Diaphanous-related formin, mDia, is an actin nucleation/polymerization factor functioning downstream of the small GTPase Rho. We found that, in addition to the Rho GTPase-mediated activation, the interaction between mDia2 and anillin is required for the localization and function of mDia2 in cytokinesis.
Abstract
Diaphanous-related formin, mDia, is an actin nucleation/polymerization factor functioning downstream of the small GTPase Rho. Although Rho is critically involved in cytokinesis, it remains elusive how Rho effectors and other regulators of cytoskeletons work together to accomplish this process. Here we focused on mDia2, an mDia isoform involved in cytokinesis of NIH 3T3 cells, and analyzed mechanisms of its localization in cytokinesis. We found that targeting of mDia2 to the cleavage furrow requires not only its binding to RhoA but also its diaphanous-inhibitory domain (DID). We then performed pulldown assays using a fragment containing the latter domain as a bait and identified anillin as a novel mDia2 interaction partner. The anillin-binding is competitive with the diaphanous autoregulatory domain (DAD) of mDia2 in its autoinhibitory interaction. A series of RNA interference and functional rescue experiments has revealed that, in addition to the Rho GTPase-mediated activation, the interaction between mDia2 and anillin is required for the localization and function of mDia2 in cytokinesis.
INTRODUCTION
Cytokinesis is a step of cell division that physically separates a dividing cell into two. In many types of cells, this process is critically driven by the actomyosin-based contraction of the contractile ring (Balasubramanian et al., 2004; Pollard, 2009). Previous studies have revealed that, as sister chromosomes separate, many molecules involved in cytokinesis are recruited to the middle region of mitotic spindle, i.e., the central spindle and also to the equatorial cell cortex, and those molecules regulate the contractile ring formation for constriction (Eggert et al., 2006). However, it still remains unknown how these molecules interact each other and how they work together to accomplish this process. Among such molecules, the small GTPase Rho functions as an essential molecular switch linking nuclear division and cytokinesis (Mabuchi et al., 1993; Piekny et al., 2005). Rho is spatiotemporally activated in anaphase to telophase by guanine nucleotide exchange factor (GEF), Ect2 (Tatsumoto et al., 1999; Kimura et al., 2000; Barr and Gruneberg, 2007). The GTP-bound, activated form of Rho then acts on downstream effectors to elicit its function. These effector molecules include ROCK/Rho-kinase, mammalian homolog of Drosophila diaphanous (mDia), and citron kinase, each of which functions as an important regulator of cytokinesis (Madaule et al., 1998; Ishizaki et al., 2000; Watanabe et al., 2008). mDia belongs to the formin family of proteins and induces actin filaments by catalyzing actin nucleation and polymerization (Goode and Eck, 2007; Paul and Pollard, 2009). There are three mDia isoforms, mDia1(diap1), mDia2(diap3), mDia3(diap2) (Higgs and Peterson, 2005). An mDia molecule has multiple domains, GBD (GTPase-binding domain) and DID/ARR (diaphanous-inhibitory domain/armadillo-repeat region) in the N-terminus, FH (formin homology) domains, FH1 and FH2, in the middle, and DAD (diaphanous auto-regulatory domain) in the C-terminus (Lammers et al., 2005; Otomo et al., 2005; Rose et al., 2005). The FH2 domain binds to the barbed end of an actin filament and catalyzes actin nucleation and polymerization, and the FH1 domain accelerates actin elongation of the FH2 domain through binding to the actin monomer-binding protein, profilin (Paul and Pollard, 2009). Diaphanous-related formin (DRF) proteins including mDia are regulated by the DID-DAD interaction, which inhibits the ability of the FH2 to nucleate actin assembly (Li and Higgs, 2003; Seth et al., 2006; Wallar et al., 2006). Although the biochemical properties of each domain have thus been characterized, little is known about the mechanisms how mDia is localized and regulated. We previously showed that, among mDia isoforms, mDia2 selectively localizes to the cleavage furrow and induces F-actin scaffold for the contractile ring in the cytokinesis of several cell types (Watanabe et al., 2008). However, it is neither known how mDia2 is specifically targeted and activated spatiotemporally at the site of cell division nor how each domain of mDia2 other than FH domains contributes to this process. To examine these issues, we have used RNA interference (RNAi) and a series of GFP-mDia2 constructs and analyzed the critical region of mDia2 for the localization in cytokinesis. We have also performed pulldown assays to screen molecules responsible for its localization and identified anillin as a novel mDia2 interaction partner in cytokinesis. We have further performed a series of functional RNAi rescue experiments. Here, we show that, in addition to the Rho GTPase-mediated activation, the interaction between mDia2 and anillin is required for the localization and function of mDia2 in cytokinesis.
MATERIALS AND METHODS
Materials
Short interfering double-stranded RNA oligomer (siRNA) for mDia2, siRNAmDia2#1 (Watanabe et al., 2008), was used for RNAi for mDia2. Three different Stealth Select RNAi siRNAs (Invitrogen, Carlsbad, CA), siRNAanillin#1 (MSS229296), #2 (MSS229295), and #3 (MSS229297), corresponding to nucleotide sequences of 2890–2914, 2785–2809, and 675–699, respectively, were used for RNAi for anillin (NM_028390). Stealth Select RNAi siRNA, siRNAEct2 (MSS203770), corresponding to nucleotide sequence of 215–239 was used for RNAi for Ect2 (NM_007900.2). Stealth RNAi negative control duplexes (Invitrogen) were used for control RNAi. Enhanced green fluorescent protein (pEGFP)-mDia2, GFP-mDia2 411-1171, GFP-mDia2 1028–1171, GFP-mDia2 1028–1156 were described previously (Watanabe et al., 2008; Miki et al., 2009). pEGFP-anillin was prepared by PCR amplification and restriction enzyme digestion as follows. ATCC human cDNA clone 681838 (BC070066.1; Manassas, VA) was subcloned into pBluescript. The plasmid was digested with its internal BamHI site and the inserted HindIII site after the stop codon sequence, and the digested fragment was subcloned into pEGFP-C3 (Clontech, Palo Alto, CA). Series of other mDia constructs or anillin constructs were prepared by PCR amplification of corresponding sequences and restriction enzyme digestion as shown in Table S1.
Primary antibodies (Abs) used were mouse DM1A monoclonal Ab (mAb) to α-tubulin from Sigma-Aldrich (Saint Louis, MO), rat mAb to α-tubulin from Chemicon (Temecula, CA), mouse 26C4 mAb to RhoA from (Santa Cruz, CA), mouse mAb to glutathione S-transferase (GST) from Nacalai Tesque, mouse mAb to GFP from Roche (Mannheim, Germany), and rabbit polyclonal Ab to GFP from MBL (Nagoya, Japan). Rabbit polyclonal antibodies to mDia2, anti-mDia2 N1, and anti-mDia2 C1 were described previously (Watanabe et al., 2008). Rabbit polyclonal Abs to anillin and septin 7 were generated in rabbits against a His-tagged full-length protein of human anillin (BC070066.1) and a synthetic peptide of C-terminal human septin 7 (CNSSRTLEKNKKKGKIF), respectively, in the Kinoshita's laboratory. The antibody to anillin was purified from antiserum by affinity chromatography.
Cell Culture and Transfection
NIH 3T3 cells, HeLa cells, and HT1080 cells were maintained in DMEM (Invitrogen) supplemented with 10% FCS at 37°C with an atmosphere containing 10% CO2. HEK293F (FreeStyle 293-F) cells (Invitrogen) were maintained in FreeStyle medium (Invitrogen) on an orbital shaker platform rotating at 37°C with an atmosphere containing 8% CO2. Plasmid and RNAi transfection experiments in asynchronous or synchronized cells were performed using Lipofectamine LTX Reagent (Invitrogen) and Lipofectamine RNAiMAX Reagent (Invitrogen), respectively, as described previously (Watanabe et al., 2008). Transfection of plasmids into HEK293F cells were performed as follows. We first added 12.5 μg of plasmid DNA or 12.5 μl of FreeStyle MAX Reagent (Invitrogen) each to 200 μl of OptiPro SFM (Invitrogen), and the two solutions were then mixed and incubated for 10 min at room temperature. The mixture was then added to 1.0 × 107 cells in 10 ml of the culture medium and incubated for 24 h before harvesting cells for pull down and immunoprecipitation assays.
Transfection of plasmids into NIH 3T3 cells with Neon Transfection System (Invitrogen) were performed according to the manufacturer's protocol with two pulses of 1400 V and 20 ms.
The RNAi rescue experiments in Figure 6 and Figure S4, A–B, were performed as follows. NIH 3T3 cells were seeded and cultured in DMEM containing 2 mM thymidine for 16 h, and the cells were then washed twice with phosphate-buffered saline (PBS). The cells were subjected to RNAi transfection with siRNAanillin#1, and were cultured for 8 h. The medium was then replaced again with the culture medium containing 2 mM thymidine and the cells were cultured for another 12 h. The cells were trypsinized and transfected with plasmids encoding either siRNAanillin#1-resistant full-length anillin-GFP or siRNAanillin#1-resistant anillin (115-1086)-GFP using Neon Transfection System (Invitrogen) and further cultured for 4 h in the medium containing 2 mM thymidine. The cells were then washed once with PBS and three times with the fresh medium for thymidine removal and cultured first for 4 h in the culture medium alone and then for 4 h in the medium containing 20 ng/ml nocodazole (Biomol, Plymouth Meeting, PA). The cells were then washed free of nocodazole as described above and further incubated for 25 min before being fixed for immunofluorescence. The RNAi rescue experiments in Figure 7 were performed as follows. NIH 3T3 cells were cultured with the thymidine medium for 16 h as described above, and the cells were washed with PBS and cultured with fresh culture medium for 4 h. The cells were then subjected to RNAi transfection with siRNAanllin#1 and cultured for 4 h and then further cultured for 10 h with the thymidine medium. The cells were trypsinized and transfected with plasmids encoding various siRNAanillin#1-resistant anillin constructs using Neon Transfection System (Invitrogen) and further cultured for 4 h in the thymidine medium. The cells were then washed and cultured for 10 h in the fresh culture medium before being fixed for immunofluorescence.
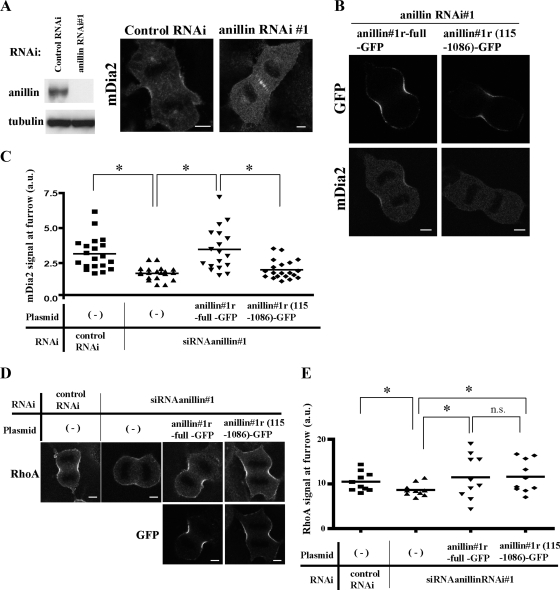
Figure 6.
The binding of anillin to mDia2 is required for the localization of mDia2 at the cleavage furrow. (A) Depletion of anillin diminished the localization of mDia2 at the cleavage furrow. The specific depletion of endogenous anillin is confirmed by immunoblotting (left). Immunofluorescence staining of mDia2 in NIH 3T3 cells treated for 36 h with control siRNA or siRNA specific for anillin (right). Bar, 5 μm. (B) Expression of the RNAi-resistant anillin-GFP with the mDia2 binding domain rescued the loss of mDia2 at the cleavage furrow. NIH 3T3 cells were synchronized and transfected with siRNAanillin#1 and with either RNAi-resistant anillin#1r-full-GFP or anillin#1r-(115–1086)-GFP. The cells were then stained for either GFP or mDia2. Bar, 5 μm. (C) Quantitative analysis of mDia2 signal at the cleavage furrow. Experiments were performed as described in B. The signal intensities of mDia2 at the cleavage furrow were quantified using MetaMorph software as described in Materials and Methods. The results are from two independent experiments, in each of which n > 9 cells were examined. *p <0.05. (D) Expression of the RNAi-resistant anillin-GFP restored the decrease of RhoA at the cleavage furrow. Experiments were performed as described in B. The cells were then stained for either RhoA (top panels) or GFP (bottom panels). Bar, 5 μm. (E) The signal intensities of RhoA at the cleavage furrow were quantified as described in Materials and Methods. The results are from two independent experiments, in each of which n > 5 cells were examined. *p < 0.05. n.s., not significant (p > 0.05).
Figure 7.
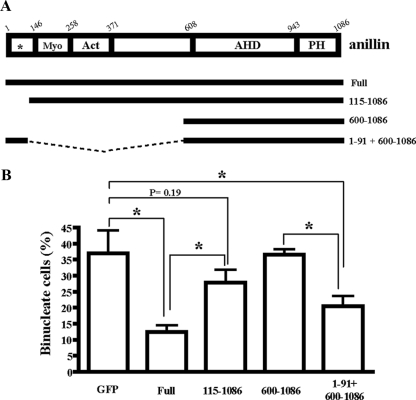
The binding of anillin to mDia2 is required for cytokinesis. (A) A schematic representation of the anillin structure and anillin-GFP constructs. Conserved regions and previously described domains are indicated; Myo, myosin-binding domain (aa146-258); Act, actin-binding and -bundling region (aa 258-371); AHD, anillin homology domain (aa 608-943); and PH, pleckstrin homology domain (aa943-1086). Asterisk indicates the nuclear localization Signal (NLS). (B) NIH 3T3 cells were synchronized with thymidine and sequentially transfected with siRNAanillin#1 and then with either empty GFP vector or a series of RNAi-resistant anillin#1r-GFP constructs (full length, 115-1086, 600-1086, or 1-91 + 600-1086). The transfected cells were washed and released for 10 h, and the cells were fixed and subjected to fluorescence staining for determining the number of binucleate cells in each population. Values are shown as percentage of binucleate cells expressing GFP per total cells expressing GFP. Error bars, SD *p < 0.05. The results are from three independent experiments, in each of which n > 200 cells were examined.
Immunofluorescence
NIH 3T3 cells or HT1080 cells were plated onto a coverslip in a 35-mm culture dish for fluorescence microscopy. The cells were fixed with trichloroacetic acid (TCA) and stained as described previously (Watanabe et al., 2008). Immunostaining was performed using the following antibodies: anti-mDia2 C1 or N1 (1:400), anti-RhoA (1:500), anti-septin 7 (1:400), anti-tubulin (1:1000), and anti-GFP (1:1000) Abs. Secondary antibodies used were as described previously (Watanabe et al., 2008). Staining was examined with a Leica SP5 confocal imaging system (Plan-Apo 63/1.40 NA; Wetzler, Germany). Figure 1A (left) shows build-up images of dividing cells that were obtained from a collection of 20–25 Z sections of 0.5-μm step intervals from the bottom to the top of the cells, and all of other images use single plane images. The images were analyzed by the Leica built-in and MetaMorph software (Molecular Devices, Downingtown, PA). Quantification of signal intensities at the cleavage furrow in Figures 3B and 6, C and E, and Figure S4B was performed using the “line scan” function in MetaMorph and Microsoft Excel (Redmond, WA) spreadsheet software. An image of the middle section of each mitotic cell was used for analysis. The background signals outside the cell were averaged and subtracted from each image before analysis. Images of 10–20 mitotic cells in each treatment were obtained from two different experiments, and line scans with a width of 3 pixels and a length of 40 pixels were performed perpendicular to the longitudinal axis at the narrowest position of the furrow. The averaged intensities corresponding to the cleavage furrow cortex were obtained as “cleavage furrow signals,” and the values were standardized by dividing them by averaged intensity values of the 20 × 20-pixel region in the cytoplasm of each image.
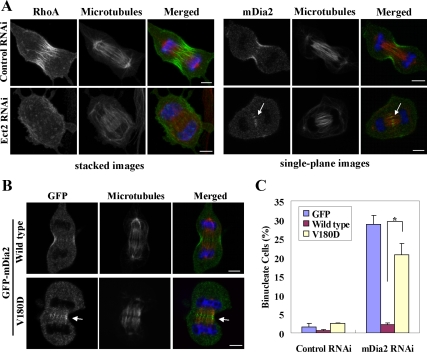
Figure 1.
Rho-dependent control of mDia2 localization at cleavage furrow during cytokinesis (A) Attenuation of the localization of RhoA and mDia2 at the cleavege furrow by depletion of Rho GEF Ect2. NIH 3T3 cells were transfected with siRNA for Ect2 and synchronized with thymidine, and at 24 h after transfection the cells were released into DMEM containing 10% FCS for 6.5 h. The cells were fixed and stained for indicated molecules (green), tubulin (red), and DNA (blue). Left, images show stacks of focal planes as described in Materials and Methods. Right, each image shows a single focal plane. An arrow indicates the accumulation of mDia2 at the central spindle. Bar, 5 μm. (B) Effects of the Rho-binding–defective mutation (V180D) on the localization of mDia2 at cleavage furrow. NIH 3T3 cells were transfected with GFP-mDia2 wild type or GFP-mDia2 containing the mutation into Val180 (V180D), and the cells were fixed and stained as shown in A. An arrow indicates the accumulation of GFP-mDia2 at the central spindle. Bar, 5 μm. (C) A failure to rescue the binucleate phenotype by expression of RNAi-resistant GFP-mDia2 V180D. NIH 3T3 cells were transfected with pEGFP, pEGFP-mDia2r#1 full-length wild type, or pEGFP-mDia2r#1 full-length V180D, and, after 24 h the cells were subjected to control or mDia2 RNAi for 48 h. Fluorescence staining was carried out as described above for determining the number of binucleate cells in each population. Values are shown as percentage of binucleate cells expressing GFP per total cells expressing GFP. Error bars, SD * p < 0.01. The results are from three independent experiments, in each of which n > 200 cells were examined.
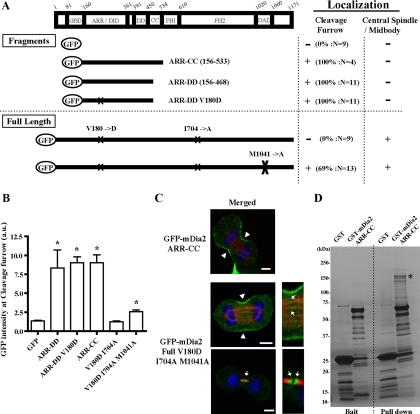
Figure 3.
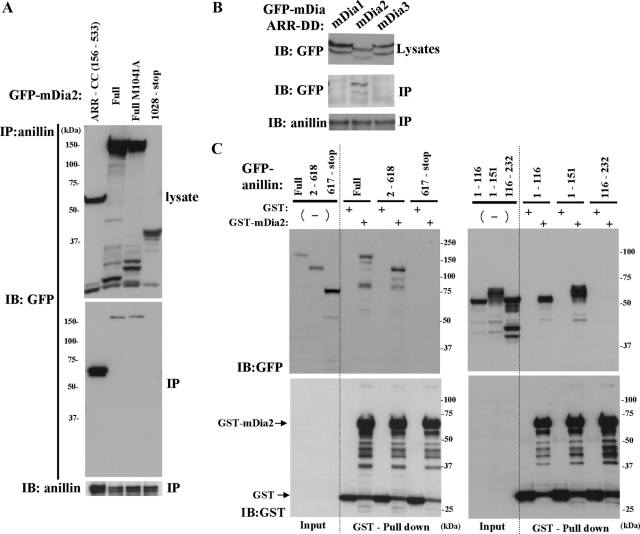
Localization and pulldown analysis of Rho-binding defective mDia2 in cytokinesis. (A) A schematic representation of the constructs and the localization analysis of GFP-fused N-terminal fragments of mDia2 lacking GBD (the top section) and full-length mDia2 containing the Rho-binding-defective mutation (V180D) (the bottom section). X indicates the positions of mutations. Experiments and analyses were carried out as described in Figure 2. M1041A, a mutation in DAD core. The localization of each GFP construct at the cleavage furrow was also examined by the quantitative analysis of GFP intensity at the cleavage furrow as shown in Figure 3B. The maximum intensity of the control GFP-expressing cells examined was used as a standard value for the judgment. The number of cells exhibiting the localization at the cleavage furrow was divided by the total number of cells examined and shown as percentages. (B) Quantitative analysis of the localization of series of GFP-mDia2 at the cleavage furrow. Experiments were performed as described in Figure 3A. The signal intensities of GFP at the cleavage furrow were quantified using MetaMorph software as described in the Materials and Methods. The number of cells examined is shown in Figure 3A. The results are from two to three independent experiments. Error bars, SEM *p < 0.05 versus GFP. (C) Localization of the N-terminal fragment of mDia2 lacking GBD (top panel) and the full-length mDia2 containing the mutations of V180D and M1041A (bottom panels). NIH 3T3 cells were transfected with a series of GFP-mDia2 mutants, and the cells were fixed and stained for GFP (green), microtubules (red), and DNA (blue). Bar, 5 μm. Arrowheads indicate the localization of mDia2 at the cleavage furrow. Arrows indicate the localization of mDia2 at the central spindle or midbody. (D) Pulldown analysis of mDia2 binding partners by the ARR-CC fragment of mDia2. Mitotically synchronized HeLa cell lysates were applied to glutathione-Sepharose beads conjugated with GST or GST-mDia2 ARR-CC (150–533). Bound proteins were eluted by a buffer containing 20 mM glutathione and analyzed by SDS-PAGE and silver staining. An asterisk indicates the band identified as anillin by mass spectrometry analysis.
Expression and Purification of Recombinant Proteins
GST-RhoA, GST-mDia1, GST-mDia2, GST-mDia3, or GST-anillin expression plasmids were introduced into Escherichia coli strain BL21 (DE3) (Novagen, Madison, WI). Expression was induced by the addition of 0.1 μM IPTG, and the cells were incubated for 16 h at 20°C. The cells were harvested by centrifugation, resuspended in a buffer (25 mM Tris-HCl, pH 7.5, 50 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol) containing Protease Inhibitor mixture (Nacalai Tesque, Kyoto, Japan), and sonicated. The homogenate was centrifuged, and the supernatant obtained was incubated with glutathione-Sepharose 4 fast flow (GSH) beads (GE Healthcare, Waukesha, WI). After washing with the buffer containing 500 mM NaCl, the GST-fusion proteins were eluted from the beads with an elution buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 20 mM GSH). The eluted proteins were dialyzed against PBS or HBS-EP Buffer (10 mM HEPES pH7.5, 3 mM EDTA, 150 mM NaCl, 0.005% Tween20) containing 1 mM dithiothreitol in Slide-A-Lyzer dialysis cassettes (Pierce, Rockford, IL). For direct binding assay and fluorescence polarization assay, the GST portion was cleaved off from the fusion protein by incubation with PreScission Protease (GE Healthcare). Recombinant RhoA was expressed and purified as described previously (Dvorsky et al., 2004).
Fluorescence Polarization Assay
Polarization measurements were performed using an EnVision plate reader (Perkin Elmer, Waltham, MA) with a polarization filter sets (FITC FP label; excitation wavelength 480 nm, emission wavelength 535 nm) at room temperature in PBS containing 1 mM dithiothreitol. An mDia2 DAD peptide containing residues 1036–1061 labeled with FITC fluorophore at the N-terminus was synthesized by Invitrogen. After the baseline fluorescence was obtained with the FITC-labeled DAD fragment at 100 nM concentration, a threefold or sixfold excess of mDiaN (N-termini of mDia: mDia1 residues 131-570, mDia2 residues 150-533, and mDia3 residues 166-542, respectively) were added, rapidly mixed and fluorescence was measured with an interval of 10 s. Finally, the changes in polarization after the addition of 480 nM, 2.4 μM, and 4.8 μM anillinN (N-terminus of anillin: residues 1-91) were recorded.
Surface Plasmon Resonance (SPR) Measurements
Protein interaction analyses were performed using Biacore T100 (GE Healthcare). A purified GST-anillin (1–91) was captured onto a CM5 sensor chip (GE Healthcare) using a GST kit for fusion capture (GE Healthcare). Subsequent binding experiments were carried out in HBS-EP Buffer. The sensor chip was regenerated in 10 mM glycine-HCl (pH 2.0). Dissociation constants (Kd) were determined by plotting and fitting the steady state affinity response against concentrations using a BIAcore T100 evaluation software.
GST Pulldown, Immunoprecipitation, and Mass Spectrometry Analysis
The RhoA pulldown assay in Figure S1 was performed as follows. Thirty μM RhoA was loaded with 150 μM guanosine 5′-[γ-thio]triphosphate (GTP-γ-S) (Sigma) in 400 μl of a buffer containing 25 mM Tris-HCl, pH 7.5, 100 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol, 0.05% Tween20, and 10 mM EDTA at 30°C for 30 min. Eighty μg of GTP-γ-S-loaded or GDP-loaded RhoA were incubated with 20 μl of GSH beads and 40 μg of GST-mDia2 (71-533) wild type or the mutant V180D in 660 μl of a buffer containing 25 mM Tris-HCl, pH 7.5, 100 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol, 0.05% Tween 20, and 2 mM EDTA at 4°C for 2 h. The beads were washed three times with 500 μl of the buffer and the protein complex was analyzed by SDS-PAGE and either Coomassie staining or immunoblotting.
For the experiments in Figure 4, HEK293F cells expressing different GFP-tagged proteins were harvested by centrifugation and resuspended in a HEPES buffer (20 mM HEPES, pH 7.5, 50 mM NaCl, 5 mM MgCl2, 0.1% TritonX-100, and 1 mM dithiothreitol) containing Protease Inhibitor mixture (Nacalai Tesque) and PhosStop (Roche). The resuspended cells were sonicated and then centrifuged. The supernatant was precleared with GSH beads or nProteinA Sepharose fast flow beads (GE Healthcare) for pulldown and immunoprecipitation assay, respectively. For pulldown assays in Figure 4C, the precleared supernatant was incubated with GSH beads conjugated with either GST or GST-mDia2 proteins for 2 h at 4°C. The beads were washed three times with the HEPES buffer and eluted with the elution buffer containing 20 mM GSH. The eluted proteins were analyzed by SDS-PAGE and immunoblotting. For immunoprecipitation in Figure 4, A and B, the precleared supernatant was incubated with either 3 μg of control IgG or anti-anillin antibody for 30 min at 4°C. Twenty μl of nProteinA Sepharose fast flow beads (GE Healthcare) were added to each of the mixture and further incubated for 1.5 h at 4°C. The beads were then washed three times with the HEPES buffer and eluted with the Laemmli sample buffer. The eluted samples were analyzed by SDS-PAGE and immunoblotting.
Figure 4.
Coimmunoprecipitation analysis of anillin and mDia2. (A and B) Anillin specifically immunoprecipitates mDia2. (A) GFP-mDia2 constructs were expressed in HEK293F cells, and the cell extracts were immunoprecipitated with an anti-anillin antibody and were analyzed by immunoblotting with anti-GFP or anti-anillin antibodies. M1041A: a mutatin in DAD core. (B) GFP-ARR-DD of mDia1, mDia2, or mDia3 construct was expressed in HEK293F cells, and the cell extracts were precipitated with anti-anillin antibodies and analyzed by immunoblotting with anti-GFP or anti-anillin antibodies. (C) mDia2 binds to the N-terminal region of anillin. GFP-anillin constructs were transfected in HEK293F cells, and the cell extracts were incubated with immobilized GST or GST-mDia2 ARR-CC and bound proteins were analyzed by immunoblotting with anti-GFP or anti-GST antibodies.
For the experiments in Figure 3D, 3 × 107 HeLa cells were plated and synchronized in anaphase or telophase and collected as described previously (Oceguera-Yanez and Narumiya, 2006). The cells were then resuspended in the HEPES buffer and sonicated as described above. After being precleared, the equal amounts of the supernatant were incubated with GSH beads conjugated with GST or GST-mDia2 (150–533) for 2 h at 4°C. The beads were washed three times with the HEPES buffer, and the protein complexes were eluted with the elution buffer and analyzed by SDS-PAGE and silver staining. The specific bands detected were gel-sliced and digested with trypsin, and the samples were subsequently analyzed by MALDI mass spectrometry (Shevchenko et al., 1996).
Statistical Analysis
Graph data are presented as mean ± SD or SE of the means and were analyzed by Student's t test. A p value of <0.05 was considered statistically significant.
RESULTS
Binding to Rho is Required for the mDia2 Localization at Cleavage Furrow
To analyze the mechanism of mDia2 localization in cytokinesis, we first examined the requirement of Rho for the mDia2 localization, because mDia2 binds to RhoA (Alberts et al., 1998). During cytokinesis, RhoA is activated by guanine nucleotide exchange facter (GEF), Ect2 (Yuce et al., 2005; Nishimura and Yonemura, 2006). We therefore inhibited the Rho-signaling by depletion of Ect2 and analyzed the localization of mDia2 in cell division. NIH 3T3 cells were transfected with siRNA for Ect2, and the cells were synchronized in mitotic phase and stained for RhoA and mDia2 (Figure 1A). In control RNAi cells, signals of RhoA and mDia2 were observed at the cleavage furrow at the equatorial cell cortex. In Ect2-RNAi cells, the signals of RhoA at the equatorial cell cortex decreased as previously reported (Yuce et al., 2005; Nishimura and Yonemura, 2006) (Figure 1A left), and the signals of mDia2 at the cleavage furrow diminished, and instead accumulated at the interdigitating microtubules of the central spindle (Figure 1A right and S2A-B). These results indicate that mDia2 localizes to the cleavage furrow in a Rho activity-dependent manner. Therefore, we next used a Rho-binding defective mutant of GFP-mDia2, and examined whether the Rho-binding activity of mDia2 is required for the localization of mDia2 in cytokinesis. A mutation into V161 of mDia1 has been shown to render mDia1 RhoA-binding defective (Otomo et al., 2005; Seth et al., 2006). We first confirmed in vitro that a mutation into V180D of mDia2 (analogous to the aforementioned mDia1 V161D mutant) also rendered mDia2 RhoA-binding defective as mDia1 (Figure S1). We found that GFP-mDia2 V180D did not localize at the cleavage furrow, but accumulated at the central spindle (Figure 1B). In addition, this mutant could not effectively rescue the cytokinesis failure induced by depletion of mDia2 (Figure 1C). These results together suggest that the binding between RhoA and mDia2 is essential for the localization and function of mDia2 in cytokinesis.
Distinct Regions of mDia2 are Required for Multiple Localizations in Cytokinesis
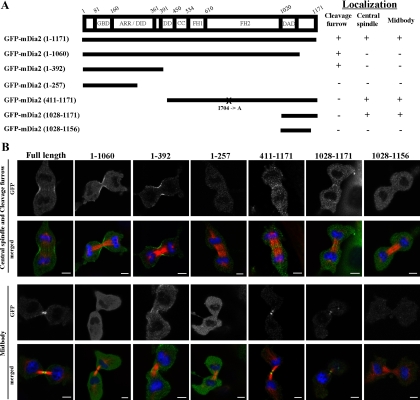
In the analysis shown in Figure 1 and also in our previous study, we have observed multiple cellular localizations of mDia2 during cell division, such as cleavage furrow, central spindle, and midbody (Watanabe et al., 2008). To determine regions of mDia2 responsible for each localization, we constructed a series of GFP-fused deletion mutants of mDia2. The presence of an N-terminal GFP tag does not perturb protein function, as GFP-mDia2 fully rescued cytokinesis failure by mDia2-RNAi (Figure 1C). These constructs were transfected in NIH 3T3 cells and the localizations of each GFP fusion proteins were examined. We found that GFP-fused N-terminal fragments of mDia2 containing GBD and ARR/DID domains such as GFP-mDia2 (1-392) localized at the cleavage furrow but not at the central spindle and midbody. On the other hand, C-terminal fragments such as GFP-mDia2 (411-1171) and GFP-mDia2 (1028-1171) localized at the central spindle and midbody but not at the cleavage furrow (Figure 2). These results suggest that distinct regions are required for the localizations of mDia2 in cell division.
Figure 2.
Analysis of the localization of mDia2 deletion mutants during cell division. (A) A schematic representation of the constructs and summary of the localization analysis. Experiments were carried out as described in B. More than 20 cells from two or three experiments were analyzed to evaluate the localization at the cleavage furrow, central spindle, and midbody. FH1 or FH2, formin homology domain 1 or 2, respectively; DID, diaphanous inhibitory domain; DAD, diaphanous auto-regulatory domain, GBD, GTPase-binding domain; ARR, Armadillo repeat domain; DD, dimerization domain. X indicates the position of mutation. I704A, the actin-polymerization defective mutation (Watanabe et al., 2008). (B) Localization of each GFP-mDia2 deletion mutants during cell division in NIH 3T3 cells. NIH 3T3 cells were transfected with a series of GFP-mDia2 deletion mutants, and the cells were fixed and stained for GFP (green), microtubules (red), and DNA (blue). Bar, 5 μm.
Identification of Anillin as an mDia2-binding Protein
Although we showed that the RhoA-binding activity is essential for the localization and function of mDia2 in cytokinesis (Figure 1, B and C), we wondered if, in addition to the RhoA-binding activity, additional factors also contribute to selective localization of mDia2 at the cleavage furrow, because all mDia isoforms, mDia1-3, bind to RhoA (Watanabe et al., 1997; Alberts et al., 1998; Yasuda et al., 2004; Rose et al., 2005), but only mDia2 localizes at the cleavage furrow (Watanabe et al., 2008). To examine this issue, we prepared GFP-mDia2 N-terminal constructs lacking GTPase-binding domain (GBD) and analyzed the localization during cell division. We found that GFP-fused N-terminal fragments of mDia2 lacking GBD domain (Figure 3A) still localized at the cleavage furrow (Figure 3, B and C). We next wondered if a Rho-binding defective mutant can localize to the cleavage furrow when it is rendered open. To this end, we introduced a mutation in the DAD core M1041 in the V180D full-length GFP-mDia2 mutant to suppress auto-inhibition (Wallar et al., 2006). We also introduced another mutation in the FH2 I704, which rendered the protein defective in actin polymerization. Introduction of I704 mutation alone could not localize the V180D protein to the cleavage furrow. However, when this is rendered open by M1041A mutation, this full-length open mutant was found to localize at the cleavage furrow (Figure 3, A-C), confirming the presence of an additional domain and mechanism for mDia2 localization at the cleavage furrow. These results prompted us to identify factors responsible for the selective localization of mDia2. To this end, we prepared GST-mDia2 ARR-coiled coil (CC), a GST-fusion protein of recombinant N-terminal fragment of mDia2 lacking GBD, and performed GST pulldown analysis. GST or GST-mDia2 ARR-CC proteins bound to GSH Sepharose beads were incubated with lysates of cells synchronized in mitosis, and the resultant protein complexes were precipitated and resolved by SDS-PAGE. A 150 kDa protein was reproducibly and specifically eluted from the GST-mDia2-bound beads. This protein was analyzed by mass spectrometry after tryptic digestion of gel slices, and identified as anillin (Figure 3D). To confirm the interaction between anillin and mDia2, we performed immunoprecipitation (IP) experiments with an anti-anillin antibody. A series of GFP-fused mDia constructs were transfected in HEK293 cells and its association with endogenous anillin was analyzed. The full-length and the N-terminal part (aa156-533), but not the C-terminal part (aa1028-stop) of GFP-mDia2 were coimmunoprecipitated with anillin (Figure 4A). We also examined the binding between anillin and mDia proteins, and found that the ARR-dimerization domain (DD; aa156-468) of GFP-mDia2 but the corresponding domain of neither GFP-mDia1 nor GFP-mDia3 was coimmunoprecipitated with anillin (Figure 4B). To characterize the domains of anillin responsible for association with mDia2, we next generated a series of GFP-fused anillin mutants and performed GST pulldown experiments in HEK293 cells. N-terminal parts (aa1–116, aa1-151), but not middle (aa116-232) or C-terminal parts (aa617-stop) of anillin were pulled down by GST-mDia2 ARR-CC (Figure 4C). These results together suggest that the N-terminal region of anillin containing aa1-116 specifically interacts with the N-terminal domains of mDia2. The mDia-binding domain of anillin was further narrowed down to aa 1-91 (see below).
Anilllin Directly Binds mDia2 and the Binding is Competitive with the DAD Domain Toward the Autoinbitory Interaction of mDia2
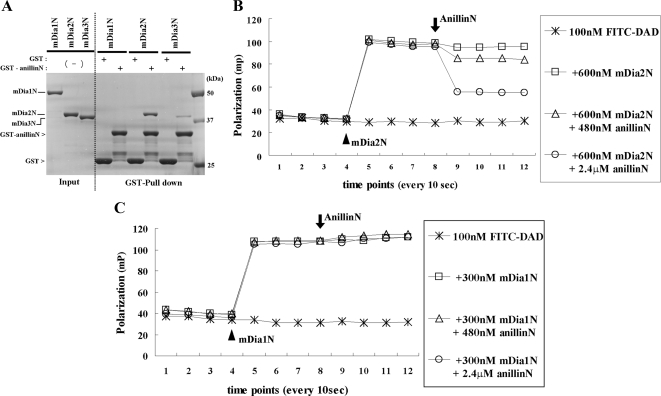
The fact that anillin and mDia2 are both actin-binding proteins led us to further characterize the direct interaction between these two proteins and the binding specificity among mDia isoforms. The recombinant N-terminal part of anillin (1-91) (anillinN) was prepared as a GST-fused protein, and the GSH bead-conjugating GST or GST-anillinN was incubated with equal amounts of recombinant N-terminal ARR-CC fragments of mDia1, mDia2, and mDia3 proteins with the same boundary (mDia1N to mDia3N). The GST-anillinN most efficiently pulled down mDia2N among mDia isoforms. mDia3N was pulled down to much less extent and no precipitation was found for mDia1N (Figure 5A). The affinity between GST-anillinN and mDia2N or mDia3N was also analyzed and measured using surface plasmon resonance on a Biacore T100 instrument (Figure S3A). The dissociation constants (Kds) of the binding GST-anillinN to mDia2N and mDia3N were determined to be 236 nM and 1.02 μM, respectively. As DAD can bind to the ARR-CC region of mDia2, we next wondered if a ternary complex between mDia2N, anillinN, and DAD can be formed or whether the binding is mutually exclusive. To examine this issue, we performed fluorescence polarization assay (Rose et al., 2005). Polarization of a fluorescently labeled 25-mer DAD peptide (aa1036-1061) was strongly increased after the addition of an excess amount of mDia2N, owing to formation and a decreased mobility of the higher molecular mass DAD-mDia2N complex. The addition of anillinN reduced polarization dose-dependently, indicating the release of the fluorescent peptide from the mDia2N complex (Figure 5B). However, anillinN did not cause the release of DAD from mDia1N (Figure 5C), and the release is much less evident from mDia3N (Figure S3B), further supporting the preferential binding of anillin to mDia2. These results together suggest that anillin directly binds mDia2 and that the binding is competitive with the DAD domain of mDia2 in its autoinhibitory interaction.
Figure 5.
Selective binding of anillin to mDia2 and its competition with DAD binding in the autoinbitory interaction of mDia2. (A) Selective binding of anillin to mDia2. Recombinant mDia proteins (mDiaN: mDia ARR-CC), 6.6 μg, were incubated with GSH-Sepharose beads and 20 μg of either GST or GST-anillin (1-91) and analyzed by SDS-PAGE and Coomassie staining. (B and C) Fluorescence polarization analysis of the competition between anillin and the DAD domain of mDia2 in binding to the ARR-CC domain of mDia2 (B) and mDia1 (C). FITC-labeled mDia2-DAD peptides (1036–1051) were sequentially mixed with indicated concentrations of recombinant mDia and anillin, and the fluorescence polarization were measured with a Perkin Elmer EnVision multilabel plate reader. Note that the increase of polarization signal indicates decreased molecular movements of fluorescence labeled small molecules upon specific bindings to proteins. An arrowhead indicates the addition of each mDia ARR/DID-CC, and an arrow indicates the addition of anillinN. The results represent the average value from two independent experiments.
The Anillin Binding Is Required for the mDia2 Localization at the Cleavage Furrow
To study the role of anillin in the localization of mDia2, we transfected NIH 3T3 cells with siRNA specific for anillin for 36 h and stained the cells for mDia2 (Figure 6A). The specific depletion of anillin at the time of examination was confirmed by Western blotting (Figure 6A, left). In anillin-RNAi cells, signals of mDia2 were not observed at the cleavage furrow, but accumulated at the central spindle (Figure 6A, right). We confirmed this finding by using three nonoverlapping siRNAs for anillin, siRNAanillin#1, #2, and #3 (data not shown). We also found that anillin RNAi induced the loss of mDia2 at the cleavage furrow in human HT1080 cells (Figure S2C), supporting that the anillin's function to localize mDia2 is conserved in other cell types. To further validate that the mislocalization of mDia2 is caused by loss of interaction between mDia2 and anillin, we attempted to rescue the phenotype by transfecting a RNAi-resistant full-length or N-terminally deleted construct of anillin-GFP, anillin#1r-full-GFP and anillin#1r-(115-1086)-GFP, respectively. As anillin is an essential scaffold protein at the cleavage furrow (Field et al., 2005; D'Avino, 2009), we first examined whether these two constructs are functional by analyzing their localization and localization of the known anillin-binding partner, septin 7. Both anillin-GFP properly localized at the cleavage furrow and rescued the disappearance of septin 7 at the cleavage furrow, indicating that the both anillin-GFP are functional proteins (Figure S4, A and B). We found that the disappearance of the mDia2 signal at the cleavage furrow induced by anillin RNAi was recovered by the expression of anillin#1r-full-GFP, but not by anillin#1r-(115-1086)-GFP (Figure 6B). The localization of mDia2 at the cleavage furrow in each cell population was further verified by quantitative fluorescence intensity measurements of the mDia2 signal (Figure 6C). We also found that anillin RNAi also partially attenuated the localization of RhoA at the cleavage furrow, but that expression of either anillin#1r-full-GFP or anillin#1r-(115-1086)-GFP corrected this attenuation (Figure 6, D and E). These results together suggest that, in addition to the RhoA-mediated activation, the binding between anillin and mDia2 is required for the localization of mDia2 at the cleavage furrow.
The Binding of Anillin to mDia2 Is Required for Cytokinesis
To investigate the importance of the binding between mDia2 and anillin in cytokinesis, we synchronized NIH 3T3 cells and cotransfected siRNAanillin#1 and anillin#1r-GFP with or without the mDia2-binding region and measured the percentage of the binucleate cells in each cell population after the progression through the mitotic phase. We first confirmed that each of the expressed series of anillin-GFP localized consistently at subcellular structures such as nucleus, ectopic cortical foci, and stress fibers as previously reported. (Figure S5; Oegema et al., 2000; Suzuki et al., 2005). We found that the cytokinesis failure induced by the depletion of anillin is efficiently rescued by the expression of anillin#1r-GFP full-length, but not by the anillin#1r-(115-1086)-GFP that lacks the mDia2 binding domain (Figure 7). We next used anillin#1r-(600-1086)-GFP. This construct is a C-terminal fragment of anillin-GFP lacking further the myosin- and actin-binding domains, and used as a nonfunctional construct in the anillin-RNAi rescue experiments (Piekny and Glotzer, 2008). This C-terminal fragment consistently localized at the cleavage furrow as reported (Oegema et al., 2000), but the expression of this fragment in anillin-RNAi cells did not rescue the cytokinesis failure phenotype (Figure 7). We then fused the N-terminal mDia2-binding region (aa1-91) to the above nonfunctional construct, anillin#1r-(600-1086)-GFP, and attempted to examine the function of the mDia2-binding region in anillin (Figure 7A). We found that the expression of anillin#1r-(1-91 + 600-1086)-GFP significantly rescued the anillin-RNAi–mediated cytokinesis failure more effectively than GFP or anillin#1r-(600–1086)-GFP (Figure 7B). We further confirmed that the expression of anillin#1r-(1-91 + 600-1086)-GFP restored the localization of mDia2 at the cleavage furrow (data not shown). These results together suggest that the interaction between mDia2 and anillin is required for proper execution of cytokinesis.
DISCUSSION
Here, we found that the inhibition of the Rho-signaling abolished mDia2 localization at the cleavage furrow and induced its accumulation at the central spindle, and further that the Rho-binding activity of mDia2 is essential for its function in execution of cytokinesis (Figure 1). We also found that the ARR/DID and CC domain, the N-terminal region of mDia2 without GBD alone, can localize at the cleavage furrow (Figure 3). We performed a pulldown assay using the fragment containing this domain and identified anillin as a novel binding partner of mDia2 (Figure 3). The anillin directly binds to mDia2 and its binding was competitive with DAD toward the autoinhibitory interaction of mDia2 (Figure 5). Finally, we found that the interaction between anillin and mDia2 was not only required for the mDia2 localization at the cleavage furrow (Figure 6), but also important for cytokinesis (Figure 7). Our findings have thus clarified the localization mechanisms of mDia2 in cytokinesis and further revealed a heretofore unnoticed interaction of an actin nucleater, mDia2, and an essential scaffold in cytokinesis, anillin. We discussed these issues in detail below.
First, although the formin family of proteins have been characterized as a major component of cytokinesis in animal cells (Castrillon and Wasserman, 1994; Swan et al., 1998; Watanabe et al., 2008), its localization and regulation mechanisms during this process have not yet been revealed. Here we have used siRNA for RhoGEF Ect2 and found that the inhibition of the Rho-signaling diminished the mDia2 localization at the cleavage furrow and induced its accumulation at the central spindle (Figure 1A). We also found that the cytokinesis failure induced by mDia2 RNAi was not effectively rescued by the expression of the Rho-binding-defective mutant of mDia2 (Figure 1C). These results suggest that targeting and function of mDia2 in cytokinesis are indeed regulated by Rho, which is consistent with the previous report showing the regulation of other Diaphanous-related formin (DRF) proteins by RhoGTPases (Seth et al., 2006). The mDia2 localization at the central spindle and midbody indicates associations of mDia2 with microtubules. Previous reports suggested that mDia2 directly or indirectly binds to tips of microtubules in interphase via FH domains (Wen et al., 2004; Bartolini et al., 2008). However, we found that the C-terminal region of mDia2 and not those suggested previously is required for the localization at the central spindle and midbody (Figure 2). As for the central spindle localization, previous studies also reported that one of other Rho effectors involved in cytokinesis, citron kinase, was transferred to the cleavage furrow in an Rho-dependent manner and localized at the central spindle on Rho inactivation (Eda et al., 2001), and that the citron kinase at the central spindle made a complex with KIF14 and PRC1 at the central spindle (Gruneberg et al., 2006). It is thus tempting to speculate that mDia2 responds to Rho-signaling and is transferred from the central spindle to the cleavage furrow, and this mechanism may be common among several Rho effectors.
Second, we found that Rho and the Rho-binding do not solely determine the mDia2 localization in cytokinesis. Notably, we found that GFP-mDia2 lacking the GBD still localized at the cleavage furrow (Figure 3, A–C). These constructs have ARR/DID and DD domains that are also called Formin homology 3 (FH3) domain and are important for the localization of several formin family proteins (Carnahan and Gould, 2003; Brandt et al., 2007; Rincon et al., 2009), suggesting that the FH3 domains of each formin family protein contributes to its localization and that such localization is driven by binding of this domain to context-dependent interaction partners. We performed pulldown assays using the mDia2 fragment containing the above region, and identified anillin as a binding partner (Figure 3D). Anillin is essential in cytokinesis in most cell types, which functions as a cross-linker of other cytoskeltal components in the contractile ring (D'Avino, 2009). Both anillin and formin family proteins have been studied as important components of the contractile ring in cytokinesis (Eggert et al., 2006), but their physical interaction has not been revealed. Here, we have revealed the physical interaction between anillin and one of the formin family proteins, mDia2 (Figures 4 and 5). The N-terminal region of anillin, which directly binds to mDia2 (Figures 5 and 7A), is well conserved in anillin of various vertebrates, but has not been characterized before.
Third, we found that this N-terminal fragment of anillin can compete in binding to the DID domain of mDia2 with DAD of mDia2 (Figure 5B). This indicates a possibility that mDia2 can be activated by anillin through the disruption of DID–DAD interaction. However, we suggest that mDia2 binds to anillin after active Rho binds to mDia2 to release the autoinhibitory interaction and stabilizes the open conformation, since the Rho-binding-defective mDia2 did not localize at the cleavage furrow and the addition of a mutation into DAD region, which structurally opens mDia, induced the accumulation of mDia2 at the cleavage furrow (Figure 3, A and C, bottom panels). Our results are thus similar to the targeting mechanism of mDia1 suggested by Brandt et al. (2007), who showed that IQGAP1 binds to mDia1 after the RhoA-mediated release of autoinhibition of mDia1 and regulates phagocytotic processes. The specific interaction between mDia and mDia binding partners and their involvement in the activation of the mDia proteins are thus key issues in controlling mDia function. Structural analyses of the interaction between mDia proteins and each interaction partner will be useful to fully understand how mDia activity is controlled by each binding partner.
Finally, we provide the evidence that the interaction between anillin and mDia2 is important in cytokinesis (Figures 6 and 7). We found in the anillin-RNAi rescue experiments using the anillin lacking the mDia2-binding region that mDia2 did not localize at the cleavage furrow even if RhoA localize as in control-RNAi cells (Figure 6, B–D), and that the anillin lacking the mDia2-binding region did not rescue the cytokinesis failure as full-length anillin (Figure 7). These results suggest that localization of RhoA at the cleavage furrow is not enough for the localization of mDia2 at the cleavage furrow, and that, in addition to it, the binding between mDia2 and anillin is required for the localization of mDia2 and execution of cytokinesis. Because anillin binds several cytoskeletal proteins and functions as an essential scaffold in cytokinesis (D'Avino, 2009), our findings suggest that mDia2 is also anchored in the anillin scaffold so that it can function effectively within this scaffold. In fact, the cytokinesis phenotypes of mDia2-RNAi cells and that of anillin-RNAi cells were similar in that the both cells showed the unstabilized contractile ring and abnormal contraction during cytokinesis (Straight et al., 2005; Watanabe et al., 2008). It is thus interesting to speculate that anillin may regulate actomyosin-ring dynamics locally by linking the active mDia2 and the contractile ring components such as actin and myosin. Based on these findings, we propose a model. When Rho is activated at the equatorial cortex of the dividing cells, mDia2 is first targeted there by activated Rho. Anillin then binds to the DID domain of this open form of mDia2 and stabilizes its localization and activation. The stabilized mDia2 can provide F-actin scaffold for the contraction of the contractile ring in cytokinesis.
Supplementary Material
ACKNOWLEDGMENTS
We thank K. Nonomura, M. Takagi, and T. Arai of our department for assistance. We also thank C. Field (Harvard University) for stimulating discussions, and Professor N. Watanabe (Tohoku University) for helpful suggestions and comments. We are grateful to Professor S. Iwata and Dr. T. Kobayashi (Kyoto University, JST, ERATO, Iwata Human Receptor Crystallography Project) for providing us a BIAcore facility. S.W. was supported by JSPS (Japan Society for the Promotion of Science) Research fellowships. This work was supported by a Grant-in-Aid for Specially Promoted Research from the Ministry for Education, Culture, Sports, Science, and Technology.
Abbreviations used:
- mDia
mammalian homolog of diaphanous
- DID
diaphanous-inhibitory domain
- DAD
diaphanous autoregulatory domain
- GEF
guanine nucleotide exchange factor
- GBD
GTPase-binding domain
- ARR
armadillo-repeat region
- FH domain
formin homology domain
- DD
dimerization domain
- CC
coiled coil.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-04-0324) on July 21, 2010.
REFERENCES
- Alberts A. S., Bouquin N., Johnston L. H., Treisman R. Analysis of RhoA-binding proteins reveals an interaction domain conserved in heterotrimeric G protein beta subunits and the yeast response regulator protein Skn7. J. Biol. Chem. 1998;273:8616–8622. doi: 10.1074/jbc.273.15.8616. [DOI] [PubMed] [Google Scholar]
- Balasubramanian M. K., Bi E., Glotzer M. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr. Biol. 2004;14:R806–18. doi: 10.1016/j.cub.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Barr F. A., Gruneberg U. Cytokinesis: placing and making the final cut. Cell. 2007;131:847–860. doi: 10.1016/j.cell.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Bartolini F., Moseley J. B., Schmoranzer J., Cassimeris L., Goode B. L., Gundersen G. G. The formin mDia2 stabilizes microtubules independently of its actin nucleation activity. J. Cell Biol. 2008;181:523–536. doi: 10.1083/jcb.200709029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt D. T., Marion S., Griffiths G., Watanabe T., Kaibuchi K., Grosse R. Dia1 and IQGAP1 interact in cell migration and phagocytic cup formation. J. Cell Biol. 2007;178:193–200. doi: 10.1083/jcb.200612071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnahan R. H., Gould K. L. The PCH family protein, Cdc15p, recruits two F-actin nucleation pathways to coordinate cytokinetic actin ring formation in Schizosaccharomyces pombe. J. Cell Biol. 2003;162:851–862. doi: 10.1083/jcb.200305012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillon D. H., Wasserman S. A. Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development. 1994;120:3367–3377. doi: 10.1242/dev.120.12.3367. [DOI] [PubMed] [Google Scholar]
- D'Avino P. P. How to scaffold the contractile ring for a safe cytokinesis—lessons from Anillin-related proteins. J. Cell Sci. 2009;122:1071–1079. doi: 10.1242/jcs.034785. [DOI] [PubMed] [Google Scholar]
- Dvorsky R., Blumenstein L., Vetter I. R., Ahmadian M. R. Structural insights into the interaction of ROCKI with the switch regions of RhoA. J. Biol. Chem. 2004;279:7098–7104. doi: 10.1074/jbc.M311911200. [DOI] [PubMed] [Google Scholar]
- Eda M., Yonemura S., Kato T., Watanabe N., Ishizaki T., Madaule P., Narumiya S. Rho-dependent transfer of Citron-kinase to the cleavage furrow of dividing cells. J. Cell Sci. 2001;114:3273–3284. doi: 10.1242/jcs.114.18.3273. [DOI] [PubMed] [Google Scholar]
- Eggert U. S., Mitchison T. J., Field C. M. Animal cytokinesis: from parts list to mechanisms. Annu. Rev. Biochem. 2006;75:543–566. doi: 10.1146/annurev.biochem.74.082803.133425. [DOI] [PubMed] [Google Scholar]
- Field C. M., Coughlin M., Doberstein S., Marty T., Sullivan W. Characterization of anillin mutants reveals essential roles in septin localization and plasma membrane integrity. Development. 2005;132:2849–2860. doi: 10.1242/dev.01843. [DOI] [PubMed] [Google Scholar]
- Goode B. L., Eck M. J. Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- Gruneberg U., Neef R., Li X., Chan E. H., Chalamalasetty R. B., Nigg E. A., Barr F. A. KIF14 and citron kinase act together to promote efficient cytokinesis. J. Cell Biol. 2006;172:363–372. doi: 10.1083/jcb.200511061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs H. N., Peterson K. J. Phylogenetic analysis of the formin homology 2 domain. Mol. Biol. Cell. 2005;16:1–13. doi: 10.1091/mbc.E04-07-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki T., Uehata M., Tamechika I., Keel J., Nonomura K., Maekawa M., Narumiya S. Pharmacological properties of Y-27632, a specific inhibitor of rho-associated kinases. Mol. Pharmacol. 2000;57:976–983. [PubMed] [Google Scholar]
- Kimura K., Tsuji T., Takada Y., Miki T., Narumiya S. Accumulation of GTP-bound RhoA during cytokinesis and a critical role of ECT2 in this accumulation. J. Biol. Chem. 2000;275:17233–17236. doi: 10.1074/jbc.C000212200. [DOI] [PubMed] [Google Scholar]
- Lammers M., Rose R., Scrima A., Wittinghofer A. The regulation of mDia1 by autoinhibition and its release by Rho*GTP. EMBO J. 2005;24:4176–4187. doi: 10.1038/sj.emboj.7600879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Higgs H. N. The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr. Biol. 2003;13:1335–1340. doi: 10.1016/s0960-9822(03)00540-2. [DOI] [PubMed] [Google Scholar]
- Mabuchi I., Hamaguchi Y., Fujimoto H., Morii N., Mishima M., Narumiya S. A rho-like protein is involved in the organisation of the contractile ring in dividing sand dollar eggs. Zygote. 1993;1:325–331. doi: 10.1017/s0967199400001659. [DOI] [PubMed] [Google Scholar]
- Madaule P., Eda M., Watanabe N., Fujisawa K., Matsuoka T., Bito H., Ishizaki T., Narumiya S. Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature. 1998;394:491–494. doi: 10.1038/28873. [DOI] [PubMed] [Google Scholar]
- Miki T., Okawa K., Sekimoto T., Yoneda Y., Watanabe S., Ishizaki T., Narumiya S. mDia2 shuttles between the nucleus and the cytoplasm through the importin-α/β- and CRM1-mediated nuclear transport mechanism. J. Biol. Chem. 2009;284:5753–5762. doi: 10.1074/jbc.M806191200. [DOI] [PubMed] [Google Scholar]
- Nishimura Y., Yonemura S. Centralspindlin regulates ECT2 and RhoA accumulation at the equatorial cortex during cytokinesis. J. Cell Sci. 2006;119:104–114. doi: 10.1242/jcs.02737. [DOI] [PubMed] [Google Scholar]
- Oceguera-Yanez F., Narumiya S. Measurement of activity of rho GTPases during mitosis. Methods Enzymol. 2006;406:332–345. doi: 10.1016/S0076-6879(06)06024-1. [DOI] [PubMed] [Google Scholar]
- Oegema K., Savoian M. S., Mitchison T. J., Field C. M. Functional analysis of a human homologue of the Drosophila actin binding protein anillin suggests a role in cytokinesis. J. Cell Biol. 2000;150:539–552. doi: 10.1083/jcb.150.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otomo T., Otomo C., Tomchick D. R., Machius M., Rosen M. K. Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol. Cell. 2005;18:273–281. doi: 10.1016/j.molcel.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Paul A. S., Pollard T. D. Review of the mechanism of processive actin filament elongation by formins. Cell Motil. Cytoskelet. 2009;66:606–617. doi: 10.1002/cm.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekny A., Werner M., Glotzer M. Cytokinesis: welcome to the Rho zone. Trends Cell Biol. 2005;15:651–658. doi: 10.1016/j.tcb.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Piekny A. J., Glotzer M. Anillin is a scaffold protein that links RhoA, actin, and myosin during cytokinesis. Curr. Biol. 2008;18:30–36. doi: 10.1016/j.cub.2007.11.068. [DOI] [PubMed] [Google Scholar]
- Pollard T. D. Mechanics of cytokinesis in eukaryotes. Curr. Opin. Cell Biol. 2009;22:50–56. doi: 10.1016/j.ceb.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincon S. A., Ye Y., Villar-Tajadura M. A., Santos B., Martin S. G., Perez P. Pob1 participates in the Cdc42 regulation of fission yeast actin cytoskeleton. Mol. Biol. Cell. 2009;20:4390–4399. doi: 10.1091/mbc.E09-03-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose R., Weyand M., Lammers M., Ishizaki T., Ahmadian M. R., Wittinghofer A. Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature. 2005;435:513–518. doi: 10.1038/nature03604. [DOI] [PubMed] [Google Scholar]
- Seth A., Otomo C., Rosen M. K. Autoinhibition regulates cellular localization and actin assembly activity of the diaphanous-related formins FRLalpha and mDia1. J. Cell Biol. 2006;174:701–713. doi: 10.1083/jcb.200605006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Straight A. F., Field C. M., Mitchison T. J. Anillin binds nonmuscle myosin II and regulates the contractile ring. Mol. Biol. Cell. 2005;16:193–201. doi: 10.1091/mbc.E04-08-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki C., Daigo Y., Ishikawa N., Kato T., Hayama S., Ito T., Tsuchiya E., Nakamura Y. ANLN plays a critical role in human lung carcinogenesis through the activation of RHOA and by involvement in the phosphoinositide 3-kinase/AKT pathway. Cancer Res. 2005;65:11314–11325. doi: 10.1158/0008-5472.CAN-05-1507. [DOI] [PubMed] [Google Scholar]
- Swan K. A., Severson A. F., Carter J. C., Martin P. R., Schnabel H., Schnabel R., Bowerman B. cyk-1, a C. elegans FH gene required for a late step in embryonic cytokinesis. J. Cell Sci. 1998;111(Pt 14):2017–2027. doi: 10.1242/jcs.111.14.2017. [DOI] [PubMed] [Google Scholar]
- Tatsumoto T., Xie X., Blumenthal R., Okamoto I., Miki T. Human ECT2 is an exchange factor for Rho GTPases, phosphorylated in G2/M phases, and involved in cytokinesis. J. Cell Biol. 1999;147:921–928. doi: 10.1083/jcb.147.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallar B. J., Stropich B. N., Schoenherr J. A., Holman H. A., Kitchen S. M., Alberts A. S. The basic region of the diaphanous-autoregulatory domain (DAD) is required for autoregulatory interactions with the diaphanous-related formin inhibitory domain. J. Biol. Chem. 2006;281:4300–4307. doi: 10.1074/jbc.M510277200. [DOI] [PubMed] [Google Scholar]
- Watanabe N., Madaule P., Reid T., Ishizaki T., Watanabe G., Kakizuka A., Saito Y., Nakao K., Jockusch B. M., Narumiya S. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 1997;16:3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Ando Y., Yasuda S., Hosoya H., Watanabe N., Ishizaki T., Narumiya S. mDia2 induces the actin scaffold for the contractile ring and stabilizes its position during cytokinesis in NIH 3T3 cells. Mol. Biol. Cell. 2008;19:2328–2338. doi: 10.1091/mbc.E07-10-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y., Eng C. H., Schmoranzer J., Cabrera-Poch N., Morris E. J., Chen M., Wallar B. J., Alberts A. S., Gundersen G. G. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat. Cell Biol. 2004;6:820–830. doi: 10.1038/ncb1160. [DOI] [PubMed] [Google Scholar]
- Yasuda S., Oceguera-Yanez F., Kato T., Okamoto M., Yonemura S., Terada Y., Ishizaki T., Narumiya S. Cdc42 and mDia3 regulate microtubule attachment to kinetochores. Nature. 2004;428:767–771. doi: 10.1038/nature02452. [DOI] [PubMed] [Google Scholar]
- Yuce O., Piekny A., Glotzer M. An ECT2-centralspindlin complex regulates the localization and function of RhoA. J. Cell Biol. 2005;170:571–582. doi: 10.1083/jcb.200501097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.