The BNIP-2 and Cdc42GAP Homology (BCH) domain from p50RhoGAP sequesters RhoA from inactivation by the adjacent GAP domain and it confers unique Rho-binding profile from that of GAP domain. This suppression is further augmented by an intramolecular interaction, adding to a new paradigm for regulating p50RhoGAP signaling.
Abstract
The BNIP-2 and Cdc42GAP homology (BCH) domain is a novel regulator for Rho GTPases, but its impact on p50-Rho GTPase-activating protein (p50RhoGAP or Cdc42GAP) in cells remains elusive. Here we show that deletion of the BCH domain from p50RhoGAP enhanced its GAP activity and caused drastic cell rounding. Introducing constitutively active RhoA or inactivating GAP domain blocked such effect, whereas replacing the BCH domain with endosome-targeting SNX3 excluded requirement of endosomal localization in regulating the GAP activity. Substitution with homologous BCH domain from Schizosaccharomyces pombe, which does not bind mammalian RhoA, also led to complete loss of suppression. Interestingly, the p50RhoGAP BCH domain only targeted RhoA, but not Cdc42 or Rac1, and it was unable to distinguish between GDP and the GTP-bound form of RhoA. Further mutagenesis revealed a RhoA-binding motif (residues 85-120), which when deleted, significantly reduced BCH inhibition on GAP-mediated cell rounding, whereas its full suppression also required an intramolecular interaction motif (residues 169-197). Therefore, BCH domain serves as a local modulator in cis to sequester RhoA from inactivation by the adjacent GAP domain, adding to a new paradigm for regulating p50RhoGAP signaling.
INTRODUCTION
Rho small GTPases such as Rho, Rac, and Cdc42 play vital roles in regulating actin cytoskeleton organization and cell morphology during cell migration, division, apoptosis, and tissue development. Activation of Cdc42 and Rac1 stimulates the formation of filopodia and lamellipodia, respectively, whereas RhoA activation stimulates the formation of stress fibers (Hall, 1998; Etienne-Manneville and Hall, 2002). Their activities are tightly regulated by at least three major classes of regulatory proteins, namely the guanine nucleotide exchange factors (GEFs), the Rho GTPase-activating proteins (GAPs), and the guanine nucleotide dissociation inhibitors (GDIs; Dovas and Couchman, 2005; Bernards and Settleman, 2007; Bos et al., 2007). Aberrant activity of Rho and their regulators are known to affect cell proliferation and transformation, motility, invasion, tumorigenesis, and pathogen infection (Sahai and Marshall, 2002; Jaffe and Hall, 2005; Munter et al., 2006; Vega and Ridley, 2008). A balanced control on Rho activity is therefore crucial for maintaining normal physiology and understanding their mechanism of action, and precise levels of control remains a key challenge.
RhoGAPs function as negative regulators by activating the intrinsic Rho GTPase activity, converting the active GTP-bound state to the inactive GDP-bound state (Moon and Zheng, 2003; Bernards and Settleman, 2005). The human genome encodes more than 80 RhoGAPs with distinctive arrays of protein domain/motifs (Tcherkezian and Lamarche-Vane, 2007). These domain/motifs could potentially act as regulator for either their GAP activity, subcellular localization, or connecting to various signaling networks via protein–protein interactions. These possibilities, however, await more systematic analyses. The p50RhoGAP (or Cdc42GAP) is a 50-kDa protein that functions biochemically as a GAP for Cdc42 and Rho (Barfod et al., 1993; Ridley et al., 1993; Lancaster et al., 1994; Wang et al., 2005; Wang et al., 2006). Expression of p50RhoGAP is up-regulated in Waldenstrom macroglobulinemia (Hatjiharissi et al., 2007), whereas the expression of its homolog BPGAP1/ARHGAP8 is up-regulated in primary colorectal tumors (Johnstone et al., 2004) and cervical cancer (Song et al., 2008). Recent studies showed that p50RhoGAP is involved in regulating cell migration (Wang et al., 2006; Szczur et al., 2006; Shen et al., 2008) and muscle cell differentiation (Kang et al., 2008), whereas its homozygous knockout embryos and newborn mice displayed reduced organ and body size, owing to increased spontaneous Jun N-terminal kinase (JNK)-mediated apoptosis (Wang et al., 2005, 2006, 2007). Despite being one of the earliest RhoGAPs identified where the biochemical and structural properties of the GAP domain of p50RhoGAP are already well defined (Barrett et al., 1997; Rittinger et al., 1997; Nassar et al., 1998), little is known on the modulation of the GAP function inside the cells. In particular, the impact of other protein modules such as the BNIP-2 and Cdc42GAP homology (BCH) domain that is located N-terminus proximal to the GAP domain remains largely undetermined. Recent studies showed that the BCH domain contributes to autoinhibition in vitro (Moskwa et al., 2005) and is required for its endosomal localization as well as binding to Rab5 and Rab11 (Sirokmány et al., 2006).
Recent work by us and others show that the BCH domains could regulate cell dynamics by engaging specific Rho small GTPases, their immediate regulators, and other cellular targets. For instance, the BCH domain of BNIP-2 induces cell protrusions by targeting Cdc42 (Low et al., 1999, 2000; Zhou et al., 2005) and promotes muscle differentiation by coupling the myogenic Cdo receptor signaling to p38α/β MAPK activation (Kang et al., 2008). In comparison, the homologous BCH domain in BNIP-Sα targets RhoA and displaces p50RhoGAP via its heterophilic interaction, leading to RhoA activation, cell rounding, and apoptosis (Zhou et al., 2002, 2006). Consistently, the BCH domain of another member, the BNIP-XL, inhibits cellular transformation by preventing oncogenic Lbc RhoGEF from activating RhoA after binding to these two proteins separately (Soh and Low, 2008). Extension to this, the BCH domain of BPGAP1/ARHGAP8, a homolog of p50RhoGAP, induces pseudopodia and cell migration by coupling to the GAP domain and proline-rich motif (Shang et al., 2003). This BCH domain also stimulates ERK (extracellular signal–regulated kinase) signaling, through a yet unknown mechanism (Lua and Low, 2005). Further, the BCH domain of BNIP-H (Caytaxin) interacts with the kidney-type glutaminase (KGA; Buschdorf et al., 2006) and peptidyl-prolyl isomerase Pin1 (Buschdorf et al., 2008) to regulate neurite outgrowth.
All these findings point to the possibility that the BCH domain of p50RhoGAP could similarly play an important role in modulating its biological function inside the cells. Here we present the evidence that the BCH domain on p50RhoGAP serves as a local modulator to sequester RhoA from being inactivated by the adjacent GAP domain. Acting in concert with an intramolecular interaction, such sequestration of substrate in cis provides a novel mechanism for regulating the local activity of p50RhoGAP toward Rho. This could have important bearings on previously unappreciated function of BCH or BCH-like domains in other RhoGAPs or RhoGEFs.
MATERIALS AND METHODS
Cell Culture and Transfection
Human 293T cells were grown in RPMI-1640 medium, and HeLa cells were grown in DMEM (high glucose) supplemented with 10% (vol/vol) fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin (all from Hyclone Laboratories, Logan, UT). Cells in six-well plates were transfected with TransIT-LT1 (Mirus Bio, Madison, WI) or Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol.
Construction of Expression Plasmids
Full-length cDNA of p50RhoGAP was cloned into a hemagglutinin (HA)-tagged, green fluorescent protein (GFP)- and FLAG-tagged expression vector, pXJ40 (Dr. E. Manser, IMCB, Singapore). FLAG-Rho-GDIα plasmid was provided by Dr. Amy L. Wilson-Delfosse (Case Western Reserve University School of Medicine; Gibson et al., 2004). Constructions used in cellular localization study were as follows: yellow fluorescent protein (YFP)-Golgi, red fluorescent protein–endoplasmic reticulum (RFP-ER; Clontech, Mountain View, CA), glycosylphosphatidylinositol (GPI)-GFP and lysosomal-associated membrane protein (Lamp1)-GFP (from A. Galmiche, Institut für Medizinische Strahlenkunde und Zellforschung, University of Würzburg, Würzburg, Germany). Deletion mutants were generated by PCR using specific primers facilitated by restriction sites. For each construct, several clones were chosen and sequenced using the ABI PRISM BigDye Terminator Cycle Sequencing kit (Applied Biosystems. Foster City, CA) to the entirety in both directions to confirm their identity. All plasmids were purified using Qiagen miniprep kit (Chatsworth, CA) for use in transfection experiments. Escherichia coli strain DH5α was used as host for propagation of the clones.
Bioinformatics
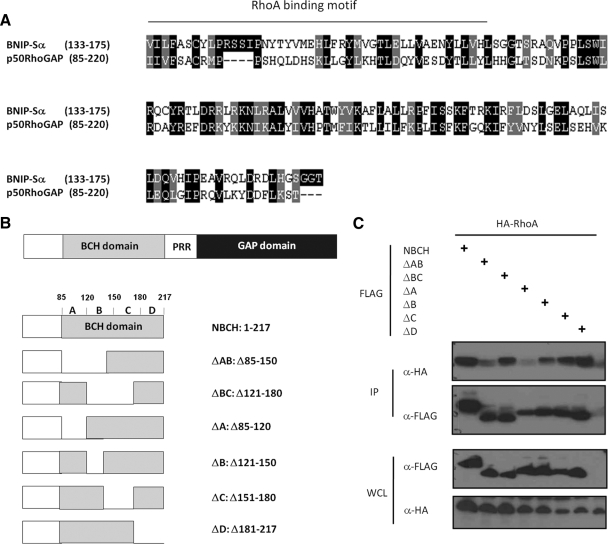
To search for putative p50RhoGAP homolog in Schizosaccharomyces pombe that contain BCH domains, the peptide sequence of full-length p50RhoGAP was used to carry out BLAST searches in S. pombe GeneDB using BLASTP (http://www.genedb.org/genedb/pombe/). The percentage of similarity between S. pombe RhoGAP and p50RhoGAP was identified by bl2seq (http://blast.ncbi.nlm.nih.gov/bl2seq/wblast2.cgi). To identify any putative RBD/motif (RBM) within the p50RhoGAP BCH domain, the sequence of p50RhoGAP BCH domain (amino acids 85-217) was used for alignment with known RBD domain of BNIP-Sα using the ClustalW (http://www2.ebi.ac.uk/clustalw/). Outputs of the multiple sequence alignment were displayed with BOXSHADE 3.21 (http://www.ch.embnet.org).
Immunoprecipitation Studies and Western Blot Analyses
Control cells or cells transfected with expression plasmids were lysed in lysis buffer (150 mM sodium chloride, 50 mM Tris, pH 7.3, 0.25 mM EDTA, 1% [wt/vol] sodium deoxycholate, 1% [vol/vol] Triton X-100, 0.2% sodium fluoride, 0.1% sodium orthovanadate, and a mixture of protease inhibitors from Roche Applied Sciences, Indianapolis, IN). Lysates were immunoprecipitated (IPed) with anti-FLAG M2 beads (Sigma, St. Louis, MO), and the associated proteins were separated on SDS-PAGE and probed with anti-HA (for cotransfection experiments) Samples were run in SDS/PAGE gels and analyzed by Western blotting with anti-HA (Zymed, South San Francisco, CA) or anti-FLAG (Sigma).
Immunofluorescence and Direct Fluorescence Studies
Cells were seeded on coverslips in a six-well plate and transfected with various expression constructs for 16–20 h and then stained for immunofluorescence detection using confocal fluorescence microscopy or directly visualized for cells expressing GFP-tagged proteins as previously described (Zhou et al., 2002). FLAG-tagged proteins were detected with monoclonal anti-FLAG followed by Texas Red– or FITC dye–conjugated goat anti-mouse IgG (Invitrogen). Filamentous actin was detected by rhodamine-phalloidin (Molecular Probes, Eugene, OR), and microtubule was detected by anti-tubulin (Sigma) followed by Alexa Fluor 488–conjugated goat anti-mouse IgG (Invitrogen). In certain cases as specified, intensity of signal detected by confocal microscopy was used to determine the relative expression levels of tagged proteins across various regions of cell populations as indicated by the lines and arrows. The images were collected using a Zeiss 510 Meta laser-scanning microscope equipped with a 60× lens (Thornwood, NY). The detector gain was first optimized by sampling various regions of the coverslip and then was fixed for each specified channel. Once set, the detector gain value was kept constant throughout the image acquisition process. As a result, signal intensities from identical channels of different images could be used for quantitative measurements.
Microscopic Analysis of Transferrin Uptake
HeLa cells on coverslips were incubated in extracellular H-medium (145 mM NaCl, 5 mM KCl, 1 mM MgCl2, 0,8 mM CaCl2, 10 mM HEPES, 5 mM glucose, pH 7.4) for 20 min at 37°C and then medium was replaced with H-medium containing 10 μg/ml Alexa 568-transferrin (Molecular Probes) for the indicated times.
Rho Inhibitor Treatment
HeLa cells on coverslips were incubated with 1 μg/ml the exoenzyme C3 transferase from Clostridium botulinum (Cytoskeleton, Denver, CO) for 4 h, followed by staining with rhodamine-phalloidin (Molecular Probes) and confocal microscopy analysis.
RhoA activity Assays
Assays for the active (GTP-bound) form of RhoA was performed as described previously (Zhou et al., 2006). Briefly, cells were cotransfected for 24 h with RhoA in the absence or presence of p50RhoGAP or mutants. Cells were then lysed and subjected to pulldown assays with glutathione S-transferase (GST) fusion of the RBD of rhotekin, which would bind and detect active RhoA in vivo (plasmid kindly provided by Dr. Simone Schoenwaelder, Monash University, Australia). Bound RhoA was separated on SDS-PAGE gels and subjected to Western blot analysis with the FLAG antibody.
In Vitro GTP- and GDP-loading Assays
A total of 15 μg of GST-RhoA fusion protein were preloaded with 10 mM GDP or GTPγS (Sigma) in binding buffer (25 mM NaCl, 20 mM Tris-HCl, pH 7.5, 0.1 mM DTT, and 50 mM EDTA) at 30°C for 10 min. The reaction was stopped with MgCl2 to a final concentration of 20 mM (Brill et al., 1996). The beads were incubated with cell lysates at 4°C in GAP lysis buffer (50 mM HEPES pH 7.4, 150 mM NaCl, 1.5 mM MgCl2, 5 mM EGTA, 10% glycerol, 1% Triton X-100, 5 mM sodium orthovanadate, and a mixture of protease inhibitors). Samples were run in SDS-PAGE gels and analyzed by Western blotting.
RESULTS
The BCH Domain of p50RhoGAP Inhibits GAP-induced Cell Rounding
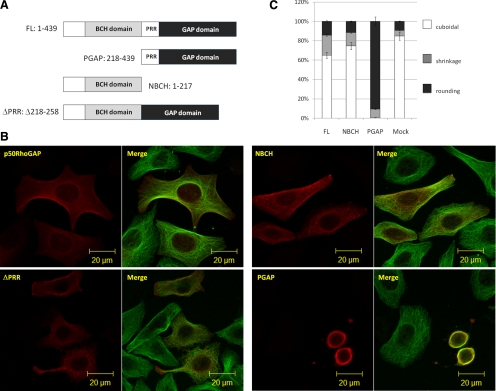
To address the impact of other protein modules/motifs on the cellular function of p50RhoGAP, human cervical epithelial HeLa cells were transfected with either the wild type or its three mutants, namely a ΔPRR mutant lacking the proline-rich region (amino acids 218-258), the NBCH (N-terminus containing the BCH domain, but lacking proline region; amino acids 1–217) or the PGAP (proline-containing carboxyl end, harboring the GAP domain; amino acids 218-439; Figure 1A). Their effects on cell morphology were compared and quantified by indirect immunofluorescence microscopy after costaining with anti-tubulin antibodies to visualize microtubules (Figure 1B). Figure 1C shows that only 15% of the cells transfected with full-length p50RhoGAP appeared round, whereas the majority of them still remained cuboidal or began to show cell retraction/shrinkage. In contrast, 90% of the cells transfected with PGAP already exhibited drastic cell rounding as shown in Figure 1B. Such effects on morphology were not due to variations in the protein expression because all their protein levels were identical (Supplementary Figure S1). To further examine the threshold of regulation by the GAP domain, we analyzed its expression levels and showed that even at very low expression levels, the PGAP domain was able to potently induce drastic cell rounding. In contrast, p50RhoGAP would increase the extents of cell rounding only when it was highly expressed (Supplementary Figure S2). This observation indicates that the N-terminal NBCH region could exert an inhibitory effect toward the otherwise very potent activity of the C-terminal GAP domain. The inhibitory effect was not due to the proline-containing sequence because cells expressing the ΔPRR mutant still displayed normal morphology. In comparison, the presence of the BCH domain in NBCH-transfected cells did not alter the overall cell morphology. This effect is different from the potent cell-rounding effect induced by the BCH domain of BNIP-Sα (Zhou et al., 2006), which was localized in punctate structures (Supplementary Figure S3), whereas p50RhoGAP BCH domain was predominantly localized to cell periphery (see Figure 2D). These results therefore support the specific and novel role of the adjacent BCH domain in suppressing the GAP activity of p50RhoGAP inside the cells.
Figure 1.
The BCH domain of p50RhoGAP inhibits GAP-induced cell rounding. (A) Schematic diagram of p50RhoGAP and its mutants: N-terminus without GAP domain (NBCH, amino acids 1-217), C-terminus without BCH domain (PGAP, amino acids 218-439), and a mutant without the proline-rich region, PPR (ΔPPR, Δ218-258). (B) HeLa cells were transfected with plasmids encoding FLAG-tagged full-length p50RhoGAP, NBCH, PGAP, or ΔPPR mutants. Cells were then fixed after 16–20 h and subjected to confocal fluorescence microscopy as described in Materials and Methods. Morphological changes and cytoskeletal rearrangements were revealed by indirect immunostaining with Alexa Fluor 488–conjugated goat anti-mouse IgG against anti-tubulin for microtubules and with FLAG antibody for expressed FLAG-tagged proteins. (C) For quantitative analysis, the ratio of cuboidal, protrusion/shrinkage, and round cells was scored with at least 150 transfected cells counted per sample per experiment. Data are means ± SD (n = 3).
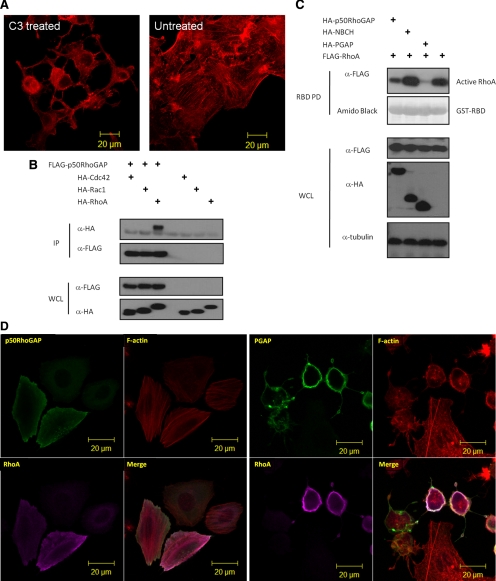
Figure 2.
The GAP domain of p50RhoGAP induces cell rounding by inactivating Rho GTPases (A) HeLa cells were treated with Rho inhibitor C3 Transferase followed by rhodamine-conjugated phalloidin staining and confocal fluorescence microscopy analysis. (B) Cells were transfected with FLAG-p50RhoGAP in the presence or absence of HA-tagged expression constructs of Cdc42, Rac1, and RhoA. Lysates were immunoprecipitated (IP) with anti-FLAG beads, and the associated proteins were separated on SDS-PAGE, blotted, and probed with HA antibody. Expression of FLAG-p50RhoGAP and HA-tagged Cdc42, Rac1, and RhoA were verified by Western blot analyses for the whole cell lysates (WCL) using anti-FLAG (third panel) and anti-HA (bottom panel), respectively. The bound GTPase was detected by anti-HA (top panel), and equal loading of IP beads were verified by anti-FLAG (second panel). (C) To determine the Rho GTPase activity, HeLa cells were transfected with FLAG-tagged wild-type RhoA in the presence and absence of HA-tagged p50RhoGAP, NBCH, or PGAP mutants. Cell were lysed and incubated with GST fusion of the Rho-binding domain of rhotekin immobilized on beads as described in Materials and Methods Bound active RhoA were resolved on SDS-PAGE and detected by immunoblotting with FLAG-antibody (top panel). Equal loading of GST fusion proteins is shown in the second panel. (D) HeLa cells were transfected with plasmids encoding HA-RhoA alone or with FLAG-tagged full-length p50RhoGAP, PGAP, or NBCH mutants. Cells were then fixed after 20 h and subjected to confocal fluorescence microscopy as described in Materials and Methods The actin filaments were detected by direct costaining with rhodamine-conjugated phalloidin. (E) HeLa cells were cotransfected with HA-tagged PGAP mutant and wild-type or constitutively active RhoA-G14V. Cells were then fixed and images analyzed by confocal fluorescence microscopy after direct staining with rhodamine-conjugated phalloidin for actin filaments.
The GAP Domain of p50RhoGAP Induces Cell Rounding by Inactivating RhoA
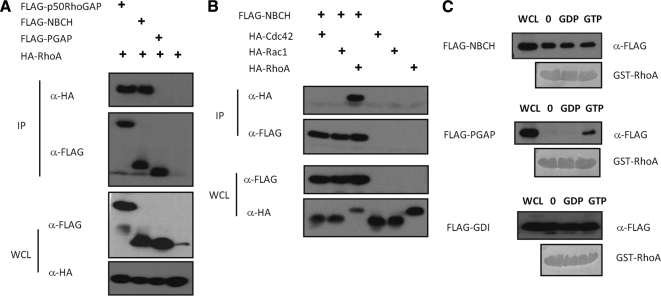
To explore the molecular basis underlying the new inhibitory role of the BCH domain in p50RhoGAP, we needed to first establish the basis of cell rounding induced by the GAP domain. Previous studies showed that p50RhoGAP could interact with Cdc42 and RhoA in vitro (Barfod et al., 1993; Lancaster et al., 1994) and also inactivated these two Rho GTPases in cells (Ridley et al., 1993; Wang et al., 2005; Wang et al., 2006). In HeLa cells, it has been reported that p50RhoGAP targets RhoA, but not Cdc42 (Moskwa et al., 2005; Sirokmány et al., 2006). Interestingly, once treated with Rho inhibitor C3 transferase from C. botulinum, HeLa cells displayed similar cytoskeleton collapse and cell rounding as induced by GAP domain (Figure 2A). This observation prompted us to examine whether cell rounding and cytoskeleton disruption induced by the GAP domain of p50RhoGAP, and its regulation by the adjacent BCH domain as shown above, would involve RhoA inactivation. First, we examined the interactions between p50RhoGAP and RhoA, Cdc42, or Rac1 in HeLa cells. These Rho GTPases were expressed in the same HA-epitope to allow direct comparison of their binding affinity. Figure 2B shows that RhoA but not Cdc42 or Rac1 was preferentially coIPed with p50RhoGAP. Next, RhoA activity assays also showed a marked reduction in the level of active RhoA in the presence of the single GAP domain when compared with full-length p50RhoGAP (Figure 2C and Supplementary Figure S4). Consistent with these biochemical data were the confocal imaging analyses for the impact of the full-length, NBCH and PGAP fragments on RhoA-induced stress fibers and cell rounding (Figure 2D). Here, expression of the PGAP fragment without the BCH domain led to extensive cell rounding accompanied by a total collapse of the F-actin. In strong contrast, the full-length p50RhoGAP or the NBCH did not affect the formation of stress fibers induced by RhoA despite that the cell periphery projections in RhoA-expressing cells were abolished in the presence of NBCH (indicated by yellow boxes). The cell rounding induced by GAP domain could be blocked by coexpressing the constitutive active RhoA-G14V with PGAP with full restoration of stress fibers (Figure 2E; comparing yellow boxes), indicating that cell rounding indeed required inactivation of RhoA.
To ascertain that such RhoA inactivation was a direct effect from the GAP activity of p50RhoGAP and not via other indirect pathways, we went on to nullify the enzymatic function of the GAP domain by mutating several key amino acids; these include the “invariant” arginine finger R282, which is critical as a catalytic residue in trans (Leonard et al., 1998), the secondary arginine R283 (Leonard et al., 1998), and the highly conserved asparagine N391 for stabilizing the Rho effector loop (Rittinger et al., 1997). PGAP mutants carrying R282A, N391V, R282A/R283A, R282A/N391V, and R282A/R283A/N391V were each expressed in HeLa cells and showed that cells expressing either single-point mutants (R282A, N391V) or double-points mutants (R282A/R283A, R282A/N391V) still exhibited drastic cell rounding (Figure 3A). Only cells expressing the triple-point mutant R282A/R283A/N391V displayed the regular cuboidal morphology indicating the complete loss of its GAP activity (Figure 3A), as evidenced by the Rho activity assay (Figure 3B) and its inability to resolve RhoA-induced stress fibers (Supplementary Figure S5).
Figure 3.
Three essential residues for GAP domain in inducing cell rounding. (A) HeLa cells were transfected with HA-tagged PGAP or the various mutants indicated, fixed after 20 h, and subjected to confocal fluorescence microscopy with anti-HA and Alexa Fluor 488 dye–conjugated goat anti-rabbit IgG as described in Materials and Methods. (B) To determine the Rho GTPase activity, HeLa cells were transfected with FLAG-tagged RhoA alone or with various HA-tagged PGAP or its various mutants. Cell were lysed and incubated with GST fusion of the Rho-binding domain of rhotekin immobilized on beads as described in Materials and Methods. Bound active RhoA were resolved on SDS-PAGE and detected by immunoblotting with FLAG-antibody (top panel). Equal loading of GST fusion proteins is shown in the second panel.
These results therefore confirm that p50RhoGAP interacts with and inactivates RhoA in the cells, resulting in extensive cytoskeletal collapse and cell rounding in a process mediated by the catalytic GAP domain. Most significantly, the adjacent BCH domain could suppress such a potent GAP activity.
Suppression of p50RhoGAP Activity Does Not Require Its Endosomal Localization or Intramolecular Interaction Alone
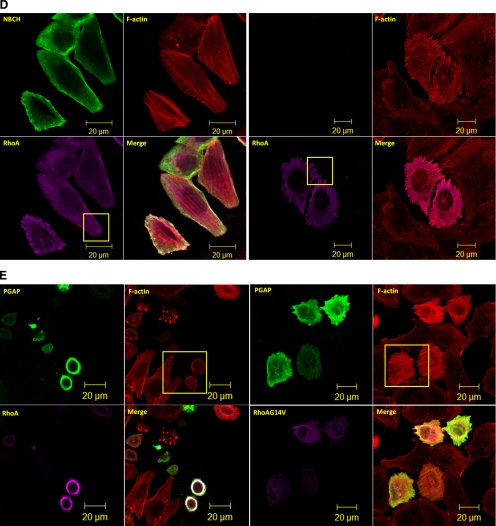
It was recently reported that the BCH domain, which shares very limited sequence homology to the Sec14p domain, is required for the intramolecular interaction with the GAP domain (Moskwa et al., 2005) and the endosomal localization of p50RhoGAP (Sirokmány et al., 2006). These observations offer two potential mechanisms for BCH domain-mediated GAP inhibition. First, the two previously reported intramolecular interaction regions (IIR 1: 1–48 and IIR 2: 169–197, Moskwa et al., 2005) could sterically inhibit GAP function via the BCH–GAP intramolecular interaction. Figure 4 shows that expression of two mutants lacking these two intramolecular interaction regions alone (FL53 and FLΔ151-217) displayed no significant effect on cell morphology or actin network, indicating that such intramolecular interactions alone did not directly suppress the GAP function in cells. However, one of the motifs, IIR 2, turned out to augment the inhibitory effect of BCH domain as shown in our further analyses (see later).
Figure 4.
Intramolecular interaction alone does not regulate GAP activity. HeLa cells were transfected with FLAG-tagged-FL53 or FLΔ151-217 mutants and subjected to confocal fluorescence microscopy after direct staining with rhodamine-conjugated phalloidin for actin filaments.
Alternatively, the BCH domain could restrict p50RhoGAP to specific cellular environment where other extrinsic factors could directly or indirectly inhibit its function. To examine this further, the endosomal marker Alexa-568–coupled transferrin and a series of cellular organelle markers were used to monitor the subcellular distribution of p50RhoGAP. Supplementary Figure S6A shows the partial colocalization of p50RhoGAP with transferring-positive endosomes, but not with LAMP1 (lysosome), mitotracker (mitochondria), and the GPI-anchored protein compartment, indicating that a fraction of this GAP was localized in endosomes. Next, we substituted the BCH domain of p50RhoGAP with a specific endosomal module from the endosome localizing protein, sorting nexin3 (SNX3; Xu et al., 2001). Should an extrinsic factor be required to inhibit the GAP function there, this substitution would retain the suppression effect. Interestingly, this SNX3-PGAP mutant still caused cell rounding (Supplementary Figure S6B), although the hybrid protein still displayed endosomal localization (Supplementary Figure S6C; arrows). This result demonstrates that the endosomal localization of p50RhoGAP per se is not responsible for bringing about extrinsic factors to inhibit its GAP function there.
The BCH Domain of p50RhoGAP Displays a Unique Rho-binding Profile
The data presented so far suggest that the intrinsic property of BCH domain itself is critical in inhibiting the RhoGAP activity. Because our earlier work had demonstrated that various BCH domains could target small GTPases, it is conceivable that the BCH domain of p50RhoGAP could also bind RhoA, consequently preventing the GAP domain from acting on the same target. To test this hypothesis, we first determined the interaction between the NBCH fragment (that harbors the BCH domain) with Rho GTPases. Figure 5A shows that both full-length p50RhoGAP and NBCH could interact with RhoA whereas binding of RhoA to PGAP remained undetectable despite the potent GAP activity toward RhoA (Figure 2C). This was attributed to their high turnover and transient nature of enzyme-substrate relationship. Moreover, similar to the full-length p50RhoGAP (Figure 2B), the BCH domain specifically interacted with RhoA, but not with Cdc42 or Rac1 (Figure 5B).
Figure 5.
The BCH domain of p50RhoGAP displays distinctive RhoA binding profile. (A) HEK293T cells were transfected with plasmid encoding HA-RhoA alone or with FLAG-p50RhoGAP, NBCH or PGAP. Lysates were immunoprecipitated (IP) with anti-FLAG beads, and the associated proteins were detected with HA antibody (top panel). Expression of FLAG-tagged proteins and HA-RhoA were verified by Western blot analyses by anti-FLAG (third panel) and anti-HA (bottom panel), respectively. Equal loading of IP beads were verified by anti-FLAG (second panel). (B) Cells were transfected with plasmid encoding FLAG-NBCH in the presence or absence of HA-tagged Cdc42, Rac1, and RhoA. Lysates were immunoprecipitated (IP) with anti-FLAG beads, and the associated proteins were separated on SDS-PAGE, and probed with HA antibody (top panel). Expression of FLAG-tagged NBCH and HA-tagged GTPases were verified by Western blot analyses using anti-FLAG (third panel) and anti-HA (bottom panel), respectively. Equal loading of IP beads were verified by anti-FLAG (second panel). (C) HEK293T lysates expressing FLAG-NBCH, PGAP fragments or Rho-GDIα were incubated with unloaded GST-RhoA, GST-RhoA preloaded with GDP, or GTPγS as described in Materials and Methods. Bound proteins and whole cell lysates (WCL) input were analyzed with anti-FLAG and equal loading of GST beads verified by amido black staining.
The presence of BCH domain and RhoGAP domain as two Rho-targeting domain in tandem raises an interesting issue as to how Rho, as their common substrate, would be recognized and how the dynamic nature of the complex regulated. To gain some insights to this process, we examined how the BCH domain of p50RhoGAP could differentiate between the GTP-bound active Rho from the GDP-bound inactive Rho. To determine if the binding toward RhoA is nucleotide-dependent, GST-RhoA was unloaded or loaded with GDP or GTPγS (a nonhydrolyzable GTP analogue), and incubated with lysates expressing NBCH in a series of parallel experiments using two different controls (i.e., the PGAP fragment of p50RhoGAP which is expected to recognize the GTP-bound form; Barrett et al., 1997; Rittinger et al., 1997; Nassar et al., 1998) and the Rho-GDI, which recognizes Rho independently of the nucleotides (Hancock and Hall, 1993; Zalcman et al., 1996; Nomanbhoy and Cerione, 1996). Unlike PGAP region that preferentially bound GTPγS-RhoA, the BCH domain shows no preference for the nucleotide-free, GDP-, or GTPγS-bound form of RhoA (Figure 5C), a profile shared also by the Rho-GDI.
Taken together, all these data demonstrate that the BCH and GAP domains, despite binding to a common target, display unique binding characteristics that could be of importance in regulating the function of Rho inside the cells.
The BCH-like Domain of S. pombe That Does Not Bind Mammalian RhoA Cannot Functionally Substitute for the BCH Domain of p50RhoGAP
To further test the hypothesis that RhoA-binding ability was indeed responsible for the inhibitory effect, we first substituted the p50RhoGAP BCH domain with the homologous BCH domain of the RhoGAP from S. pombe. The yeast BCH-like domain was chosen because the evolutionary relationship between human and yeast is distant enough that the yeast BCH domain is unable to bind human RhoA. Bioinformatics analysis shows that the S. pombe homolog shares 43% similarity with p50RhoGAP full length and 51% similarity in their BCH domains (Supplementary Figure S7A). The S. pombe BCH domain was fused with the PGAP fragment (pBCHhPG; Supplementary Figure S7B) and lost the RhoA-binding ability as determined by coIP study (Supplementary Figure S7C). Once introduced into HeLa cells, pBCHhPG mutant was able to inactivate RhoA (Supplementary Figure S7D) and induced drastic cell rounding (Supplementary Figure S7B), similar to the earlier observation by PGAP that had its entire adjacent BCH domain removed. This result indicates that binding of RhoA and therefore its sequestration could play a vital role in the suppression of GAP-induced cell rounding.
The BCH Domain of p50RhoGAP Contains a Novel Rho-binding Motif
To further establish the “substrate sequestration” model, a p50RhoGAP mutant lacking only the RhoA-binding ability in its BCH domain is therefore required. To help identify such potential RhoA-binding sites, analysis of primary sequences of p50RhoGAP BCH domain with another RhoA-binding BCH domain from BNIP-Sα (Zhou et al., 2002) was first carried out. Such analysis revealed a putative RBM at the positions 85–125 of p50RhoGAP BCH domain (Figure 6A). Based on the secondary structure prediction, this BCH domain was further subdivided into four subregions (i.e., regions A, residues 85-120, putative RBM; B, residues 121-150; C, residues 151-180; and D, residues 181-217). Next, two NBCH mutants lacking the putative RBM (Δ85-120, Δ85-150) were generated. Four other constructs (Δ121-150, Δ121-180, Δ151-180, and Δ181-217) with deletion regions falling outside the putative RBM yet covering the other segments of the entire p50RhoGAP BCH domain were also prepared (Figure 6B). Cells were cotransfected with an HA-RhoA expression plasmid together with the FLAG-tagged NBCH wild type or the deletion mutants as described above. CoIP assays showed that the two mutants (ΔA and ΔAB) lacking the predicted RBM led to severe loss of binding to RhoA (Figure 6C). Other mutants, however, still retained their binding to RhoA. As a further control for their structural integrity, these two non-RhoA-binding mutants still retained their ability to form the homophilic interaction (Supplementary Figure S8). The region spanning amino acid residues 85-120 of the BCH domain therefore contains a novel RhoA-binding motif.
Figure 6.
Identification of a RhoA-binding motif within the BCH domain of p50RhoGAP. (A) Multiple sequence alignments of BCH domains from Homo sapiens BNIP-Sα (AY078983) and p50RhoGAP/Cdc42GAP (Q07960) using ClustalW and formatted using BOXSHADE. Identical residues are shaded black whereas similar or conserved ones are in gray. The region corresponding to the previously described Rho-binding motif in BNIP-Sα (Zhou et al., 2006) was underlined. (B) Schematic diagram of NBCH domain and its mutants. (C) Cells were cotransfected with HA-tagged RhoA and FLAG-tagged NBCH wild type or mutants as depicted in Figure 6B. Lysates were immunoprecipitated (IP) with anti-FLAG beads, and the associated proteins were separated on SDS-PAGE, blotted, and probed with HA antibody. Expression of FLAG-NBCH constructs and HA- RhoA were verified by Western blot analyses of the whole cell lysates (WCL) using anti-FLAG (third panel) and anti-HA (bottom panel), respectively. The bound RhoA was detected by anti-HA (top panel), and equal loading of IP beads were verified by anti-FLAG (second panel).
Full Suppression of p50RhoGAP Activity by BCH Domain Requires RhoA Sequestration Acting in Concert with Its Intramolecular Interaction
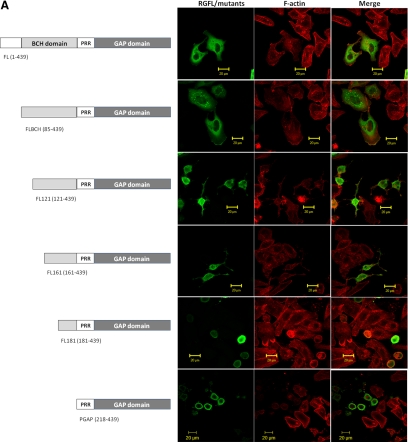
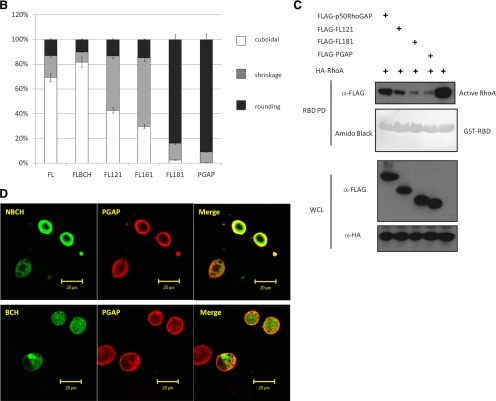
Next, we examined whether the p50RhoGAP lacking the RBM in the BCH domain could indeed lead to loss of its suppression effect on GAP activity. A series of FLAG-tagged p50RhoGAP truncation mutants were generated and their effect on cell rounding were then determined (Figure 7A). Among these mutants, FLBCH contains an entire intact BCH domain (with only the N-terminal amino acid 1-84 removed), whereas FL121 and FL161 mutants have their RBM removed. All these three mutants lack a previously identified IIR 1 (1-48, Moskwa et al., 2005), but they still retain IIR 2 (169–197, Moskwa et al., 2005). To examine whether RhoA sequestration was the only mechanism that the BCH domain employs to inhibit GAP activity, a further deletion with an impaired IIR 2 was also included, designated FL181. HeLa cells transfected with either the wild type or the mutant with an intact BCH domain (FLBCH) displayed normal cuboidal morphology (Figure 7, A and B), indicating strong inhibition of the GAP activity. Once the RBM was removed, as seen for mutants FL121 and FL161, their inhibition was significantly reduced. In FL121 and FL161 mutants (where RBM was absent), close to 60–70% of the transfected cells displayed drastic cell protrusion and rounding when compared with the FLBCH (20%) or the wild type (30%). This data demonstrated that the RhoA-binding ability indeed plays an important role in suppression of the GAP activity. However, the impact of two non-RhoA-binding mutants (FL121 and FL161) was not as potent as that from PGAP (near 100%), which had the entire BCH domain removed. In contrast, FL181, which had lost its RhoA-binding and both IIRs, led to a similar cell rounding effect (>95%) as did PGAP. Consistently, RBD assay shows enhanced GAP activity of FL121 compared with wild-type p50RhoGAP, whereas FL181 conferred the same maximal GAP activity as did PGAP (Figure 7C). To ensure that these effects on morphology were not due to variations in the protein expression, their respective expression levels were verified to be the same by Western analysis of the total cell lysates (Supplementary Figures S9) or by confocal microscopy analyses (Supplementary Figures S10). This result therefore indicates that both RhoA sequestration and intramolecular interaction (via IIR 2) act in concert to allow the BCH domain to fully suppress the activity of the adjacent GAP domain.
Figure 7.
The BCH domain of p50RhoGAP inhibits the adjacent GAP function by sequestering RhoA. (A) HeLa cells were transfected for 20 h with FLAG-tagged p50RhoGAP wild type and mutants including FLBCH, FL121, FL161, FL181, or PGAP. Cells were then fixed and incubated with FLAG monoclonal antibodies, followed by Alexa Fluor 488-conjugated goat anti-mouse IgG. Cell morphology was monitored by direct staining with rhodamine-conjugated phalloidin for actin filaments. (B) For quantitative analysis, the ratio of cuboidal, protrusion/shrinkage, and round cells was scored and at least 150 transfected cells were counted per sample per experiment. Data are means ± SD (n = 3). (C) Cells were transfected with FLAG-tagged p50RhoGAP full-length, FL121, FL181, or PGAP in the presence of HA-RhoA. After 20 h, cell were lysed and incubated with GST fusion of the Rho-binding domain of rhotekin-immobilized on beads, in order to assess the impacts of p50RhoGAP and the mutants in regulating RhoA activity as described in Materials and Methods. Bound GTPases were resolved on SDS-PAGE and detected by immunoblotting with HA-antibody (top panel). Equal loading of GST fusion proteins is shown in the second panel. (D) HeLa cells were cotransfected with HA-tagged PGAP and FLAG-tagged-NBCH or BCH domain. Cells were then fixed and analyzed with confocal fluorescence microscopy as described in Materials and Methods.
It is intriguing that, despite having the ability to bind RhoA, the BCH domain alone did not affect the overall cell morphology (although it did abolish periphery projections induced by Rho despite retaining stress fiber; see discussion later) and the Rho activity. However, when present with the GAP domain, this BCH domain greatly suppresses the adjacent GAP activity by sequestering RhoA and forming an intramolecular interaction. This phenomenon raises further speculation that the regulatory function of this BCH domain is physically coupled to the GAP domain. Consistently, coexpression of either BCH or NBCH domain with the GAP domain as two separate entities (in trans) failed to suppress any of the GAP-mediated cell rounding (Figure 7D). Such requirement for its physical linkage in cis would suggest a need for specific conformation or/and microenvironment to modulate the local pools of RhoA.
Taken together, we have demonstrated that the BCH domain of p50RhoGAP acts as a novel local modulator of GAP activity by engaging a specific RhoA-binding motif that exhibits a unique binding profile in order to sequester RhoA and prevent it from being inactivated by the adjacent RhoGAP domain. Furthermore, we show that such inhibition could work in concert with an IIS distal to RhoA-binding, thus adding a new paradigm of regulating p50RhoGAP and Rho signaling.
DISCUSSION
BCH Domain as a Regulatory Module for GTPases Signaling
Despite having a highly conserved enzymatic GAP module that inactivates Rho, many RhoGAPs exist to regulate diverse and complex cellular processes. This is mainly attributed to the ability of one RhoGAP to target more than one member of Rho GTPases and the potential of multiple RhoGAPs to regulate one single Rho. Furthermore, the existence of dynamic arrays of multiple domains in different RhoGAPs could potentially add to the repertoire of their functional regulation in space and time by engaging new interacting partners and cross-talks, directing to specific compartments or/and undergoing posttranslational modifications. RhoGAPs confer broad substrate specificity toward Rho. For example, p200RhoGAP stimulates the GTPase activities of both Rac1 and RhoA in vitro and in vivo (Moon et al., 2003) whereas hCdGAP targets Cdc42 and Rac1 (Tcherkezian et al., 2006). Similarly, p50RhoGAP also acts on Cdc42 and Rho in vitro (Barfod et al., 1993; Lancaster et al., 1994), whereas the loss of p50RhoGAP expression is linked to activation of Cdc42 inside the cells (Wang et al., 2005, 2006; Kang et al., 2008). Here, we show that p50RhoGAP instead binds preferentially to RhoA and not to Cdc42 in HeLa cells, primarily through its BCH domain. Recent studies by Sirokmány et al. (2006) also showed that in HeLa cells, p50RhoGAP has no effect on the distribution of Cdc42 and that both proteins show no colocalization. We believe that such variation in the substrate recognition is likely due to different cell types and different subcellular environments, thus providing greater functional plasticity and regulation for Rho signaling. In all cases, however, there is still a lack of evidence on how its activity could be regulated at the molecular level. By domain deletion, swapping, delineating the key RhoA-binding region within the BCH domain, and linking their interaction to cell morphological changes, we have revealed a cryptic role of the N-terminal BCH domain toward the activity of the adjacent GAP domain in p50RhoGAP, offering new insights to the regulation of Rho signaling by the multidomain RhoGAPs. Further, its preference for RhoA but not Cdc42 or Rac1 could provide a selective regulation of the GAP activity for RhoA, at least in the HeLa cells.
We have recently established the roles of BNIP-2 BCH domain in conferring Cdc42-dependent cell protrusions (Zhou et al., 2005) and p38-mediated pathways (Kang et al., 2008) and the ability of the BCH domains of BNIP-Sα and BNIP-XL in trans to counteract suppression of RhoA by p50RhoGAP (Zhou et al., 2006) and to counteract activation of RhoA by Lbc RhoGEF (Soh and Low, 2008), respectively. Unlike the above scenarios where regulation occurs via interaction of two separate molecules, our current findings provide the first evidence that BCH domain acts as an important regulatory switch for the GAP function when present together on the same protein.
Our results show that upon removal of the BCH domain the GAP domain conferred much enhanced activity toward RhoA, although no RhoA was detectable in the immmunocomplex of GAP. This is attributed to their high turnover and transient nature of enzyme–substrate interaction, which is also generally observed in other reactions such as kinases and their substrates. Consistently, the active GAP promotes extensive cell rounding that could only be abrogated when the triple catalytic mutant was introduced or blocked by constitutive active RhoG14V. In contrast, the BCH domain did not affect the Rho activity when present alone despite its strong binding to RhoA, and no impact on the overall cell morphology was observed. However, it is worth noting that this BCH domain alone did suppress RhoA-induced cellular projections, leading to a more cuboidal structure that still retained intact stress fibers. These data imply that although NBCH domain does not directly affect the RhoA activity per se, it could still modulate the impact of RhoA signaling, probably by indirectly interfering with some of its downstream effectors. However, when BCH domain was present with the GAP domain, the binding of full-length p50RhoGAP with Rho became evident but the GAP activity toward Rho was significantly reduced. Consequently, the cells did not round up.
Interestingly, despite sharing the same preference for RhoA as the BCH domains of BNIP-Sα and BNIP-XL and the ability to interact with both GDP- or GTP-bound forms of Rho, the BCH domain of p50RhoGAP, when present alone, does not significantly affect cell morphology; however, when present with the GAP domain, it acts as an potent inhibitor of GAP in cis. This highlights the absolute requirement for the p50RhoGAP BCH to be present with the GAP in totality in specific microenvironment or/and requiring certain conformation in order to exert its impact on local pools of RhoA, thus further demonstrating the versatility of different BCH domains in regulating Rho signaling in various subcellular environments. Consistent with this, its dual ability to interact with GDP- or GTP-bound form of Rho would permit a dynamic cross-regulation such that capturing the GTP-bound Rho would ensure no inactivation by GAP, whereas capturing GDP-Rho could prevent activation by GEF. Depending on which and when such GAP and GEFs are being activated, the overall outcome would be different or would be neutralized. This could also explain why overexpression of BCH domain alone does not readily lead to any noticeable effects in cells. However, it remains unclear at the molecular level as to how the binding of BCH domain to Rho would be different from other Rho-binding modules such as GAP, GEF, and GDI as well as other effector molecules. It is also worth noting that besides p50RhoGAP and its homolog BPGAP1, several BCH-like domains are also present in cis with the GEF domain in Dbl, Duo, and Trio RhoGEFs and also in neurofibromatosis type 1 (NF1) RasGAP (Zhou and Low, unpublished data). It remains an exciting prospect to determine whether these BCH domains could act to modulate the local GTPases activity by a similar or distinct mechanism from that shown by p50RhoGAP.
How Could RhoA Sequestration Be Reversed?
Our data showed that the BCH domain of p50RhoGAP could serve as a local sink to sequester RhoA and subsequently prevent it from inactivation. This could explain why previous knockout (Wang et al., 2005, 2006) and our shRNA studies (Supplementary Figure S11) show no significant elevated level of stress fibers. Thus, it remain to be seen how upon relief of sequestration, the substrate could in turn become available for the adjacent GAP domain, and vice versa. Their unique profiles in recognizing the substrate might hold keys to understanding some of the complex nature of regulation. For example, could there be a dynamic conformational switch that allows transient transfer of the substrate between the two domains? How would the cycling of GTP/GDP-bound state of Rho determine whether they would be sequestered by BCH domain, released from there, and subjected to activation by GEF or inactivation by GAP? Although the ability of BCH domain to capture either the GDP-bound or GTP-bound form as discussed above could provide a dynamic dual regulation for RhoGAP as well as RhoGEF functions, it is possible that the exchange of nucleotides of RhoA might regulate the activity of p50RhoGAP. For example, p50RhoGAP could be inhibited when bound to GDP-RhoA, but upon RhoA activation by a nearby RhoGEF and facilitated by BCH domain, the RhoA is released to allow the catalytic activation of p50RhoGAP. This hypothesis was supported by our new finding that p50RhoGAP could indeed interact with p115RhoGEF through its BCH domain (Supplementary Figure S12). Because BCH does not directly increase active Rho level by itself and that BCH binds RhoA-GDP or RhoA-GTP equally well, we believe that this domain could serve as a scaffold that link p115RhoGEF to other unknown biological functions or/and possibly be involved in reversing RhoA sequestration, allowing Rho to be inactivated by the adjacent GAP domain. This adds to the versatility of BCH domain in regulating not just directly on Rho but also the RhoGAP and RhoGEF, with the latter also observed for another BCH-containing member, BNIP-XL that targets the DH-PH domain of Lbc RhoGEF (Soh and Low, 2008).
The fact that coexpression of individual BCH domain with full-length p50RhoGAP could not enhance cell rounding and GAP activity also suggests that homophilic interaction between BCH domains of p50RhoGAP does not inhibit its ability to sequester RhoA. On the other hand, findings that phospholipids have the ability to “switch” the GTPase substrate preference of p190RhoGAP provide an attractive possibility (Ligeti et al., 2004; Ligeti and Settleman, 2006). The limited degree of homology between the BCH domain and the Sec-14p domain that functions in phosphatidylinositol transfer (Mousley et al., 2007; Schaaf et al., 2008) suggests that the BCH domain of p50RhoGAP could possess lipid-binding property and p50RhoGAP activity could be modulated via a lipid-dependant mechanism. On the other hand, it was previously proposed that Rac could relieve the intramolecular interaction of p50RhoGAP, albeit in vitro, by binding to the extreme N terminus (Moskwa et al., 2005). Similarly, it binds and activates p190B RhoGAP in cells (Bustos et al., 2008). However, based on an IP study, we did not readily detect any Rac1-p50RhoGAP interaction in HeLa cells, raising the possibility that other forms of Rac (e.g., Rac2 and Rac3) or other closely related Rho GTPases could instead be involved in reversing the sequestration by displacing RhoA from the BCH domain. However, to achieve that, such GTPases must also interact with similar motif in the BCH domain. All these possibilities await further detailed investigation.
Concerted Regulatory Mechanisms for Modulating p50RhoGAP Activity
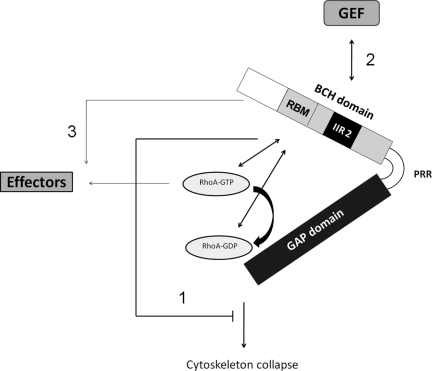
Intramolecular interaction plays critical roles in regulating Rho GTPase regulator proteins, including the RhoGEF (e.g., p115RhoGEF, Vav, proto-Dbl, and Ect2; Hart et al., 1998; Aghazadeh et al., 2000; Bi et al., 2001; Kim et al., 2005) and RhoGAP (e.g., 120RasGAP, CdGAP, oligophrenin-1, and GGAP; Drugan et al., 2000; Jenna et al., 2002; Fauchereau et al., 2003; Xia et al., 2003). In most cases, certain protein–protein interaction domains are critical for masking their catalytic domains (Jenna et al., 2002; Xia et al., 2003; Yohe et al., 2007). It has previously been reported that the N-terminus of p50RhoGAP, including part of the BCH domain (i.e., IIR 2), could mask the GAP activity via intramolecular interaction in vitro (Moskwa et al., 2005). Interestingly, the present study shows that mutants lacking either IIR 1 (outside of BCH) or 2 (within BCH) were still able to suppress the GAP function in cells. However, when the latter motif was removed together with the Rho-binding motif, the entire BCH domain became ineffective to suppress the GAP function. This demonstrated that maximal GAP inhibition by BCH domain required both RhoA sequestration as well as autoinhibition mediated by the intramolecular interaction (Figure 8). Uniquely, such regulation requires the domain being present in close proximity with the GAP domain in cis but not in trans because overexpression of BCH as a discrete entity delinked from the GAP domain does not prevent the GAP function (Figure 7C). This observation highlights the importance of concentrating the BCH domain with the GAP domain to control the local activity of Rho.
Figure 8.
Model depicting BCH domain as a local modulator to sequester RhoA from inactivation by the adjacent GAP domain and possibly as a scaffold that links the RhoGAP function to RhoGEF and other effectors. The GAP domain of p50RhoGAP inactivates RhoA and subsequently induces drastic cytoskeleton collapse and cell rounding. This cellular effect could be inhibited in cis by the N-terminal BCH domain, but not its proline-rich region (PRR), that acts via a concerted mechanism including the binding of the BCH domain to RhoA via its Rho-binding motif (RBM), thus displacing GAP domain from inhibiting RhoA, and the intramolecular interaction region 2 (IIR 2) that augments RhoA sequestration to confer complete suppression of the GAP activity (pathway 1). As the BCH domain displays distinctive Rho-binding profile from the GAP domain and is independent of the nucleotide-binding status, it could help ensure that the GTP-bound active form of Rho be effectively sequestered from inactivation by GAP. In addition, independent BCH domain could also serve as a scaffold that links RhoGEF (e.g., at least with p115RhoGEF shown in this study; pathway 2) or interfering with downstream signaling of RhoA without directly affecting Rho activity (e.g., abolishing periphery projections; pathway 3). However, how these multitude mechanisms operate, either in isolation or in concert for a dynamic regulation, awaits further investigation. See text for details.
In summary, we have provided several lines of evidence that the adjacent BCH domain on p50RhoGAP could serve as a local modulator to sequester and prevent RhoA from being inactivated by the GAP domain. Having both BCH and GAP domain in close proximity on p50RhoGAP could also ensure an efficient means for targeting and regulating local activity of RhoA. Our findings shed light to increasing evidence that BCH domain presents a dynamic functional module for GTPases signaling and therefore calling for further investigation on previously unappreciated roles of homologous BCH domains in other RhoGAPs or RhoGEFs, some of which are now underway.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Alan Hall (Memorial Sloan-Kettering Cancer Center) for the generous gifts of the original cDNAs for RhoA and p50RhoGAP, Dr. Amy L. Wilson-Delfosse (Case Western Reserve University School of Medicine) for the generous gift of the Rho-GDIα plasmid, Dr. Snezhana Oliferenko and Aleksandar Vjestica (Temasek Life Sciences Laboratory Limited, Singapore) for the S. pombe total cDNA, and Dr. Robert R. Krauss (Mount Sinai School of Medicine) for the pSilencer2.1-U6 hygro plasmids for p50RhoGAP. We also thank our reviewers for their critical suggestions. Y.T.Z is a Singapore Millennium Foundation Research Fellow. This work was supported in part by grants from the Biomedical Research Council of Singapore, Ministry of Education Singapore (Academic Research Fund T208A3121) and the Mechanobiology Institute, National University of Singapore to B.C.L., and also by the Project 111 (B06016) to S.C.L. from the State Bureau of Foreign Experts and Ministry of Education, China.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-05-0408) on July 21, 2010.
REFERENCES
- Aghazadeh B., Lowry W. E., Huang X. Y., Rosen M. K. Structural basis for relief of autoinhibition of the Dbl homology domain of proto-oncogene Vav by tyrosine phosphorylation. Cell. 2000;102:625–633. doi: 10.1016/s0092-8674(00)00085-4. [DOI] [PubMed] [Google Scholar]
- Barfod E. T., Zheng Y., Kuang W. J., Hart M. J., Evans T., Cerione R. A., Ashkenazi A. Cloning and expression of a human CDC42 GTPase-activating protein reveals a functional SH3-binding domain. J. Biol. Chem. 1993;268:26059–26062. [PubMed] [Google Scholar]
- Barrett T., Xiao B., Dodson E. J., Dodson G., Ludbrook S. B., Nurmahomed K., Gamblin S. J., Musacchio A., Smerdon S. J., Eccleston J. F. The structure of the GTPase-activating domain from p50rhoGAP. Nature. 1997;385:458–461. doi: 10.1038/385458a0. [DOI] [PubMed] [Google Scholar]
- Bernards A., Settleman J. GAPs in growth factor signalling. Growth Factors. 2005;23:143–149. doi: 10.1080/08977190500130480. [DOI] [PubMed] [Google Scholar]
- Bernards A., Settleman J. GEFs in growth factor signaling. Growth Factors. 2007;25:355–361. doi: 10.1080/08977190701830375. [DOI] [PubMed] [Google Scholar]
- Bi F., Debreceni B., Zhu K., Salani B., Eva A., Zheng Y. Autoinhibition mechanism of proto-Dbl. Mol. Cell. Biol. 2001;21:1463–1474. doi: 10.1128/MCB.21.5.1463-1474.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J. L., Rehmann H., Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Brill S., Li S., Lyman C. W., Church D. M., Wasmuth J. J., Weissbach L., Bernards A., Snijders A. J. The Ras GTPase-activating-protein-related human protein IQGAP2 harbors a potential actin binding domain and interacts with calmodulin and Rho family GTPases. Mol. Cell. Biol. 1996;16:4869–4878. doi: 10.1128/mcb.16.9.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschdorf J. P., Chew L. L., Zhang B., Cao Q., Liang F. Y., Liou Y. C., Zhou Y. T., Low B. C. Brain-specific BNIP-2-homology protein Caytaxin relocalises glutaminase to neurite terminals and reduces glutamate levels. J. Cell Sci. 2006;119:3337–3350. doi: 10.1242/jcs.03061. [DOI] [PubMed] [Google Scholar]
- Buschdorf J. P., Chew L. L., Soh U. J., Liou Y. C., Low B. C. Nerve growth factor stimulates interaction of Cayman ataxia protein BNIP-H/Caytaxin with peptidyl-prolyl isomerase Pin1 in differentiating neurons. PLoS ONE. 2008;3:e2686. doi: 10.1371/journal.pone.0002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos R. I., Forget M. A., Settleman J. E., Hansen S. H. Coordination of Rho and Rac GTPase function via p190B RhoGAP. Curr. Biol. 2008;18:1606–1611. doi: 10.1016/j.cub.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovas A., Couchman J. R. RhoGDI: multiple functions in the regulation of Rho family GTPase activities. Biochem. J. 2005;390:1–9. doi: 10.1042/BJ20050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drugan J. K., Rogers-Graham K., Gilmer T., Campbell S., Clark G. J. The Ras/p120 GTPase-activating protein (GAP) interaction is regulated by the p120 GAP pleckstrin homology domain. J. Biol. Chem. 2000;275:35021–35027. doi: 10.1074/jbc.M004386200. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- Fauchereau F., Herbrand U., Chafey P., Eberth A., Koulakoff A., Vinet M. C., Ahmadian M. R., Chelly J., Billuart P. The RhoGAP activity of OPHN1, a new F-actin-binding protein, is negatively controlled by its amino-terminal domain. Mol. Cell Neurosci. 2003;23:574–586. doi: 10.1016/s1044-7431(03)00078-2. [DOI] [PubMed] [Google Scholar]
- Gibson R. M., Gandhi P. N., Tong X., Miyoshi J., Takai Y., Konieczkowski M., Sedor J. R., Wilson-Delfosse A. L. An activating mutant of Rac1 that fails to interact with Rho GDP-dissociation inhibitor stimulates membrane ruffling in mammalian cells. Biochem. J. 2004;378:409–419. doi: 10.1042/BJ20030979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Hancock J. F., Hall A. A novel role for RhoGDI as an inhibitor of GAP proteins. EMBO J. 1993;12:1915–1921. doi: 10.1002/j.1460-2075.1993.tb05840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart M. J., Jiang X., Kozasa T., Roscoe W., Singer W. D., Gilman A. G., Sternweis P. C., Bollag G. Direct stimulation of the guanine nucleotide exchange activity of p115 RhoGEF by Galpha13. Science. 1998;280:2112–2114. doi: 10.1126/science.280.5372.2112. [DOI] [PubMed] [Google Scholar]
- Hatjiharissi E., et al. Proteomic analysis of waldenstrom macroglobulinemia. Cancer Res. 2007;67:3777–3784. doi: 10.1158/0008-5472.CAN-06-3089. [DOI] [PubMed] [Google Scholar]
- Jaffe A. B., Hall A. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Jenna S., Hussain N. K., Danek E. I., Triki I., Wasiak S., McPherson P. S., Lamarche-Vane N. The activity of the GTPase-activating protein CdGAP is regulated by the endocytic protein intersectin. J. Biol. Chem. 2002;277:6366–6373. doi: 10.1074/jbc.M105516200. [DOI] [PubMed] [Google Scholar]
- Johnstone C. N., Castellvi-Bel S., Chang L. M., Bessa X., Nakagawa H., Harada H., Sung R. K., Pique J. M., Castells A., Rustgi A. K. ARHGAP8 is a novel member of the RHOGAP family related to ARHGAP1/CDC42GAP/p50RHOGAP: mutation and expression analyses in colorectal and breast cancers. Gene. 2004;336:59–71. doi: 10.1016/j.gene.2004.01.025. [DOI] [PubMed] [Google Scholar]
- Kang J. S., Bae G. U., Yi M. J., Yang Y. J., Oh J. E., Takaesu G., Zhou Y. T., Low B. C., Krauss R. S. A Cdo-Bnip-2-Cdc42 signaling pathway regulates p38alpha/beta MAPK activity and myogenic differentiation. J. Cell Biol. 2008;182:497–507. doi: 10.1083/jcb.200801119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. E., Billadeau D. D., Chen J. The tandem BRCT domains of Ect2 are required for both negative and positive regulation of Ect2 in cytokinesis. J. Biol. Chem. 2005;280:5733–5739. doi: 10.1074/jbc.M409298200. [DOI] [PubMed] [Google Scholar]
- Lancaster C. A., Taylor-Harris P. M., Self A. J., Brill S., van Erp H. E., Hall A. Characterization of rhoGAP. A GTPase-activating protein for rho-related small GTPases. J. Biol. Chem. 1994;269:1137–1142. [PubMed] [Google Scholar]
- Leonard D. A., Lin R., Cerione R. A., Manor D. Biochemical studies of the mechanism of action of the Cdc42-GTPase-activating protein. J. Biol. Chem. 1998;273:16210–16215. doi: 10.1074/jbc.273.26.16210. [DOI] [PubMed] [Google Scholar]
- Ligeti E., Settleman J. Regulation of RhoGAP specificity by phospholipids and prenylation. Methods Enzymol. 2006;406:104–117. doi: 10.1016/S0076-6879(06)06009-5. [DOI] [PubMed] [Google Scholar]
- Ligeti E., Dagher M. C., Hernandez S. E., Koleske A. J., Settleman J. Phospholipids can switch the GTPase substrate preference of a GTPase-activating protein. J. Biol. Chem. 2004;279:5055–5508. doi: 10.1074/jbc.C300547200. [DOI] [PubMed] [Google Scholar]
- Low B. C., Lim Y. P., Lim J., Wong E. S., Guy G. R. Tyrosine phosphorylation of the Bcl-2-associated protein BNIP-2 by fibroblast growth factor receptor-1 prevents its binding to Cdc42GAP and Cdc42. J. Biol. Chem. 1999;274:33123–33130. doi: 10.1074/jbc.274.46.33123. [DOI] [PubMed] [Google Scholar]
- Low B. C., Seow K. T., Guy G. R. The BNIP-2 and Cdc42GAP homology domain of BNIP-2 mediates its homophilic association and heterophilic interaction with Cdc42GAP. J. Biol. Chem. 2000;275:37742–37751. doi: 10.1074/jbc.M004897200. [DOI] [PubMed] [Google Scholar]
- Lua B. L., Low B. C. Activation of EGF receptor endocytosis and ERK1/2 signaling by BPGAP1 requires direct interaction with EEN/endophilin II and a functional RhoGAP domain. J. Cell Sci. 2005;118:2707–2721. doi: 10.1242/jcs.02383. [DOI] [PubMed] [Google Scholar]
- Moon S. Y., Zang H., Zheng Y. Characterization of a brain-specific Rho GTPase-activating protein, p200RhoGAP. J. Biol. Chem. 2003;278:4151–4159. doi: 10.1074/jbc.M207789200. [DOI] [PubMed] [Google Scholar]
- Moon S. Y., Zheng Y. Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 2003;13:13–22. doi: 10.1016/s0962-8924(02)00004-1. [DOI] [PubMed] [Google Scholar]
- Moskwa P., Paclet M. H., Dagher M. C., Ligeti E. Autoinhibition of p50 Rho GTPase-activating protein (GAP) is released by prenylated small GTPases. J. Biol. Chem. 2005;280:6716–6720. doi: 10.1074/jbc.M412563200. [DOI] [PubMed] [Google Scholar]
- Mousley C. J., Tyeryar K. R., Vincent-Pope P., Bankaitis V. A. The Sec14-superfamily and the regulatory interface between phospholipid metabolism and membrane trafficking. Biochim. Biophys. Acta. 2007;1771:727–736. doi: 10.1016/j.bbalip.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munter S., Way M., Frischknecht F. Signaling during pathogen infection. Sci. STKE. 2006;2006:re5. doi: 10.1126/stke.3352006re5. [DOI] [PubMed] [Google Scholar]
- Nassar N., Hoffman G. R., Manor D., Clardy J. C., Cerione R. A. Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP. Nat. Struct. Biol. 1998;5:1047–1052. doi: 10.1038/4156. [DOI] [PubMed] [Google Scholar]
- Nomanbhoy T. K., Cerione R. A. Characterization of the interaction between RhoGDI and Cdc42Hs using fluorescence spectroscopy. J. Biol. Chem. 1996;271:10004–10009. doi: 10.1074/jbc.271.17.10004. [DOI] [PubMed] [Google Scholar]
- Ridley A. J., Self A. J., Kasmi F., Paterson H. F., Hall A., Marshall C. J., Ellis C. rho family GTPase activating proteins p190, bcr and rhoGAP show distinct specificities in vitro and in vivo. EMBO J. 1993;12:5151–5160. doi: 10.1002/j.1460-2075.1993.tb06210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittinger K., Walker P. A., Eccleston J. F., Smerdon S. J., Gamblin S. J. Structure at 1.65 A of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature. 1997;389:758–762. doi: 10.1038/39651. [DOI] [PubMed] [Google Scholar]
- Sahai E., Marshall C. J. RHO-GTPases and cancer. Nat. Rev. Cancer. 2002;2:133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- Schaaf G., et al. Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily. Mol. Cell. 2008;29:191–206. doi: 10.1016/j.molcel.2007.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang X., Zhou Y. T., Low B. C. Concerted regulation of cell dynamics by BNIP-2 and Cdc42GAP homology/Sec14p-like, proline-rich, and GTPase-activating protein domains of a novel Rho GTPase-activating protein, BPGAP1. J. Biol. Chem. 2003;278:45903–45914. doi: 10.1074/jbc.M304514200. [DOI] [PubMed] [Google Scholar]
- Shen Y., et al. Nudel binds Cdc42GAP to modulate Cdc42 activity at the leading edge of migrating cells. Dev. Cell. 2008;14:342–353. doi: 10.1016/j.devcel.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Sirokmány G., Szidonya L., Kaldi K., Gaborik Z., Ligeti E., Geiszt M. Sec14 homology domain targets p50RhoGAP to endosomes and provides a link between Rab and Rho GTPases. J. Biol. Chem. 2006;281:6096–6105. doi: 10.1074/jbc.M510619200. [DOI] [PubMed] [Google Scholar]
- Soh U. J., Low B. C. BNIP2 extra long inhibits RhoA and cellular transformation by Lbc RhoGEF via its BCH domain. J. Cell Sci. 2008;121:1739–1749. doi: 10.1242/jcs.021774. [DOI] [PubMed] [Google Scholar]
- Song J. Y., Lee J. K., Lee N. W., Jung H. H., Kim S. H., Lee K. W. Microarray analysis of normal cervix, carcinoma in situ, and invasive cervical cancer: identification of candidate genes in pathogenesis of invasion in cervical cancer. Int. J. Gynecol. Cancer. 2008;18:1051–1059. doi: 10.1111/j.1525-1438.2007.01164.x. [DOI] [PubMed] [Google Scholar]
- Szczur K., Xu H., Atkinson S., Zheng Y., Filippi M. D. Rho GTPase CDC42 regulates directionality and random movement via distinct MAPK pathways in neutrophils. Blood. 2006;108:4205–4213. doi: 10.1182/blood-2006-03-013789. [DOI] [PubMed] [Google Scholar]
- Tcherkezian J., Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biol. Cell. 2007;99:67–86. doi: 10.1042/BC20060086. [DOI] [PubMed] [Google Scholar]
- Tcherkezian J., Triki I., Stenne R., Danek E. I., Lamarche-Vane N. The human orthologue of CdGAP is a phosphoprotein and a GTPase-activating protein for Cdc42 and Rac1 but not RhoA. Biol. Cell. 2006;98:445–456. doi: 10.1042/BC20050101. [DOI] [PubMed] [Google Scholar]
- Vega F. M., Ridley A. J. Rho GTPases in cancer cell biology. FEBS Lett. 2008;582:2093–2101. doi: 10.1016/j.febslet.2008.04.039. [DOI] [PubMed] [Google Scholar]
- Wang L., Yang L., Burns K., Kuan C. Y., Zheng Y. Cdc42GAP regulates c-Jun N-terminal kinase (JNK)-mediated apoptosis and cell number during mammalian perinatal growth. Proc. Natl. Acad. Sci. USA. 2005;102:13484–13489. doi: 10.1073/pnas.0504420102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Yang L., Debidda M., Witte D., Zheng Y. Cdc42 GTPase-activating protein deficiency promotes genomic instability and premature aging-like phenotypes. Proc. Natl. Acad. Sci. USA. 2007;104:1248–1253. doi: 10.1073/pnas.0609149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Yang L., Filippi M. D., Williams D. A., Zheng Y. Genetic deletion of Cdc42GAP reveals a role of Cdc42 in erythropoiesis and hematopoietic stem/progenitor cell survival, adhesion, and engraftment. Blood. 2006;107:98–105. doi: 10.1182/blood-2005-05-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia C., Ma W., Stafford L. J., Liu C., Gong L., Martin J. F., Liu M. GGAPs, a new family of bifunctional GTP-binding and GTPase-activating proteins. Mol. Cell. Biol. 2003;23:2476–2488. doi: 10.1128/MCB.23.7.2476-2488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Hortsman H., Seet L., Wong S. H., Hong W. SNX3 regulates endosomal function through its PX-domain-mediated interaction with PtdIns(3)P. Nat. Cell Biol. 2001;3:658–666. doi: 10.1038/35083051. [DOI] [PubMed] [Google Scholar]
- Yohe M. E., Rossman K. L., Gardner O. S., Karnoub A. E., Snyder J. T., Gershburg S., Graves L. M., Der C. J., Sondek J. Auto-inhibition of the Dbl family protein Tim by an N-terminal helical motif. J. Biol. Chem. 2007;282:13813–13823. doi: 10.1074/jbc.M700185200. [DOI] [PubMed] [Google Scholar]
- Zalcman G., Closson V., Camonis J., Honore N., Rousseau-Merck M. F., Tavitian A., Olofsson B. RhoGDI-3 is a new GDP dissociation inhibitor (GDI): identification of a non-cytosolic GDI protein interacting with the small GTP-binding proteins RhoB and RhoG. J. Biol. Chem. 1996;271:30366–30374. doi: 10.1074/jbc.271.48.30366. [DOI] [PubMed] [Google Scholar]
- Zhou Y. T., Soh U. J., Shang X., Guy G. R., Low B. C. The BNIP-2 and Cdc42GAP homology/Sec14p-like domain of BNIP-Sα is a novel apoptosis-inducing sequence. J. Biol. Chem. 2002;277:7483–7492. doi: 10.1074/jbc.M109459200. [DOI] [PubMed] [Google Scholar]
- Zhou Y. T., Guy G. R., Low B. C. BNIP-2 induces cell elongation and membrane protrusions by interacting with Cdc42 via a unique Cdc42-binding motif within its BNIP-2 and Cdc42GAP homology domain. Exp. Cell Res. 2005;303:263–274. doi: 10.1016/j.yexcr.2004.08.044. [DOI] [PubMed] [Google Scholar]
- Zhou Y. T., Guy G. R., Low B. C. BNIP-Salpha induces cell rounding and apoptosis by displacing p50RhoGAP and facilitating RhoA activation via its unique motifs in the BNIP-2 and Cdc42GAP homology domain. Oncogene. 2006;25:2393–2408. doi: 10.1038/sj.onc.1209274. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.