A and 9A polybasic motif mutants to sucrose loaded PC vesicles containing 10 mol % PA.
Inhibition of Phospholipase D Promotes Nuclear Import of Lipin1β
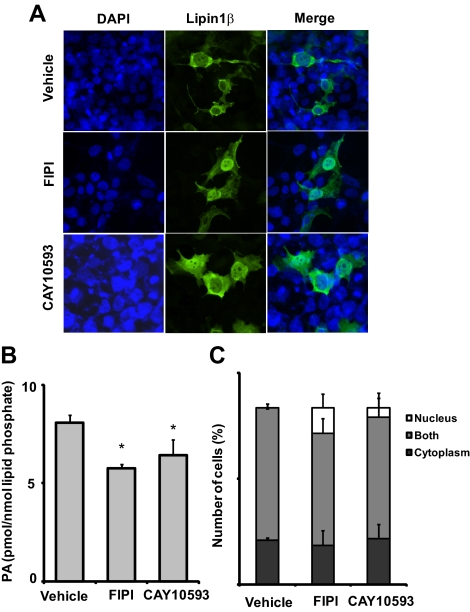
Our finding that the lipin1β polybasic motif is a critical determinant of both nuclear localization and selective binding to PA raises the possibility that nuclear localization and membrane association of lipins could be regulated in a coordinated manner by this lipid. We attempted to evaluate association of lipin1 with membranes by subcellular fractionation. Although we observed partial localization of lipin 1β with the endoplasmic reticulum marker calnexin in fixed cells (Figure 1), after hypotonic lysis and differential centrifugation in our hands lipin1 was predominantly soluble. This may indicate that the affinity of the interaction with membranes is too low to be preserved significantly post lysis. As reported by others (e.g., Bou Khalil et al., 2009), a small (>10%) fraction of the total lipin1β was recovered in a microsomal fraction. Mutation of the polybasic motif sequences did not alter recovery of lipin1β in this fraction, suggesting that other types of interactions are involved in microsomal targeting of the protein (not shown). We therefore explored effects of manipulations designed to alter cellular PA levels on the subcellular localization of lipin1β monitored by indirect immunofluorescence. Recent studies using halopemide derivatives as potent selective chemical inhibitors reveal that phosphatidylcholine specific phospholipase D (PLD) is a significant source of PA in cells grown in serum (Scott et al., 2009; Su et al., 2009). We found that treatment of HEK293 cells with empirically determined optimally effective concentrations of two such halopemide-based PLD inhibitors, FIPI (which inhibits both PLD1 and PLD2) and CAY10593 (which exhibits selectivity for PLD2) produced 30 and 20% decreases in total cellular PA levels measured by HPLC tandem mass spectrometry, respectively. This decrease in cellular PA levels was accompanied by the appearance of cells exhibiting exclusive localization of lipin1β to the nucleus (Figure 5). The PLD1 and PLD2 targets of FIPI are largely localized to cytoplasmic membrane organelles including the plasma membrane, Golgi apparatus, and endoplasmic reticulum (Stace and Ktistakis, 2006; Morris, 2007), the latter of which appears to be an important site of lipin1β action in phospholipid metabolism (Bou Khalil et al., 2009). These results suggest that elevated levels of PA on cytoplasmic membranes antagonize nuclear localization of lipin1β. Control experiments using GFP reporters fused to the SV40 T-antigen nuclear localization sequence indicated that treatment of cells with these PLD inhibitors did not have generalized effects on nuclear integrity and nuclear import (not shown).
Figure 5.
Nuclear localization of lipin1 is promoted by pharmacological inhibition of phospholipase D. (A) HEK 293 cells expressing wild-type lipin1β and the 4A and 9A polybasic motif mutants were treated with 1.5 μM of the dual PLD1/PLD2 inhibitor FIPI or 50 μM of the PLD2 selective inhibitor CAY10593 for 4h. Subcellular localization of lipin1β (green) was visualized by indirect immunofluorescence and compared with the nuclear marker DAPI (blue). (B) Total PA levels in cells incubated with vehicle or the indicated PLD inhibitors at the concentrations used for the experiment shown in panel A and total PA levels (the sum of 16 abundant PA species) quantitated by HPLC ESI MS/MS. (C) Percent of total number of transfected cells, which lipin1β was localized in cytoplasm, nucleus, or both cytoplasm and nucleus (mean ± SD of at least 100 cells counted in 10 distinct fields).
The Lipin 1 Polybasic Motif Is Critical for Lipin1β Function in Phospholipid and Neutral Lipid Metabolism
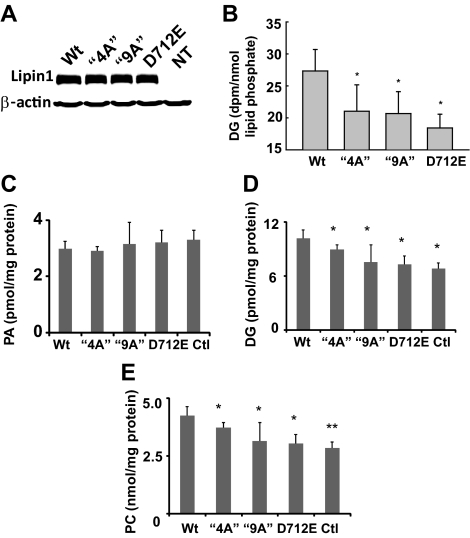
We used lentivirus vectors to overexpress wild-type lipin1β and the lipin1β 4A and 9A variants in cultured liver cells (Figure 6A). Using a radiolabeling approach we found that, in comparison to cells expressing the inactive lipin1β D712E mutant, overexpression of wt lipin1β, but not the 4A and 9A mutants, resulted in significant production of the lipin1β product DG (Figure 6 B), suggesting that although the 4A and 9A mutants are enzymatically active in vitro they are not functional in cellular lipid metabolism. The lipin1 substrate PA represents a family of structurally related molecules with distinct radyl chain lengths, linkages, and saturations (Stace and Ktistakis, 2006). Furthermore both PA and the product of lipin catalyzed PA hydrolysis, DG, can be produced by additional cellular pathways. DG is a precursor for synthesis of the major cellular glycerophospholipid PC, raising the possibility that a more detailed analysis might reveal selective effects of manipulations of lipin1β on particular species or pools of these lipids. Accordingly, we used HPLC tandem mass spectrometry assays to quantitate changes in molecular species of PA and DG in cells overexpressing wild-type catalytically inactive 4A and 9A lipin1β mutants. Overexpression of wild-type l lipin1β or the various lipin 1β mutants had no significant effect on PA levels in these cells (Figure 6C). As observed with the radiolabeling studies, overexpression of wild-type lipin1β significantly elevated levels of a series of molecular species of DG and the major glycerophospholipid product derived from this pathway, PC, while the lipin1β 4A and 9A mutants did not. These results identify a critical role for the lipin1β polybasic motif in expression of lipin1β catalytic activity linked to the production of key intermediates required for the synthesis of glycerophospholipids and triglycerides.
Figure 6.
Effects of overexpression of lipin1β wt and mutants on diacylglycerol (DG), phosphatidylcholine (PC), and phosphatidic acid (PA) levels. (A) HA-tagged wt lipin1β and the indicated mutants were overexpressed in HepG2 cells and detected by Western blotting. (B) HepG2 cells expressing wt lipin1β and the indicated mutants were radiolabeled with [3H]palmitic acid for 24 h. DG levels were determined after separation by TLC, and quantitation by scintillation counting. (C–E) Levels of 16 abundant molecular species of PA, DG, and PC were determined in HepG2 cells overexpressing lipin1β wt, and mutants incubated with 1 μM palmitic acid were measured by HPLC ESI MS/MS. Data are means ± SD of at least three independent determinations.
Requirement of Catalytic Activity and the Polybasic Motif for Lipin1β Function in Adipogenic Differentiation
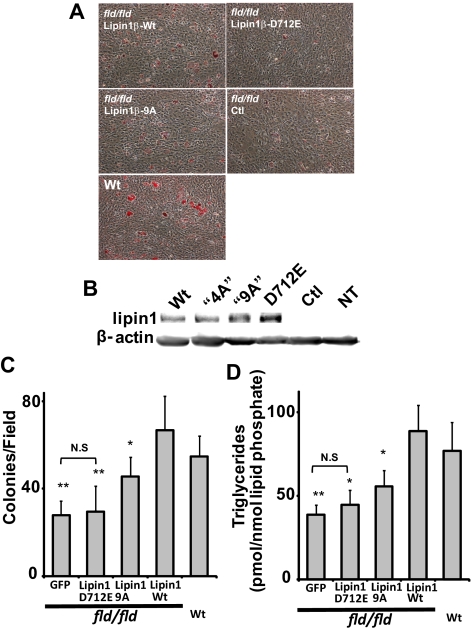
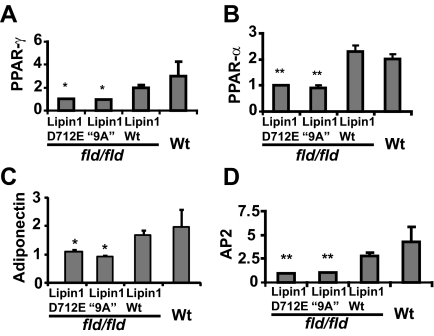
Embryo fibroblasts cultured from mice (MEFs) that are homozygous for the lipin1 fld allele are defective in their capacity for adipogenic differentiation. This function can be restored by retroviral expression of lipin1 in these cells (Phan et al., 2004; Peterfy et al., 2005). To investigate the requirement for the lipin1 polybasic motif in this process, we examined effects of lentivirus-mediated overexpression of wild-type catalytically inactive and 9A variants of lipin1β on adipogenic differentiation of these cells measured by oil red-O staining (Figure 7). We found that the differentiation response of fld/fld MEFs was weak, but the small number of colonies of differentiated cells was substantially increased by overexpression of wild-type lipin1β to an extent that was comparable to differentiation observed with fibroblasts from wild-type mouse embryos. Although these recombinant lipin proteins were expressed to a similar extent as determined by Western blotting, the ability of the lipin1β 9A mutant to restore adipogenesis in these cells detected by quantitation of oil red-O stained colonies was attenuated to 55% of the number of colonies observed in cells expressing wild-type lipin1β. TG accumulation in cells expressing the lipin 1β 9A mutant in comparison to fld/fld MEFs expressing wild-type lipin 1β was attenuated to a slightly greater extent (65%). Adipogenic differentiation requires coordinated expression of a set of adipocyte specific genes and acquisition of the ability to synthesize and accumulate triglycerides. To begin to discriminate the relative roles for the metabolic and transcriptional functions of lipin1β in this process, we examined the ability of the catalytically inactive lipin1β D712E mutant to restore adipogenic differentiation in fld/fld MEFs. Although also strongly expressed, inactive mutant did not increase the number of oil red-O–stained adipocyte colonies over control or increase TG levels in the population of cells. We therefore conclude that proper expression of lipin1β catalytic activity is critical for complementation of the adipogenesis defect of fld/fld MEFs. While not active in the overexpression assays shown in Figure 6, increases in oil red-O–stained colonies and TG accumulation indicate that overexpression of the lipin1 9A mutant can restore conversion of PA to DG for TG and phospholipid synthesis to some extent in these cells. These observations could reflect compensatory increases in expression of other lipin isoforms or the possibility that this mutant exhibits partial catalytic activity in this setting. We also examined the ability of wild-type lipin1β and the lipin1β D712E and “9A” mutants to promote expression of adipocyte-specific genes (including genes that are known to be PPAR-α responsive) when expressed in fld/fld MEFs (Figure 8). We found that expression of wild-type lipin1β increased expression of a subset of these genes when fld/fld MEFs were incubated in differentiation medium. However statistically significant increases in expression of these genes were not observed in cells expressing the lipin1β 9A mutant. The ability of the lipin1β 9A mutant to effect a partial rescue of adipogenic differentiation in the apparent absence of statistically significant increases in expression of other genes known to be important for this process (notably PPARγ) is paradoxical and might be explained by sufficiency of the basal levels of gene expression in fld/fld MEFs for adipogenesis or by modest but functionally significant increases in adipogenic gene expression in the differentiating subpopulation of cells that are not detected by our assays. Regarding the latter possibility, while nuclear localization of this mutant is substantially decreased as discussed above, lipin 1 may contain additional weaker determinants of nuclear localization (Figure 1). Taken together our results indicate that both the transcriptional and metabolic functions of lipin 1 are required for full complementation of the adipogenic differentiation of fld/fld MEFs. Of particular relevance to this issue we found that consistent with its inability to restore adipogenic differentiation in fld/fld MEFs (Figure 7), the catalytically inactive lipin1β D712E mutant was unable to restore adipogenic gene expression in these cells (Figure 8), suggesting that the enzyme activity of lipin1β resulting in localized changes in a PA or a PA metabolite might be necessary for the transcriptional coactivator function of lipin1β.
Figure 7.
The polybasic motif plays an important role in complementation of the adipogenesis defect of fld/fld mouse embryo fibroblasts by lipin1β. (A) Oil red-O staining (red) of differentiated wt and fld/fld mouse embryo fibroblasts expressing wild-type lipin1β, the indicated lipin1β mutants, or a GFP control using lentivirus vectors at day 6 postinduction of differentiation. (B) Expression of lipin 1 variants was quantitated by Western blotting. (C) Adipogenic differentiation was quantitated by counting the number of oil red-O–stained colonies per field. (D) Adipogenic differentiation was quantitated by measuring total TG levels (the sum of 36 abundant TG species) using HPLC ESI MS/MS. Data shown are means ± SD of three or more independent determinations. Statistically significant differences between cells expressing wt lipin 1 and the indicated lipin 1 mutants are indicated by asterisks. Note that TG accumulation or the number of differentiated colonies was not significantly different (N.S.) between cells expressing GFP or Lipin1 D712E.
Figure 8.
The polybasic motif is required for complementation of the adipogenic gene transcription defect of fld/fld mouse embryo fibroblasts by lipin1β. RT-PCR was used to quantify PPAR-γ (A), PPAR-α (B), adiponectin (C), and aP2 mRNA expression (D) in MEFs isolated from wt mice, and fld/fld mice infected with lipin1β wt, lipin 1β D712E, and lipin1β “9A” lentivirus. Samples for RNA analysis were prepared from cells at 6 d postinduction of differentiation. Values are normalized (= 1.0) to RNA levels in control virus–infected fld/fld MEFs.
DISCUSSION
Phosphatidic acid has both metabolic and signaling functions (Stace and Ktistakis, 2006; Itoh et al., 2009; Nishikimi et al., 2009). In comparison to its precursors and metabolites, steady state PA levels are low in growing cells despite the considerable metabolic flux through the cellular PA pool required to sustain phospholipid synthesis (Ivanova et al., 2009). In addition to production de novo, PA levels can be acutely elevated in cells by the activity of DG kinases and PLD, which are coupled to agonist-stimulated pathways that promote PA-mediated signaling events (Stace and Ktistakis, 2006). Taken together, these findings focus attention on the role of PA as an intracellular lipid signal which has driven efforts to identify PA binding proteins that recognize and respond to temporal and spatial alterations in PA levels. In comparison to recognized domains that interact selectively with other lipid species such as phosphoinositides, a PA binding modular protein domain has not been defined. Instead, these studies have unearthed a disparate variety of proteins that interact with and are regulated by PA. A common feature of these proteins is the presence of sequences of basic amino acids, a so-called polybasic motif, often flanked by hydrophobic amino acids that has been presumed to participate in electrostatically mediated interactions with the PA headgroup (Stace and Ktistakis, 2006; Itoh et al., 2009; Nishikimi et al., 2009). The results reported here indicate that lipin1β exhibits a pronounced binding selectivity for PA in two different assay systems, including bilayer membranes containing low molar fractions of PA. Mutational studies localize an important determinant of this binding to a polybasic motif sequence that is well conserved among the three mammalian lipin isoforms. Studies using model peptides of similar overall net positive charge suggest that affinity of the lipin1β polybasic motif sequence for PA alone would be insufficient to account for the observed binding affinity and high selectivity with which lipin1β binds to PA in model membranes indicating that other mechanisms (for example secondary and tertiary structural determinants) likely contribute to the apparent affinity and selectivity with which the purified protein interacts with PA (Murray et al., 1998). Our finding that when fused to GFP the 9-aa lipin polybasic motif sequence directs the protein to the nucleus with apparently negligible association with cellular membranes is also consistent with this idea. An additional consideration is suggested by the observation that the preparations of purified wild-type and polybasic motif mutants of lipin1β used for our studies appear to be exclusively oligomeric, possibly a tetramer (Mr ∼500 KDa) when analyzed by size exclusion chromatography (H.R. and A.J.M., unpublished observations). Clearly this issue warrants further investigation, but if the individual polybasic motif sequences are functional in these putative lipin1 oligomers this could explain why PA induces such a marked increase in binding affinity of the protein for model membranes. Functional interactions between these binding sites could also play a role in the positive cooperative effect of PA on catalytic activity of yeast and mammalian lipins in assays with detergent solubilized substrates reported by several groups (Han et al., 2006; Donkor et al., 2007). While we found that lipin1β polybasic motif mutants were catalytically active in vitro, unlike wild-type lipin1β, they were unable to promote increases in levels of DG and PC when overexpressed in cells. In addition to membrane recruitment, interaction of the polybasic motif mutant with PA may also be important in orienting the enzyme with substrates in cellular membranes for proper expression of catalytic activity in a manner that is not preserved in assays using detergent solubilized substrates.
Our results also identify the lipin1β polybasic motif as a critical and necessary primary determinant of nuclear localization and imply that lipin1β exchange between the nucleus and cytoplasm is dynamic. Pharmacological manipulations that decrease cellular PA levels by inhibition of cytoplasmic PLD enzymes promote nuclear localization of lipin1β, suggesting that nuclear cytoplasmic shuttling of the enzyme is regulated by PA levels on cellular membranes. The similarity between acidic lipid binding polybasic motifs and nuclear localization sequences has been noted by others. The yeast sterile 5 scaffold contains a polybasic motif that functions as both a membrane anchor and nuclear localization sequence (Winters et al., 2005). The mammalian small GTPase REM also contains an acidic lipid binding polybasic motif at the C terminus that directs the protein to the nucleus when proximal hydrophobic residues are mutated (Heo et al., 2006). The lipin1 polybasic motif is also flanked by hydrophobic residues that might contribute to membrane association in a similar manner (Figure 1). Our results raise the possibility that, like these previously reported examples, this sequence motif also serves a “dual function” membrane binding/nuclear localization sequence. If this is the case, then alterations in PA levels in cytoplasmic membranes could regulate the balance between the enzymatic role of lipin1β in phospholipid and triglyceride synthesis and the transcriptional function which is clearly important for adipocyte differentiation through expression of PPAR-α responsive genes. In support of this idea, we found that pharmacological inhibition of phospholipase D leading to decreases in cellular PA levels promoted nuclear localization of lipin 1β.
To begin to investigate the importance of the N-terminal polybasic motif for lipin 1 function, we examined the ability of lipin1β polybasic motif mutants to complement the adipogenic defect of lipin1-deficient fld/fld MEFs. We found that expression of wild-type lipin1β in these cells restored differentiation measured by oil red-O staining, TG accumulation, and expression of several adipocyte markers. For reasons that are presently unclear, this finding differs from a prior report that lipin 1α but not lipin 1β was able to complement this defect in fld/fld MEFs (Peterfy et al., 2005). By contrast, we did not observe significant increases in expression of these markers in cells expressing the lipin1β “9A” mutant implying that nuclear localization is important for these effects. Evaluation of adipogenic differentiation of lipin 1 expressing fld/fld MEFs evaluated by oil red-O staining and TG accumulation again identified a critical role for the polybasic motif. However, in comparison to the catalytically inactive lipin1β D712E mutant, which was completely inactive in this assay, the reduced number oil red-O–stained colonies of lipin1β “9A” expressing cells suggest that, while we were unable to detect effects of overexpression of this variant on cellular levels of DG and PA in overexpression experiments, it has some enzymatic activity in this particular setting or could perhaps elicit compensatory changes in expression of the lipin 2 and lipin 3 isoforms. Most surprisingly, the catalytically inactive lipin1 D712E mutant, which exhibited wild-type nuclear localization patterns in our hands and has been reported to have unimpaired PGC1-α binding and can fully activate PPAR-α response elements in a reporter transcription assay (Finck et al., 2006), was also unable to restore adipogenic gene expression in fld/fld MEFs, raising the possibility that a direct or indirect product of the enzyme is important for the transcriptional effects. The inability of the lipin1β “9A” mutant to significantly increase adipogenic gene expression may also be indicative of a role for nuclear localized lipin1β enzyme activity in this process. On the other hand, mutation of a conserved serine residue in the C terminus of lipin1 and lipin2 has been reported to impair catalysis but not transcriptional coactivator function, again measured using reporter gene response element assays (Donkor et al., 2009). These findings may point to fundamental differences between reporter gene response element assays conducted in transfected cells and our study in which we examined restoration of gene transcription by lentivirus-mediated expression of lipin1β variants in lipin1-deficient MEFs. An analysis of the effects of the manipulations of lipin1β expression we report using mass spectrometry based “lipidomic” approaches to identify candidate lipin1β-generated metabolites that enhance or enable the transcriptional coactivator function may be warranted.
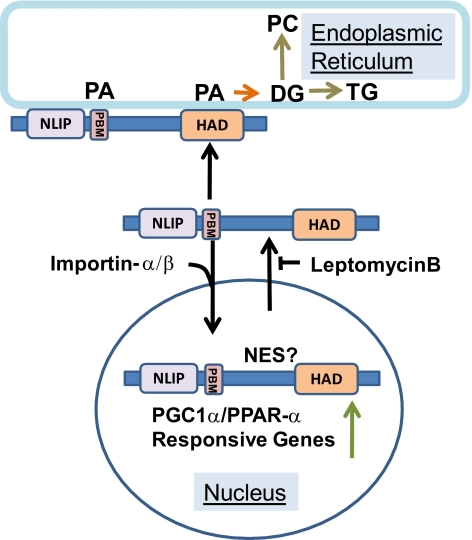
Several ongoing and recent studies have identified additional factors that contribute to an increasingly complex picture of lipin1 regulation in metabolism and nuclear gene transcription. These include posttranslational modifications such as phosphorylation and sumoylation (Harris et al., 2007; Liu and Gerace, 2009) and protein–protein interactions (Peterfy et al., 2009). While more work will be needed to determine how these processes impact on the phenomena reported here, our study provides new insights into sequence determinants of nuclear cytoplasmic trafficking of lipin 1 and the interaction of this enzyme with phospholipids including its subtrate PA. Our studies suggest a mechanism by which the lipin1 substrate, PA, could alter the balance of lipin1 localization between cellular membranes and the nucleus. PA-regulated nuclear cytoplasmic shuttling might therefore provide a mechanism to coordinate mammalian lipin1 function in synthesis of glycerolipids and transcriptional regulation (Figure 9) in an analogous (but mechanistically distinct) manner to that which has been elegantly demonstrated for the budding yeast lipin homolog smp2/PAP1p (Carman and Henry, 2007).
Figure 9.
Regulation of nucleo-cytoplasmic lipin1β traffic by phosphatidic acid. Our results suggest a model in which lipin1β actively shuttles between the nucleus and cytoplasm. This process is controlled by a nuclear import sequence identified here as the polybasic motif and putative nuclear export signal localized elsewhere in the C terminus of the protein. Interactions between lipin1β and PA mediated by the polybasic motif antagonize the nuclear import process and are important for proper expression of lipin1β catalytic activity required for synthesis of glycerophospholipids. NLIP is the conserved lipin N-terminal homology domain, HAD is the lipin catalytic domain with homology to halo acid dehalogenases and PMB is the polybasic motif. NES designates a putative nuclear export sequence.
ACKNOWLEDGMENTS
This research was supported by National Institutes of Health grants GM50388 and P20RR021954 and was conducted using resources provided by the Lexington KY Department of Veterans Affairs Medical Center. H.R. is the recipient of an American Heart Association Post Doctoral Fellowship.
Abbreviations used:
- DG
nDiacylglycerol
- FIPI
n5-Fluoro-2-indolyl des-chlorohalopemide
- HPLC ESI MS/MS
nhigh performance liquid chromatography electrospray ionization tandem mass spectrometry
- MEF
nmouse embryo fibroblast
- PA
nPhosphatidic acid
- PC
nPhosphatidylcholine
- PI4,5P2
nPhosphatidylinositol 4,5-bisphosphate.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-01-0073) on July 21, 2010.
REFERENCES
- Bou Khalil M., Sundaram M., Zhang H. Y., Links P. H., Raven J. F., Manmontri B., Sariahmetoglu M., Tran K., Reue K., Brindley D. N., Yao Z. The level and compartmentalization of phosphatidate phosphatase-1 (lipin-1) control the assembly and secretion of hepatic VLDL. J. Lipid Res. 2009;50:47–58. doi: 10.1194/jlr.M800204-JLR200. [DOI] [PubMed] [Google Scholar]
- Buser C. A., McLaughlin S. Ultracentrifugation technique for measuring the binding of peptides and proteins to sucrose-loaded phospholipid vesicles. Methods Mol. Biol. 1998;84:267–281. doi: 10.1385/0-89603-488-7:267. [DOI] [PubMed] [Google Scholar]
- Carman G. M., Han G. S. Phosphatidic acid phosphatase, a key enzyme in the regulation of lipid synthesis. J. Biol. Chem. 2009;284:2593–2597. doi: 10.1074/jbc.R800059200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman G. M., Henry S. A. Phosphatidic acid plays a central role in the transcriptional regulation of glycerophospholipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 2007;282:37293–37297. doi: 10.1074/jbc.R700038200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkor J., Sariahmetoglu M., Dewald J., Brindley D. N., Reue K. Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J. Biol. Chem. 2007;282:3450–3457. doi: 10.1074/jbc.M610745200. [DOI] [PubMed] [Google Scholar]
- Donkor J., Zhang P., Wong S., O'Loughlin L., Dewald J., Kok B. P., Brindley D. N., Reue K. A conserved serine residue is required for the phosphatidate phosphatase activity but not the transcriptional coactivator functions of lipin-1 and lipin-2. J. Biol. Chem. 2009;284:29968–29978. doi: 10.1074/jbc.M109.023663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du G., Morris A. J., Sciorra V. A., Frohman M. A. G-protein-coupled receptor regulation of phospholipase D. Methods Enzymol. 2002;345:265–274. doi: 10.1016/s0076-6879(02)45022-7. [DOI] [PubMed] [Google Scholar]
- Finck B. N., Gropler M. C., Chen Z., Leone T. C., Croce M. A., Harris T. E., Lawrence J. C., Jr., Kelly D. P. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell metabolism. 2006;4:199–210. doi: 10.1016/j.cmet.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Han G. S., Wu W. I., Carman G. M. The Saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 2006;281:9210–9218. doi: 10.1074/jbc.M600425200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T. E., Huffman T. A., Chi A., Shabanowitz J., Hunt D. F., Kumar A., Lawrence J. C., Jr. Insulin controls subcellular localization and multisite phosphorylation of the phosphatidic acid phosphatase, lipin 1. J. Biol. Chem. 2007;282:277–286. doi: 10.1074/jbc.M609537200. [DOI] [PubMed] [Google Scholar]
- Heo W. D., Inoue T., Park W. S., Kim M. L., Park B. O., Wandless T. J., Meyer T. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 2006;314:1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Hasegawa J., Tsujita K., Kanaho Y., Takenawa T. The tyrosine kinase Fer is a downstream target of the PLD-PA pathway that regulates cell migration. Sci. Signal. 2009;2:ra52. doi: 10.1126/scisignal.2000393. [DOI] [PubMed] [Google Scholar]
- Ivanova P. T., Milne S. B., Myers D. S., Brown H. A. Lipidomics: a mass spectrometry based systems level analysis of cellular lipids. Curr. Opin. Chem. Biol. 2009;13:526–531. doi: 10.1016/j.cbpa.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- la Cour T., Kiemer L., Molgaard A., Gupta R., Skriver K., Brunak S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 2004;17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- Lange A., Mills R. E., Lange C. J., Stewart M., Devine S. E., Corbett A. H. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G. H., Gerace L. Sumoylation regulates nuclear localization of lipin-1alpha in neuronal cells. PLoS One. 2009;4:e7031. doi: 10.1371/journal.pone.0007031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris A. J. Regulation of phospholipase D activity, membrane targeting and intracellular trafficking by phosphoinositides. Biochem. Soc. Symp. 2007:247–257. doi: 10.1042/BSS0740247. [DOI] [PubMed] [Google Scholar]
- Murray D., Hermida-Matsumoto L., Buser C. A., Tsang J., Sigal C. T., Ben-Tal N., Honig B., Resh M. D., McLaughlin S. Electrostatics and the membrane association of Src: theory and experiment. Biochemistry. 1998;37:2145–2159. doi: 10.1021/bi972012b. [DOI] [PubMed] [Google Scholar]
- Nishikimi A., Fukuhara H., Su W., Hongu T., Takasuga S., Mihara H., Cao Q., Sanematsu F., Kanai M., Hasegawa H., Tanaka Y., Shibasaki M., Kanaho Y., Sasaki T., Frohman M. A., Fukui Y. Sequential regulation of DOCK2 dynamics by two phospholipids during neutrophil chemotaxis. Science. 2009;324:384–387. doi: 10.1126/science.1170179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara L., Han G. S., Peak-Chew S., Grimsey N., Carman G. M., Siniossoglou S. Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase. J. Biol. Chem. 2006;281:34537–34548. doi: 10.1074/jbc.M606654200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamuklar Z., Federico L., Liu S., Umezu-Goto M., Dong A., Panchatcharam M., Fulerson Z., Berdyshev E., Natarajan V., Fang X., van Meeteren L. A., Moolenaar W. H., Mills G. B., Morris A. J., Smyth S. S. Autotaxin/lysopholipase D and lysophosphatidic acid regulate murine hemostasis and thrombosis. J. Biol. Chem. 2009;284:7385–7394. doi: 10.1074/jbc.M807820200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterfy M., Harris T. E., Fujita N., Reue K. Insulin-stimulated interaction with 14–3-3 promotes cytoplasmic localization of lipin-1 in adipocytes. J. Biol. Chem. 2009 doi: 10.1074/jbc.M109.072488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterfy M., Phan J., Reue K. Alternatively spliced lipin isoforms exhibit distinct expression pattern, subcellular localization, and role in adipogenesis. J. Biol. Chem. 2005;280:32883–32889. doi: 10.1074/jbc.M503885200. [DOI] [PubMed] [Google Scholar]
- Peterfy M., Phan J., Xu P., Reue K. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 2001;27:121–124. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- Phan J., Peterfy M., Reue K. Lipin expression preceding peroxisome proliferator-activated receptor-gamma is critical for adipogenesis in vivo and in vitro. J. Biol. Chem. 2004;279:29558–29564. doi: 10.1074/jbc.M403506200. [DOI] [PubMed] [Google Scholar]
- Phan J., Reue K. Lipin, a lipodystrophy and obesity gene. Cell Metab. 2005;1:73–83. doi: 10.1016/j.cmet.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Reue K. The lipin family: mutations and metabolism. Curr. Opin. Lipidol. 2009;20:165–170. doi: 10.1097/MOL.0b013e32832adee5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciorra V. A., Rudge S. A., Prestwich G. D., Frohman M. A., Engebrecht J., Morris A. J. Identification of a phosphoinositide binding motif that mediates activation of mammalian and yeast phospholipase D isoenzymes. EMBO J. 1999;18:5911–5921. doi: 10.1093/emboj/18.21.5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott S. A., Selvy P. E., Buck J. R., Cho H. P., Criswell T. L., Thomas A. L., Armstrong M. D., Arteaga C. L., Lindsley C. W., Brown H. A. Design of isoform-selective phospholipase D inhibitors that modulate cancer cell invasiveness. Nat. Chem. Biol. 2009;5:108–117. doi: 10.1038/nchembio.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stace C. L., Ktistakis N. T. Phosphatidic acid- and phosphatidylserine-binding proteins. Biochim. Biophys. Acta. 2006;1761:913–926. doi: 10.1016/j.bbalip.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Su W., Yeku O., Olepu S., Genna A., Park J. S., Ren H., Du G., Gelb M. H., Morris A. J., Frohman M. A. 5-Fluoro-2-indolyl des-chlorohalopemide (FIPI), a phospholipase D pharmacological inhibitor that alters cell spreading and inhibits chemotaxis. Mol. Pharmacol. 2009;75:437–446. doi: 10.1124/mol.108.053298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W. S., Imai A., Sugiyama M., Furui T., Tamaya T., Saio M., Morris A. J. Translocation of lysophosphatidic acid phosphatase in response to gonadotropin-releasing hormone to the plasma membrane in ovarian cancer cell. Am. J. Obstet. Gynecol. 2004;191:143–149. doi: 10.1016/j.ajog.2004.01.038. [DOI] [PubMed] [Google Scholar]
- Winters M. J., Lamson R. E., Nakanishi H., Neiman A. M., Pryciak P. M. A membrane binding domain in the ste5 scaffold synergizes with gbetagamma binding to control localization and signaling in pheromone response. Mol. Cell. 2005;20:21–32. doi: 10.1016/j.molcel.2005.08.020. [DOI] [PubMed] [Google Scholar]