Abstract
The peroxisome proliferator-activated receptors (PPARs) are central regulators of fat metabolism, energy homeostasis, proliferation, and inflammation. The three PPAR subtypes, PPARα, β/δ, and γ activate overlapping but also very different target gene programs. This review summarizes the insights into PPAR subtype-specific transactivation provided by genome-wide studies and discusses the recent advances in the understanding of the molecular mechanisms underlying PPAR subtype specificity with special focus on the regulatory role of AF-1.
1. Introduction
The peroxisome proliferator-activated receptors (PPARs) constitute a subgroup of the nuclear receptor (NR) family. The founding member of the family, PPARα, was identified in 1990 and named by its ability to become activated by chemicals known to induce peroxisome proliferation in rodents [1]. Subsequently, the two other PPAR subtypes, PPARβ/δ and PPARγ, were identified by homology screens [2, 3]. The three PPAR subtypes are encoded by distinct genes located on different chromosomes (reviewed by [4]). Alternate promoter usage and splicing give rise to two different protein isoforms from the PPARγ gene called PPARγ1 and PPARγ2, with the latter containing 30 additional amino acids at the N-terminus (Swiss-Prot http://www.expasy.org/). All three PPAR subtypes can be activated by a large variety of fatty acids and fatty acid metabolites, such as hydroxylated eicosanoids, prostaglandins, and leukotrienes, and by many synthetic compounds. PPARα is specifically activated by fibrates and other hypolipidemic drugs, whereas PPARγ is activated by the insulin-sensitizing, antidiabetic thiazolidinedione drugs [4].
The PPARs play important regulatory roles in numerous cellular processes related to metabolism, inflammation, differentiation, proliferation, and atherosclerosis (reviewed by [5, 6]). The three subtypes display dissimilar patterns of tissue distribution and activate both overlapping and distinct sets of target genes. Most notably, whereas PPARα [7, 8] and -β/δ [9] are potent activators of genes involved in lipid oxidation, PPARγ stands out by its additional ability to activate lipogenic genes and adipocyte differentiation [10, 11]. In fact, PPARγ is obligate for adipocyte differentiation and is sufficient to transform many nonadipogenic cell lines into adipocyte-like cells [12, 13]. As a reflection of these subtype-specific properties, PPARα and PPARβ/δ are highly expressed in tissues with high β-oxidation rates such as liver, muscle, heart, and brown adipose tissue. By contrast PPARγ is highly expressed in adipose tissue, and PPARγ2 is fat selective, whereas the PPARγ1 isoform is expressed at low levels in several tissues, including colon, spleen, liver, and muscles. PPARβ/δ is the most ubiquitously expressed subtype with the highest levels found in the intestines and keratinocytes (see [14] and reviewed by [5]).
Like most NRs, the PPAR protein structure consist of four domains: The N-terminal A/B-domain containing the ligand-independent activation function 1 (AF-1), the C-domain, which is the DNA-binding domain (DBD), the D-domain, also called the hinge region, and finally the E-domain, commonly referred to as the ligand binding domain (LBD). The E-domain contains the ligand-dependent AF-2, which is highly dependent on the C-terminal helix 12. While the A/B -and D-domains are only poorly conserved between the PPAR subtypes, the C -and E-domains share a high degree of sequence and structural homology (reviewed by [4]). In fact, the C-domains are completely interchangeable between the PPAR subtypes and appear to have no effect on specificity [15, 16]. The PPARs bind DNA as obligate heterodimers with members of the retinoid X receptor (RXR) family of nuclear receptors to modified direct repeat 1 elements (DR1) with the consensus sequence 5′-AACTAGGNCA A AGGTCA-3′. The PPARs occupy the 5′ extended half site of the binding site [17].
Given the important role of the PPARs in regulation of metabolism, inflammation, differentiation, and cellular growth, a large number of specific and potent synthetic ligands have been generated. This has spurred a huge interest in understanding the molecular mechanisms of PPAR transactivation and in genome-wide approaches to identify new PPAR target genes. These studies have provided the field with important insights into how different ligands modulate the transactivation capacity of the PPARs and to what extent the individual PPAR domains are involved in ensuring subtype-specificity by enabling or preventing transactivation of specific subsets of target genes. This paper focuses on the recent advances in understanding PPAR subtype-specific transactivation as seen from a molecular and a genome-wide perspective. In particular, the regulatory role of the AF-1 in maintaining PPAR subtype-specificity and transactivation capacity is reviewed.
2. Molecular Mechanisms of PPAR Subtype Specific Transactivation
The ability of the individual PPAR subtypes to induce very different cellular fates is intriguing because they share a high degree of sequence- and structural homology and activate overlapping sets of target genes. Nevertheless, the PPARs maintain a high degree of subtype-specificity when expressed in a given cell line at comparable levels [11, 18, 19] and adenoviral expression of PPARγ1 in mouse liver leads to induction of several genes which are not readily activated by PPARα, that is, genes involved in lipid accumulation and adipogenesis [20]. These results indicate that although the chromatin setting ultimately determines the accessibility of the PPAR response elements, intrinsic properties of the individual PPAR subtypes are key determinants of the gene programs that can be activated. Mechanisms maintaining subtype-specificity are of significant general interest because subtype selective gene regulation is a recurrent theme among transcription factor families, and therefore several attempts have been made to address this issue. These studies reported that the PPAR subtypes differ only little in their ability to transactivate artificial promoter reporter constructs in transient transfections [21–24] and display limited specificity in their binding to naked DNA containing target gene PPAR response elements (PPREs) in mobility shift assays, although poorly conserved PPREs preferentially bind PPARγ:RXR heterodimers [25]. By contrast, work from our laboratory has shown that in the endogenous chromatin setting, the binding level of a particular PPAR subtype to a given genomic PPRE generally correlates with its potential to transactivate the corresponding target gene, although exceptions clearly exist [11].
2.1. The LBD and AF-2
Because of the obvious therapeutic potential for modulating PPAR transactivation through the administration of ligands that bind to the E-domain, the cofactor interaction surfaces and the molecular mechanisms underlying activation of the AF-2 have been extensively studied. The recent publication of an almost complete structure of the DNA bound PPARγ:RXRα heterodimer [26] represents a major breakthrough in the understanding of the positioning of the PPARγ and RXR domains relative to each other and their interactions. Unfortunately, the PPARγ and RXRα A/B-domains could not be crystallized, most likely due to their high mobility and lack of internal structure. However, the overall impression from this study is that the PPARγ LBD is the centerpiece of the complex, around which all other domains from both PPARγ and RXRα are arranged [26]. This accentuates previous descriptions of extensive interdomain cross-talk in the PPARs [27, 28].
The PPARγ LBD, is composed of 13 α-helices and a small four-stranded β-sheet that forms a large (approx. 1300 Å3) T-shaped hydrophobic ligand-binding pocket typical for the promiscuous NRs, such as PPARs and the pregnane X receptor (PXR), that bind many different ligands with low affinity [29]. The ligand-binding pocket of PPARβ/δ is narrower than those of PPARγ and PPARα, which appears to be a major determinant of ligand binding, as it prohibits binding of TZDs and severely decreases the affinity of PPARβ/δ towards fibrate ligands due to the bulky acid and alkyl groups on these compounds [30]. PPARα contains the most lipophilic ligand binding pocket, which potentially explains why PPARα, as opposed to PPARβ/δ and PPARγ, binds a variety of saturated fatty acids [31]. An additional layer of ligand binding specificity is imposed by the size and charge of the amino acids lining the ligand binding pocket [30]. Ligand-binding affects the stability of the PPARs with liganded PPARα being transiently stabilized [32] while proteasomal degradation of PPARγ is increased upon ligand binding [33].
The structural basis for AF-2-mediated transcriptional activity is a ligand-induced conformational change in the LBD, causing the most C-terminal helix 12 to fold up against the core [30]; thereby generating an activation surface, often described as a charge clamp, onto which coactivators can dock. Many coactivators bind to the NR E-domains through a motif with the consensus sequence LXXLL, which facilitates direct interaction with the charge clamp. In contrast, corepressors often have a LXXXIXXX(I/L) motif that interacts with an overlapping surface but is unable to fit into the charge clamp (reviewed by [34]). The variations in the primary sequences of the PPAR LBDs results in slight differences in the cofactor interaction surfaces and ligand-induced conformations of these domains [35, 36] and PPAR subtype-specificity is thought to be partly imposed by differential affinity of the receptors towards the individual cofactors [37, 38]. Thus, the cofactor expression pattern in a specific cellular context may favor transcriptional activation of one PPAR subtype over the others.
Probably, the transcriptional activity of the PPARs should not be regarded as determined by a static positioning of helix 12 in either the “on” or “off” position. Rather, it appears that ligand-binding shifts the equilibrium of the different helix 12 positions in the receptor population towards the more active conformations (reviewed by [39]) [40]. Helix 12 is absolutely necessary for the activity of AF-2, and PPAR mutants with helix 12 partly or completely deleted are dominant negative inhibitors of PPAR signaling [41, 42]. Interestingly, the requirement for helix 12 does not appear to equalize a requirement for ligand-binding. Recently, it has been demonstrated that a natural PPARγ mutant that is impervious to activation by virtually all known agonists has intact adipogenic potential [43]. Indeed, the PPARs display high basal transcriptional activity that can be explained by the AF-1 in the A/B-domain and the presence of endogenous ligands. In addition, ligand-independent recruitment of coactivators to the AF-2 has been observed in overexpression and in vitro studies, indicating that in addition to the AF-1, the AF-2 also contributes to ligand-independent transactivation. Perhaps, this is possible because the shift in the positioning of helix 12 upon ligand-binding is not as pronounced for the PPARs as for the steroid NRs [44, 45].
2.2. The Elusive Structure of the PPAR A/B-Domain
Compared to the AF-2, the mechanisms for AF-1-mediated transcriptional activity are less well-understood despite several publications pointing to an important role for the A/B-domains in maintaining PPAR subtype-specificity [15, 16, 46]. The fact that it so far has proven impossible to crystallize a NR A/B-domain indicates that these domains are poorly structured, a notion that has been confirmed by experiments using deuterium exchange mass spectrometry [26], circular dichroism spectroscopy, nuclear magnetic resonance spectroscopy [47, 48], or limited proteolysis [49, 50]. It has been suggested that secondary structure formation is an important step in AF-1-mediated transactivation and both the PPARα [51] and glucocorticoid receptor (GR) [47] AF-1 display α-helical characteristics in the presence of trifluoroethanol, a strong α-helix stabilizing agent. Furthermore, mutational analyses of the PPARα [51], GR [52, 53], and hepatocyte nuclear factor 4 (HNF-4) [54] AF-1 domains have shown that conservation of the hydrophobic amino acids, that potentially could be involved in α-helix formation is especially important for the transcriptional activity, while mutation of individual acidic amino acids made less of an impact, suggesting that α-helix formation is an important step in AF-1-mediated gene activation. Interestingly, it was recently shown that mitogen-activated protein kinase (MAPK) phosphorylation of serine 211 in the glucocorticoid receptor A/B-domain induces formation of secondary or tertiary structure in this region which facilitates interaction between the AF-1 and coregulators thereby leading to enhanced transcriptional activity [55]. Another model proposes that, as individual coactivators offer different surfaces for unstructured activation-domains to fold up on, distinct conformations could be induced by the different coactivators, thereby resulting in differential transcriptional activity or specificity [56]. Several of the cofactors reported to interact with NR A/B-domains have no sequence or known structural homology, and this model offers an attractive explanation for how that is possible.
2.3. PPAR Transcriptional Activity Is Regulated by Modification of the A/B-Domain
It is well-established that posttranslational modifications of the PPAR A/B-domains influence the transcriptional activity of both the AF-1 and AF-2 through various mechanisms. The PPARα and –γ A/B-domains, but not the PPARβ/δ A/B-domain, are modified by phosphorylation. MAPK- phosphorylation of serine 12 and 21 in the PPARα A/B-domain, enhances the transcriptional activity by transiently increasing receptor stability through reduced ubiquitination [32]. Oppositely, phosphorylation of serine 76 by glycogen synthase kinase 3 (GSK3) leads to increased ubiquitination and degradation of PPARα [93]. MAPK mediated phosphorylation of serine 82 in the PPARγ1 A/B-domain (corresponding to serine 112 in PPARγ2) has been shown to inhibit both ligand-dependent and independent transactivation [94], the former by decreasing the ligand-binding affinity of the receptor [27]. Interestingly, it was recently published that the phosphorylated PPARγ AF-1 domain is bound by the peptidyl-prolyl isomerase Pin1, whereby both polyubiquitination and the transcriptional activity of PPARγ is inhibited, possibly due to the decreased turnover rate of the receptor [95]. Oppositely, it has also been reported that S112 phosphorylation of PPARγ2 by a constitutively active MAPK kinase (MEK) [96] or by cyclin-dependent kinase 9 (Cdk 9) residing in the positive transcription elongation factor b complex (P-TEFb) results in increased transcriptional activity [71]. Thus, it appears that the cellular and/or molecular context determines the transcriptional effect of PPARγ A/B-domain phosphorylation. Mice homozygous for the S112A mutation are indistinguishable from the wild types on a normal diet, but they are significantly more glucose tolerant in the setting of diet-induced obesity [97], an effect analogous to the outcome of PPARγ activation by TZD treatment. In line with phosphorylation of S112 decreasing the insulin sensitizing actions of PPARγ, humans carrying a P115Q mutation, that blocks phosphorylation of serine 114 (corresponding to serine 112 in mice), are extremely obese but are also less insulin-resistant than expected based on their degree of obesity [98]. In addition to affecting the activity of PPARγ through regulation of MAPK, MEK has also been reported to interact directly with PPARγ and promote its nuclear export [99]. Recently, it was furthermore reported that phosphorylation of serine 16 and 21 of PPARγ by Casein-kinase-II likewise promotes shuttling of PPARγ from the nucleus to the cytosol [100]. Besides phosphorylation, PPARγ transactivation is also repressed by conjugation of small ubiquitin-related modifier (SUMO) to lysine 107 in the A/B-domain [101].
2.4. The PPAR A/B-Domain Is Involved in the Recruitment of Cofactors
In addition to regulating the PPAR transcriptional activity by affecting receptor stability, cellular compartmentalization, and perhaps interdomain communication in response to the posttranslational modification status, the PPAR A/B-domains are involved in recruiting a handful of cofactors. The PPARγ AF-1 is the most well-described of the three PPAR A/B-domains and the coactivators Tat-interacting protein 60 (Tip60) [82] and PPARγ coactivator-2 (PGC-2) [15] but also the corepressor tribbles homolog 3 (TRB3) [92] are recruited to PPARγ exclusively through binding to the A/B-domain. Both PPARα and PPARγ have been shown to bind the histone acetyl transferase (HAT) coactivators p300 and CREBP-binding protein (CBP) through interaction surfaces in both the A/B-and E-domains in GST-pulldown studies [61]. The significance of the A/B-domain interaction was previously unknown, but we have recently shown that recruitment of p300 and CBP is compromised by deletion of the PPARγ A/B-domain specifically on the PPREs of the target genes that required AF-1 activity to become fully activated [46]. The SWI/SNF chromatin remodeling complex BRG1-associated factor 60c (BAF60c) subunit which interacts directly with PPARγ likewise appear to have the potential to bind both the A/B- and E-domains, but the AF-1 interaction is stronger and ligand-independent [58]. PPARα has been shown to be coactivated by binding of the target gene product bifunctional enzyme (BFE) to the A/B-domain [57], and the ubiquitin ligase murine double minute 2 (MDM2) is bound by the N-terminal of all three PPAR subtypes [69]. In addition, we have recently demonstrated that the Mediator complex is tethered to the PPARγ A/B-domain through the MED14 subunit and that MED14 is required for full transcriptional activation of PPARγ subtype-specific genes by PPARγ [102]. A complete list of the cofactors currently known to interact with the PPARs is shown in Table 1.
Table 1.
Cofactors regulating PPAR activity.
| Coactivator | Enzymatic activity | Interaction | Reference |
|---|---|---|---|
| Bifunctional enzyme (BFE) | Dehydrogenase | α (A/B) | [57] |
| BRG1-associated factor 60c (BAF60c) | None | γ (A/B) | [58] |
| Coactivator-associated arginine methyltransferase 1 (CARM1) | HMT | SRC-1-3 | [59] |
| Constitutive coactivator of PPARγ (CCPG) | None | γ (D) | [60] |
| CREBP- binding protein (CBP) | HAT | α,γ (A/B, LBD, SRC) | [61] |
| Hydrogen peroxide-inducible clone 5 protein (Hic5) | None | γ | [62] |
| LIM domain-only protein (LMO4) | None | γ (D, LBD) | [63] |
| Lipin 1 | None | α, PGC-1 | [64] |
| Mediator subunit 1 (MED1) | None | α, β/δ,γ (LBD) | [65] |
| Mediator subunit 14 (MED14) | None | γ (A/B) | [66] |
| Multiple Endocrine Neoplasia type 1 (MEN1) | None | γ (LBD) | [67] |
| Multiprotein bridging factor 1 (MBF-1) | None | γ (D, LBD) | [68] |
| Murine double minute 2 (MDM2) | Ubiquitin ligase | α, β/δ (A/B) | [69] |
| p300 | HAT | α,γ (A/B, LBD, SRC) | [61] |
| Poly ADP-ribose polymerase 2 (PARP-2) | ADP-ribose polymerase | γ | [70] |
| Positive transcription elongation factor b complex (P-TEFb) | Kinase | γ | [71] |
| PPARα-interacting complex 285 (PRIC285) | DNA helicase | α, β/δ, γ (DBD) | [72, 73] |
| PPARα-interacting complex 320 (PRIC320) | DNA helicase | α | [74] |
| PPAR-interacting protein (PRIP) | None | α,γ (LBD) | [75] |
| PPARγ coactivator 1α (PGC-1α) | None | α,γ (DBD) | [76] |
| PPARγ coactivator 2 (PGC-2) | None | γ (A/B) | [15] |
| PR domain containing 16 (PRDM16) | None | α,γ | [77] |
| PRIP-interacting protein with methyltransferase domain (PIMT) | HMT | PRIP, CBP, MED1 | [78] |
| Protein arginine N-methyltransferase 2 (PRMT2) | HMT? | γ | [79] |
| Steroid receptor coactivator-1 (SRC-1) | HAT | α, β/δ, γ (LBD) | [80] |
| Steroid receptor coactivator-2 (SRC-2) | HAT | α,γ (LBD) | [81] |
| Steroid receptor coactivator-3 (SRC-3) | HAT | α, β/δ,γ (LBD) | [81] |
| Tat interactive protein (Tip60) | HAT | γ (A/B) | [82] |
| Thyroid hormone receptor interacting protein 3 (TRIP3) | None | γ (LBD) | [83] |
|
| |||
| Corepressor | Enzyme activity | Interaction | Reference |
|
| |||
| Histone deacetylase 1 (HDAC1) | HDAC | NCoR, SMRT | [84] |
| Histone deacetylase 3 (HDAC3) | HDAC | NCoR, SMRT | [85, 86] |
| Insulin-like growth factor-binding protein-3 (IGFBP-3) | None | γ | [87] |
| Nuclear receptor corepressor 1 (NCoR) | None | α, β/δ, γ | [88] |
| Receptor-interacting protein 140 (RIP140) | None | α,γ (LBD) | [89] |
| Scaffold attachment factor B1 (SAFB1) | None | α, β/δ, γ | [90] |
| Silencing mediator of retinoid and thyroid receptors (SMRT) | None | α, β/δ, γ | [88] |
| Silent mating type information regulation 2 homolog 1 (SIRT1) | HDAC | NCoR, PGC-1 | [91] |
| Tribbles homolog 3 (TRB3) | None | γ (A/B) | [92] |
Histone methyltransferase (HMT), Histone acetyltransferase (HAT), and Histone deactylase (HDAC).
2.5. The A/B-Domains Play an Important Role in Maintaining PPAR Subtype-Specificity
Because the A/B-domain is the least conserved region among the PPARs, it has long been suspected that subtype-specificity, and target gene selectivity is completely or partly mediated through this domain. This hypothesis has been investigated by deleting the A/B-domain or by constructing chimeric PPARs where domains have been swapped between the subtypes.
Despite the undisputed observation that the PPARα and -γ A/B-domains are potent transactivators when expressed as GAL4-fusion proteins [51] there has been controversy regarding the physiological importance of the activity of the PPAR A/B-domains. Deletion of the A/B-domain was reported to have no effect [82, 103] or to significantly decrease PPAR-mediated expression from a reporter plasmid in transient transfections [51]. Interestingly, deletion of the A/B-domain affected the transcriptional activity of PPARα differentially depending on the target gene promoter used in the reporter construct. One study employed the acyl-CoA oxidase promoter and found that the A/B-domain contributed significantly to the transcriptional activity of PPARα [51], while another study showed that luciferase expression driven by the cytochrome P450 4A6 promoter was completely unaffected by deletion of the AF-1 [103]. We have recently on a global scale shown that deletion of the PPARγ A/B-domain selectively decreases the transactivation potential of PPARγ on the highly subtype-specific target genes. We found that out of 257 PPARγ-induced genes only 25 are dependent on the PPARγ A/B-domain to become fully activated in the presence of the TZD rosiglitazone. The A/B domain dependent genes are the highly PPARγ selective target genes many of which are involved in lipid storage. Notably, in the absence of synthetic agonist, transactivation of this subgroup of genes in particular relies almost exclusively on the PPARγ A/B-domain [46].
The importance of the PPAR A/B-domains in maintaining subtype-specificity has been indicated by several studies showing that these domains are not interchangeable. Thus, although both the α and γ A/B-domains contain potent activation functions [51], they cannot functionally substitute for each other as evidenced by the observation that a chimera consisting of the PPARα A/B-domain and the PPARγ CDE-domains is able to transactivate the PPARγ selective target genes similar to that of PPARγCDE [46]. However, the A/B-domains of PPARγ and PPARα can impose a partial subtype-specific activation in the context of a noncognate receptor. Spiegelman and coworkers showed that swapping the PPARγ A/B-domain on to the nonadipogenic PPARβ/δCDE conferred adipogenic potential to this receptor subtype [15]. Subsequently, Tontonoz and coworkers reported that the PPAR A/B-domains function to restrict target gene activation in the context of the cognate receptor and showed that A/B-domain deleted PPARβ/δ (PPARβ/δCDE) can induce adipogenesis. This study thereby raised the question as to what degree the adipogenic potential of the PPARγ A/B-domain-PPARβ/δCDE chimera used in the Spiegelman study arose by the addition of the PPARγ A/B-domain or the lack of the PPARβ/δ A/B-domain [13, 16]. In agreement with the A/B-domains conferring subtype-specificity to the PPARs in part by limiting nonselective target gene activation, we have shown that compared to the full length receptors, the A/B-domain deleted PPARαCDE and -γCDE are far better transactivators of the noncognate highly subtype selective PPAR target genes normally activated by the opposite subtype. However, reminiscent of the Spiegelman data we also found that addition of the PPARα A/B-domain greatly enhances the ability of PPARγCDE to activate a PPARα specific target gene [46]. Thus, it appears that the A/B-domains contribute to maintaining PPAR subtype-specificity by both potentiating activation of the highly subtype selective target genes, and by restricting nonselective target gene activation exclusively in the context of the cognate CDE domains. By contrast, the A/B-domain plays only a minor role in the activation of the target genes shared between the subtypes (Figure 1).
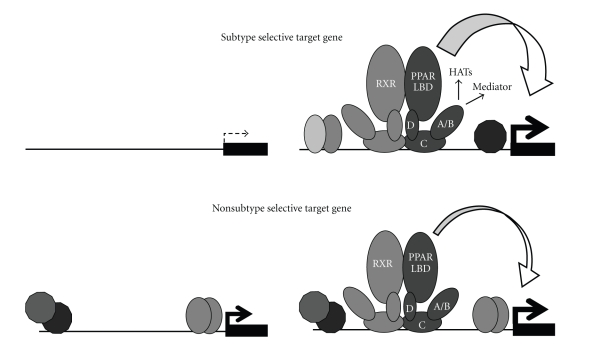
Figure 1.
The PPAR A/B-domains potentiate transactivation of the highly subtype selective target genes. Illustration of the differential requirement for the PPAR A/B-domains in the transactivation of highly subtype selective and non-subtype selective target genes. The PPAR subtype specific target genes are generally expressed at very low levels in the absence of PPARs, but expression is dramatically increased upon introduction of full length exogenous PPAR. The activities of the PPAR A/B-domains are necessary to obtain this potent induction of the highly subtype specific target genes, presumably by facilitating recruitment and tethering of histone acetylase complexes (HATs) and the Mediator complex. Conversely, non-subtype selective PPAR target genes are usually already expressed at high levels in the absence of PPARs, and their expression levels are only increased by a few fold in response to ectopic PPAR expression. The PPAR A/B-domains appear to be dispensable for transactivation of this group of target genes.
3. Genome-Wide Approaches to Mapping PPAR Subtype-Specific Transactivation
The PPAR transcriptome in cells and tissues has been mapped by ectopically expressing a particular subtype and/or treating with a specific agonist and mapping the changes in gene expression using array technology. More recently the combination of chromatin immunoprecipitation (ChIP) with microarray analysis (ChIP-chip), high throughput sequencing (ChIP-seq), or with pair end-tagging sequencing (ChIP-PET) has allowed the mapping of the PPAR cistrome in cells and tissues following various treatments. These global techniques have led to important insights into the role of the different PPAR subtypes in the regulation of metabolism and differentiation and into the action of PPAR agonists.
3.1. Expression Array Studies
Although microarray studies of NIH-3T3 fibroblast overexpressing PPARβ/δ have confirmed that this PPAR subtype rightfully is recognized as being an inducer of genes involved in energy expenditure and β-oxidation of fatty acids [16] it appears that at least in insulin-resistant (db/db) mice, activation of this pathway by PPARδ-specific agonists is limited to the muscles. Administration of PPARβ/δ agonist ameliorates both muscle and liver insulin resistance in db/db mice by lowering the hepatic glucose output, increasing glucose disposal, and inhibiting fatty acid release from the adipose tissue. Surprisingly however, the expression arrays only detected induction of carnitine palmitoyltransferase 1 (Cpt1), a key gene in fatty acid β-oxidation, in the muscles, whereas the pathways responsible for the increased glucose disposal appeared to be hepatic fatty acid synthesis and pentose phosphate shunt that generates NADPH to provide reducing power for lipid synthesis [104]. Consistent with previous reports [105], PPARβ/δ agonist treatment did lead to increased β-oxidation rates in the muscles, suggesting that PPARβ/δ promote insulin sensitivity by consuming glucose through induction of hepatic fat-production in combination with a counterbalancing fat burning in muscle [104]. A recent study comparing hepatic gene regulation between wild type and PPARβ/δ -or PPARα knockout mice showed that while PPARα expression is highly upregulated during fasting, P p a r β/δ mRNA is downregulated. In accordance with this finding, the differences in gene expression between the wild type and PPARβ/δ knockout mice were more pronounced in the fed state but surprisingly a relatively large subset of genes were downregulated in a PPARβ/δ-dependent manner during the fast. Interestingly, there is only limited overlap between the hepatic genes regulated in a PPARα or PPARβ/δ-dependent manner in the fed state, while a large proportion of the target genes appear to be regulated by both subtypes in the fasted state. It is evident that some of the differential effects on liver gene expression in the two knockout mouse models may be due to indirect effects imposed by other tissues; however, in agreement with the general perception of PPARβ/δ being an inhibitor of inflammation, pathway analyses showed that several gene sets involved in these processes were enriched in the knockout mice in the fed state. In the fasted state, the electron transport and oxidative phosphorylation pathways were decreased and in both metabolic states deletion of PPARβ/δ resulted in downregulation of genes involved in lipoprotein metabolism and carbohydrate metabolism, which included glycogen metabolism, glycolysis, gluconeogenesis, and the pentose phosphate pathway [106]. In agreement with the conclusion from the db/db mouse study indicating that PPARβ/δ is an important regulator of glucose disposal and utilization [104], these changes in gene expression resulted in significantly increased fasting plasma glucose levels in the PPARβ/δ knockout mice [106]. Besides studies of the PPARα knockout mice [106, 107], other approaches to determine the PPARα transcriptome includes overexpression studies in fibroblasts [16], exposure of hepatoma cell lines to synthetic PPARα agonist [108], and in vivo examinations of the alterations in the gene expression pattern of mouse and monkey liver [109, 110] and mouse intestine [111] in response to agonist treatment. These studies unanimously report that PPARα is the major inducer of β-oxidation in these tissues. In addition, it has also been reported that PPARα function to repress amino acid catabolism [112].
The PPARγ transcriptome of the adipogenic 3T3-L1 cell line has been characterized in several expression array studies because of the high endogenous expression level of PPARγ and the observation that white adipose tissue is essential for the insulin sensitizing effects of the TZD PPARγ agonists [113, 114]. Studies using these agonist to drive the differentiation of preadipocytes show that the TZDs are potent activators of adipogenesis and induce or enhance the expression of adipocyte specific markers and genes involved in lipid storage and transport, but also lipid hydrolysis and oxidation [115, 116]. Interestingly however, when mature adipocytes are exposed to TZDs for a few hours up to a couple of days, it leads to decreased expression of both PPARγ itself and of lipid storage and adipokine genes while fatty acid activation and degradation is induced [116, 117]. The global effects of TZD treatment on gene expression has also been investigated in macrophages [118], colon cancer cells [119], aorta [120], and dendritic cells [121] with regulation of genes involved in lipid metabolism and inflammation being a consistent finding in these studies.
Other approaches to annotate the PPARγ transcriptome include analysis of NIH-3T3 fibroblasts transduced with retrovirus encoding PPARγ2 [16, 122] and adenoviral overexpression of PPARγ1 in the liver of PPARα knockout mice for 2–6 days [20]. The latter resulted in hepatic steatosis, thus underscoring the lipogenic potential of PPARγ. In order to increase the chances of identifying genes targeted directly by PPARγ as opposed to genes being differentially expressed as a consequence of secondary regulation, we made use of acute adenoviral expression of the PPARs in NIH-3T3 fibroblasts, which have very low levels of endogenous PPARs [11]. This system allows us to induce rapid expression of the PPARs and subsequently evaluate the immediate effects on target gene activity at the mRNA level within 8 h after transduction, whereby secondary effects (e.g., induction of endogenous PPARs) on gene expression were minimized. By combining this cellular system with expression array analysis, we found that ectopic PPARγ2 expression in combination with TZD treatment acutely activated a large number of known and novel target genes in the NIH-3T3 cells. Both expression of genes involved in lipid anabolic and catabolic pathways were induced but the net outcome was lipid accumulation [46]. These results corroborate previous findings from our lab that when expressed at adipocyte levels, PPARγ2 is a strong transactivator capable of activating most PPAR target genes, even if other subtypes expressed at the same level would be better activators [11].
3.2. ChIP-Chip and ChIP-Seq Studies
The generation of genome-wide profiles of PPAR, RXR, and cofactor occupancy using the ChIP-chip, ChIP-seq, and ChIP-PET technologies has significantly increased the understanding of PPAR-mediated transactivation. First, these studies have shown that most target genes have multiple PPAR binding sites not only in the proximal promoter region but also in introns and at distant positions up- and downstream of the gene. Notably, about 50% of all PPAR:RXR-binding sites are found in introns. This distributions of binding sites reflects that of most other nuclear receptors [123–125] and of the several other transcription factors [126–128]. While these studies provide invaluable help in determining the position of binding sites, they also complicate the picture of functional PPREs and underscore the weaknesses of traditional promoter and enhancer characterization, where often only the proximal promoter or genomic sequences immediately upstream of the transcription start site are cloned in front of a luciferase reporter.
The first PPAR cistrome to be published was that of PPARγ in differentiating and mature 3T3-L1 adipocytes as mapped by ChIP-seq [129] and ChIP-chip [130]. Subsequently, the PPARγ cistrome in 3T3-L1 cells has also been mapped by others using ChIP-chip [131], ChIP-seq [132], and ChIP-PET [133]. The mappings revealed between 2730 and 7000 genomic PPARγ binding sites, depending on the method and false discovery rate employed. Notably, all genes encoding proteins involved in fatty acid handling and storage as well as lipolysis were found to have adjacent PPARγ:RXR binding sites, but surprisingly this was also true for most genes encoding enzymes involved in steroid metabolism [133], glycolysis, the pentose phosphate pathway, and the TCA cycle [129, 131]. Interestingly, a significant overlap between PPARγ:RXR binding sites and binding sites of CCAT/enhancer binding protein (C/EBP) α and -β was found [129, 130] indicating that the cooperativity between PPARγ and members of the C/EBP family in the regulation of adipocyte gene expression (reviewed previously [134]) takes place on a much larger scale than previously anticipated.
A recent study profiling the PPARγ cistrome in primary mouse macrophages found that there was only very limited overlap between the genomic sites bound by the receptor in this cell type and in adipocytes. Interestingly, the transcription factor PU.1, which is involved in monocyte development and not expressed in adipocytes, was enriched at the macrophage specific sites. This study thus addresses the intriguing question of how cell type-specific PPARγ transactivation is achieved at the level of chromatin binding and suggests that tissue-specific factors may play a role in facilitating PPARγ binding to the tissue selective binding sites [132].
So far, genome-wide cistromes are not available for other PPAR subtypes, but recently PPARα binding sites in a human hepatoma cell line was mapped by ChIP-chip using an array covering promoter regions from 7.5 kb upstream to 2.5 kb downstream of the transcription start site. This study showed increased binding of PPARα to 4220 genomic regions in response to agonist treatment [135]. The group of genes assigned to these binding sites that were upregulated as determined by microarray analysis, clustered as being involved in sterol and lipid biosynthetic pathways, which is surprising given the general perception of PPARα as an inducer of lipid degradation. The downregulated genes were involved in innate and humoral immune response, which is consistent with the well-described anti-inflammatory activity of PPARα (Reviewed previously [5]).
4. Summary and Perspectives
As described in this paper, several molecular mechanisms conferring subtype-specificity to the PPARs or leading to preferential activation of a specific PPAR subtype in a certain cellular context have been elucidated. (1) The PPAR subtypes bind to the individual genomic PPREs with differential affinity. (2) The PPARs are activated by different ligands. (3) The PPAR subtypes display differential affinity towards various cofactors. (4) PPAR transcriptional activity is modulated by subtype-specific posttranslational modifications. (5) The PPAR A/B-domains potentiate subtype-specific activation of target genes while restricting nonselective target gene activation.
Most likely, PPAR subtype-specificity is maintained through the concerted effects of the regulatory mechanisms exerted by the individual PPAR domains or communicating PPAR and RXR domains. However, the current knowledge strongly suggests that the relative importance of these contributions is differential and that especially the A/B-domains are important mediators of PPAR subtype-specificity. Future studies should aim at pinpointing the exact sections of the A/B-and E-domains, and potentially the D-domain, that are involved in maintaining PPAR subtype-specificity and to fully elucidate the molecular mechanisms underlying this activity.
References
- 1.Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347(6294):645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 2.Kliewer SA, Forman BM, Blumberg B, et al. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(15):7355–7359. doi: 10.1073/pnas.91.15.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dreyer C, Krey G, Keller H, Givel F, Helftenbein G, Wahli W. Control of the peroxisomal β-oxidation pathway by a novel family of nuclear hormone receptors. Cell. 1992;68(5):879–887. doi: 10.1016/0092-8674(92)90031-7. [DOI] [PubMed] [Google Scholar]
- 4.Escher P, Wahli W. Peroxisome proliferator-activated receptors: insight into multiple cellular functions. Mutation Research. 2000;448(2):121–138. doi: 10.1016/s0027-5107(99)00231-6. [DOI] [PubMed] [Google Scholar]
- 5.Kersten S, Desvergne B, Wahli W. Roles of PPARS in health and disease. Nature. 2000;405(6785):421–424. doi: 10.1038/35013000. [DOI] [PubMed] [Google Scholar]
- 6.Keshamouni VG, Han S, Roman J. Peroxisome proliferator-activated receptors in lung cancer. PPAR Research. 2007;2007:10 pages. doi: 10.1155/2007/90289. Article ID 90289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gulick T, Cresci S, Caira T, Moore DD, Kelly DP. The peroxisome proliferator-activated receptor regulates mitochondrial fatty acid oxidative enzyme gene expression. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(23):11012–11016. doi: 10.1073/pnas.91.23.11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor α mediates the adaptive response to fasting. Journal of Clinical Investigation. 1999;103(11):1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y-X, Lee C-H, Tiep S, et al. Peroxisome-proliferator-activated receptor δ activates fat metabolism to prevent obesity. Cell. 2003;113(2):159–170. doi: 10.1016/s0092-8674(03)00269-1. [DOI] [PubMed] [Google Scholar]
- 10.Mueller E, Drori S, Aiyer A, et al. Genetic analysis of adipogenesis through peroxisome proliferator-activated receptor γ isoforms. Journal of Biological Chemistry. 2002;277(44):41925–41930. doi: 10.1074/jbc.M206950200. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen R, Grøntved L, Stunnenberg HG, Mandrup S. Peroxisome proliferator-activated receptor subtype- and cell-type-specific activation of genomic target genes upon adenoviral transgene delivery. Molecular and Cellular Biology. 2006;26(15):5698–5714. doi: 10.1128/MCB.02266-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim SW, Her SJ, Kim SY, Shin CS. Ectopic overexpression of adipogenic transcription factors induces transdifferentiation of MC3T3-E1 osteoblasts. Biochemical and Biophysical Research Communications. 2005;327(3):811–819. doi: 10.1016/j.bbrc.2004.12.076. [DOI] [PubMed] [Google Scholar]
- 13.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell. 1994;79(7):1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 14.Escher P, Braissant O, Basu-Modak S, Michalik L, Wahli W, Desvergne B. Rat PPARs: quantitative analysis in adult rat tissues and regulation in fasting and refeeding. Endocrinology. 2001;142(10):4195–4202. doi: 10.1210/endo.142.10.8458. [DOI] [PubMed] [Google Scholar]
- 15.Castillo G, Brun RP, Rosenfield JK, et al. An adipogenic cofactor bound by the differentiation domain of PPARγ . The EMBO Journal. 1999;18(13):3676–3687. doi: 10.1093/emboj/18.13.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hummasti S, Tontonoz P. The peroxisome proliferator-activated receptor N-terminal domain controls isotype-selective gene expression and adipogenesis. Molecular Endocrinology. 2006;20(6):1261–1275. doi: 10.1210/me.2006-0025. [DOI] [PubMed] [Google Scholar]
- 17.IJpenberg A, Jeannin E, Wahli W, Desvergne B. Polarity and specific sequence requirements of peroxisome proliferator-activated receptor (PPAR)/retinoid X receptor heterodimer binding to DNA. A functional analysis of the malic enzyme gene PPAR response element. Journal of Biological Chemistry. 1997;272(32):20108–20117. doi: 10.1074/jbc.272.32.20108. [DOI] [PubMed] [Google Scholar]
- 18.Ravnskjaer K, Boergesen M, Rubi B, et al. Peroxisome proliferator-activated receptor α (PPARα) potentiates, whereas PPARγ attenuates, glucose-stimulated insulin secretion in pancreatic β-cells. Endocrinology. 2005;146(8):3266–3276. doi: 10.1210/en.2004-1430. [DOI] [PubMed] [Google Scholar]
- 19.Brun RP, Tontonoz P, Forman BM, et al. Differential activation of adipogenesis by multiple PPAR isoforms. Genes and Development. 1996;10(8):974–984. doi: 10.1101/gad.10.8.974. [DOI] [PubMed] [Google Scholar]
- 20.Yu S, Matsusue K, Kashireddy P, et al. Adipocyte-specific gene expression and adipogenic steatosis in the mouse liver due to peroxisome proliferator-activated receptor γ1 (PPARγ1) overexpression. Journal of Biological Chemistry. 2003;278(1):498–505. doi: 10.1074/jbc.M210062200. [DOI] [PubMed] [Google Scholar]
- 21.Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors α and δ . Proceedings of the National Academy of Sciences of the United States of America. 1997;94(9):4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helledie T, Antonius M, Sorensen RV, et al. Lipid-binding proteins modulate ligand-dependent trans-activation by peroxisome proliferator-activated receptors and localize to the nucleus as well as the cytoplasm. Journal of Lipid Research. 2000;41(11):1740–1751. [PubMed] [Google Scholar]
- 23.Kassam A, Hunter J, Rachubinski RA, Capone JP. Subtype- and response element-dependent differences in transactivation by peroxisome proliferator-activated receptors α and γ . Molecular and Cellular Endocrinology. 1998;141(1-2):153–162. doi: 10.1016/s0303-7207(98)00085-9. [DOI] [PubMed] [Google Scholar]
- 24.Schoonjans K, Peinado-Onsurbe J, Lefebvre A-M, et al. PPARα and PPARγ activators direct a distinct tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. The EMBO Journal. 1996;15(19):5336–5348. [PMC free article] [PubMed] [Google Scholar]
- 25.Juge-Aubry C, Pernin A, Favez T, et al. DNA binding properties of peroxisome proliferator-activated receptor subtypes on various natural peroxisome proliferator response elements: importance of the 5′-flanking region. Journal of Biological Chemistry. 1997;272(40):25252–25259. doi: 10.1074/jbc.272.40.25252. [DOI] [PubMed] [Google Scholar]
- 26.Chandra V, Huang P, Hamuro Y, et al. Structure of the intact PPAR-γ-RXR-α nuclear receptor complex on DNA. Nature. 2008;456(7220):350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shao D, Rangwala SM, Bailey ST, Krakow SL, Reginato MJ, Lazar MA. Interdomain communication regulating ligand binding by PPAR-γ . Nature. 1998;396(6709):377–380. doi: 10.1038/24634. [DOI] [PubMed] [Google Scholar]
- 28.Deeb SS, Fajas L, Nemoto M, et al. A Pro12Ala substitution in PPARγ2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nature Genetics. 1998;20(3):284–287. doi: 10.1038/3099. [DOI] [PubMed] [Google Scholar]
- 29.Nolte RT, Wisely GB, Westin S, et al. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ . Nature. 1998;395(6698):137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 30.Xu HE, Lambert MH, Montana VG, et al. Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(24):13919–13924. doi: 10.1073/pnas.241410198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu HE, Lambert MH, Montana VG, et al. Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Molecular Cell. 1999;3(3):397–403. doi: 10.1016/s1097-2765(00)80467-0. [DOI] [PubMed] [Google Scholar]
- 32.Blanquart C, Barbier O, Fruchart J-C, Staels B, Glineur C. Peroxisome proliferator-activated receptor α (PPARα) turnover by the ubiquitin-proteasome system controls the ligand-induced expression level of its target genes. Journal of Biological Chemistry. 2002;277(40):37254–37259. doi: 10.1074/jbc.M110598200. [DOI] [PubMed] [Google Scholar]
- 33.Hauser S, Adelmant G, Sarraf P, Wright HM, Mueller E, Spiegelman BM. Degradation of the peroxisome proliferator-activated receptor γ is linked to ligand-dependent activation. Journal of Biological Chemistry. 2000;275(24):18527–18533. doi: 10.1074/jbc.M001297200. [DOI] [PubMed] [Google Scholar]
- 34.Rosenfeld MG, Glass CK. Coregulator codes of transcriptional regulation by nuclear receptors. Journal of Biological Chemistry. 2001;276(40):36865–36868. doi: 10.1074/jbc.R100041200. [DOI] [PubMed] [Google Scholar]
- 35.Jørgensen C, Krogsdam A-M, Kratchmarova I, et al. Opposing effects of fatty acids and acyl-CoA esters on conformation and cofactor recruitment of peroxisome proliferator-activated receptors. Annals of the New York Academy of Sciences. 2002;967:431–439. doi: 10.1111/j.1749-6632.2002.tb04299.x. [DOI] [PubMed] [Google Scholar]
- 36.Stanley TB, Leesnitzer LM, Montana VG, et al. Subtype specific effects of peroxisome proliferator-activated receptor ligands on corepressor affinity. Biochemistry. 2003;42(31):9278–9287. doi: 10.1021/bi034472c. [DOI] [PubMed] [Google Scholar]
- 37.Koppen A, Houtman R, Pijnenburg D, Jeninga EH, Ruijtenbeek R, Kalkhoven E. Nuclear receptor-coregulator interaction profiling identifies TRIP3 as a novel peroxisome proliferator-activated receptor γ cofactor. Molecular and Cellular Proteomics. 2009;8(10):2212–2226. doi: 10.1074/mcp.M900209-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oberkofler H, Esterbauer H, Linnemayr V, Strosberg AD, Krempler F, Patsch W. Peroxisome proliferator-activated receptor (PPAR) γ coactivator-1 recruitment regulates PPAR subtype specificity. Journal of Biological Chemistry. 2002;277(19):16750–16757. doi: 10.1074/jbc.M200475200. [DOI] [PubMed] [Google Scholar]
- 39.Wärnmark A, Treuter E, Wright APH, Gustafsson J-Å. Activation functions 1 and 2 of nuclear receptors: molecular strategies for transcriptional activation. Molecular Endocrinology. 2003;17(10):1901–1909. doi: 10.1210/me.2002-0384. [DOI] [PubMed] [Google Scholar]
- 40.Hu X, Lazar MA. The CoRNR motif controls the recruitment of compressors by nuclear hormone receptors. Nature. 1999;402(6757):93–96. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- 41.Berger J, Patel HV, Woods J, et al. A PPARγ mutant serves as a dominant negative inhibitor of PPAR signaling and is localized in the nucleus. Molecular and Cellular Endocrinology. 2000;162(1-2):57–67. doi: 10.1016/s0303-7207(00)00211-2. [DOI] [PubMed] [Google Scholar]
- 42.Masugi J, Tamori Y, Kasuga M. Inhibition of adipogenesis by a COOH-terminally truncated mutant of PPARγ2 in 3T3-L1 cells. Biochemical and Biophysical Research Communications. 1999;264(1):93–99. doi: 10.1006/bbrc.1999.1488. [DOI] [PubMed] [Google Scholar]
- 43.Walkey CJ, Spiegelman BM. A functional peroxisome proliferator-activated receptor-γ ligand-binding domain is not required for adipogenesis. Journal of Biological Chemistry. 2008;283(36):24290–24294. doi: 10.1074/jbc.C800139200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tudor C, Feige JN, Pingali H, et al. Association with coregulators is the major determinant governing peroxisome proliferator-activated receptor mobility in living cells. Journal of Biological Chemistry. 2007;282(7):4417–4426. doi: 10.1074/jbc.M608172200. [DOI] [PubMed] [Google Scholar]
- 45.Molnár F, Matilainen M, Carlberg C. Structural determinants of the agonist-independent association of human peroxisome proliferator-activated receptors with coactivators. Journal of Biological Chemistry. 2005;280(28):26543–26556. doi: 10.1074/jbc.M502463200. [DOI] [PubMed] [Google Scholar]
- 46.Bugge A, Grøntved L, Aagaard MM, Borup R, Mandrup S. The PPARγ2 A/B-domain plays a gene-specific role in transactivation and cofactor recruitment. Molecular Endocrinology. 2009;23(6):794–808. doi: 10.1210/me.2008-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dahlman-Wright K, Baumann H, McEwan IJ, et al. Structural characterization of a minimal functional transactivation domain from the human glucocorticoid receptor. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(5):1699–1703. doi: 10.1073/pnas.92.5.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wärnmark A, Wikström A, Wright APH, Gustafsson J-Å, Härd T. The N-terminal regions of estrogen receptor α and β are unstructured in vitro and show different TBP binding properties. Journal of Biological Chemistry. 2001;276(49):45939–45944. doi: 10.1074/jbc.M107875200. [DOI] [PubMed] [Google Scholar]
- 49.Birnbaumer M, Schrader WT, O’Malley BW. Assessment of structural similarities in chick oviduct progesterone receptor subunits by partial proteolysis of photoaffinity-labeled proteins. Journal of Biological Chemistry. 1983;258(12):7331–7337. [PubMed] [Google Scholar]
- 50.Bain DL, Franden MA, McManaman JL, Takimoto GS, Horwitz KB. The N-terminal region of the human progesterone A-receptor. Structural analysis and the influence of the DNA binding domain. Journal of Biological Chemistry. 2000;275(10):7313–7320. doi: 10.1074/jbc.275.10.7313. [DOI] [PubMed] [Google Scholar]
- 51.Hi R, Osada S, Yumoto N, Osumi T. Characterization of the amino-terminal activation domain of peroxisome proliferator-activated receptor α. Importance of α-helical structure in the transactivating function. Journal of Biological Chemistry. 1999;274(49):35152–35158. doi: 10.1074/jbc.274.49.35152. [DOI] [PubMed] [Google Scholar]
- 52.Almlöf T, Gustafsson J-Å, Wright APH. Role of hydrophobic amino acid clusters in the transactivation activity of the human glucocorticoid receptor. Molecular and Cellular Biology. 1997;17(2):934–945. doi: 10.1128/mcb.17.2.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Almlof T, Wright APH, Gustafsson J-A. Role of acidic and phosphorylated residues in gene activation by the glucocorticoid receptor. Journal of Biological Chemistry. 1995;270(29):17535–17540. doi: 10.1074/jbc.270.29.17535. [DOI] [PubMed] [Google Scholar]
- 54.Kistanova E, Dell H, Tsantili P, Falvey E, Cladaras C, Hadzopoulou-Cladaras M. The activation function-1 of hepatocyte nuclear factor-4 is an acidic activator that mediates interactions through bulky hydrophobic residues. Biochemical Journal. 2001;356(2):635–642. doi: 10.1042/0264-6021:3560635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garza AMS, Khan SH, Kumar R. Site-specific phosphorylation induces functionally active conformation in the intrinsically disordered N-terminal activation function (AF1) domain of the glucocorticoid receptor. Molecular and Cellular Biology. 2010;30(1):220–230. doi: 10.1128/MCB.00552-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hermann S, Berndt KD, Wright AP. How transcriptional activators bind target proteins. Journal of Biological Chemistry. 2001;276(43):40127–40132. doi: 10.1074/jbc.M103793200. [DOI] [PubMed] [Google Scholar]
- 57.Juge-Aubry CE, Kuenzli S, Sanchez J-C, Hochstrasser D, Meier CA. Peroxisomal bifunctional enzyme binds and activates the activation function-1 region of the peroxisome proliferator-activated receptor α . Biochemical Journal. 2001;353(2):253–258. doi: 10.1042/0264-6021:3530253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Debril M-B, Gelman L, Fayard E, Annicotte J-S, Rocchi S, Auwerx J. Transcription factors and nuclear receptors interact with the SWI/SNF complex through the BAF60c subunit. Journal of Biological Chemistry. 2004;279(16):16677–16686. doi: 10.1074/jbc.M312288200. [DOI] [PubMed] [Google Scholar]
- 59.Yadav N, Cheng D, Richard S, et al. CARM1 promotes adipocyte differentiation by coactivating PPARγ . EMBO Reports. 2008;9(2):193–198. doi: 10.1038/sj.embor.7401151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li D, Kang Q, Wang D-M. Constitutive coactivator of peroxisome proliferator-activated receptor (PPARγ), a novel coactivator of PPARγ that promotes adipogenesis. Molecular Endocrinology. 2007;21(10):2320–2333. doi: 10.1210/me.2006-0520. [DOI] [PubMed] [Google Scholar]
- 61.Gelman L, Zhou G, Fajas L, Raspé E, Fruchart J-C, Auwerx J. p300 Interacts with the N- and C-terminal part of PPARγ2 in a ligand-independent and -dependent manner, respectively. Journal of Biological Chemistry. 1999;274(12):7681–7688. doi: 10.1074/jbc.274.12.7681. [DOI] [PubMed] [Google Scholar]
- 62.Drori S, Girnun GD, Tou L, et al. Hic-5 regulates an epithelial program mediated by PPARγ . Genes and Development. 2005;19(3):362–375. doi: 10.1101/gad.1240705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schock SC, Xu J, Duquette PM, et al. Rescue of neurons from ischemic injury by peroxisome proliferator-activated receptor-γ requires a novel essential cofactor LMO4. Journal of Neuroscience. 2008;28(47):12433–12444. doi: 10.1523/JNEUROSCI.2897-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Finck BN, Gropler MC, Chen Z, et al. Lipin 1 is an inducible amplifier of the hepatic PGC-1α/PPARα regulatory pathway. Cell Metabolism. 2006;4(3):199–210. doi: 10.1016/j.cmet.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 65.Zhu Y, Qi C, Jain S, Rao MS, Reddy JK. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. Journal of Biological Chemistry. 1997;272(41):25500–25506. doi: 10.1074/jbc.272.41.25500. [DOI] [PubMed] [Google Scholar]
- 66.Grøntved L, Madsen MS, Boergesen M, Roeder RG, Mandrup S. MED14 tethers mediator to the N-terminal domain of peroxisome proliferator-activated receptor γ and is required for full transcriptional activity and adipogenesis. Molecular and Cellular Biology. 2010;30(9):2155–2169. doi: 10.1128/MCB.01238-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dreijerink KMA, Varier RA, van Beekum O, et al. The multiple endocrine neoplasia type 1 (MEN1) tumor suppressor regulates peroxisome proliferator-activated receptor γ-dependent adipocyte differentiation. Molecular and Cellular Biology. 2009;29(18):5060–5069. doi: 10.1128/MCB.01001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brendel C, Gelman L, Auwerx J. Multiprotein bridging factor-1 (MBF-1) is a cofactor for nuclear receptors that regulate lipid metabolism. Molecular Endocrinology. 2002;16(6):1367–1377. doi: 10.1210/mend.16.6.0843. [DOI] [PubMed] [Google Scholar]
- 69.Gopinathan L, Hannon DB, Peters JM, Vanden Heuvel JP. Regulation of peroxisome proliferator-activated receptor-α by MDM2. Toxicological Sciences. 2009;108(1):48–58. doi: 10.1093/toxsci/kfn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bai P, Houten SM, Huber A, et al. Peroxisome proliferator-activated receptor (PPAR)-2 controls adipocyte differentiation and adipose tissue function through the regulation of the activity of the retinoid X receptor/PPARγ heterodimer. Journal of Biological Chemistry. 2007;282(52):37738–37746. doi: 10.1074/jbc.M701021200. [DOI] [PubMed] [Google Scholar]
- 71.Iankova I, Petersen RK, Annicotte J-S, et al. Peroxisome proliferator-activated receptor γ recruits the positive transcription elongation factor b complex to activate transcription and promote adipogenesis. Molecular Endocrinology. 2006;20(7):1494–1505. doi: 10.1210/me.2005-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Surapureddi S, Yu S, Bu H, et al. Identification of a transcriptionally active peroxisome proliferator-activated receptor α-interacting cofactor complex in rat liver and characterization of PRIC285 as a coactivator. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(18):11836–11841. doi: 10.1073/pnas.182426699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tomaru T, Satoh T, Yoshino S, et al. Isolation and characterization of a transcriptional cofactor and its novel isoform that bind the deoxyribonucleic acid-binding domain of peroxisome proliferator-activated receptor-γ . Endocrinology. 2006;147(1):377–388. doi: 10.1210/en.2005-0450. [DOI] [PubMed] [Google Scholar]
- 74.Surapureddi S, Viswakarma N, Yu S, Guo D, Rao MS, Reddy JK. PRIC320, a transcription coactivator, isolated from peroxisome proliferator-binding protein complex. Biochemical and Biophysical Research Communications. 2006;343(2):535–543. doi: 10.1016/j.bbrc.2006.02.160. [DOI] [PubMed] [Google Scholar]
- 75.Zhu Y, Kan L, Qi C, et al. Isolation and characterization of peroxisome proliferator-activated receptor (PPAR) interacting protein (PRIP) as a coactivator for PPAR. Journal of Biological Chemistry. 2000;275(18):13510–13516. doi: 10.1074/jbc.275.18.13510. [DOI] [PubMed] [Google Scholar]
- 76.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92(6):829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 77.Seale P, Bjork B, Yang W, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454(7207):961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu Y, Qi C, Cao W-Q, Yeldandi AV, Rao MS, Reddy JK. Cloning and characterization of PIMT, a protein with a methyltransferase domain, which interacts with and enhances nuclear receptor coactivator PRIP function. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(18):10380–10385. doi: 10.1073/pnas.181347498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Qi C, Chang J, Zhu Y, Yeldandi AV, Rao SM, Zhu Y-J. Identification of protein arginine methyltransferase 2 as a coactivator for estrogen receptor α . Journal of Biological Chemistry. 2002;277(32):28624–28630. doi: 10.1074/jbc.M201053200. [DOI] [PubMed] [Google Scholar]
- 80.Zhu Y, Qi C, Calandra C, Rao MS, Reddy JK. Cloning and identification of mouse steroid receptor coactivator-1 (mSRC-1), as a coactivator of peroxisome proliferator-activated receptor gamma. Gene Expression. 1996;6(3):185–195. [PMC free article] [PubMed] [Google Scholar]
- 81.Li H, Gomes PJ, Chen JD. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(16):8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van Beekum O, Brenkman AB, Grøntved L, et al. The adipogenic acetyltransferase Tip60 targets activation function 1 of peroxisome proliferator-activated receptor γ . Endocrinology. 2008;149(4):1840–1849. doi: 10.1210/en.2007-0977. [DOI] [PubMed] [Google Scholar]
- 83.Koppen A, Houtman R, Pijnenburg D, Jeninga EH, Ruijtenbeek R, Kalkhoven E. Nuclear receptor-coregulator interaction profiling identifies TRIP3 as a novel peroxisome proliferator-activated receptor γ cofactor. Molecular and Cellular Proteomics. 2009;8(10):2212–2226. doi: 10.1074/mcp.M900209-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yoo EJ, Chung JJ, Choe SS, Kim KH, Kim JB. Down-regulation of histone deacetylases stimulates adipocyte differentiation. Journal of Biological Chemistry. 2006;281(10):6608–6615. doi: 10.1074/jbc.M508982200. [DOI] [PubMed] [Google Scholar]
- 85.Knutson SK, Chyla BJ, Amann JM, Bhaskara S, Huppert SS, Hiebert SW. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. The EMBO Journal. 2008;27(7):1017–1028. doi: 10.1038/emboj.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Montgomery RL, Potthoff MJ, Haberland M, et al. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. Journal of Clinical Investigation. 2008;118(11):3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chan SSY, Schedlich LJ, Twigg SM, Baxter RC. Inhibition of adipocyte differentiation by insulin-like growth factor-binding protein-3. American Journal of Physiology. 2009;296(4):E654–E663. doi: 10.1152/ajpendo.90846.2008. [DOI] [PubMed] [Google Scholar]
- 88.Direnzo J, Söderström M, Kurokawa R, et al. Peroxisome proliferator-activated receptors and retinoic acid receptors differentially control the interactions of retinoid X receptor heterodimers with ligands, coactivators, and corepressors. Molecular and Cellular Biology. 1997;17(4):2166–2176. doi: 10.1128/mcb.17.4.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Treuter E, Albrektsen T, Johansson L, Leers J, Gustafsson J-A. A regulatory role for RIP140 in nuclear receptor activation. Molecular Endocrinology. 1998;12(6):864–881. doi: 10.1210/mend.12.6.0123. [DOI] [PubMed] [Google Scholar]
- 90.Debril M-B, Dubuquoy L, Feige J-N, et al. Scaffold attachment factor B1 directly interacts with nuclear receptors in living cells and represses transcriptional activity. Journal of Molecular Endocrinology. 2005;35(3):503–517. doi: 10.1677/jme.1.01856. [DOI] [PubMed] [Google Scholar]
- 91.Picard F, Kurtev M, Chung N, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ . Nature. 2004;429(6993):771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Takahashi Y, Ohoka N, Hayashi H, Sato R. TRB3 suppresses adipocyte differentiation by negatively regulating PPARγ transcriptional activity. Journal of Lipid Research. 2008;49(4):880–892. doi: 10.1194/jlr.M700545-JLR200. [DOI] [PubMed] [Google Scholar]
- 93.Burns KA, Vanden Heuvel JP. Modulation of PPAR activity via phosphorylation. Biochimica et Biophysica Acta. 2007;1771(8):952–960. doi: 10.1016/j.bbalip.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Adams M, Reginato MJ, Shao D, Lazar MA, Chatterjee VK. Transcriptional activation by peroxisome proliferator-activated receptor γ is inhibited by phosphorylation at a consensus mitogen-activated protein kinase site. Journal of Biological Chemistry. 1997;272(8):5128–5132. doi: 10.1074/jbc.272.8.5128. [DOI] [PubMed] [Google Scholar]
- 95.Fujimoto Y, Shiraki T, Horiuchi Y, et al. Proline cis/trans-isomerase Pin1 regulates peroxisome proliferator-activated receptor γ activity through the direct binding to the activation function-1 domain. Journal of Biological Chemistry. 2010;285(5):3126–3132. doi: 10.1074/jbc.M109.055095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang B, Berger J, Zhou G, et al. Insulin- and mitogen-activated protein kinase-mediated phosphorylation and activation of peroxisome proliferator-activated receptor γ . Journal of Biological Chemistry. 1996;271(50):31771–31774. doi: 10.1074/jbc.271.50.31771. [DOI] [PubMed] [Google Scholar]
- 97.Rangwala SM, Rhoades B, Shapiro JS, et al. Genetic modulation of PPARγ phosphorylation regulates insulin sensitivity. Developmental Cell. 2003;5(4):657–663. doi: 10.1016/s1534-5807(03)00274-0. [DOI] [PubMed] [Google Scholar]
- 98.Ristow M, Müller-Wieland D, Pfeiffer A, Krone W, Kahn CR. Obesity associated with a mutation in a genetic regulator of adipocyte differentiation. The New England Journal of Medicine. 1998;339(14):953–959. doi: 10.1056/NEJM199810013391403. [DOI] [PubMed] [Google Scholar]
- 99.Burgermeister E, Chuderland D, Hanoch T, Meyer M, Liscovitch M, Seger R. Interaction with MEK causes nuclear export and downregulation of peroxisome proliferator-activated receptor γ . Molecular and Cellular Biology. 2007;27(3):803–817. doi: 10.1128/MCB.00601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.von Knethen A, Tzieply N, Jennewein C, Brüne B. Casein-kinase-II-dependent phosphorylation of PPARγ provokes CRM1-mediated shuttling of PPARγ from the nucleus to the cytosol. Journal of Cell Science. 2010;123(2):192–201. doi: 10.1242/jcs.055475. [DOI] [PubMed] [Google Scholar]
- 101.Yamashita D, Yamaguchi T, Shimizu M, Nakata N, Hirose F, Osumi T. The transactivating function of peroxisome proliferator-activated receptor γ is negatively regulated by SUMO conjugation in the amino-terminal domain. Genes to Cells. 2004;9(11):1017–1029. doi: 10.1111/j.1365-2443.2004.00786.x. [DOI] [PubMed] [Google Scholar]
- 102.Grøntved L, Madsen MS, Boergesen M, Roeder RG, Mandrup S. MED14 tethers mediator to the N-terminal domain of peroxisome proliferator-activated receptor γ and is required for full transcriptional activity and adipogenesis. Molecular and Cellular Biology. 2010;30(9):2155–2169. doi: 10.1128/MCB.01238-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hsu M-H, Palmer CNA, Song W, Griffin KJ, Johnson EF. A carboxyl-terminal extension of the zinc finger domain contributes to the specificity and polarity of peroxisome proliferator-activated receptor DNA binding. Journal of Biological Chemistry. 1998;273(43):27988–27997. doi: 10.1074/jbc.273.43.27988. [DOI] [PubMed] [Google Scholar]
- 104.Lee C-H, Olson P, Hevener A, et al. PPARδ regulates glucose metabolism and insulin sensitivity. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(9):3444–3449. doi: 10.1073/pnas.0511253103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tanaka T, Yamamoto J, Iwasaki S, et al. Activation of peroxisome proliferator-activated receptor δ induces fatty acid β-oxidation in skeletal muscle and attenuates metabolic syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(26):15924–15929. doi: 10.1073/pnas.0306981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sanderson LM, Boekschoten MV, Desvergne B, Müller M, Kersten S. Transcriptional profiling reveals divergent roles of PPARα and PPARβ/δ in regulation of gene expression in mouse liver. Physiological Genomics. 2010;41(1):42–52. doi: 10.1152/physiolgenomics.00127.2009. [DOI] [PubMed] [Google Scholar]
- 107.Anderson SP, Dunn C, Laughter A, et al. Overlapping transcriptional programs regulated by the nuclear receptors peroxisome proliferator-activated receptor α, retinoid X receptor, and liver X receptor in mouse liver. Molecular Pharmacology. 2004;66(6):1440–1452. doi: 10.1124/mol.104.005496. [DOI] [PubMed] [Google Scholar]
- 108.Vanden Heuvel JP, Kreder D, Belda B, et al. Comprehensive analysis of gene expression in rat and human hepatoma cells exposed to the peroxisome proliferator WY14,643. Toxicology and Applied Pharmacology. 2003;188(3):185–198. doi: 10.1016/s0041-008x(03)00015-2. [DOI] [PubMed] [Google Scholar]
- 109.Yamazaki K, Kuromitsu J, Tanaka I. Microarray analysis of gene expression changes in mouse liver induced by peroxisome proliferator-activated receptor α agonists. Biochemical and Biophysical Research Communications. 2002;290(3):1114–1122. doi: 10.1006/bbrc.2001.6319. [DOI] [PubMed] [Google Scholar]
- 110.Cariello NF, Romach EH, Colton HM, et al. Gene expression profiling of the PPAR-alpha agonist ciprofibrate in the cynomolgus monkey liver. Toxicological Sciences. 2005;88(1):250–264. doi: 10.1093/toxsci/kfi273. [DOI] [PubMed] [Google Scholar]
- 111.Bünger M, van den Bosch HM, van der Meijde J, Kersten S, Hooiveld GJEJ, Müller M. Genome-wide analysis of PPARα activation in murine small intestine. Physiological Genomics. 2007;30(2):192–204. doi: 10.1152/physiolgenomics.00198.2006. [DOI] [PubMed] [Google Scholar]
- 112.Kersten S, Mandard S, Escher P, et al. The peroxisome proliferator-activated receptor α regulates amino acid metabolism. The FASEB Journal. 2001;15(11):1971–1978. doi: 10.1096/fj.01-0147com. [DOI] [PubMed] [Google Scholar]
- 113.Okuno A, Tamemoto H, Tobe K, et al. Troglitazone increases the number of small adipocytes without the change of white adipose tissue mass in obese Zucker rats. Journal of Clinical Investigation. 1998;101(6):1354–1361. doi: 10.1172/JCI1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chao L, Marcus-Samuels B, Mason MM, et al. Adipose tissue is required for the antidiabetic, but not for the hypolipidemic, effect of thiazolidinediones. Journal of Clinical Investigation. 2000;106(10):1221–1228. doi: 10.1172/JCI11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Albrektsen T, Frederiksen KS, Holmes WE, Boel E, Taylor K, Fleckner J. Novel genes regulated by the insulin sensitizer rosiglitazone during adipocyte differentiation. Diabetes. 2002;51(4):1042–1051. doi: 10.2337/diabetes.51.4.1042. [DOI] [PubMed] [Google Scholar]
- 116.Gerhold DL, Liu F, Jiang G, et al. Gene expression profile of adipocyte differentiation and its regulation by peroxisome proliferator-activated receptor-γ agonists. Endocrinology. 2002;143(6):2106–2118. doi: 10.1210/endo.143.6.8842. [DOI] [PubMed] [Google Scholar]
- 117.Wang P, Renes J, Bouwman F, Bunschoten A, Mariman E, Keijer J. Absence of an adipogenic effect of rosiglitazone on mature 3T3-L1 adipocytes: increase of lipid catabolism and reduction of adipokine expression. Diabetologia. 2007;50(3):654–665. doi: 10.1007/s00125-006-0565-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hodgkinson CP, Ye S. Microarray analysis of peroxisome proliferator-activated receptor γ-induced changes in gene expression in macrophages. Biochemical and Biophysical Research Communications. 2003;308(3):505–510. doi: 10.1016/s0006-291x(03)01416-5. [DOI] [PubMed] [Google Scholar]
- 119.Gupta RA, Brockman JA, Sarraf P, Willson TM, DuBois RN. Target genes of peroxisome proliferator-activated receptor γ in colorectal cancer cells. Journal of Biological Chemistry. 2001;276(32):29681–29687. doi: 10.1074/jbc.M103779200. [DOI] [PubMed] [Google Scholar]
- 120.Keen HL, Ryan MJ, Beyer A, et al. Gene expression profiling of potential PPARγ target genes in mouse aorta. Physiological Genomics. 2004;18:33–42. doi: 10.1152/physiolgenomics.00027.2004. [DOI] [PubMed] [Google Scholar]
- 121.Szatmari I, Töröcsik D, Agostini M, et al. PPARγ regulates the function of human dendritic cells primarily by altering lipid metabolism. Blood. 2007;110(9):3271–3280. doi: 10.1182/blood-2007-06-096222. [DOI] [PubMed] [Google Scholar]
- 122.Åkerblad P, Månsson R, Lagergren A, et al. Gene expression analysis suggests that EBF-1 and PPARγ2 induce adipogenesis of NIH-3T3 cells with similar efficiency and kinetics. Physiological Genomics. 2005;23(2):206–216. doi: 10.1152/physiolgenomics.00015.2005. [DOI] [PubMed] [Google Scholar]
- 123.Carroll JS, Meyer CA, Song J, et al. Genome-wide analysis of estrogen receptor binding sites. Nature Genetics. 2006;38(11):1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 124.Gao H, Fält S, Sandelin A, Gustafsson J-Å, Dahlman-Wright K. Genome-wide identification of estrogen receptor α-binding sites in mouse liver. Molecular Endocrinology. 2008;22(1):10–22. doi: 10.1210/me.2007-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Thomas AM, Hart SN, Kong B, Fang J, Zhong X-B, Guo GL. Genome-wide tissue-specific farnesoid X receptor binding in mouse liver and intestine. Hepatology. 2010;51(4):1410–1419. doi: 10.1002/hep.23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lim C-A, Yao F, Wong JJ-Y, et al. Genome-wide mapping of RELA(p65) binding identifies E2F1 as a transcriptional activator recruited by NF-κB upon TLR4 activation. Molecular Cell. 2007;27(4):622–635. doi: 10.1016/j.molcel.2007.06.038. [DOI] [PubMed] [Google Scholar]
- 127.Wederell ED, Bilenky M, Cullum R, et al. Global analysis of in vivo Foxa2-binding sites in mouse adult liver using massively parallel sequencing. Nucleic Acids Research. 2008;36(14):4549–4564. doi: 10.1093/nar/gkn382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Frietze S, Lan X, Jin VX, Farnham PJ. Genomic targets of the KRAB and SCAN domain-containing zinc finger protein 263. Journal of Biological Chemistry. 2010;285(2):1393–1403. doi: 10.1074/jbc.M109.063032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nielsen R, Pedersen TÅ, Hagenbeek D, et al. Genome-wide profiling of PPARγ:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes and Development. 2008;22(21):2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lefterova MI, Zhang Y, Steger DJ, et al. PPARγ and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes and Development. 2008;22(21):2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wakabayashi K-I, Okamura M, Tsutsumi S, et al. The peroxisome proliferator-activated receptor γ/retinoid X receptor α heterodimer targets the histone modification enzyme PR-Set7/Setd8 gene and regulates adipogenesis through a positive feedback loop. Molecular and Cellular Biology. 2009;29(13):3544–3555. doi: 10.1128/MCB.01856-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lefterova MI, Steger DJ, Zhuo D, et al. Cell-specific determinants of peroxisome proliferator-activated receptor γ function in adipocytes and macrophages. Molecular and Cellular Biology. 2010;30(9):2078–2089. doi: 10.1128/MCB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hamza MS, Pott S, Vega VB, et al. De-novo identification of PPARγ/RXR binding sites and direct targets during adipogenesis. PLoS ONE. 2009;4(3, article e4907) doi: 10.1371/journal.pone.0004907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.MacDougald OA, Mandrup S. Adipogenesis: forces that tip the scales. Trends in Endocrinology and Metabolism. 2002;13(1):5–11. doi: 10.1016/s1043-2760(01)00517-3. [DOI] [PubMed] [Google Scholar]
- 135.van der Meer DLM, Degenhardt T, Väisänen S, et al. Profiling of promoter occupancy by PPARα in human hepatoma cells via ChIP-chip analysis. Nucleic Acids Research. 2010;38(9):2839–2850. doi: 10.1093/nar/gkq012. [DOI] [PMC free article] [PubMed] [Google Scholar]