Abstract
Arabidopsis thaliana knockout lines for the plant-specific eukaryotic translation initiation factors eIFiso4G1 (i4g1) and eIFiso4G2 (i4g2) genes have been obtained. To address the potential for functional redundancy of these genes, homozygous double mutant lines were generated by crossing individual knockout lines. Both single and double mutant plants were analyzed for changes in gross morphology, development, and responses to selected environmental stressors. Single gene knockouts appear to have minimal effect on morphology, germination rate, growth rate, flowering time, or fertility. However, double mutant i4g1/i4g2 knockout plants show reduced germination rates, slow growth rates, moderate chlorosis, impaired fertility and reduced long term seed viability. Double mutant plants also exhibit altered responses to dehydration, salinity, and heat stress. The i4g2 and i4g1/i4g2 double mutant has reduced amounts of chlorophyll a and b suggesting a role in the expression of chloroplast proteins. General protein synthesis did not appear to be affected as the levels of gross protein expression did not appear to change in the mutants. The lack of a phenotype for either of the single mutants suggests there is considerable functional overlap. However, the strong phenotypes observed for the double mutant indicates that the individual gene products may have specialized roles in the expression of proteins involved in plant growth and development.
Electronic supplementary material
The online version of this article (doi:10.1007/s11103-010-9670-z) contains supplementary material, which is available to authorized users.
Keywords: Translation initiation, Initiation factor, eIF4G, eIFiso4G, Protein synthesis, Plant translation
Introduction
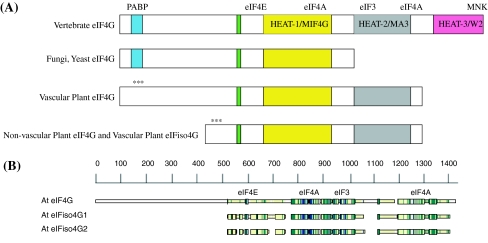
Eukaryotic translation initiation is a complicated process requiring the interaction of multiple initiation factors (Pestova et al. 2007). Cellular eukaryotic mRNAs contain a 5′ methylated cap structure (m7GpppN), which serves as the binding site for the eukaryotic translation initiation factor 4F (eIF4F) cap-binding complex. eIF4F consists of a small cap-binding subunit (eIF4E) and a larger protein subunit (eIF4G) that facilitates interactions with other initiation factors (Hernandez and Vazquez-Pianzola 2005; Prévôt et al. 2003). eIF4G is comprised of multiple domains (see Fig. 1a). In addition to an N-terminal domain that recruits poly(A)-binding protein (PABP) and an eIF4E binding domain, there are up to three C-terminal HEAT domains (MIF4G, MA3, W2) (Bellsolell et al. 2006; Korneeva et al. 2000; LeFebvre et al. 2006; Marcotrigiano et al. 2001; Marintchev and Wagner 2004, 2005; Oberer et al. 2005; Schutz et al. 2008). The MIF4G HEAT domain is the most highly conserved domain found in all forms of eIF4G and interacts with eIF4A (Marcotrigiano et al. 2001; Marintchev and Wagner 2004; Schutz et al. 2008). The MA3 HEAT domain is retained in vertebrates and plants (Marintchev and Wagner 2005); however, the W2 HEAT domain is only present in vertebrates (Marintchev and Wagner 2005). The structures of the HEAT domains of animal and yeast eIF4G have been determined and consist of multiple α-helices that form a right-handed solenoid typical of the HEAT family and interact with other initiation factors or proteins during initiation (Bellsolell et al. 2006; Korneeva et al. 2000; LeFebvre et al. 2006; Marcotrigiano et al. 2001; Marintchev and Wagner 2004; Marintchev and Wagner 2005; Schutz et al. 2008).
Fig. 1.
a eIF4G and eIFiso4G Domain Organization. The organization of eIF4G from vertebrates, fungi/yeast and plants have in common the eIF4E binding domain and core HEAT domain for binding eIF4A and eIF3 (MIF4G). Fungi and yeast have lost the second and third HEAT domains that include the second eIF4A binding domain and MNK binding domain (Hernandez and Vazquez-Pianzola 2005; Marintchev and Wagner 2005; Schutz et al. 2008). Plant eIF4G and eIFiso4G have retained only the first and second HEAT domains. The PABP binding domain for plant eIF4G or eIFiso4G (***) has not been experimentally determined. b Amino Acid Sequence Comparison. The amino acid sequences for Arabidopsis thaliana eIF4G (At3g60240) and the two eIFiso4G (At5g57870, At2g24050) proteins were aligned using CLUSTALW2 (Larkin et al. 2007) and visualized with MACAW (v.2.0.5) (Schuler et al. 1991). The approximate binding domains for eIF4E/iso4E, eIF4A and eIF3 are indicated. The actual sequence alignments are shown in Supplemental Data Figure B. The PABP binding domain (***) for plant eIF4G or eIFiso4G has not been experimentally determined
Higher plants are unique in that they encode a second eIF4F complex, eIFiso4F, comprised of distinct isoforms of both the cap-binding protein eIF4E (eIFiso4E) and the scaffolding protein eIF4G (eIFiso4G) (Browning et al. 1992; Browning 1996). eIFiso4G lacks a significant amount of N-terminal sequence compared to eIF4G (Fig. 1), but is highly conserved in the two HEAT domains (MIF4G, MA3) responsible for eIF4E, eIF4A, and eIF3 binding. eIFiso4F is 3–5 fold more abundant than eIF4F in wheat germ extracts, maize root tips, and cauliflower florets (Browning et al. 1990, 1992), suggesting that it may be the primary complex used to initiate general translation in most plant cells, though both eIF4F and eIFiso4F are capable of initiating translation in vitro (Gallie and Browning 2001). However, the ability of eIFiso4F to support initiation of some mRNAs is not equal to eIF4F (Mayberry et al. 2009), suggesting that there are features of some mRNAs that allow them to interact preferentially with eIFiso4F or eIF4F. The eIF4F complex more efficiently translates mRNAs that contain secondary structure in the 5′ untranslated region (UTR), and is also more adept at initiating translation of uncapped monocistronic and dicistronic messages (Gallie and Browning 2001).
Mutations in eIFiso4G have been found to confer resistance to rice yellow mottle virus (RYMV) and map to a domain involved in eIF4A binding (Albar et al. 2006). Resistance to potyviruses has also been reported for eIFiso4G mutants (Nicaise et al. 2007). A mutation in eIF4G (cum2) is also associated with virus resistance to cucumber mosaic virus (CMV) replication in Arabidopsis (Yoshii et al. 2004). In addition, the cap binding proteins, eIF4E and eIFiso4E, which form complexes with eIF4G and eIFiso4G respectively, are also virus resistance genes for various viruses which suggests an integral relationship between the host translation initiation factors and different types of plant virus replication (Gallois et al. 2010; Robaglia and Caranta 2006).
A. thaliana encodes three isoforms of the eIF4F/eIFiso4F large subunit; eIF4G (At3g60240), eIFiso4G1 (At5g57870), and eIFiso4G2 (At2g24050) and four isoforms of the small subunit cap-binding protein; eIF4E1 (At4g18040), eIF4E2 (At1g29590), eIF4E3 (At1g29550) and eIFiso4E (At5g35620). The cap binding proteins, eIFiso4E and eIF4E1, share about 41% identity at the amino acid sequence level and both have a similar molecular mass of approximately 24 kDa. The eIF4E2 and eIF4E3 isoforms are very similar to eIF4E1 amino acid sequence (54 and 56% identity respectively) and appear to be gene duplication events on chromosome 1 (95% identity to each other); however, eIF4E2 or eIF4E3 are not expressed at significant levels according to various transcription profiling databases suggesting specific tissue or developmental roles. Reduced levels of eIF4E and eIFiso4E expression in tobacco were shown to have effects on plant growth and polysome formation (Combe et al. 2005).
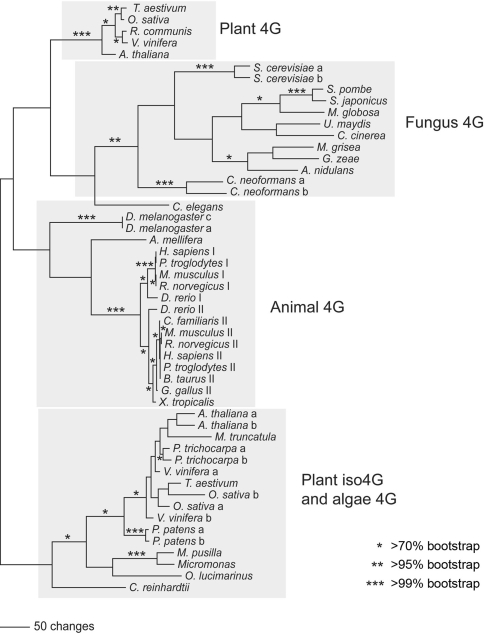
The eIFiso4G isoforms differ in molecular mass from the eIF4G (86 kDa and 83 kDa v. 168 kDa, respectively) and have ~27% amino acid identity (41% similarity) to eIF4G. The eIFiso4G1 and eIFiso4G2 isoforms share ~57% amino acid identity (72% similarity) in the conserved domains with each other. A schematic of the alignment of amino acid sequences for eIF4G and the two forms of eIFiso4G is shown in Fig. 1b. The HEAT-1 (MIF4G) domain is conserved across all the major taxonomic groups (Fig. 1a), and this region was used as the basis for a phylogenetic analysis of the eIF4G and eIFiso4G isoforms (Fig. 2). The phylogenetic analysis was conducted using parsimony, pairwise distance, and maximum likelihood methods using PAUP* (Swofford 2000), with bootstrapping (Felsestein 1985) used to assess support for the results. Well supported branches of the tree did not differ significantly among these analyses (Fig. 2 is based on the parsimony results). Branches that are supported by >70, >95, and >99% of the bootstrap proportions are marked in Fig. 2; branches supported by >70% bootstrap proportions are considered significantly supported (Hillis and Bull 1993).
Fig. 2.
eIF4G/eIFiso4G Phylogeny. Amino acid sequences from the conserved HEAT-1/MIF4G domain were aligned for 50 eIF4G or eIFiso4G sequences from plants (vascular and non-vascular), animals, and fungi using the program MAFFT (Katoh and Toh 2008). A phylogenetic analysis of these aligned sequences (see Supplementary data Fig. C) was then conducted using PAUP* (Swofford 2000), under the parsimony, minimum evolution, and maximum likelihood criteria (the parsimony results are shown here), with bootstrapping used to assess support. Asterisks indicate >70% (*), >95% (**), and >99% (***) bootstrap support
The plant, fungus, and animal eIF4G sequences, as well as the plant eIFiso4G and nonvascular plant eIF4G sequences, generally clustered in discrete monophyletic groups. The only exception was the C. elegans eIF4G sequence, which sometimes grouped weakly with the fungus eIF4G sequences (Fig. 2). This tree is consistent with an ancient gene duplication that gave rise to the eIFiso4G and eIFiso4G-like genes in plants, sometime before the diversification of plants, animals, and fungi. Both genes have been retained in higher plants, although the eIFiso4G gene appears to have been lost in the ancestor of animals and fungi.
It is clear that the functional core of eIF4G is shared by eIFiso4G; however, the N-terminus of eIF4G is highly variable and reflects divergence in the evolutionary process. Plant, animal, fungal and yeast eIF4G have an extended N-terminus; however, there is little similarity between these N-terminal regions from animals, plants, fungi or yeast suggesting independent evolution. Even the N-terminal amino acid sequence similarity between wheat and Arabidopsis eIF4G is quite low. The function of the N-terminal region of eIF4G remains unknown and likely evolved independently in each organism for regulatory purposes.
Regulation of eIF4F/eIFiso4F activity in plants differs significantly from other eukaryotic mechanisms. 4E-binding proteins, a major class of eIF4E regulatory proteins in other eukaryotes (Raught and Gingras 1999, 2007) have yet to be reported in plants. The 4E-BP families encoded by other higher eukaryotes function to sequester eIF4E, providing a layer of regulation on protein synthesis by limiting formation of the eIF4F complex (Raught and Gingras 2007). AtLOX2 (Arabidopsis Lipoxygenase 2), and AtBT3b, (the β subunit of basic transcription factor 3, part of the nascent-polypeptide associated complex) were shown to interact with Arabidopsis eIFiso4E using the yeast two hybrid system (Freire et al. 2000; Freire 2005). However, the biological significance of these interactions has yet to be determined, but suggests the possibility of regulation through sequestration of cap-binding proteins using non-canonical factors. Another type of initiation complex regulation appears to be absent in plants, phosphorylation by MNK1 of eIF4E. Plant eIF4G isoforms lack the C-terminal HEAT-3/W2 domain observed in animal eIF4Gs. In animal eIF4Gs, the HEAT-3/W2 domain serves as a site for interaction with MNK1 (MAP kinase activated protein kinase) which phosphorylates eIF4E when in complex with eIF4G (Buxade et al. 2008). The apparent absence of conserved 4E-BPs, and the lack of a conserved MNK1 kinase binding domain in plant eIF4G isoforms suggests that plants may regulate initiation of translation through alternative mechanisms.
To determine the independent or overlapping function in plants of the two forms of eIFiso4G, gene knockouts were obtained and the resulting phenotypes observed. The overall health of the plants was monitored by cataloging changes in basic morphology or development. The responses to environmental stresses (hydration, increased salinity, heat stress) were also monitored. The loss of either subunit, eIFiso4G1 or eIFiso4G2, resulted in almost imperceptible changes in phenotype consistent with functional redundancy; however, the loss of both forms, resulted in a severe growth phenotype, chlorosis due to less chlorophyll, and changes in fertility and seed stability. These results suggest that at least one of the paralogs for eIFiso4G are required for appropriate expression of some genes that may participate in the regulation of plant growth and development. This phenomenon may represent a synergistic effect of the loss of two overlapping gene functions as the phenotype of the double mutant is greater than that of either parent (Perez-Perez et al. 2009).
Results
Screening for eIF4G isoform mutants
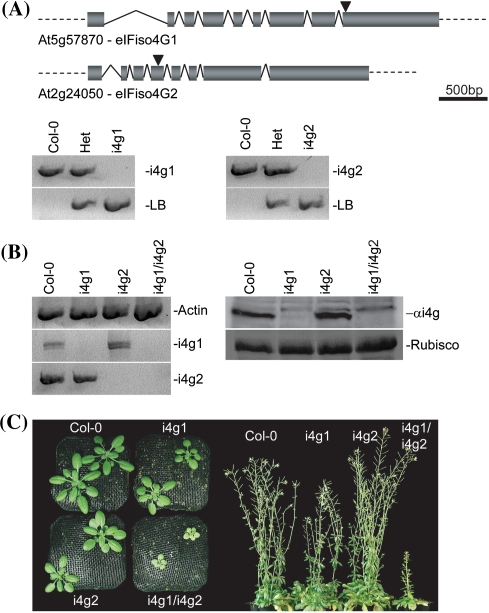
To examine the effects of individual eIFiso4G knockouts, transgenic T-DNA disruption lines were obtained for At5g57870 (eIFiso4G1) and At2g24050 (eIFiso4G2) from the Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu/cgi-bin/tdnaexpress) through TAIR. SALK_009905 (i4g1) has a T-DNA insertion in exon 9 of the predicted At5g57870 open reading frame and SALK_076633 (i4g2) has a T-DNA insertion in exon 4 of At2g24050 (Fig. 3a). To ensure that any observed phenotypic effects were due to disruption of the target genes, and not due to accompanying secondary mutations, the individual lines were back crossed once to wild-type Columbia (Col-0) plants and resulting progeny were allowed to self pollinate to create segregating populations. Individual homozygous knockout plants were identified via PCR (Fig. 3a) and carried through an additional generation following self-pollination. The resulting F3 progeny were used for further analysis. Knockout confirmation was determined by RT–PCR and western blotting (Fig. 3b). Homozygous i4g1 or i4g2 plant lines lack detectable mRNA for the respective gene that is missing. eIFiso4G1 is the more highly expressed gene since the knockout in this gene shows very little apparent eIFiso4G2 protein. In contrast, the i4g2 knockout shows similar levels of eIFiso4G protein to wild type. This difference is not due to lack of cross-reactivity of the antibody raised to At eIFiso4G1 with eIFiso4G2. E. coli expressed recombinant AteIFiso4G2 cross reacts with the rabbit antibody to recombinant AteIFiso4G1 (data not shown). Analysis of expression databases also indicates that the amount of eIFiso4G2 mRNA is considerably lower than eIFiso4G1 under most environmental conditions, growth states or tissue types [see Supplemental Figure A, eFP (Winter et al. 2007) and Supplemental Table A, PARE (German et al. 2008)]. However, eIFiso4G2 mRNA expression appears higher than eIFiso4G1 in late stage embryo development, uninucleate microphore, and the apical section of the torpedo in the embryo (see Supplemental Figure A). These results suggest that eIFiso4G2 may be necessary only at certain times in development or in particular tissues.
Fig. 3.
Arabidopsis eIFiso4G knockout lines. a Genomic screening. i4g1 lines carry a pROK2 T-DNA insertion in exon 9. i4g2 lines contain a pROK2 T-DNA insertion in exon 4. PCR screening of genomic DNA with gene specific primers yields a 928 bp band which is absent in homozygous i4g1 plants, but present in both wild-type Col-0 and heterozygous plants. Screening with a combination of pROK2 specific and gene specific primers yields a band of 601 bp in i4g1 and heterozygous plants that is absent in Col-0. PCR screening of genomic DNA with gene specific primers yields a 915 bp band which is absent in homozygous i4g2 plants, but present in both wild-type Col-0 and heterozygous plants. Screening with a combination of pROK2 specific and gene specific primers yields a band of 753 bp in i4g2 and heterozygous plants that is absent in Col-0. b Confirmation of eIFiso4G isoform knockout by RT–PCR and western blotting. mRNA populations from wild-type and mutant plants were probed for actin, eIFiso4G1, and eIFiso4G2 message (left panel). Western blotting of total plant extracts probed with antibody to AteIFiso4G1 (right panel upper). Coomassie stained Rubisco is shown for a total protein loading comparison (right panel lower). c Phenotypic effects. (Left panel) Overhead comparison of plants at 21 days post sowing. (Right panel) Profile of plant lines at ~56 days post sowing
To create i4g1/i4g2 double mutant plants, homozygous individual mutants were cross-pollinated to each other and the resulting F1 progeny were allowed to self-pollinate to create a segregating population. Initial i4g1/i4g2 segregating populations yielded no double homozygotes (60 total plants), though 23 heterozygous/homozygous individuals were identified, in both gene combinations with near equal frequency. Heterozygous/homozygous individuals were allowed to self-pollinate, and multiple independent double homozygous individuals were identified in the resulting F3 segregating populations. These confirmed double homozygous plants were allowed to self-pollinate and the progeny were used for all subsequent experiments.
The double homozygous plants were backcrossed to each respective parent and the parent phenotype was restored (data not shown).
Effects of eIFiso4G gene knockout(s)
To determine the effects of eIFiso4G gene knockouts on general morphology, wild type Col-0, i4g1 or i4g2 plant lines were examined for general differences in germination rate, leaf production and shape, root length, pigmentation, inflorescence production, fertility, and general health. Single gene knockouts appear phenotypically similar to wild-type Columbia (Col-0) plants (Fig. 3c), with no obvious morphological differences or developmental abnormalities, and appear in general good health. In contrast, i4g1/i4g2 double mutants have impaired germination rates, grow and develop slower, exhibit mild chlorosis, with reduced secondary inflorescence and reduced fertility (Fig. 3c). In general they are in poor health relative to their wild-type and single eIFiso4G knockout counterparts, suggesting a synergistic effect of the two genes (Perez-Perez et al. 2009).
eIFiso4G double mutants exhibit changes in leaf production and growth
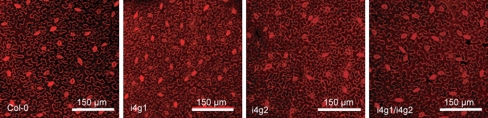
To examine the development of leaves in mutant plants, Col-0, i4g1, i4g2, and i4g1/i4g2, were grown under standard conditions following stratification. Rosette leaves that were larger than 1 mm in width were counted at weekly intervals starting at 3 weeks post stratification. i4g1/i4g2 double mutant plants consistently produced roughly 90% the number of rosette leaves relative to Col-0 at each time point, while i4g1 or i4g2 plants were relatively unaffected (Table 1). Leaf length was measured from the tip to the narrowest point at the base at the each time point. In addition to a reduced number of rosette leaves, i4g1/i4g2 double mutant plants produce leaves that are significantly smaller than Col-0 plants (Fig. 3c). Both i4g1 and i4g2 single mutant plants produced leaves similar in size to, or even slightly larger than Col-0 plants. To determine whether that decreased size in the double mutant plants is due to a decrease in cell density or accompanied by changes in cell morphology, leaves from 21 days soil grown plants were stained with propidium iodide and examined by confocal laser microscopy at 20× magnification (Fig. 4). Epidermal cells were examined in 450 μm × 450 μm squares for obvious changes in size and shape and the number of cells were counted within the square. i4g1/i4g2 double mutant epidermal cells have no discernable change in shape, but have an average cell density of 87.8 ± 6.1 cells relative to their Col-0 counterparts, which have an average cell density of 151.7 ± 10.3 cell. i4g1 and i4g2 plants were similar to Col-0 plants, with cell densities of 147.3 ± 6.9 and 148.8 ± 6.5 respectively.
Table 1.
Analysis of growth of leaves, roots and flowering time
| Leaf growth | |||
|---|---|---|---|
| Plant line | >1 mm Rosette leaves | SD | % Wild type |
| 21 Days | |||
| Col-0 | 8.1 | 1.0 | 100 |
| i4g1 | 7.3 | 1.1 | 90 |
| i4g2 | 8.2 | 1.1 | 101 |
| i4g1/i4g2 | 6.9 | 1.2 | 85 |
| 28 Days | |||
| Col-0 | 10.2 | 1.0 | 100 |
| i4g1 | 9.8 | 1.0 | 96 |
| i4g2 | 10.2 | 1.2 | 100 |
| i4g1/i4g2 | 9.3 | 1.0 | 91 |
| 35 Days | |||
| Col-0 | 11.5 | 1.3 | 100 |
| i4g1 | 11.1 | 1.0 | 96 |
| i4g2 | 10.9 | 1.5 | 95 |
| i4g1/i4g2 | 10.4 | 1.5 | 91 |
| Root growth | |||
|---|---|---|---|
| Plant line | Primary root length (mm) | SD | % Wild type |
| 3 Days | |||
| Col-0 | 12.7 | 1.8 | 100 |
| i4g1 | 12.4 | 1.4 | 97 |
| i4g2 | 11.8 | 1.4 | 92 |
| i4g1/i4g2 | 10.7 | 1.8 | 84 |
| 5 Days | |||
| Col-0 | 21.5 | 2.4 | 100 |
| i4g1 | 21.3 | 1.9 | 99 |
| i4g2 | 20.9 | 1.6 | 97 |
| i4g1/i4g2 | 16.6 | 3.4 | 77 |
| 7 Days | |||
| Col-0 | 28.5 | 3.5 | 100 |
| i4g1 | 28.4 | 2.4 | 100 |
| i4g2 | 27.6 | 1.9 | 97 |
| i4g1/i4g2 | 22.2 | 3.5 | 78 |
| 10 Days | |||
| Col-0 | 48.5 | 4.6 | 100 |
| i4g1 | 47.4 | 3.7 | 98 |
| i4g2 | 46.4 | 3.0 | 96 |
| i4g1/i4g2 | 29.7 | 4.5 | 61 |
| 12 Days | |||
| Col-0 | 62.9 | 6.0 | 100 |
| i4g1 | 62.8 | 5.1 | 100 |
| i4g2 | 61.9 | 3.3 | 98 |
| i4g1/i4g2 | 37.7 | 5.3 | 60 |
| Flowering | |||
|---|---|---|---|
| Plant line | Floral bud appearance (Days) | SD | % Wild type |
| Col-0 | 42.3 | 1.4 | 100 |
| i4g1 | 42.4 | 2.8 | 100 |
| i4g2 | 40.2 | 2.4 | 95 |
| i4g1/i4g2 | 52.4 | 2.9 | 124 |
| Plant line | Rosette leaves >1 mm at flowering | SD | % Wild type |
|---|---|---|---|
| Col-0 | 11.4 | 1.0 | 100 |
| i4g1 | 10.7 | 0.9 | 94 |
| i4g2 | 11.1 | 1.3 | 97 |
| i4g1/i4g2 | 11.1 | 0.9 | 98 |
SD standard deviation
Fig. 4.
Confocal imagery of epidermal cells. Propidium iodide stained epidermal cells of 21 day old leaves at 20× magnification. A 450 μm × 450 μm section is shown
eIFiso4G double mutants exhibit changes in root growth
To examine changes in root growth rates, plants were grown aseptically on a vertical bias. Primary root length was measured at 2–3 day intervals for 12 days post germination. i4g1/i4g2 double mutant root length was consistently ~80% of wild-type for the first 7 days, and tapered to roughly 60% by 12 days (Table 1). In contrast, i4g1 and i4g2 single mutant plants exhibit no change in primary root length relative to Col-0 plants (Table 1). In addition to a shortened primary root, i4g1/i4g2 double mutant plants exhibit reduced secondary lateral rooting, and reduced root hair density when examined under microscopy (data not shown).
eIFiso4G double mutants exhibit delayed flowering and reduced fertility
To determine the flowering rates for each of the mutant lines, plants were grown in soil under standard conditions and the number of days that had passed prior to emergence of the primary inflorescence was recorded, as were the number of rosette leaves present at the time of flowering. i4g1/i4g2 double mutant flowering was delayed by approximately 10 days relative to Col-0 plants, or roughly 24% (Table 1). The number of rosette leaves present at the time of flowering was similar to that of Col-0, i4g1, and i4g2 single mutant plants, suggesting that the emergence of the primary inflorescence occurs at a similar stage of development. In contrast to the relatively small delay seen in emergence of the primary inflorescence (Table 1), a significant differential exists in the duration of the plant lifecycle. The typical lifecycle observed for Col-0, i4g1, and i4g2 plants grown under standard conditions is 10–12 weeks, while the life cycle for the i4g1/i4g2 double mutant extends to 20–24 weeks, with considerable reduction in both secondary inflorescence and seed production. Following emergence of the primary inflorescence, elongation of the primary bolt is slowed relative to wild-type and single mutant plants, producing a shortened primary bolt with little to no secondary inflorescence. Siliques that form are smaller and contain fewer seeds, though seeds produced are of a size and shape that is indistinguishable from Col-0 seed. Pollen production appears limited based on increased difficulty in using the i4g1/i4g2 as a pollen donor in out-crosses.
To examine the effects these inflorescence shortcomings have on overall fertility, 10 plants for each mutant line were grown under normal conditions, and allowed to self-fertilize. The seed produced from each plant was collected, weighed, and averaged. Seed production for i4g1, i4g2, and i4g1/i4g2 double mutants relative to Col-0 plants was 62, 104, and 16% respectively (Table 2). Because the seeds produced by the i4g1/i4g2 double mutant are of similar size and shape to those produced by Col-0 (data not shown), the reduction in seed harvest weight is likely due to a simple reduction in the amounts of secondary inflorescence in combination with a reduction in fertility rather than anything abnormal about the seeds themselves.
Table 2.
Analysis of viability and fertility
| Viability | |||
|---|---|---|---|
| Plant line | % Viable @ 4 days | ||
| Fresh seed | |||
| Col-0 | 98 | ||
| i4g1 | 97 | ||
| i4g2 | 97 | ||
| i4g1/i4g2 | 77 | ||
| Year old seed | |||
| Col-0 | 98 | ||
| i4g1 | 97 | ||
| i4g2 | 98 | ||
| i4g1/i4g2 | 23 | ||
| Fertility | |||
|---|---|---|---|
| Plant line | Average individual seed production (mg/plant) | SD | % Wild type |
| Col-0 | 66.8 | 24.3 | 100 |
| i4g1 | 41.6 | 12.3 | 62 |
| i4g2 | 69.8 | 28.3 | 104 |
| i4g1/i4g2 | 10.8 | 8.0 | 16 |
SD standard deviation
To examine changes in germination rates, Col-0 and mutant seeds were grown under aseptic conditions and examined for emergence of seedlings following 4 days of growth at standard conditions. Germination rates were similar to wild-type for single mutant plants, but noticeably reduced for the double mutant (Table 2). Germination rates in the double mutant decreased significantly as the seed increased in age, falling precipitously to roughly a third of the original germination rate following long term storage (12+ months) at room temperature (Table 2). The reduced germination rate of fresh seed and dramatic reduction in germination rates for stored seeds suggests potential abnormalities in the seed coat, or possibly in embryo development.
Analysis of expressed seed proteins in the i4g1/i4g2 mutant by two dimensional gels did not show any obvious differences in protein expression suggesting that overall protein synthesis in seeds is not affected, nor did electron microscopy of seeds show any obvious abnormalities in the seed structure (data not shown). The pigmentation of the i4g1/i4g2 seeds appeared to be lighter than those of Col-0 or the parents suggesting there might be differences in the level of seed pigment synthesis (data not shown).
eIFiso4G2 and eIFiso4G double mutants exhibit changes in chlorophyll profiles
In addition to their reduced stature, the leaves on i4g1/i4g2 plants exhibit mild chlorosis almost immediately following germination, with a comparatively pale and yellowish hue relative to Col-0 and single mutant plants. An analysis of chlorophyll production determined that single mutant i4g1 plants are largely unchanged relative to wild-type Col-0 (102% chlorophyll a, 100% chlorophyll b), while i4g2 and i4g1/i4g2 plants produce reduced levels of chlorophyll a (83 and 66% respectively) and chlorophyll b (81 and 76% respectively) (Table 3). Mature i4g1/i4g2 leaves also exhibit premature browning and senescence (not shown).
Table 3.
The eIFiso4G1/eIFiso4G2 double mutant produces reduced levels of chlorophyll
| Mutant | Chlorophyll a (μg/mg FW) |
Chlorophyll a % wild type |
Chlorophyll b (μg/mg FW) |
Chlorophyll b % wild type |
Ratio chlorophyll a/b |
|---|---|---|---|---|---|
| Col-0 | 0.90 | 100 | 0.25 | 100 | 3.60 |
| i4g1 | 0.91 | 102 | 0.25 | 100 | 3.64 |
| i4g2 | 0.75 | 83 | 0.20 | 81 | 3.75 |
| i4g1/i4g2 | 0.59 | 66 | 0.19 | 76 | 3.11 |
Leaves from 6 to 10 plants were pooled to average out any plant-to-plant variation. The data are representative of several repeats
Wild type and i4g1/i4g2 double mutant light harvesting complexes were analyzed by “green gel” electrophoresis (Horie et al. 2009). The amount of chlorophyll in the i4g1/i4g2 mutant appeared lower, but light and dark treated plants showed no discernable differences in the light harvesting complex composition (see Supplemental Figure D) suggesting that the overall translation of the proteins in light harvesting complexes was not altered.
eIFiso4G double mutants have reduced tolerance to dehydration
To examine effects on tolerance to dehydration, aseptically grown 21 day old plants were weighed and then transferred to dry Whatman 3 M filter paper and left at room temperature for 24 h without watering. Tolerance to dehydration was determined by comparing the fresh weight and the dry weight following 24 h on filter paper. Following the treatment, the double mutant plants retained the least amount of their original fresh weight at 7%. In contrast, wild-type plants retained nearly twice as much at 13% of their original fresh weight. Single mutant plants behaved more similarly to Col-0 plants, retaining 13 and 11% of their original fresh weight (Table 4). However, this difference may also be due to the smaller size of the double mutant plants making them more susceptible to dehydration.
Table 4.
Analysis of dehydration
| Plant line | Weight (mg) | SD | % Parent |
|---|---|---|---|
| 0 h | |||
| Col-0 | 426.5 | 126.0 | NA |
| i4g1 | 415.5 | 77.9 | NA |
| i4g2 | 467.6 | 108.8 | NA |
| i4g1/i4g2 | 155.9 | 89.8 | NA |
| 24 h | |||
| Col-0 | 56.5 | 21.0 | 13 |
| i4g1 | 55.1 | 24.3 | 13 |
| i4g2 | 50.8 | 14.6 | 11 |
| i4g1/i4g2 | 11.5 | 6.4 | 7 |
SD standard deviation
eIFiso4G double mutants have reduced tolerance to salinity
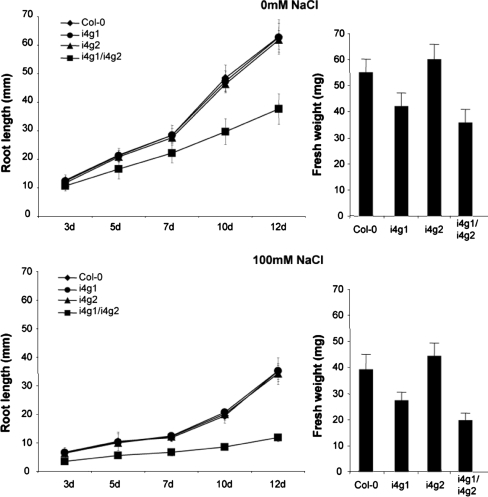
To examine the effects of salinity on root growth and fresh weight, 4 day old aseptically grown plants were transplanted to new media supplemented with 0 mM or 100 mM NaCl. Plants used to examine salinity effects on root growth were grown on a vertical bias for an additional 12 days and root length measurements were taken every 2–3 days. i4g1/i4g2 double mutant plants grown on 100 mM NaCl responded with reduced growth rates relative to Col-0, i4g1, and i4g2 plants with root lengths ranging from 56.5 to 33.7% of the Col-0 roots by 12 days (Fig. 5). Root length for i4g1/i4g2 plants grown on 100 mM NaCl were reduced to 31.4% of plants grown on 0 mM NaCl by 12 days, compared to roughly 55% for Col-0 and single mutant plants (55.8, 56.1, and 55.3% for Col-0, i4g1, and i4g2 respectively). Plants used to examine global salinity effects were grown aseptically for 4 days and then transferred to fresh media supplemented with 0 mM or 100 mM NaCl and grown for an additional 7 days prior to weighing. i4g1/i4g2 double mutant plants appear to be more susceptible to 100 mM NaCl, growing to roughly 55% of their 0 mM NaCl fresh weight, compared to 65–73% for Col-0 and single mutant plants.
Fig. 5.
Salinty responses. Root length and fresh weight was measured following transfer of 4 day old seedlings to new media containing 0 mM or 100 mM NaCl. Aseptically grown seedlings that were transplanted onto 100 mM NaCl media were weighed and compared to seedlings transferred to 0 mM NaCl media
eIFiso4G double mutants show no response to heat shock
To analyze thermotolerance, hypocotyl lengths in wild-type and eIFiso4G mutants were measured following treatment at different temperatures. Seedlings were germinated aseptically and grown at 22°C for 2 days. The seedlings were then subjected to continuous 22°C or 37°C for 90 min, or 45°C for 90 min, or pretreatment at 37°C prior to treatment at 45°C. The seedlings were then grown in the dark for 3 days at 22°C, and the hypocotyl length was measured. No statistical difference was observed between Col-0 and individual eIFiso4G mutants at any of the temperature treatments (Fig. 6), indicating that the individual mutant plants are not impaired in their ability to respond to high temperatures. The hypocotyl lengths of i4g1/i4g2 double mutant plants were constant in length regardless of the temperature treatment applied, suggesting a lack of any temperature related response or that the i4g1/i4g2 double mutant plants are already preconditioned to heat stress under normal growth conditions perhaps due to alternations in gene expression at the translation level.
Fig. 6.
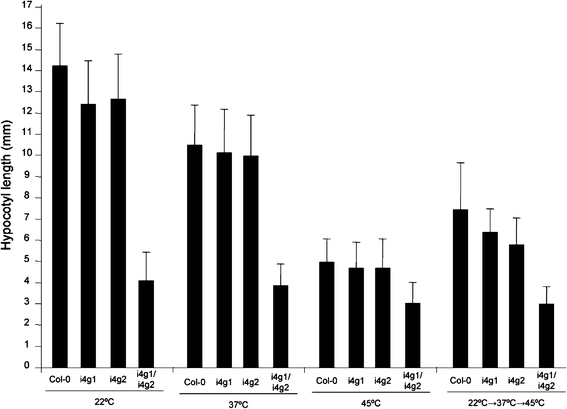
Thermotolerance. Hypocotyl lengths were measured following treatment at elevated temperatures. Plants were treated at room temperature, 37°C for 90 min, 45°C for 90 min, or 45°C for 60 min following pretreatment at 37°C for 90 min and 22°C for 60 min
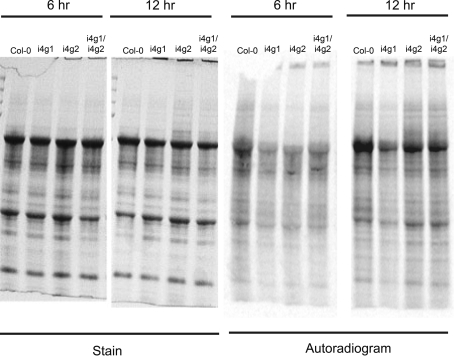
Incorporation of [35S]-methionine in vivo
The ability of the i4g1, i4g2 and i4g1/i4g2 mutants compared to wild type Col0 plants to incorporate [35S]-methionine was measured in vivo. The label was incorporated for 6 and 12 h and the proteins analyzed by SDS PAGE. No gross differences were observed between any of the plant extracts (see Fig. 7). These results, combined with the analysis of seed proteins by two-dimensional gels and green gel analysis, suggest that there are not gross changes in the overall rate of translation of highly expressed proteins in the i4g1/i4g2 double mutant. The phenotype is more likely due to lowered gene expression of a small number of proteins possibly involved in regulatory mechanisms.
Fig. 7.
Incorporation of [35S]-methionine in vivo. Leaves of seedlings (12 days) were labeled with [35S]-methionine for 6 or 12 h (room temperature, continuous light) as described in Experimental Procedures
Discussion
Though the process of translation initiation is very similar in all eukaryotes, the translational machinery and regulatory mechanisms that plants employ may differ from those used by animals and fungi. The eIF4G isoforms in plants differ from what is observed in yeast and animals. For example, yeast has two genes for eIF4G that are similar in molecular mass and functionally overlap (Clarkson et al. 2010; Goyer et al. 1993). Animals have two forms of eIF4G (eIF4GI, eIF4GII) that are similar in molecular weight, but appear to be have alternative transcription start sites, different translation start sites and differences in degradation by viral proteases (Byrd et al. 2002, 2005; Caron et al. 2004; Coldwell and Morley 2006; Gradi et al. 1998a, b; Svitkin et al. 1999) suggesting alternative regulation and activities for these isoforms. The presence in plants of multiple eIF4G isoforms with differing molecular mass and limited homology (eIF4G vs. eIFiso4G) suggests the possibility of functional specialization. The eIFiso4G isoforms are plant specific, and so it would stand to reason that they may be required for translation of mRNAs for plant specific functions, including photosynthesis, cellulose biosynthesis, flowering and seed production, and response to environmental stressors due to the sessile nature of plants. An interesting result for these mutants regarding potyvirus resistance was previously reported (Nicaise et al. 2007). The i4g1 mutant is resistant to PPV and LMV, but not to ClYVV or TuMV. The i4g2 mutant was susceptible to all 4 viruses. In contrast, the double mutant, i4g1/i4g2 was resistant to PPV as was the parent i4g1, but the double mutant was also resistant to TuMV for which neither parent showed resistance (Nicaise et al. 2007). Specific point mutations in the VPg of TuMV can overcome the virus resistance conferred by the double i4g1/i4g2 mutant or an eIFiso4E mutant suggesting specific interactions of the VPg with the eIFiso4F components (Gallois et al. 2010).
We have examined the morphological effects of single and double eIFiso4G isoform knockouts and found that individual isoform knockouts are phenotypically similar to wild-type plants when grown under normal conditions, while double mutant knockout plants are significantly impaired, resulting in plants that are stunted with reduced viability. This suggests there is some functional redundancy between the two eIFiso4G isoforms, and an essential function for eIFiso4F complexes during normal plant development and reproduction. The data suggest possible roles in the translation of proteins or regulatory proteins involved in stress responses, coordinated temporal or tissue specific expression, photosynthesis, or flowering and seed production.
Conditions of thermal stress (heat shock) result in the translational repression of non-heat shock messages due to a loss of cap-dependent translation (Gallie et al. 1995; Key et al. 1981; Pitto et al. 1992). Despite repression of general protein synthesis, heat shock associated mRNAs are efficiently translated, presumably through cap-independent mechanisms directed by their 5′UTR sequences (Gallie et al. 1995; Pitto et al. 1992). Both eIFiso4G and eIF4G show increased protein expression levels following thermal stress, while their mRNA levels remain constant, either through an increase in translation efficiency for their own messages or a decrease in protein turnover (Gallie et al. 1998). Protein expression levels for eIF4G/eIFiso4G partnering cap-binding proteins (eIF4E and eIFiso4E respectively) do not show a corresponding increase in expression following heat shock, suggesting that cap recognition during heat stress is not important and only eIF4G and/or eIFiso4G are primarily required for cap-independent translation. The observation that protein expression levels for eIF4G isoforms change during heat stress lead us to test the ability of i4 g, i4g2 and i4g1/i4g2 mutants to respond to simple environmental pressures. Double mutant plants showed less tolerance when challenged with salinity or dehydration while single mutant plants respond similarly to wild-type plants, suggesting a requirement for eIFiso4G isoforms in response to stress. The i4g1/i4g2 double mutant did not respond to heat stress in a measurable way and behaves as though constitutively heat shocked.
The presence of multiple initiation complex isoforms may regulate protein synthesis initiation throughout the plant life cycle. Protein expression patterns for eIF4G and eIFiso4G do change through the course of plant development. During germination, both factors are detectable in 2–3 day old wheat embryos, though only eIFiso4G is detectable in endosperm tissue (Gallie et al. 1998). eIFiso4G is readily detectable through 7 days post germination in both root and leaf tissues, while eIF4G expression levels fall below detectable limits by 5 days post germination. During seed development, levels of eIFiso4G expression are highest in early development (8–10 days post flowering), and steadily decline through late development (20–25 days post flowering), while eIF4G expression patterns are the exact opposite (Gallie et al. 1998). Determining whether there are patterns of tissue specific expression for each eIFiso4G isoform remains a major goal for future work.
In this study we observe that i4g1/i4g2 double mutant plants are compromised in their ability to produce the photosynthesis related pigment chlorophyll. Both chlorophyll a and chlorophyll a are significantly reduced in the double mutant plant, and somewhat reduced in the i4G2 single mutant plants. This reduction in chlorophyll makes the double mutant plants appear moderately chlorotic, and suggests that energy production through photosynthesis may be impaired. However, no difference was observed in the protein composition of the light harvesting complexes. Double mutant plants also exhibit pronounced premature senescence in their primary rosette leaves long before their life cycle is completed. Flowering time and seed viability are also drastically affected in i4g1/i4g2 double mutant plants. Long-term seed viability is significantly reduced, with germination rates falling to 23% within a year. That observation, coupled with the observation that i4g1/i4g2 double mutant seedlings are more susceptible to dehydration suggests there may be a problem with the seed coat and hence increased susceptibility to excessive dehydration.
In conclusion, the eIFiso4G isoforms clearly have a role in the normal growth and development of plants with roles in both vegetative growth and reproduction. eIFiso4G presumably also plays a role in production of proteins involved in photosynthesis, as chlorophyll production is affected. Although a significant phenotype is associated with the i4g1/i4g2 double mutant, there do not appear to be gross changes in the ability to translate most proteins, suggesting that the phenotype may be due to loss or reduction of proteins that are expressed as low levels such as transcription factors or other regulatory proteins. eIFiso4G isoforms show considerable functional redundancy as single mutant plants display little difference to wild-type plants under the growth conditions examined, while double mutant plants show pronounced phenotypes suggesting a synergistic effect (Perez-Perez et al. 2009). These results suggest that there is an important role(s) for the two eIFiso4G isoforms in gene expression. Determination of expression patterns and genome wide expression of translationally active mRNAs in the i4g1/i4g2 mutant compared to Col-0 are in progress and will provide considerable insight into the biology of the observed phenotypes.
Experimental procedures
Plant material and growth conditions
Arabidopsis thaliana ecotype Columbia-0 (Col-0) was used for all wild-type controls (Lehle Seeds). The homozygous knockout lines for At5g57870 (i4g1) and At2g24050 (i4g2) mutants were isolated from the SALK lines 009905 and 076633, respectively. Homozygous i4g1/i4g2 double mutant plants were generated through crossing followed by self fertilization. All plants were grown at 22°C with a 16 h light period following stratification for 7 days in wet soil at 4°C unless otherwise noted. For plants grown under aseptic conditions, seeds were sterilized with a solution of 20% bleach, and 1% SDS solution, rinsed (5×) with sterile H2O, resuspended in sterile 0.01% agar and placed onto 1× Murashige and Skoog (MS) nutrient agar (0.8%) containing 2.0% sucrose with a sterile pipette. Plates were left at 4°C in the dark for 7 days and then grown under standard conditions.
Screening of the SALK libraries
Floral tissue was collected from individual plants and genomic DNA isolated (Dellaporta et al. 1983). PCR analysis of genomic DNA was carried out using a combination of primers: for i4G1, LF 009905, 5′-TCATCACATTGTTCAGGTTAACACC-3′, RT009905, 5′-TTCGCTCAACTTGGGACCACT-3′, and LBb1, 5′GCGTGGACCG CTTGCTGCAACT-3′, and for i4G2, 076633LF, 5′-TCAACCTTCAAACAC AAAAGCTGA-3′, 076633RT, 5′- AACCCTTTTCCCCGTCAAGGT-3′ and LBb1.
The combination of 009905LF and 009905RT produces a fragment of 928 bp and the combination of 009905RT and LBb1 produces a fragment of 601 bp. The combination of 076633LF and 076633RT produces a fragment of 915 bp and the combination of 0776633LF and LBb1 produces a fragment of 753 bp. The gene specific primer/TDNA insert primer PCR fragments were sequenced for verification of insertion location.
RT PCR
First strand cDNA was generated from flower derived total RNA using oligo dT(20) as a primer and Superscript II reverse transcriptase (Invitrogen). First strand cDNA was amplified with the following primer sets to screen for mRNA presence: At5g57870 ×9F, 5′-CTATAACCGAAATACATACTGAGGCAGAG-3′ and At5g57870 ×9R, 5′-ATGTCTGCAGCCTGTGAGGCCAACAAGG-3′ produces a 964 bp fragment. At2g24050 ×1F, 5′-ATGCAACAACAAGGTGAACCATCGG-3′ and At2g24050 ×6R, 5′-GCTCGTACTTCTCCGGAGTAAGC-3′ produces a 574 bp fragment. At3g18780 ×3F, 5′-GTGCTGGATTCTGGTGATGGTGTGTC-3′ and At3g18780 ×3R, 5′-CTGGAATGTGCTGAGGGAAGCAAGAATG-3′ produces a 607 bp fragment.
Western blotting
Four plants per line of Col-0, i4g1, i4g2, and i4g1/i4g2 plants were grown in soil under standard conditions for 28 days. Above soil tissue was pooled for each representative line, ground into a fine powder under liquid nitrogen, and resuspended in 1× protein dissociation buffer (62.5 mm Tris–HCl pH 6.8, 2% SDS, 5% β-ME) supplemented with Complete™ protease inhibitor cocktail (Roche). Fragments were resolved on a 5% stacking/12.5% resolving acrylamide gel and transferred to nitrocellulose via semi-dry transfer as previously described (Browning et al. 1990). The membrane was blocked using 5% milk in HNAT (10 mM Hepes–KOH, pH 7.6, 150 mM NaCl, 0.2% bovine serum albumin, 0.02% Tween 80) and probed with a 1:2,000 dilution of rabbit primary antibody raised to recombinant Arabidopsis eIFiso4G1 (Mayberry et al. 2009) in 5% milk in HNAT for 2 h at room temperature. The secondary antibody was goat anti-rabbit HRP (Kirkegaard Perry) at a 1:5,000 dilution in 5% milk in HNAT for 1 h at room temperature and visualized with chemiluminescent substrate (Pierce Chemical).
Leaf, root, and flower analysis
Twenty-four plants per line of Col-0, i4g1, i4g2, and i4g1/i4g2 plants were soil grown under standard conditions following a 1 week stratification at 4º in wet soil. Rosette leaves greater than 1 mm in width were counted in weekly intervals beginning at 3 weeks post removal from 4°C. Twenty rosette leaves were measured from the tip to the base with a digital caliper at each time interval. Root analysis was performed on plants aseptically grown on 1× MS media supplemented with 2% sucrose. Plants were germinated on horizontal plates for 2 days and then moved to a vertical bias. Primary root lengths were measured with a digital caliper at 2–3 day intervals post germination. The plants used to measure rosette leaves were also used to determine the time to flowering. Flowering time was determined by the emergence of primary inflorescence tissue
Fertility, and germination rate testing
Ten plants per line of Col-0, i4g1, i4g2 and i4g1/i4g2 plants were soil grown under standard conditions and allowed to self-pollinate and go to seed. After drying, the seeds from individual plants were collected and weighed. Germination rates were determined by observation of seedling emergence at 4 days under aseptic growth conditions on 1× MS media supplemented with 2.0% sucrose.
Confocal microscopy
Whole plant seedlings or rosette leaf samples from 28d plants were floated on a 50 mM solution of propidium iodide for 5–10 min. Root samples were directly mounted to slides in distilled water under a glass coverslip. Leaf samples were sectioned into approximately 1 mm squares prior to mounting. Samples were excited at 488 nm and examined with a Leica SP2 AOBS confocal laser microscope at 20× magnification. 6 leaves for each representative line were used to count epidermal cells in a defined area.
Chlorophyll measurements
Chlorophyll a and b were measured spectrophotometrically as described (Jeffrey and Humphrey 1975). Leaf samples were ground in liquid nitrogen and extracted with 90% (v/v) acetone. The absorbance at 664 and 647 nm was determined and used to calculate chlorophyll a and b content by the equations: Chl a = 11.93A664 − 1.93A647 and Chl b = 20.36A647 − 5.50A664, respectively. Each experiment was repeated 2–3 times and representative results presented.
Thermotolerance, dehydration, and salinity assays
Plants were grown under aseptic conditions on 1× MS nutrient agar with 2.0% sucrose and grown for 3 days prior to treatment. Thermotolerance assays were conducted as described (Whitham et al. 2000). Plants for dehydration analysis were grown under aseptic conditions on 1× MS nutrient agar with 2.0% sucrose for 21 days at 22°C with a 16 h photoperiod and transferred to dry 3 M Whatman filter paper for 24 h. Dehydration tolerance was determined by the percentage of fresh tissue weight remaining following treatment (Novillo et al. 2004). Aseptically grown 4 day old seedlings were transferred to new 1× MS media (2% sucrose) supplemented with 0 mM or 100 mM NaCl. Plants used for determining effects on new root growth were grown for an additional 12 days on a vertical bias and were measured at 3, 5, 7, 9, and 12 days post transfer. Plants used for determining effects on fresh weight were grown for an additional 7 days under standard conditions prior to weighing.
Incorporation of [35S]-methionine in vivo
Seedlings (12 day) on sterile MS plates were labeled with a 1:1 mixture of [35S]-methionine (PerkinElmer NEG009A) and 0.02% Tween 20 (Gilmour et al. 1988). Briefly, a 2 μl drop of the [35S]-methionine/Tween 20 mixture was placed on each leaf surface. The plates were covered and plants exposed to continuous light at room temperature during the 6–12 h labeling period. The seedlings were removed from the plates and weighed. A proportional amount of 2× SDS gel loading buffer containing protease inhibitors (Roche Complete tablet) was added to give a tissue/buffer ratio of 0.38 mg tissue/μl of buffer. The seedlings were ground with plastic pestle in a 1.5 ml microfuge tube, heated for 4 min at 95°C, ground again and centrifuged at room temperature for 10 min. The supernatant removed and 20 μl applied to a 4–12% SDS PAGE (Invitrogen). The gel was stained with coomassie brilliant blue, dried and exposed to a phosphorimager screen for 24 h.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by grants from NSF (MCB0214996) to K.S.B and DBI-0820047 to D.R.G.. The mutants and phenotype data from this report will be part of a NSF Arabidopsis 2010 project on translation initiation factors (DBI-0820047) to K.S.B. and D.R.G and others. A. Aertker and J. Tran were summer NSF REU recipients for K.S.B.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- eIF
Eukaryotic initiation factor
- HEAT domain
Huntington, elongation factor3, subunit of protein phosphatase 2A, target of rapamycin
References
- Albar L, Bangratz-Reyser M, Hebrard E, Ndjiondjop MN, Jones M, Ghesquiere A. Mutations in the eIF(iso)4G translation initiation factor confer high resistance of rice to Rice yellow mottle virus. Plant J. 2006;47:417–426. doi: 10.1111/j.1365-313X.2006.02792.x. [DOI] [PubMed] [Google Scholar]
- Bellsolell L, Cho-Park PF, Poulin F, Sonenberg N, Burley SK. Two structurally atypical HEAT domains in the C-terminal portion of human eIF4G support binding to eIF4A and Mnk1. Structure. 2006;14:913–923. doi: 10.1016/j.str.2006.03.012. [DOI] [PubMed] [Google Scholar]
- Browning KS. The plant translational apparatus. Plant Mol Bio. 1996;32:107–144. doi: 10.1007/BF00039380. [DOI] [PubMed] [Google Scholar]
- Browning KS, Humphreys J, Hobbs W, Smith GB, Ravel JM. Determination of the amounts of the protein synthesis initiation and elongation factors in wheat germ. J Biol Chem. 1990;265:17967–17973. [PubMed] [Google Scholar]
- Browning KS, Webster C, Roberts JKM, Ravel JM. Identification of an isozyme form of protein synthesis initiation factor 4F in plants. J Biol Chem. 1992;267:10096–10100. [PubMed] [Google Scholar]
- Buxade M, Parra-Palau JL, Proud CG. The Mnks: MAP kinase-interacting kinases (MAP kinase signal-integrating kinases) Front Biosci. 2008;13:5359–5373. doi: 10.2741/3086. [DOI] [PubMed] [Google Scholar]
- Byrd MP, Zamora M, Lloyd RE. Generation of multiple isoforms of eukaryotic translation initiation factor 4GI by use of alternate translation initiation codons. Mol Cell Biol. 2002;22:4499–4511. doi: 10.1128/MCB.22.13.4499-4511.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd MP, Zamora M, Lloyd RE. Translation of eukaryotic translation initiation factor 4GI (eIF4GI) proceeds from multiple mRNAs containing a novel cap-dependent internal ribosome entry site (IRES) that is active during poliovirus infection. J Biol Chem. 2005;280:18610–18622. doi: 10.1074/jbc.M414014200. [DOI] [PubMed] [Google Scholar]
- Caron S, Charon M, Cramer E, Sonenberg N, Dusanter-Fourt I. Selective modification of eukaryotic initiation factor 4F (eIF4F) at the onset of cell differentiation: recruitment of eIF4GII and long-lasting phosphorylation of eIF4E. Mol Cell Biol. 2004;24:4920–4928. doi: 10.1128/MCB.24.11.4920-4928.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson BK, Gilbert WV, Doudna JA. Functional overlap between eIF4G isoforms in Saccharomyces cerevisiae. PLoS ONE. 2010;5:e9114. doi: 10.1371/journal.pone.0009114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coldwell MJ, Morley SJ. Specific isoforms of translation initiation factor 4GI show differences in translational activity. Mol Cell Biol. 2006;26:8448–8460. doi: 10.1128/MCB.01248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combe JP, Petracek ME, van EG, Meulewaeter F, Twell D. Translation initiation factors eIF4E and eIFiso4E are required for polysome formation and regulate plant growth in tobacco. Plant Mol Biol. 2005;57:749–760. doi: 10.1007/s11103-005-3098-x. [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21. doi: 10.1007/BF02712670. [DOI] [Google Scholar]
- Felsestein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- Freire MA. Translation initiation factor (iso) 4E interacts with BTF3, the beta subunit of the nascent polypeptide-associated complex. Gene. 2005;345:271–277. doi: 10.1016/j.gene.2004.11.030. [DOI] [PubMed] [Google Scholar]
- Freire MA, Tourneur C, Granier F, Camonis J, El Amrani A, Browning KS, Robaglia C. Plant lipoxygenase 2 is a translation initiation factor-4E- binding protein. Plant Mol Biol. 2000;44:129–140. doi: 10.1023/A:1006494628892. [DOI] [PubMed] [Google Scholar]
- Gallie DR, Browning KS. eIF4G functionally differs from eIFiso4G in promoting internal initiation, cap-independent translation, and translation of structured mRNAs. J Biol Chem. 2001;276:36951–36960. doi: 10.1074/jbc.M103869200. [DOI] [PubMed] [Google Scholar]
- Gallie DR, Caldwell C, Pitto L. Heat shock disrupts cap and poly(A) tail function during translation and increases mRNA stability of introduced reporter mRNA. Plant Physiol. 1995;108:1703–1713. doi: 10.1104/pp.108.4.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR, Le H, Tanguay RL, Browning KS. Translation initiation factors are differentially regulated in cereals during development and following heat shock. Plant J. 1998;14:715–722. doi: 10.1046/j.1365-313x.1998.00175.x. [DOI] [Google Scholar]
- Gallois JL, Charron C, Sanchez F, Pagny G, Houvenaghel MC, Moretti A, Ponz F, Revers F, Caranta C, German-Retana S. Single amino acid changes in the turnip mosaic virus viral genome-linked protein (VPg) confer virulence towards Arabidopsis thaliana mutants knocked out for eukaryotic initiation factors eIF(iso)4E and eIF(iso)4G. J Gen Vir. 2010;91:288–293. doi: 10.1099/vir.0.015321-0. [DOI] [PubMed] [Google Scholar]
- German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, De PE, Lu C, Schroth G, Meyers BC, Green PJ. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Hajela RK, Thomashow MF. Cold acclimation in Arabidopsis thaliana. Plant Phys. 1988;87:745–750. doi: 10.1104/pp.87.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyer C, Altmann M, Lee HS, Blanc A, Deshmukh M, Woolford JL, Jr, Trachsel H, Sonenberg N. TIF4631 and TIF4632: two yeast genes encoding the high- molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor 4F) contain an RNA recognition motif-like sequence and carry out an essential function. Mol Cell Biol. 1993;13:4860–4874. doi: 10.1128/mcb.13.8.4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradi A, Imataka H, Svitkin YV, Rom E, Raught B, Morino S, Sonenberg N. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradi A, Svitkin YV, Imataka H, Sonenberg N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc Natl Acad USA. 1998;95:11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez G, Vazquez-Pianzola P. Functional diversity of the eukaryotic translation initiation factors belonging to eIF4 families. Mech Dev. 2005;122:865–876. doi: 10.1016/j.mod.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Hillis DM, Bull JJ. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol. 1993;42:182–192. [Google Scholar]
- Horie Y, Ito H, Kusaba M, Tanaka R, Tanaka A. Participation of chlorophyll b reductase in the initial step of the degradation of light-harvesting chlorophyll a/b-protein complexes in Arabidopsis. J Biol Chem. 2009;284:17449–17456. doi: 10.1074/jbc.M109.008912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey SW, Humphrey GF. New spectrophotometric equations for determining chlorophylls a, b, c1 and c2 in higher plants, algae, and natural phytoplankton. Biochem Physiol Pflanz. 1975;167:191–194. [Google Scholar]
- Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Key JL, Lin CY, Chen YM. Heat shock proteins of higher plants. Proc Natl Acad USA. 1981;78:3526–3530. doi: 10.1073/pnas.78.6.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneeva NL, Lamphear BJ, Hennigan FLC, Rhoads RE. Mutually cooperative binding of eukaryotic translation initiation factor (eIF) 3 and eIF4A to human eIF4G–1. J Biol Chem. 2000;275:41369–41376. doi: 10.1074/jbc.M007525200. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- LeFebvre AK, Korneeva NL, Trutschl M, Cvek U, Duzan RD, Bradley CA, Hershey JW, Rhoads RE. Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. J Biol Chem. 2006;281:22917–22932. doi: 10.1074/jbc.M605418200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J, Lomakin IB, Sonenberg N, Pestova TV, Hellen CUT, Burley SK. A conserved HEAT domain within elF4G directs assembly of the translation initiation machinery. Mol Cell. 2001;7:193–203. doi: 10.1016/S1097-2765(01)00167-8. [DOI] [PubMed] [Google Scholar]
- Marintchev A, Wagner G. Translation initiation: structures, mechanisms and evolution. Q Rev Biophys. 2004;37:197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- Marintchev A, Wagner G. eIF4G and CBP80 Share a common origin and similar domain organization: implications for the structure and function of eIF4G. Biochem. 2005;44:12265–12272. doi: 10.1021/bi051271v. [DOI] [PubMed] [Google Scholar]
- Mayberry LK, Allen ML, Dennis MD, Browning KS. Evidence for variation in the optimal translation initiation complex: plant eIF4B, eIF4F, and eIF(iso)4F differentially promote translation of mRNAs. Plant Physiol. 2009;150:1844–1854. doi: 10.1104/pp.109.138438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicaise V, Gallois JL, Chafiai F, Allen LM, Schurdi-Levraud V, Browning KS, Candresse T, Caranta C, Le GO, German-Retana S. Coordinated and selective recruitment of eIF4E and eIF4G factors for potyvirus infection in Arabidopsis thaliana. FEBS Lett. 2007;581:1041–1046. doi: 10.1016/j.febslet.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Novillo F, Alonso JM, Ecker JR, Salinas J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc Natl Acad Sci USA. 2004;101:3985–3990. doi: 10.1073/pnas.0303029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberer M, Marintchev A, Wagner G. Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes Dev. 2005;19:2212–2223. doi: 10.1101/gad.1335305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Perez JM, Candela H, Micol JL. Understanding synergy in genetic interactions. Trends Genet. 2009;25:368–376. doi: 10.1016/j.tig.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Lorsch JR, Hellen CUT. The mechanism of translation initiation in eukaryotes. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational control in biology and medicine. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2007. pp. 87–128. [Google Scholar]
- Pitto L, Gallie DR, Walbot V. Role of the leader sequence during thermal repression of translation in maize, tobacco, and carrot protoplasts. Plant Physiol. 1992;100:1827–1833. doi: 10.1104/pp.100.4.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévôt D, Darlix JL, Ohlmann T. Conducting the initiation of protein synthesis: the role of eIF4G. Biol Cell. 2003;95:141–156. doi: 10.1016/S0248-4900(03)00031-5. [DOI] [PubMed] [Google Scholar]
- Raught B, Gingras AC. eIF4E activity is regulated at multiple levels. Int J Biochem Cell B. 1999;31:43–57. doi: 10.1016/S1357-2725(98)00131-9. [DOI] [PubMed] [Google Scholar]
- Raught B, Gingras A-C. Signaling to translation initiation. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational control in biology and medicine. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2007. pp. 369–400. [Google Scholar]
- Robaglia C, Caranta C. Translation initiation factors: a weak link in plant RNA virus infection. Trends Plant Sci. 2006;11:40–45. doi: 10.1016/j.tplants.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Schuler GD, Altschul SF, Lipman DJ. A workbench for multiple alignment construction and analysis. Proteins Struc Func Gen. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- Schutz P, Bumann M, Oberholzer AE, Bieniossek C, Trachsel H, Altmann M, Baumann U. Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein–protein interactions. Proc Natl Acad Sci USA. 2008;105:9564–9569. doi: 10.1073/pnas.0800418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin YV, Gradi A, Imataka H, Morino S, Sonenberg N. Eukaryotic initiation factor 4GII (eIF4GII), but not eIF4GI, cleavage correlates with inhibition of host cell protein synthesis after human rhinovirus infection. J Virol. 1999;73:3467–3472. doi: 10.1128/jvi.73.4.3467-3472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford (2000) PAUP*. Phylogenetic analysis using parsimony (*and Other Methods)
- Whitham SA, Anderberg RJ, Chisholm ST, Carrington JC. Arabidopsis RTM2 gene is necessary for specific restriction of tobacco etch virus and encodes an unusual small heat shock-like protein. Plant Cell. 2000;12:569–582. doi: 10.1105/tpc.12.4.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii M, Nishikiori M, Tomita K, Yoshioka N, Kozuka R, Naito S, Ishikawa M. The Arabidopsis cucumovirus multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J Virol. 2004;78:6102–6111. doi: 10.1128/JVI.78.12.6102-6111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.