Overexpression of Rheb activates mTOR signaling via a PI3K/PKB-independent mechanism and is sufficient to induce skeletal muscle hypertrophy. The hypertrophic effects of Rheb are driven through a rapamycin-sensitive (RS) mechanism, mTOR is the RS element that confers the hypertrophy and the kinase activity of mTOR is necessary for this event.
Abstract
It has been widely proposed that signaling by mammalian target of rapamycin (mTOR) is both necessary and sufficient for the induction of skeletal muscle hypertrophy. Evidence for this hypothesis is largely based on studies that used stimuli that activate mTOR via a phosphatidylinositol 3-kinase (PI3K)/protein kinase B (PKB)-dependent mechanism. However, the stimulation of signaling by PI3K/PKB also can activate several mTOR-independent growth-promoting events; thus, it is not clear whether signaling by mTOR is permissive, or sufficient, for the induction of hypertrophy. Furthermore, the presumed role of mTOR in hypertrophy is derived from studies that used rapamycin to inhibit mTOR; yet, there is very little direct evidence that mTOR is the rapamycin-sensitive element that confers the hypertrophic response. In this study, we determined that, in skeletal muscle, overexpression of Rheb stimulates a PI3K/PKB-independent activation of mTOR signaling, and this is sufficient for the induction of a rapamycin-sensitive hypertrophic response. Transgenic mice with muscle specific expression of various mTOR mutants also were used to demonstrate that mTOR is the rapamycin-sensitive element that conferred the hypertrophic response and that the kinase activity of mTOR is necessary for this event. Combined, these results provide direct genetic evidence that a PI3K/PKB-independent activation of mTOR signaling is sufficient to induce hypertrophy. In summary, overexpression of Rheb activates mTOR signaling via a PI3K/PKB-independent mechanism and is sufficient to induce skeletal muscle hypertrophy. The hypertrophic effects of Rheb are driven through a rapamycin-sensitive (RS) mechanism, mTOR is the RS element that confers the hypertrophy, and the kinase activity of mTOR is necessary for this event.
INTRODUCTION
Skeletal muscles make up 40–50% of the body's mass, and they are not only the motors that drive locomotion, but they also play a crucial role in whole body metabolism (Lee et al., 2000; Izumiya et al., 2008). Accordingly, it has been well recognized that the maintenance of skeletal muscle mass contributes significantly to disease prevention and issues associated with the quality of life (Seguin and Nelson, 2003).
Skeletal muscle mass is known to be regulated by a variety of different stimuli, including mechanical loads, neural activity, cytokines, growth factors, and hormones; however, the molecular mechanisms that regulate these changes remain poorly defined (Frost and Lang, 2007; Sandri, 2008). Nevertheless, our knowledge of these mechanisms is advancing, and recent studies have revealed that adaptive changes in muscle mass are often correlated with the activity of a protein kinase called the mammalian target of rapamycin (mTOR). For example, hypertrophy resulting from increased mechanical loading and β2-andrenergic agonists is associated with an increase in mTOR signaling, whereas atrophy resulting from disuse, glucocorticoids, and food deprivation is associated with a decrease in mTOR signaling (Wu et al.; Baar and Esser, 1999; Hornberger et al., 2001; Lewis et al., 2006; Kline et al., 2007).
Several studies have not only shown that mTOR signaling is correlated with changes in muscle mass, but in many cases, it also has been suggested that signaling through mTOR is necessary for these changes to occur. For example, hypertrophy induced by mechanical loading, insulin-like growth factor 1 (IGF-1) and constitutively active protein kinase B (c.a.-PKB) has been shown to be significantly, if not completely, blocked by the mTOR inhibitor rapamycin (Bodine et al., 2001; Park et al., 2005; Sandri, 2008). Because rapamycin is considered to be a specific inhibitor of mTOR, many have concluded that mTOR is the rapamycin-sensitive element that confers the hypertrophic effects of these stimuli (Davies et al., 2000; Bodine, 2006; Nader, 2007). However, like most pharmacological inhibitors, rapamycin can exert nonspecific (mTOR-independent) actions. For example, rapamycin can bind and sequester the FKBP12 protein, and the FKBP12 protein has been shown to play an important role in the function of the ryanodine receptor and signaling by members of the transforming growth factor-β superfamily (e.g., myostatin) (Avila et al., 2003; Wang and Donahoe, 2004; Osman et al., 2009). Furthermore, systemic administration of rapamycin would be expected to inhibit mTOR signaling in all cells throughout the body, and therefore, it is not clear if the anti-hypertrophic effects of rapamycin are due specifically to inhibition of mTOR signaling in skeletal muscle cells. Hence, it can be argued that there is strong evidence to support the conclusion that signaling through a rapamycin-sensitive mechanism is necessary for the hypertrophic effects of stimuli such as IGF-1, c.a.-PKB, and mechanical loads, but there is very little direct evidence that mTOR is the rapamycin-sensitive element, in skeletal muscle, that confers the hypertrophic response (Park et al., 2005).
As mentioned, mTOR is a protein kinase, and mTOR has been shown to phosphorylate a variety of molecules implicated in the regulation of muscle mass. The specific substrates that mTOR phosphorylates depends on whether mTOR is bound to the proteins raptor or rictor (Efeyan and Sabatini, 2010). When bound to raptor, mTOR forms a rapamycin-sensitive signaling complex called mTORC1, and signaling by mTORC1 has been reported to be directly activated by the Ras homologue enriched in brain (Rheb) (Yang et al., 2006; Sato et al., 2009). When activated by Rheb, mTORC1 can phosphorylate substrates such as the threonine 389 residue of the ribosomal S6 kinase (p70S6k) (Yang et al., 2006). In contrast, when mTOR is bound to rictor it forms the mTORC2 complex, and mTORC2 phosphorylates distinct substrates from that of mTORC1 (Efeyan and Sabatini, 2010). One of the most commonly recognized mTORC2 substrates is the serine 473 residue of protein kinase B (PKB), and unlike signaling by mTORC1, signaling by the mTORC2 is not activated by Rheb or inhibited by rapamycin (Hresko and Mueckler, 2005; Sarbassov et al., 2005; Efeyan and Sabatini, 2010). Thus, if mTOR is the rapamycin-sensitive element that confers the hypertrophic effects of stimuli such as IGF-1, c.a.-PKB, and mechanical loads, then this is probably due to signaling by mTOR when it is in the mTORC1 complex.
During the past two decades it has become clear that very low concentrations of rapamycin (2–5 nM) block mTORs ability to phosphorylate mTORC1 substrates such as p70S6k (Price et al., 1992). Consequently, it has been widely assumed that rapamycin exerts its growth inhibitory actions by blocking the ability of mTOR to phosphorylate downstream substrates (i.e., mTOR regulates growth through a kinase-dependent mechanism). However, even moderately high concentrations of rapamycin (1 μM) do not inhibit mTOR's intrinsic kinase activity (Isotani et al., 1999; Oshiro et al., 2004). Hence, a rapamycin-sensitive event does not necessarily imply an mTOR kinase dependent event. For example, rapamycin has been shown to inhibit myogenesis, and expression of either a rapamycin-resistant mutant of mTOR (RR-mTOR) or rapamycin-resistant kinase-dead mutant of mTOR (RRKD-mTOR) can rescue myogenesis from the inhibitory effects of rapamycin (Erbay and Chen, 2001). In other words, myogenesis occurs through a rapamycin-sensitive mechanism and mTOR is the rapamycin-sensitive element that confers myogenesis, but surprisingly, mTOR kinase activity is not required for this event. This important observation demonstrates that caution should be exercised before assuming that the growth inhibitory effects of rapamycin are due to inhibition of mTOR's ability to phosphorylate downstream substrates. To date, we are not aware of any in vivo studies that have determined whether mTOR kinase activity is necessary for the hypertrophic effects of stimuli such as IGF-1, c.a.-PKB and mechanical loads; thus, the role of mTOR kinase activity in these events remains to be defined.
Signaling through mTOR has not only been implicated as being necessary but also has been implicated as being sufficient for the induction of skeletal muscle hypertrophy. For example, IGF-1 has been shown to induce mTOR signaling through a pathway involving phosphatidylinositol 3-kinase (PI3K) and protein kinase B (PKB) [IGF-1 → PI3K/PKB → mTOR] (Rommel et al., 2001; Laviola et al., 2007). IGF-1 also has been shown to induce myotube hypertrophy, and this effect is significantly blocked by rapamycin (Rommel et al., 2001; Park et al., 2005). Furthermore, overexpression of c.a.-PKB activates mTOR signaling and induces muscle fiber hypertrophy through a rapamycin-sensitive mechanism (Bodine et al., 2001; Pallafacchina et al., 2002; Izumiya et al., 2008). Together, these observations have led many to conclude that the activation of mTOR signaling is sufficient to induce hypertrophy (Bodine et al., 2001; Nader, 2007). However, recent studies have shown that the activation of PI3K/PKB by IGF-1 and c.a.-PKB also can affect a variety of mTOR-independent growth regulatory molecules such as the glycogen synthase kinase 3β, tuberin, and the FOXO transcription factors (Frost and Lang, 2007; Sandri, 2008; Hamilton et al., 2009). Hence, it is very possible that signaling by mTOR is permissive, rather than sufficient, for the induction of hypertrophy. In fact, the activation of mTOR signaling can induce a negative feedback loop that impairs signaling by PKB, and based on this point, it has recently been argued that a PI3K/PKB-independent activation of mTOR may not be sufficient for the induction of skeletal muscle hypertrophy (Hamilton et al., 2009). Thus, the overall goal of this study was to determine whether a PI3K/PKB-independent activation of mTOR signaling, in skeletal muscle, is sufficient to induce hypertrophy, and determine whether mTOR kinase activity is necessary for this event.
MATERIALS AND METHODS
Materials
Insulin (Humalog) was purchased from Eli Lilly (Indianapolis, IN). Rabbit anti-laminin antibodies and wortmannin were purchased from Sigma-Aldrich (St. Louis, MO). Anti-total p70S6k, anti-green fluorescent protein (GFP), anti-total mTOR, anti-phospho-mTOR(2481), anti-total PKB, anti-phospho-PKB(473), anti-phospho-PKB(308), anti-Rheb, anti-total S6, and anti-phospho-S6(Ser235/236) antibodies were purchased from Cell Signaling Technology (Danvers, MA). Anti-phospho-p70S6k(389), tetramethylrhodamine B isothiocyanate (TRITC)-conjugated anti-rabbit, and fluorescein isothiocyanate (FITC)-conjugated anti-rat antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Peroxidase-conjugated anti-rabbit and peroxidase-conjugated anti-rat antibodies were purchased from Vector Laboratories (Burlingame, CA). Rat anti-hemagglutinin (HA) antibodies were purchased from Roche (Madison, WI).
Plasmid Constructs and Purification
pEGFP-C3 (GFP) was purchased from Clontech (Mountain View, CA). pRK5-myc-p70S6K-glutathione transferase (GST p70S6k) has been described previously (Miyazaki and Esser, 2009) and was kindly provided by Dr. Karyn Esser (University of Kentucky, Lexington, KY). pCDNA3-HA-Rheb (HA-Rheb) has been described previously (Li et al., 2004) and was a generous gift from Dr. Kun-Liang Guan (University of California–San Diego, La Jolla, CA). The dual-luciferase bicistronic reporter of cap-dependent translation has been described previously (Carter and Sarnow, 2000) and was kindly provided by Dr. Sunnie Thompson (University of Alabama, Birmingham, AL). All plasmid DNA was grown in DH5α Escherichia coli, purified with an Endofree plasmid kit (QIAGEN, Valencia, CA), and resuspended in sterile phosphate-buffered saline (PBS).
Cell Culture and Transfection
C2C12 myoblasts were cultured in growth media consisting of high glucose DMEM (Hyclone Laboratories, Logan, UT) supplemented with antibiotics and antimycotics (100 U/ml penicillin G, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin) and 10% fetal bovine serum (Invitrogen, Carlsbad, CA). Lipofectamine 2000 (Invitrogen) was used to transfect 425,000 suspended C2C12 myoblasts with 4 μg of GFP or 4 μg of HA-Rheb, with or without 0.2 μg of GST p70S6k. The transfected myoblasts were plated on one well of a six-well dish and grown for 48 h in antibiotic/antimycotic-free growth media. After 48 h, the myoblasts were switched to serum-free high-glucose DMEM for 2 h before being subjected to experimental treatments. All cell culture experiments were performed in a humidified 95% air, 5% CO2 incubator at 37°C.
Animals
All animal experiments in this study followed protocols approved by the Animal Care and Use Committee at the University of Wisconsin–Madison. FVB mice with human skeletal actin driven expression of a FLAG-tagged rapamycin-resistant (Ser2035Thr) mutant of mTOR (RR-mTOR) or a FLAG tagged rapamycin-resistant kinase dead (Ser2035Thr/Asp2357Glu) mutant of mTOR (RRKD-mTOR) have been described previously (Ge et al., 2009). These transgenic mice were bred with wild-type FVB mice and the transgenic offspring were maintained as hemizygotes. Genotypes were confirmed with tail snips by PCR, and 8- to 10-wk-old female offspring were used.
Skeletal Muscle Transfection (Electroporation)
Mice were anesthetized with 100 mg/kg ketamine plus 10 mg/kg xylazine, and a small incision was made through the skin covering the tibialis anterior (TA) muscle. A 27-gauge needle was used to inject plasmid DNA solution (2.5 μg/μl GFP or HA-Rheb, with or without 0.17 μg/μl GST p70S6k) into the proximal (6 μl) and distal (6 μl) ends of the muscle belly. After the injections, electric pulses were applied through two stainless steel pin electrodes (1-cm gap; Harvard Apparatus, Holliston, MA) laid on top of the proximal and distal myotendinous junctions. Eight 20-ms square-wave electric pulses at a frequency of 1 Hz were delivered with an ECM 830 electroporation unit (BTX; Harvard Apparatus) at a field strength of 180 V/cm. After the electroporation procedure, the incision was closed with Vetbond surgical glue (Henry Schein, Melville, NY).
Rapamycin Injections
Rapamycin was purchased from LC laboratories (Woburn, MA) and was dissolved in dimethyl sulfoxide (DMSO) to generate a 5 μg/μl stock solution. The appropriate volume of the stock solution needed to inject mice with 1 mg/kg body weight was dissolved in 200 μl of PBS. For the vehicle control condition, mice were injected with an equivalent amount of DMSO dissolved in 200 μl of PBS. Immediately after the electroporation procedure, vehicle or rapamycin solutions were administered via intraperitoneal injections, and these injections were repeated every 24 h for up to 7 d.
Sample Preparation for Immunoprecipitations and Western Blot Analysis
On collection, skeletal muscles were immediately frozen in liquid nitrogen. Frozen muscles were homogenized with a Polytron homogenizer for 20 s in ice-cold buffer A (40 mM Tris, pH 7.5, 1 mM EDTA, 5 mM EGTA, 0.5% Triton X-100, 25 mM β-glycerophosphate, 25 mM NaF, 1 mM Na3VO4, 10 μg/ml leupeptin, and 1 mM phenylmethylsulfonyl fluoride), and the whole homogenate was used for further analysis. For cell culture experiments, myoblasts were lysed in the ice cold buffer A, centrifuged at 500 × g for 5 min, and the supernatant was used for further analysis. Sample protein concentration was determined with a DC protein assay kit (Bio-Rad Laboratories, Hercules, CA), and unless otherwise noted (e.g., immunoprecipitation), equivalent amounts of protein from each sample were dissolved in Laemmli buffer and subjected to Western blot analysis as described below.
Immunoprecipitation
Immunoprecipitation of FLAG-tagged mTOR was performed as described previously (Hornberger et al., 2007). In brief, whole muscle homogenates were centrifuged at 8200 × g for 10 min, and 500 μg of protein from the supernatant was diluted to a volume of 1 ml with fresh ice-cold buffer A. This sample was then incubated with 40 μl of EZview red ANTI-FLAG M2 agarose affinity gel beads (Sigma-Aldrich) with gentle rocking at 4°C for 2 h. After the incubation, the beads were pelleted by centrifugation at 8200 × g for 30 s and washed with fresh ice-cold buffer A. After four washes, the pellets were dissolved in 2× Laemmli buffer containing no dithiothreitol and heated to 100°C for 5 min. The beads were again pelleted by centrifugation at 8200 × g for 30 s, and the supernatant was subjected to Western blot analysis as described below.
Western Blot Analysis
Western blot analyses were performed as described previously (O'Neil et al., 2009). In brief, samples were subjected to electrophoretic separation on SDS-PAGE acrylamide gels. After electrophoretic separation, proteins were transferred to a polyvinylidene difluoride membrane, blocked with 5% powdered milk in Tris-buffered saline and 1% Tween 20 (TBST) for 1 h, and then incubated overnight at 4°C with primary antibody dissolved in TBST containing 1% bovine serum albumin (BSA). After an overnight incubation, the membranes were washed for 30 min in TBST and then probed with a peroxidase-conjugated secondary antibody for 1 h at room temperature. After 30 min of washing in TBST, the blots were developed on film using regular enhanced chemiluminescence (ECL) reagent (Pierce Chemical, Rockford, IL) or ECL Plus reagent (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). Once the appropriate image was captured, the membranes were stained with Coomassie Blue to verify equal loading in all lanes. Densitometric measurements were carried out using the public domain NIH Image program (ImageJ) developed at the National Institutes of Health and available at http://rsb.info.nih.gov/nih-image/.
Measurement of Cap-dependent Translation
TA muscles were cotransfected with 30 μg of plasmid DNA encoding either HA-Rheb or GFP and 5 μg of plasmid DNA encoding a dual-luciferase bicistronic reporter of cap-dependent translation. At 48 h after transfection, TA muscles were collected, frozen in liquid nitrogen, and stored at −80°C until further analysis. For this analysis, muscles were homogenized with a Polytron homogenizer in passive lysis buffer (Promega, Madison, WI), and Renilla and firefly luciferase activity in 25 μg of sample protein was measured with a FLUOstar Optima luminometer (BMG Labtech, Durham, NC) by using the Dual-Luciferase Reporter Assay kit (Promega) as described in the manufacturer's instructions.
Immunohistochemical Analysis
Muscles were excised and placed at resting length in ice-cold PBS containing 4% paraformaldehyde. The samples were gently rocked in this solution at 4°C for 30 min and then submerged in optimal cutting temperature compound (Tissue-Tek, Sakura, Torrance, CA) and frozen in liquid nitrogen-chilled isopentane. Cross sections (10 μm in thickness) from the midbelly of the muscle were obtained with a cryostat and fixed in −20°C acetone for 10 min. Sections were warmed to room temperature for 5 min and then rehydrated with cool steam vapors. Under gentle rocking, the rehydrated sections were incubated in PBS for 15min followed by a 20-min incubation in a solution B (PBS containing 0.5% BSA and 0.5% Triton X-100). Sections were then incubated with the indicated primary antibodies dissolved in solution B for 1 h at room temperature. Sections were washed with PBS and then incubated with the appropriate fluorophore-conjugated secondary antibodies dissolved in solution B for 1 h at room temperature. Finally, the sections were washed with PBS and mounted with Vectashield mounting media (Vector Laboratories, Burlingame, CA) and a coverslip. Transfected fibers (HA or GFP positive) and laminin, phospho-S6, or total S6 were identified in dual-fluorescent images and captured with a DS-QiMc camera on an 80i epi-fluorescence microscope (both from Nikon, Tokyo, Japan) with FITC and TRITC cubes, respectively. The monochrome images were merged with NIS Elements D image analysis software (Nikon), and the staining intensity or cross-sectional area (CSA) of 35–100 randomly selected transfected and nontransfected fibers per sample was measured by tracing the periphery of individual fibers. All analyses were performed by investigators that were unaware of the sample identification.
Statistical Analysis
All values are expressed as means + SEM. Statistical significance was determined by using analysis of variance, followed by Student–Newman–Keuls post hoc analysis or Dunn's procedure for planned comparisons. Differences between groups were considered significant if p ≤ 0.05. All statistical analyses were performed on SigmaStat software (Systat Software, San Jose, CA).
RESULTS
Overexpression of Rheb Induces mTOR Signaling through a PI3K-independent Mechanism in Skeletal Muscle Myoblasts
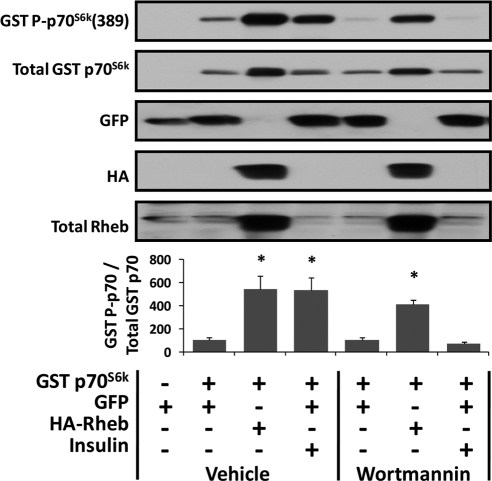
To accomplish the goal of this study, overexpression of Rheb was used as a means to induce a PI3K/PKB-independent activation of mTOR signaling. Rheb was selected for this study because previous reports have consistently shown Rheb to be one of the most proximal activators of mTOR signaling (Sun et al., 2008; Avruch et al., 2009). Furthermore, studies in nonskeletal muscle cells have shown that Rheb activates mTOR through a PI3K/PKB-independent mechanism (Garami et al., 2003; Tee et al., 2003). To confirm that this mechanism is conserved in skeletal muscle cells, C2C12 myoblasts were cotransfected with Rheb and GST-tagged p70S6k (GST p70S6k) or GFP and GST p70S6k as a control condition. At 48 h after transfection, the samples were collected and changes in the phosphorylation of GST p70S6k on the threonine 389 residue [GST p70S6k(389)] were evaluated as a marker of mTOR signaling (Hornberger et al., 2007). The results indicated that overexpression of Rheb induces a significant increase in mTOR signaling in skeletal muscle myoblasts (Figure 1).
Figure 1.
Overexpression of Rheb induces mTOR signaling through a PI3K-independent mechanism in skeletal muscle myoblasts. C2C12 myoblasts were transfected with GFP or cotransfected with a combination of GFP and GST-tagged p70S6k (GST p70S6k) or HA-Rheb and GST p70S6k. At 48 h after transfection, the myoblasts were switched to serum-free media for 2 h and then incubated with 500 nM wortmannin or the solvent vehicle (DMSO) for an additional 25 min. Where indicated, the myoblasts were stimulated with 100 nM insulin during the final 10 min of this incubation period. Cell lysates were subjected to Western blot analysis with the indicated antibodies. The ratio of GST P-p70S6K(389) to total GST p70S6K (GST P-p70/Total GST p70) was calculated and is expressed as a percentage of the drug treatment control samples (vehicle, GFP + GST p70S6K or wortmannin, GFP + GST p70S6K). Values are the mean + SEM, n = 4–6/group. *, significantly different from drug treatment control.
To determine whether the Rheb induced mTOR signaling through a PI3K-dependent mechanism, the PI3K inhibitor wortmannin was used. As a positive control from signaling through PI3K, myoblasts were stimulated with insulin in the presence or absence of wortmannin. Numerous studies have shown that insulin activates mTOR signaling through a PI3K-dependent mechanism (Avruch et al., 2006). Consistent with these studies, it was determined that wortmannin completely blocked the insulin-induced increase in mTOR signaling (Figure 1; Avruch et al., 2006). This observation verified that wortmannin successfully inhibited PI3K-dependent signaling; however, wortmannin did not inhibit the Rheb-induced increase in mTOR signaling (Figure 1). Together, these results indicate that overexpression of Rheb induces mTOR signaling through a PI3K-independent mechanism in C2C12 myoblasts.
Overexpression of Rheb Induces mTOR Signaling through a PI3K/PKB-independent Mechanism in Skeletal Muscle In Vivo
To determine whether the overexpression of Rheb could induce mTOR signaling in adult skeletal muscle in vivo, mouse TA muscles were transfected with Rheb, or GFP as a control condition, and the phosphorylation of the ribosomal S6 (S6) protein on the Ser235/236 residues (P-S6) was evaluated as a marker of mTOR signaling. As shown in Figure 2, Rheb-transfected fibers revealed a significantly greater amount of P-S6 compared with GFP-transfected fibers (Figure 2, A–G). It also was determined that Rheb-transfected fibers had a slightly, yet significantly, greater amount of total S6 protein compared with GFP-transfected fibers (Figure 2, H–N).
Figure 2.
Overexpression of Rheb induces an increase in the phosphorylation and total amount of the ribosomal S6 protein in vivo. Mouse TA muscles were transfected with GFP (A–C and H–J) or HA-Rheb (D–F and K–M). At 7 d after transfection, the muscles were collected, and cross sections from the midbelly of the muscle were subjected to immunohistochemistry for GFP or HA-Rheb and the ribosomal S6 protein phosphorylated on the Ser235/236 residues (P-S6) (A–F) or total S6 protein (H–M). Gray scale images of the signal for GFP (B and I), HA-Rheb (E and L), P-S6 (C and F), or total S6 (J and M). The staining intensity of P-S6 (G) or total S6 (N) in GFP-transfected fibers (black bars) and HA-Rheb–transfected fibers (gray bars) was expressed relative to the values obtained in nontransfected fibers from the same section and is plotted on a histogram. Values indicate the mean ± SEM, n = 3 muscles (230–240 transfected and 200 nontransfected fibers)/group. *, significantly different from the values obtained in GFP-transfected fibers, p ≤ 0.05.
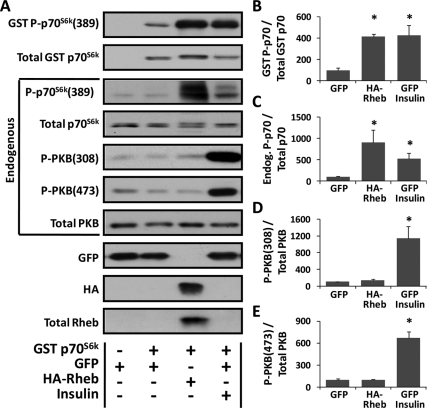
In a separate series of experiments, TA muscles were cotransfected with Rheb and GST p70S6k or GFP and GST p70S6k as a control condition. Similar to our findings in C2C12 myoblasts, the results indicated that overexpression of Rheb and insulin stimulation were both sufficient to induce an increase in the threonine 389 phosphorylation of GST p70S6k (Figure 3, A and B). Furthermore, the high transfection efficiency of our in vivo procedure, coupled with the 26-kDa size difference between GST p70S6k and endogenous p70S6k, allowed us to demonstrate that the overexpression of Rheb induced a significant increase in the threonine 389 phosphorylation of endogenous p70S6k (Figure 3, A and C). However, unlike the effects of insulin, overexpression of Rheb did not significantly alter the phosphorylation of endogenous PKB on either the threonine 308 residue [P-PKB(308)] or the serine 473 residue [P-PKB(473)] (Figure 3, A, D, and E). Numerous studies have shown that the activation of PI3K results in an increase in PKB(308) phosphorylation (Vanhaesebroeck and Alessi, 2000; Hawkins et al., 2006). Thus, the lack of an effect of Rheb on PKB(308) phosphorylation suggests that, similar to the results obtained in C2C12 myoblasts, overexpression of Rheb induces mTOR signaling through a PI3K-independent mechanism in adult skeletal muscle in vivo. Furthermore, the serine 473 residue on PKB has been reported to be directly phosphorylated by the mTORC2 complex (Hresko and Mueckler, 2005; Sarbassov et al., 2005; Guertin et al., 2006). Hence, the lack of an effect of Rheb on PKB(473) phosphorylation suggests that overexpression of Rheb did not induce mTORC2 signaling, which is consistent with previous in vitro and cell culture studies that have shown that Rheb exclusively activates signaling by the mTORC1 complex (Yang et al., 2006; Sato et al., 2009). Thus, when combined, these data indicate that overexpression of Rheb induces mTOR signaling through a PI3K/PKB-independent mechanism in adult skeletal muscle in vivo and that the effects of Rheb on mTOR seem to be specific to the mTORC1 complex.
Figure 3.
Overexpression of Rheb induces mTOR signaling through a PI3K/PKB-independent mechanism in skeletal muscle in vivo. (A–E) Mouse TA muscles were transfected with GFP or cotransfected with a combination of GFP and GST tagged p70S6k (GST p70S6k) or HA-Rheb and GST p70S6k. (A) At 48 h after transfection, the muscles were collected, and lysates were subjected to Western blot analysis with the indicated antibodies. Note that the analyses were performed on both GST p70S6k and the endogenous proteins. (B–E) In muscles cotransfected with GST p70S6k, the phosphorylated to total protein ratio for GST P-p70S6K(389) (B), endogenous P-p70S6K(389) (C), endogenous P-PKB(308) (D), and endogenous P-PKB(473) (E) was calculated and is expressed as a percentage of the values obtained in the control samples (GFP + GST p70S6k). Values are the mean + SEM, n = 3–5/group. *, significantly different from control, p ≤ 0.05.
Overexpression of Rheb Induces an Increase in Cap-dependent Translation in Skeletal Muscle In Vivo
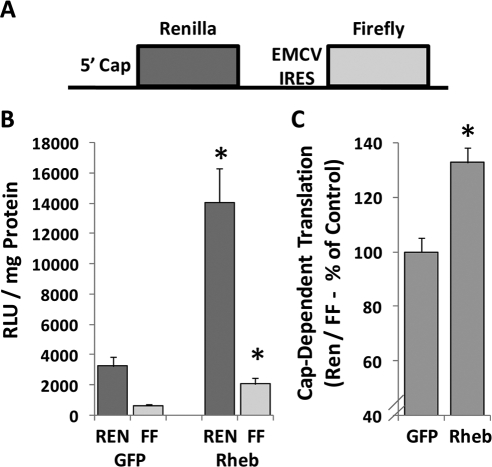
It has been proposed that activation of mTOR signaling can induce skeletal muscle hypertrophy, in part, by promoting an increase in the rate of protein synthesis (Bodine, 2006; Hamilton et al., 2009). The majority of protein synthesis is thought to occur through a cap-dependent mechanism (Mahoney et al., 2009); therefore, we set out to determine whether overexpression of Rheb could induce an increase in cap-dependent translation. In these experiments, muscles were cotransfected with GFP or Rheb and a dual-luciferase bicistronic reporter of cap-dependent translation (Carter and Sarnow, 2000). As described in Figure 4A, this reporter contains both the Renilla luciferase gene (REN) and firefly luciferase gene (FF). The expression of REN is controlled by cap-dependent translation, whereas the expression of FF is controlled by cap-independent translation via the internal ribosomal entry site (IRES) from the encephalomyocarditis virus (EMCV). The results from these experiments indicate that Rheb induced a significant increase in the expression of both the cap-dependent and cap-independent translation reporters (Figure 4B). More importantly, Rheb induced a significant increase in the ratio of the cap-dependent to cap-independent reporters, suggesting that Rheb induces an increase in cap-dependent translation (Figure 4C).
Figure 4.
Overexpression of Rheb induces an increase in cap-dependent translation in skeletal muscle in vivo. (A) Structure of the bicistronic reporter used for measuring cap-dependent translation of REN and EMCV IRES (i.e., cap-independent) translation of FF. (B and C) Mouse TA muscles were cotransfected with a combination of the bicistronic reporter and HA-Rheb or GFP as a control condition. At 48 h after transfection, the muscles were collected and luciferase activities were measured by a dual-luciferase assay. (B) Measurements of the relative light units (RLU) produced by REN and FF luciferase per milligram of total muscle protein. (C) Ratio of cap-dependent translation (REN) to cap-independent translation (FF) is expressed as a percentage of the values obtained in GFP transfected muscles. Values are the mean + SEM, n = 5–7/group. *, significantly different from the values obtained in GFP-transfected muscles.
Overexpression of Rheb Induces Skeletal Muscle Hypertrophy In Vivo
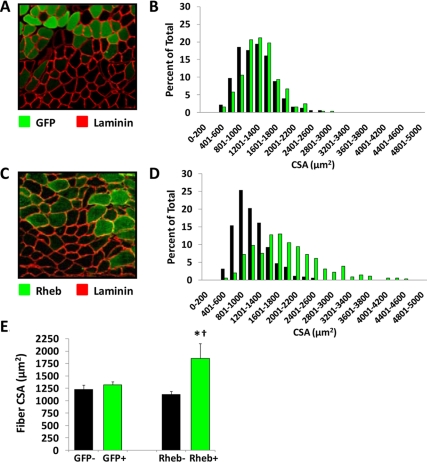
To determine whether the overexpression of Rheb is sufficient for the induction of skeletal muscle hypertrophy, mouse TA muscle were transfected with HA-Rheb or GFP as a control condition. The muscles were collected at 7 d after transfection, and the average CSA of the transfected and nontransfected fibers from each muscle was determined. The results indicated that in muscles transfected with GFP, the average CSA of the transfected and nontransfected fibers was not significantly different (Figure 5, A, B, and E). In muscles transfected with HA-Rheb, the average CSA of HA-Rheb-transfected fibers was 64% greater than that of the nontransfected fibers (Figure 5, C–E). Similar results also were obtained when muscles were transfected with a yellow fluorescent protein-tagged variant of Rheb (Supplemental Figure 1). Furthermore, the CSA of HA-Rheb-transfected fibers were significantly larger than that of GFP-transfected fibers (Figure 5E). Combined, these results indicate that overexpression of Rheb is sufficient for the induction of skeletal muscle hypertrophy. In addition, it also was determined that the average CSA of the nontransfected fibers from GFP and HA-Rheb muscles was not significantly different (Figure 5E). This observation indicates that the hypertrophic effects of Rheb were exerted in a cell autonomous manner.
Figure 5.
Overexpression of Rheb induces skeletal muscle fiber hypertrophy in vivo. Mouse TA muscles were transfected with GFP (A and B) or HA-Rheb (C and D). At 7 d after transfection, the muscles were collected, and cross sections from the midbelly of the muscle were subjected to immunohistochemistry for GFP and laminin (A) or HA-Rheb and laminin (C). (B and D) The CSA of transfected fibers (green bars) and nontransfected fibers (black bars) from GFP- (B) and HA-Rheb–transfected muscles (D) was determined and plotted on a histogram (n = 330–360 transfected and 330–360 nontransfected fibers/group). (E) Average CSA of the transfected (+) and nontransfected (−) fibers in GFP- and HA-Rheb–transfected muscles. Values are the mean + SEM, n = 4 muscles/group. *, significantly different from nontransfected fibers in HA-Rheb–transfected muscles; †, significantly different from GFP-transfected fibers, p ≤ 0.05.
Overexpression of Rheb Induces Hypertrophy via an mTOR-dependent Mechanism That Requires mTOR Kinase Activity
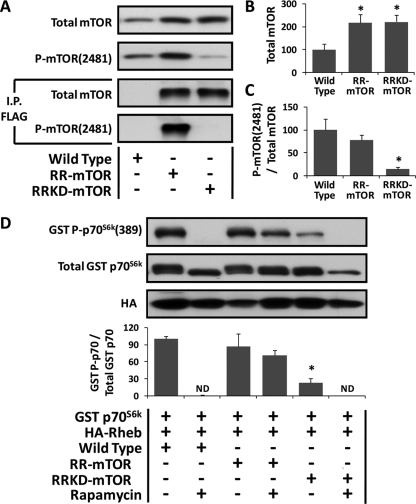
To define the role of mTOR in the Rheb-induced hypertrophic response, transgenic mice that express a FLAG-tagged rapamycin-resistant (Ser2035Thr) mutant of mTOR (RR-mTOR) or a FLAG tagged rapamycin-resistant kinase dead (Ser2035Thr/Asp2357Glu) mutant of mTOR (RRKD-mTOR) were used. These mice have been described by Ge et al. (2009), and in these mice, the expression of the transgene is under the control of the human skeletal actin promoter. As expected, expression of the transgene could not be detected in the liver or kidneys from these mice (data not shown). However, total mTOR expression in skeletal muscles from the RR-mTOR and RRKD-mTOR was significantly higher than that observed in wild-type mice (Figure 6, A and B, Note: these values result from a combination of both endogenously expressed wild-type mTOR and the transgenically expressed RR-mTOR or RRKD-mTOR). Furthermore, FLAG immunoprecipitates from skeletal muscle samples confirmed that both the RR-mTOR and RRKD-mTOR mice, but not wild-type mice, expressed a FLAG-tagged variant of mTOR. Moreover, phosphorylation of the Ser2481 residue on mTOR (an mTOR autophosphorylation site) could easily be detected on the FLAG-immunoprecipitated mTOR from RR-mTOR, but not RRKD-mTOR, mice (Figure 6A; Peterson et al., 2000). Consequently, the ratio of Ser2481 phosphorylated mTOR to total mTOR in muscles from RRKD-mTOR, but not RR-mTOR mice, was significantly reduced (Figure 6C). Combined, these results indicated that 1) the RR-mTOR and RRKD-mTOR mice possessed the appropriate tissue specific expression of the transgenes, 2) the expressed RRKD-mTOR lacked kinase activity, and 3) the ratio of kinase active mTOR to total mTOR was significantly reduced in muscles from the RRKD-mTOR mice.
Figure 6.
Confirmation of the phenotypes in RR-mTOR and RRKD-mTOR transgenic mice. (A) TA muscles from wild-type, RR-mTOR, and RRKD-mTOR mice were collected, and whole muscle lysates were subjected directly to Western blot analysis or subjected to immunoprecipiation (I.P.) for the FLAG-tag followed by Western blot analysis with the indicated antibodies. (B and C) Quantification of total mTOR (B) and the ratio of P-mTOR(2481) to total mTOR (C) in whole muscle lysates. The data are expressed as a percentage of the values obtained in wild-type muscles. *, significantly different from wild type, p ≤ 0.05. (D) TA muscles from wild-type, RR-mTOR and RRKD-mTOR mice were cotransfected with HA-Rheb and GST p70S6k. Immediately after transfection, mice were subjected to a regime of daily vehicle or 1 mg/kg rapamycin injections. At 48 h after transfection, the muscles were collected, and lysates were subjected to Western blot analysis with the indicated antibodies. The ratio of GST P-p70S6K(389) to total GST p70S6K (GST P-p70/Total GST p70) was calculated and expressed as a percentage of the values obtained in wild-type vehicle-treated samples. Values are the mean + SEM, n = 3–5/group. *, significantly different from wild-type vehicle-treated samples, p ≤ 0.05. ND, not detected.
The RR-mTOR and RRKD-mTOR transgenes both contain the Ser2035Thr mutation, and this mutation has been shown previously to render mTOR resistant to the inhibitory effects of rapamycin (Brown et al., 1995). To confirm that expression of mTOR containing the Ser2035Thr mutation conferred rapamycin resistance, TA muscles from wild-type, RR-mTOR, and RRKD-mTOR mice were cotransfected with Rheb and GST p70S6k. Immediately after the cotransfection, the mice were subjected to daily injections of rapamycin or the solvent vehicle and then assessed for changes in GST p70S6k(389) phosphorylation after 48 h. The results demonstrated that rapamycin eliminated GST p70S6k(389) phosphorylation in muscles from wild-type, but not RR-mTOR, mice (Figure 6D). Thus, these results demonstrated that phosphorylation of the threonine 389 residue on GST p70S6k is regulated by a rapamycin-sensitive mechanism and that expression of mTOR with Ser2035Thr mutation was able to rescue this event from the inhibitory actions of rapamycin. Furthermore, mTOR has been reported previously to be the kinase that phosphorylates p70S6k on the threonine 389 residue (Hornberger et al., 2007). Consistent with this conclusion, GST p70S6k(389) phosphorylation was significantly lower in vehicle-treated RRKD-mTOR mice and could not be detected in muscles from rapamycin treated RRKD-mTOR mice (Hornberger et al., 2007). Thus, the results from this series of experiments establish that the RR-mTOR and RRKD-mTOR transgenic mice conveyed the appropriate phenotypes.
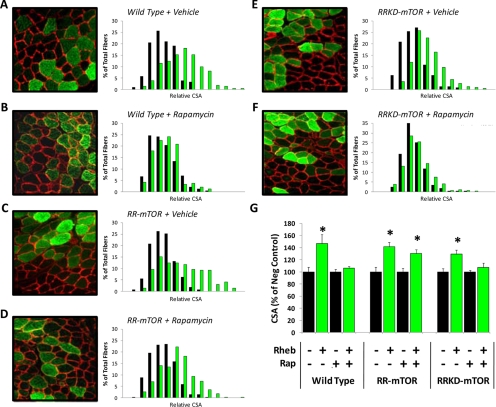
The RR-mTOR and RRKD-mTOR mice were then used to determine whether Rheb induced hypertrophy through an mTOR-dependent mechanism and whether mTOR kinase activity was necessary for this event. Specifically, TA muscles from wild-type, RR-mTOR, and RRKD-mTOR were transfected with Rheb, and then the mice were subjected to daily injections of rapamycin or the solvent vehicle as a control condition. The muscles were collected at 7 d after transfection, and the average CSA of the transfected and nontransfected fibers was determined. The results from these experiments demonstrated that rapamycin blocked the hypertrophic effects of Rheb in wild-type and RRKD-mTOR, but not RR-mTOR, mice (Figure 7). Combined, these results indicate that 1) Rheb induces hypertrophy through a rapamycin-sensitive mechanism, 2) mTOR is the rapamycin-sensitive element in skeletal muscle that confers Rheb-induced hypertrophy, and 3) mTOR kinase activity is necessary for the Rheb-induced hypertrophic response.
Figure 7.
Overexpression of Rheb induces hypertrophy via an mTOR-dependent mechanism that requires mTOR kinase activity. TA muscles from wild-type mice (A and B), RR-mTOR mice (C and D), and RRKD-mTOR mice (E and F) were transfected with HA-Rheb. Immediately after transfection, the mice were subjected to a regime of daily vehicle (A, C, and E) or 1 mg/kg rapamycin (Rap) injections (B, D, and F). At 7 d after transfection, the muscles were collected and cross-sections from the midbelly of the muscle were subjected to immunohistochemistry for HA-Rheb (green) and laminin (red). Histograms of the relative CSA for transfected fibers (green bars) and nontransfected fibers (black bars) are shown to the right of each image (n = 210–340 transfected and 210–340 nontransfected fibers/group). (G) Average relative CSA of the transfected (+) and nontransfected (−) fibers for each of the conditions described in A–F. Values are the mean + SEM, n = 4–5 muscles (210–340 fibers)/group. *, significantly different from the nontransfected fibers within a given condition, p ≤ 0.05.
DISCUSSION
The results of this study indicate that a PI3K/PKB-independent activation of mTOR signaling, in skeletal muscle, is sufficient to induce hypertrophy. This conclusion is functionally significant because it has been shown that some physiologically relevant types of growth-promoting stimuli induce mTOR signaling through a PI3K/PKB-independent mechanism. For example, mechanical loads have been shown to induce mTOR signaling through a PI3K/PKB-independent mechanism, and it has been widely proposed, although never demonstrated, that the mechanical load induced activation of mTOR signaling is sufficient to induce hypertrophy (Hornberger et al., 2004; Bodine, 2006; Hornberger et al., 2006). Based on the results of this study, it would seem that this hypothesis is correct.
The results of this study not only demonstrate that a PI3K/PKB-independent activation of mTOR signaling is sufficient to induce hypertrophy but also demonstrate that mTOR kinase activity is necessary for this event. This observation raises questions about the identity of the downstream substrates through which mTOR induces hypertrophy. Although the identity of these specific substrates remains to be fully defined, it is important to consider that hypertrophy results from a net positive change in the balance between protein synthesis and protein breakdown, and mTOR has been implicated in the regulation of both of these processes (Bodine, 2006; Mahoney et al., 2009; Jung et al., 2010). For example, mTOR has been shown to phosphorylate several proteins that control the rate of translation (Mahoney et al., 2009). Thus, the activation of mTOR signaling might promote hypertrophy, in part, by inducing the phosphorylation of substrates that enhance translational efficiency. In support of this possibility, we determined that overexpression of Rheb induced a 33% increase in ratio of cap-dependent to cap-independent translation (Figure 4).
Previous studies have not only shown that mTOR can regulate protein synthesis through the control of translation efficiency, but there is also substantial evidence that mTOR can regulate protein synthesis by controlling translational capacity (i.e., ribosome biogenesis) (Mahoney et al., 2009). For example, mTOR has been shown to bind to the promoter region of rDNA, and this occurs through a rapamycin-sensitive mechanism (Tsang et al., 2010). Furthermore, mTOR has been reported to phosphorylate several factors implicated in the regulation of rDNA transcription such as UBF and TIF-1A (Mahoney et al., 2009). Hence, activation of mTOR signaling might promote hypertrophy by inducing an increase in protein synthesis via both enhanced translational efficiency and translational capacity. In support of this possibility, we have observed that Rheb transfected fibers express a 1.7-fold greater amount of the ribosomal S6 protein compared with nontransfected fibers of the same muscle (Figure 2, F–J). Furthermore, the results in Figure 4 not only revealed that overexpression of Rheb induces an increase in the expression ratio of cap-dependent to cap-independent translation reporters but also revealed that overexpression of Rheb induced a large increase in the total expression of both reporters. This observation is consistent with an increase in translational capacity; therefore, it seems likely that a PI3K/PKB-independent activation of mTOR signaling induces hypertrophy by promoting an increase in protein synthesis via both enhanced translational efficiency and translational capacity.
As mentioned, mTOR also has been implicated in the regulation of protein degradation. For example, rapamycin has been shown to induce lysosomal protein degradation via autophagy, and this seems to be at least partly due to a decrease in the phosphorylation of two key regulators of autophagy, Atg13 and ULK1 (Jung et al., 2010). Furthermore, mTOR phosphorylates Atg13 and ULK1 through a rapamycin-sensitive mechanism, and overexpression of Rheb has been shown to induce Atg13 and ULK1 phosphorylation with a concomitant inhibition of autophagy (Wang et al., 2009; Jung et al., 2010). Thus, there are several lines of evidence that indicate that mTOR can phosphorylate and consequently inhibit substrates that regulate lysosomal protein degradation. In addition, there is also evidence that signaling by mTOR can regulate protein degradation by controlling the expression of ubiquitin ligases such as Atrogin 1 (Herningtyas et al., 2008). Based on these points, we propose that a PI3K/PKB-independent activation of mTOR induces hypertrophy by regulating the phosphorylation of substrates that promote an increase in protein synthesis (translational efficiency and capacity) and a decrease in protein degradation. Further study is needed to evaluate the relative contributions, both individually and in combination, of each of these proposed aspects of mTOR-mediated hypertrophy.
In summary, a better understanding of the molecular mechanisms that regulate muscle mass is required for the development of therapies that can attenuate or prevent the loss of muscle mass during conditions such as aging, disuse, or neuromuscular disease. In this study, we demonstrate that the overexpression of Rheb induces mTOR signaling through a PI3K/PKB-independent mechanism and that this event is sufficient to induce a robust and cell autonomous hypertrophic response. Furthermore, it was determined that the hypertrophic effects of Rheb occurred through a rapamycin-sensitive mechanism, that mTOR was the rapamycin-sensitive element in skeletal muscle that conferred the hypertrophic response, and that the kinase activity of mTOR was necessary for this event. Combined, these results strongly indicate that a PI3K/PKB-independent activation of mTOR signaling, in skeletal muscle, is sufficient to induce hypertrophy. Thus, the results of this study fill several key gaps in our knowledge of the molecular mechanisms that regulate skeletal muscle mass and also provide evidence that the development of mTOR-specific agonists could serve as a therapeutic strategy for preventing muscle atrophy.
Supplementary Material
ACKNOWLEDGMENTS
Special thanks are extended to Drs. Karyn Esser, Kun-Liang Guan, and Sunnie Thompson for providing plasmids. This work was supported by National Institutes of Health grants AR-053280 and AR-057347 (to T.A.H.) and AR-048194 (to J. C.).
Abbreviations used:
- c.a.-PKB
constitutively active protein kinase B
- CSA
cross-sectional area
- GFP
green fluorescent protein
- GST
glutathione transferase
- IGF-1
insulin-like growth factor 1
- mTOR
mammalian target of rapamycin
- p70S6k
ribosomal S6 kinase 1
- PI3K
phosphatidylinositol 3-kinase
- PKB
protein kinase B
- Rheb
Ras homologue enriched in brain
- RR-mTOR
rapamycin-resistant mammalian target of rapamycin
- RRKD-mTOR
rapamycin-resistant kinase dead mammalian target of rapamycin
- TA
tibialis anterior.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-05-0454) on July 28, 2010.
REFERENCES
- Avila G., Lee E. H., Perez C. F., Allen P. D., Dirksen R. T. FKBP12 binding to RyR1 modulates excitation-contraction coupling in mouse skeletal myotubes. J. Biol. Chem. 2003;278:22600–22608. doi: 10.1074/jbc.M205866200. [DOI] [PubMed] [Google Scholar]
- Avruch J., Hara K., Lin Y., Liu M., Long X., Ortiz-Vega S., Yonezawa K. Insulin and amino-acid regulation of mTOR signaling and kinase activity through the Rheb GTPase. Oncogene. 2006;25:6361–6372. doi: 10.1038/sj.onc.1209882. [DOI] [PubMed] [Google Scholar]
- Avruch J., Long X., Lin Y., Ortiz-Vega S., Rapley J., Papageorgiou A., Oshiro N., Kikkawa U. Activation of mTORC1 in two steps: Rheb-GTP activation of catalytic function and increased binding of substrates to raptor. Biochem. Soc. Trans. 2009;37:223–226. doi: 10.1042/BST0370223. [DOI] [PubMed] [Google Scholar]
- Baar K., Esser K. Phosphorylation of p70(S6k) correlates with increased skeletal muscle mass following resistance exercise. Am. J. Physiol. 1999;276:C120–C127. doi: 10.1152/ajpcell.1999.276.1.C120. [DOI] [PubMed] [Google Scholar]
- Bodine S. C. mTOR signaling and the molecular adaptation to resistance exercise. Med. Sci. Sports Exerc. 2006;38:1950–1957. doi: 10.1249/01.mss.0000233797.24035.35. [DOI] [PubMed] [Google Scholar]
- Bodine S. C., et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 2001;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- Brown E. J., Beal P. A., Keith C. T., Chen J., Shin T. B., Schreiber S. L. Control of p70 s6 kinase by kinase activity of FRAP in vivo. Nature. 1995;377:441–446. doi: 10.1038/377441a0. [DOI] [PubMed] [Google Scholar]
- Carter M. S., Sarnow P. Distinct mRNAs that encode La autoantigen are differentially expressed and contain internal ribosome entry sites. J. Biol. Chem. 2000;275:28301–28307. doi: 10.1074/jbc.M004657200. [DOI] [PubMed] [Google Scholar]
- Davies S. P., Reddy H., Caivano M., Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A., Sabatini D. M. mTOR and cancer: many loops in one pathway. Curr. Opin. Cell Biol. 2010;22:169–176. doi: 10.1016/j.ceb.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbay E., Chen J. The mammalian target of rapamycin regulates C2C12 myogenesis via a kinase-independent mechanism. J. Biol. Chem. 2001;276:36079–36082. doi: 10.1074/jbc.C100406200. [DOI] [PubMed] [Google Scholar]
- Frost R. A., Lang C. H. Protein kinase B/Akt: a nexus of growth factor and cytokine signaling in determining muscle mass. J. Appl. Physiol. 2007;103:378–387. doi: 10.1152/japplphysiol.00089.2007. [DOI] [PubMed] [Google Scholar]
- Garami A., Zwartkruis F. J., Nobukuni T., Joaquin M., Roccio M., Stocker H., Kozma S. C., Hafen E., Bos J. L., Thomas G. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol. Cell. 2003;11:1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- Ge Y., Wu A. L., Warnes C., Liu J., Zhang C., Kawasome H., Terada N., Boppart M. D., Schoenherr C. J., Chen J. mTOR regulates skeletal muscle regeneration in vivo through kinase-dependent and kinase-independent mechanisms. Am. J. Physiol. Cell Physiol. 2009;297:C1434–C1444. doi: 10.1152/ajpcell.00248.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin D. A., Stevens D. M., Thoreen C. C., Burds A. A., Kalaany N. Y., Moffat J., Brown M., Fitzgerald K. J., Sabatini D. M. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev. Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Hamilton D. L., MacKenzie M. G., Baar K. R. Molecular mechanisms of skeletal muscle hypertrophy: using molecular biology to understand muscle growth. In: Magalhães J., Ascensão A., editors. Muscle Plasticity—Advances in Biochemical and Physiological Research. Kerala, India: Research Signpost; 2009. pp. 45–93. [Google Scholar]
- Hawkins P. T., Anderson K. E., Davidson K., Stephens L. R. Signalling through Class I PI3Ks in mammalian cells. Biochem. Soc. Trans. 2006;34:647–662. doi: 10.1042/BST0340647. [DOI] [PubMed] [Google Scholar]
- Herningtyas E. H., Okimura Y., Handayaningsih A. E., Yamamoto D., Maki T., Iida K., Takahashi Y., Kaji H., Chihara K. Branched-chain amino acids and arginine suppress MaFbx/atrogin-1 mRNA expression via mTOR pathway in C2C12 cell line. Biochim. Biophys. Acta. 2008;1780:1115–1120. doi: 10.1016/j.bbagen.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Hornberger T. A., Chu W. K., Mak Y. W., Hsiung J. W., Huang S. A., Chien S. The role of phospholipase D and phosphatidic acid in the mechanical activation of mTOR signaling in skeletal muscle. Proc. Natl. Acad. Sci. USA. 2006;103:4741–4746. doi: 10.1073/pnas.0600678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornberger T. A., Hunter R. B., Kandarian S. C., Esser K. A. Regulation of translation factors during hindlimb unloading and denervation of skeletal muscle in rats. Am. J. Physiol. Cell Physiol. 2001;281:C179–C187. doi: 10.1152/ajpcell.2001.281.1.C179. [DOI] [PubMed] [Google Scholar]
- Hornberger T. A., Stuppard R., Conley K. E., Fedele M. J., Fiorotto M. L., Chin E. R., Esser K. A. Mechanical stimuli regulate rapamycin-sensitive signalling by a phosphoinositide 3-kinase-, protein kinase B- and growth factor-independent mechanism. Biochem. J. 2004;380:795–804. doi: 10.1042/BJ20040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornberger T. A., Sukhija K. B., Wang X. R., Chien S. mTOR is the rapamycin-sensitive kinase that confers mechanically-induced phosphorylation of the hydrophobic motif site Thr(389) in p70(S6k) FEBS Lett. 2007;581:4562–4566. doi: 10.1016/j.febslet.2007.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hresko R. C., Mueckler M. mTOR.RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3–L1 adipocytes. J. Biol. Chem. 2005;280:40406–40416. doi: 10.1074/jbc.M508361200. [DOI] [PubMed] [Google Scholar]
- Isotani S., Hara K., Tokunaga C., Inoue H., Avruch J., Yonezawa K. Immunopurified mammalian target of rapamycin phosphorylates and activates p70 S6 kinase alpha in vitro. J. Biol. Chem. 1999;274:34493–34498. doi: 10.1074/jbc.274.48.34493. [DOI] [PubMed] [Google Scholar]
- Izumiya Y., Hopkins T., Morris C., Sato K., Zeng L., Viereck J., Hamilton J. A., Ouchi N., LeBrasseur N. K., Walsh K. Fast/glycolytic muscle fiber growth reduces fat mass and improves metabolic parameters in obese mice. Cell Metab. 2008;7:159–172. doi: 10.1016/j.cmet.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C. H., Ro S. H., Cao J., Otto N. M., Kim D. H. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline W. O., Panaro F. J., Yang H., Bodine S. C. Rapamycin inhibits the growth and muscle-sparing effects of clenbuterol. J. Appl. Physiol. 2007;102:740–747. doi: 10.1152/japplphysiol.00873.2006. [DOI] [PubMed] [Google Scholar]
- Laviola L., Natalicchio A., Giorgino F. The IGF-I signaling pathway. Curr. Pharm. Des. 2007;13:663–669. doi: 10.2174/138161207780249146. [DOI] [PubMed] [Google Scholar]
- Lee R. C., Wang Z., Heo M., Ross R., Janssen I., Heymsfield S. B. Total-body skeletal muscle mass: development and cross-validation of anthropometric prediction models. Am. J. Clin. Nutr. 2000;72:796–803. doi: 10.1093/ajcn/72.3.796. [DOI] [PubMed] [Google Scholar]
- Lewis M. I., Bodine S. C., Kamangar N., Xu X., Da X., Fournier M. Effect of severe short-term malnutrition on diaphragm muscle signal transduction pathways influencing protein turnover. J. Appl. Physiol. 2006;100:1799–1806. doi: 10.1152/japplphysiol.01233.2005. [DOI] [PubMed] [Google Scholar]
- Li Y., Inoki K., Guan K. L. Biochemical and functional characterizations of small GTPase Rheb and TSC2 GAP activity. Mol. Cell Biol. 2004;24:7965–7975. doi: 10.1128/MCB.24.18.7965-7975.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney S. J., Dempsey J. M., Blenis J. Chapter 2: Cell Signaling in Protein Synthesis Ribosome Biogenesis and Translation Initiation and Elongation. Prog. Mol. Biol. Transl. Sci. 2009;90C:53–107. doi: 10.1016/S1877-1173(09)90002-3. [DOI] [PubMed] [Google Scholar]
- Miyazaki M., Esser K. A. REDD2 is enriched in skeletal muscle and inhibits mTOR signaling in response to leucine and stretch. Am. J. Physiol. Cell Physiol. 2009;296:C583–C592. doi: 10.1152/ajpcell.00464.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nader G. A. Muscle growth learns new tricks from an old dog. Nat. Med. 2007;13:1016–1018. doi: 10.1038/nm0907-1016. [DOI] [PubMed] [Google Scholar]
- O'Neil T. K., Duffy L. R., Frey J. W., Hornberger T. A. The role of phosphoinositide 3-kinase and phosphatidic acid in the regulation of mTOR following eccentric contractions. J. Physiol. 2009;587:3691–3701. doi: 10.1113/jphysiol.2009.173609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiro N., Yoshino K., Hidayat S., Tokunaga C., Hara K., Eguchi S., Avruch J., Yonezawa K. Dissociation of raptor from mTOR is a mechanism of rapamycin-induced inhibition of mTOR function. Genes Cells. 2004;9:359–366. doi: 10.1111/j.1356-9597.2004.00727.x. [DOI] [PubMed] [Google Scholar]
- Osman B., Doller A., Akool el S., Holdener M., Hintermann E., Pfeilschifter J., Eberhardt W. Rapamycin induces the TGFbeta1/Smad signaling cascade in renal mesangial cells upstream of mTOR. Cell Signal. 2009;21:1806–1817. doi: 10.1016/j.cellsig.2009.07.016. [DOI] [PubMed] [Google Scholar]
- Pallafacchina G., Calabria E., Serrano A. L., Kalhovde J. M., Schiaffino S. A protein kinase B-dependent and rapamycin-sensitive pathway controls skeletal muscle growth but not fiber type specification. Proc. Natl. Acad. Sci. USA. 2002;99:9213–9218. doi: 10.1073/pnas.142166599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I. H., Erbay E., Nuzzi P., Chen J. Skeletal myocyte hypertrophy requires mTOR kinase activity and S6K1. Exp. Cell Res. 2005;309:211–219. doi: 10.1016/j.yexcr.2005.05.017. [DOI] [PubMed] [Google Scholar]
- Peterson R. T., Beal P. A., Comb M. J., Schreiber S. L. FKBP12-rapamycin-associated protein (FRAP) autophosphorylates at serine 2481 under translationally repressive conditions. J. Biol. Chem. 2000;275:7416–7423. doi: 10.1074/jbc.275.10.7416. [DOI] [PubMed] [Google Scholar]
- Price D. J., Grove J. R., Calvo V., Avruch J., Bierer B. E. Rapamycin-induced inhibition of the 70-kilodalton S6 protein kinase. Science. 1992;257:973–977. doi: 10.1126/science.1380182. [DOI] [PubMed] [Google Scholar]
- Rommel C., Bodine S. C., Clarke B. A., Rossman R., Nunez L., Stitt T. N., Yancopoulos G. D., Glass D. J. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat. Cell Biol. 2001;3:1009–1013. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- Sandri M. Signaling in muscle atrophy and hypertrophy. Physiology. 2008;23:160–170. doi: 10.1152/physiol.00041.2007. [DOI] [PubMed] [Google Scholar]
- Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Sato T., Nakashima A., Guo L., Tamanoi F. Specific activation of mTORC1 by Rheb G-protein in vitro involves enhanced recruitment of its substrate protein. J. Biol. Chem. 2009;284:12783–12791. doi: 10.1074/jbc.M809207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguin R., Nelson M. E. The benefits of strength training for older adults. Am. J. Prev. Med. 2003;25:141–149. doi: 10.1016/s0749-3797(03)00177-6. [DOI] [PubMed] [Google Scholar]
- Sun Y., Fang Y., Yoon M. S., Zhang C., Roccio M., Zwartkruis F. J., Armstrong M., Brown H. A., Chen J. Phospholipase D1 is an effector of Rheb in the mTOR pathway. Proc. Natl. Acad. Sci. USA. 2008;105:8286–8291. doi: 10.1073/pnas.0712268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee A. R., Manning B. D., Roux P. P., Cantley L. C., Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- Tsang C. K., Liu H., Zheng X. F. mTOR binds to the promoters of RNA polymerase I- and III-transcribed genes. Cell Cycle. 2010;9:953–957. doi: 10.4161/cc.9.5.10876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaesebroeck B., Alessi D. R. The PI3K-PDK1 connection: more than just a road to PKB. Biochem. J. 2000;346:561–576. [PMC free article] [PubMed] [Google Scholar]
- Wang T., Donahoe P. K. The immunophilin FKBP 12, a molecular guardian of the TGF-beta family type I receptors. Front. Biosci. 2004;9:619–631. doi: 10.2741/1095. [DOI] [PubMed] [Google Scholar]
- Wang T., Lao U., Edgar B. A. TOR-mediated autophagy regulates cell death in Drosophila neurodegenerative disease. J. Cell Biol. 2009;186:703–711. doi: 10.1083/jcb.200904090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Zhao W., Zhao J., Zhang Y., Qin W., Pan J., Bauman W. A., Blitzer R. D., Cardozo C. REDD1 is a major target of testosterone action in preventing dexamethasone-induced muscle loss. Endocrinology. 151:1050–1059. doi: 10.1210/en.2009-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Inoki K., Kim E., Guan K. L. TSC1/TSC2 and Rheb have different effects on TORC1 and TORC2 activity. Proc. Natl. Acad. Sci. USA. 2006;103:6811–6816. doi: 10.1073/pnas.0602282103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.