Abstract
TGF-β/Smad3 promotes renal fibrosis, but the mechanisms that regulate profibrotic genes remain unclear. We hypothesized that miR-192, a microRNA expressed in the kidney may mediate renal fibrosis in a Smad3-dependent manner. Microarray and real-time PCR demonstrated a tight association between upregulation of miR-192 in the fibrotic kidney and activation of TGF-β/Smad signaling. Deletion of Smad7 promoted miR-192 expression and enhanced Smad signaling and fibrosis in obstructive kidney disease. In contrast, overexpression of Smad7 to block TGF-β/Smad signaling inhibited miR-192 expression and renal fibrosis in the rat 5/6 nephrectomy model; in vitro, overexpression of Smad7 in tubular epithelial cells abolished TGF-β1–induced miR-192 expression. Furthermore, Smad3 but not Smad2 mediated TGF-β1–induced miR-192 expression by binding to the miR-192 promoter. Last, overexpression of a miR-192 mimic promoted and addition of a miR-192 inhibitor blocked TGF-β1–induced collagen matrix expression. Taken together, miR-192 may be a critical downstream mediator of TGF-β/Smad3 signaling in the development of renal fibrosis.
TGF-β is a profibrogenic cytokine that mediates renal fibrosis positively by activating its downstream mediators called Smad2 and Smad3 but negatively by its inhibitory factor Smad71–4; however, the mechanisms as to how the Smads regulate the fibrogenic genes during renal fibrosis remain unclear.
MicroRNAs (miRNAs) are small, noncoding RNAs with approximately 22 nucleotides. The mature miRNAs can complementarily bind to the mRNA 3′ untranslated region to regulate the gene expression by translational repression or induction of mRNA degradation.5 Increasing evidence shows that TGF-β may act by regulating miRNAs to exhibit its biological effects such as epithelial-to-mesenchymal transition (EMT),5 suggesting that TGF-β regulates the expression of these miRNAs to promote EMT.
Several miRNAs, including miR-192, -194, -204, -215, and -216, are highly expressed in the kidney, as compared with other organs.6,7 In the context of renal fibrosis, expression levels of miR-192 increased significantly in glomeruli isolated from diabetic mice.8 In vitro, miR-192 is induced by TGF-β1 and mediates TGF-β–induced collagen expression in mesangial cells (MCs) by downregulating ZEB2 expression.8 In contrast, a recent study also found that TGF-β1 suppresses miR-192 expression in human tubular epithelial cells (TEC) and loss of miR-192 promotes fibrogenesis in diabetic nephropathy.9 The discrepancy in these two studies with opposite findings and understanding of miR-192 in diabetic nephropathy necessitates further investigation of the potential role of miR-192 and the mechanisms that regulate miR-192 expression during renal fibrosis under various disease conditions. Thus, this study tested the hypothesis that TGF-β1 may act by stimulating Smad3 to regulate miR-192 expression during renal fibrosis. This was tested in rodent models of obstructive and remnant kidney diseases induced in mice that lacked Smad3 or Smad7, had conditional knockout (KO) for Smad2, or overexpressed renal Smad7. In addition, TGF-β–induced miR-192 expression via the Smad3-dependent mechanism was determined in TECs overexpressing Smad7 or knocking down for Smad2 or Smad3 and in Smad2 or Smad3 KO mouse embryonic fibroblasts (MEFs).
Results
Activation of TGF-β/Smad Signaling Is Essential for Upregulation of miR-192 During Renal Fibrosis In Vivo and In Vitro
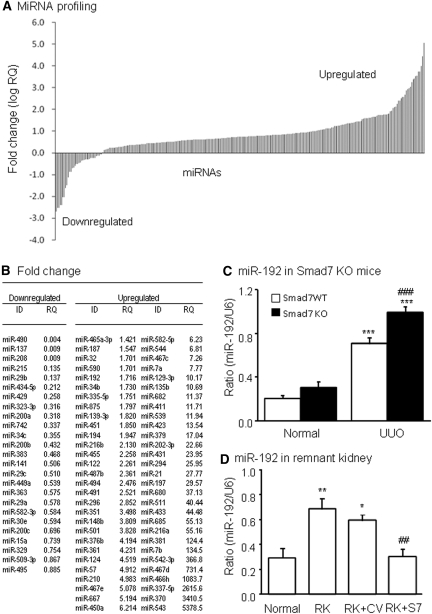
We first examined miRNA expression in obstructive kidney disease by microarray and real-time PCR. As shown in Figure 1, compared with normal kidney, 120 miRNAs including miR-192 were significantly upregulated from mouse kidneys after unilateral ureteral obstruction (UUO) for 5 days (Figure 1, A and B). This was associated with activation of TGF-β/Smad signaling and renal fibrosis, as reported previously.10 Because miR-192 has been shown to be important in diabetic glomerulosclerosis,8 we examined whether miR-192 was also important for tubulointerstitial fibrosis in the UUO kidney. Interestingly, real-time PCR revealed that deletion of Smad7, an inhibitor of TGF-β signaling, enhanced further renal miR-192 expression in obstructive kidneys when compared with Smad7 wild-type (WT) mice (Figure 1C), which, again, was associated with a further increase in TGF-β/Smad signaling and progressive renal fibrosis, as described previously.10 Similarly, in a rat model of remnant kidney disease in which progressive renal fibrosis has been shown to be TGF-β/Smad dependent,11 miR-192 expression was significantly increased (Figure 1D). In contrast, inhibition of TGF-β/Smad signaling by overexpressing renal Smad7 prevented upregulation of renal miR-192 (Figure 1D) and renal fibrosis, as described previously,10 suggesting a close link between activation of TGF-β/Smad signaling and miR-192 expression during renal fibrosis in various disease conditions.
Figure 1.
Activation of TGF-β signaling increases miR-192 expression in vivo. (A) miRNA expression profile in UUO kidneys of mice determined by TaqMan miRNA low-density array. Data are expressed as fold increase compared with normal mice. (B) List of fold changes of miRNAs in day 7 UUO kidney compared with normal mouse kidneys. (C) Real-time PCR results of miR-192 expression in Smad7 WT/KO kidneys with or without UUO at day 7. (D) Real-time PCR results of miR-192 expression in remnant kidneys (RK) treated with Smad7 gene transfer (RK+S7) or control empty vector (RK+CV) at day 28. Each bar represents the mean ± SEM for at least five mice. *P < 0.05, **P < 0.01, ***P < 0.001 versus normal mice; ##P < 0.01, ###P < 0.001 versus the WT-UUO or RK+CV. RQ, relative quantification.
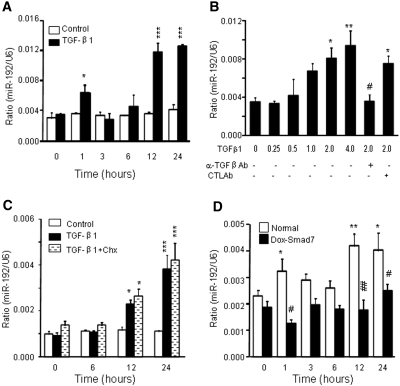
We further delineated the relationship between TGF-β/Smad signaling and miR-192 expression in a rat TEC line (NRK52E) in vitro. As shown in Figure 2, A and B, addition of TGF-β1 was able to induce miR-192 expression in a time- and dosage-dependent manner, which was blocked by a neutralizing anti–TGF-β antibody. Pretreated NRK52E cells with cycloheximide, a protein synthesis inhibitor, did not prevent miR-192 expression induced by TGF-β1 (Figure 2C), indicating that upregulation of miR-192 by TGF-β1 did not require de novo protein synthesis; however, overexpression of Smad7 in NRK52E cells virtually abolished TGF-β1–induced miR-192 expression (Figure 2D), demonstrating an essential role for TGF-β/Smad signaling in miR-192 expression.
Figure 2.
TGF-β induces miR-192 expression in a time- and dosage-dependent manner in NRK52E cells. (A) Real-time PCR shows that TGF-β1 (2 ng/ml), not PBS, induces miR-192 expression in a time-dependant manner. (B) Real-time PCR demonstrates that TGF-β1 induces miR-192 expression at 1 hour in a dosage-dependent manner, being significant at 2 ng/ml. The induction of miR-192 expression by TGF-β1 is inhibited by a neutralizing TGF-β1 antibody (α-TGF-β1 Ab) but not by an isotype control antibody (CTL Ab). (C) Real-time PCR demonstrates that cycloheximide (Chx) does not suppress TGF-β1 induction of miR-192 expression. (D) Real-time PCR analysis demonstrates that Dox-induced overexpression of Smad7 in rat TEC suppresses TGF-β–induced miR-192 expression. Each bar represents the mean ± SEM for at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 versus either the BSA control or time 0 (or dosage 0); #P < 0.05, ##P < 0.01 versus the control antibody or normal NRK52E at same time point.
TGF-β1–Induced miR-192 Expression Is Mediated by Smad3, not Smad2, In Vivo and In Vitro
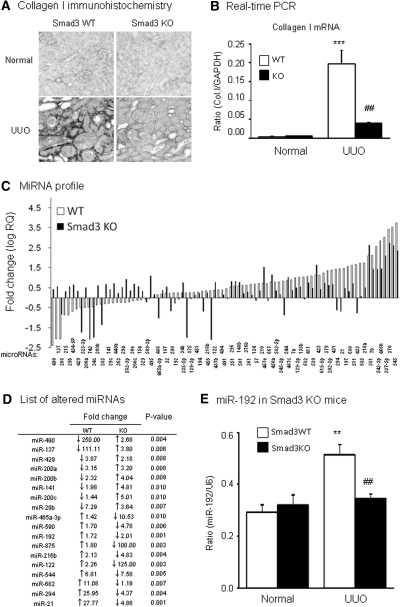
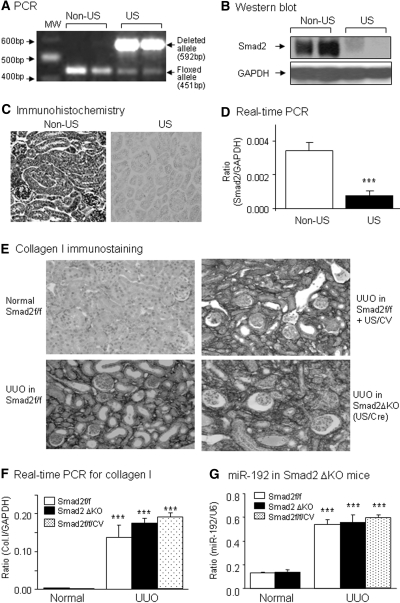
To dissect the specific role for TGF-β/Smad signaling, Smad2 or Smad3, in miR-192 expression, we examined mouse kidneys of UUO induced in Smad3 WT/KO mice or conditional Smad2 KO mice. Consistent with a previous report,12 mice null for Smad3 were protected against renal tubulointerstitial fibrosis such as collagen I matrix deposition and mRNA expression (Figure 3, A and B). By microarray and real-time PCR, compared with normal mice, miR-192 expression was significantly upregulated in the UUO kidney of Smad3 WT mice but downregulated in Smad3 KO mice (Figure 3, C through E). To determine the role of Smad2 in miR-192 expression, we conditionally deleted Smad2 from the Smad2 floxed/floxed mice using a ultrasound-microbubble–mediated Cre recombinase technique.13 As shown in Figure 4A, PCR detected that >90% of floxed Smad2 allele was deleted from the kidney by the ultrasound-microbubble–mediated Cre recombinase, which was confirmed at the mRNA and protein expression levels by real-time PCR, Western blot, and immunohistochemistry with anti-Smad2 antibodies (Figure 4, B through D). In contrast to the results found in Smad3 KO mice, real-time PCR and immunohistochemistry detected that conditional deletion of Smad2 produced a nonprotective effect on tubulointerstitial fibrosis such as collagen I expression after UUO (Figure 4, E and F), which was associated with high levels of miR-192 expression in the fibrotic kidney with or without conditional Smad2 KO (Figure 4G).
Figure 3.
Smad3 mediates miR-192 expression during renal fibrosis in a mouse model of UUO. (A and B) Immunohistochemistry (A) and real-time PCR (B) results demonstrate that mice null for Smad3 are protected against renal tubulointerstitial fibrosis as demonstrated by inhibition of collagen I expression and accumulation in day 7 UUO kidney. (C) miRNA expression profile in the UUO kidney of WT and Smad3 KO mice determined by TaqMan miRNA low-density array. Data are expressed as the fold increase against normal kidneys. (D) List of altered miRNAs showing significant differences in the UUO kidney of WT and Smad3 KO mice. miR-192 increases in ligated kidney of WT mice but reduces in Smad3 KO mice. (E) Real-time PCR results of miR-192 expression in Smad3 KO kidneys. Real-time PCR analysis demonstrates that deletion of Smad blocks miR-192 expression. Each bar represents the mean ± SEM for at least five mice. **P < 0.01, ***P < 0.001 versus normal mice; ##P < 0.01 versus WT-UUO kidney. RQ, relative quantification. Magnification, ×250.
Figure 4.
Conditional deletion of Smad2 produces no inhibitory effect on miR-192 expression and renal fibrosis in a mouse model of UUO. (A through D) Characterization of conditional Smad2 KO mice by ultrasound (US)-microbubble–mediated Cre recombination. (A) PCR detects that >90% of the floxed Smad2 (451 bp) is deleted from the kidney of Smad2 f/f mice when compared with the deleted allele (592 bp). Western blot (B), immunohistochemistry (C), and real-time PCR (D) show that ultrasound-mediated Cre/lox recombination results in a substantial deletion of Smad2 from the kidneys. (E and F) Immunohistochemistry (E) and real-time PCR (F) demonstrate that deletion of Smad2 from the kidneys (Smad2ΔKO) does not protect against renal tubulointerstitial fibrosis such as collagen I (Col I) expression in day 7 UUO kidneys (G) Real-time PCR results of miR-192 expression in Smad2 floxed/floxed (Smad2f/f) kidneys, Smad2ΔKO kidneys (Smad2f/f mice treated with US-mediated Cre recombinase), and Smad2f/f kidneys treated with control vector (Smad2f/f/CV). Real-time PCR analysis demonstrates that specific deletion of Smad2 in kidney did not block miR-192 expression. Each bar represents the mean ± SEM for at least five mice. ***P < 0.001 versus normal mice. MW, molecular weight. Magnification, ×250.
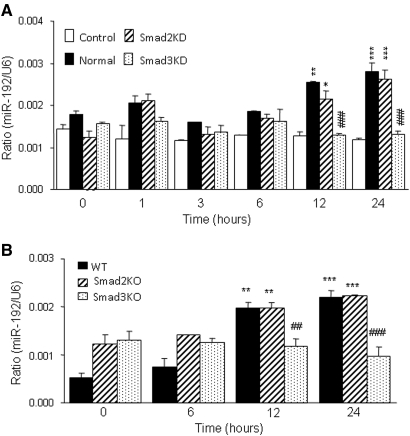
The essential role for Smad3, not Smad2, in regulating miR-192 expression was further determined in vitro in NRK52E TEC cell line in which Smad2 or Smad3 expression were specifically knocked down by the small interfering RNA technique, as described in our previous studies.13–15 As shown in Figure 5A, TGF-β1–induced miR-192 expression was significantly blocked in Smad3 but not in Smad2 knockdown TECs. This observation was further confirmed in Smad2 or Smad3 KO MEF cells.16 MEFs lacking Smad3, not Smad2, were protected against TGF-β1–induced miR-192 mRNA expression (Figure 5B).
Figure 5.
TGF-β–induced miR-192 expression is Smad3 dependent but Smad2 independent. (A) Real-time PCR analysis demonstrates that knockdown of Smad3 but not Smad2 in TEC blocks TGF-β–induced miR-192 expression. (B) Real-time PCR analysis demonstrates that deletion of Smad3 but not Smad2 in MEF blocks TGF-β–induced miR-192 expression. Each bar represents the mean ± SEM for at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 versus either the BSA control or time 0 (or dose 0); ##P < 0.01, ###P < 0.001 versus the TGF-β1–treated samples of normal TECs or WT MEF.
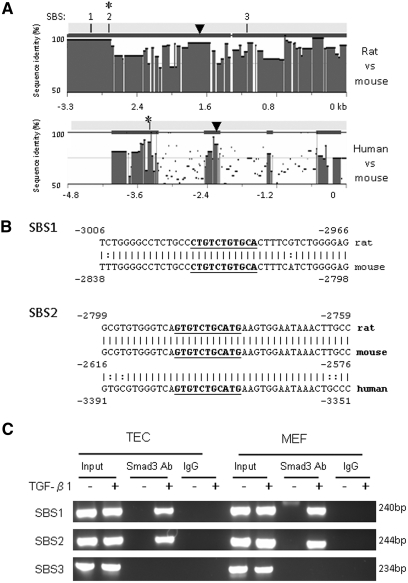
To delineate whether Smad3 specifically binds miR-192 promoter in response to TGF-β1, we performed in silico analysis (rVista 2.0, http://rvista.dcode.org/)17 to search for potential Smad-binding sites (SBSs) in miR-192 promoter. Studies by Sun et al.7 revealed a noncoding region with high cross-species conservation located at 2158 bp upstream of the miR-192 on human chromosome 11. Within this region, there is a potential transcription factor–binding site for Ets-1.7 In this study, we found not only an Ets-1 site but also three conserved SBSs at −800 to −3500 bp in the 5′ end of miR-192 in mouse and rat genomes (Figure 6A); however, only SBS2 is conserved in human miR-192 promoter (Figure 6, A and B). The presence of these potential SBSs suggests that Smad3 may interact with these binding sites to induce miR-192 expression. To confirm this finding, we used a chromatin immunoprecipitation (ChIP) assay to determine the interaction of Smad3 with the miR-192 promoter region in TECs and MEFs. As shown in Figure 6C, the antibody against Smad3 could successfully immunoprecipitate the DNA fragments from both TECs and MEFs containing the potential SBS1 and SBS2 but not SBS3 (Figure 6C), supporting that Smad3 could physically interact with the miR-192 promoter region.
Figure 6.
Smad3 binds to conserved SBSs in miR-192 promoter. (A) Sequence analysis by rVista 2.0 program shows the degree of sequence conservation of miR-192 promoter and reveals three conserved SBSs located at −3500 to −2000 bp from the miR-192 mature sequence in rat and mouse genomes and one conserved SBS located at −3600 to −3000 bp from the miR-192 mature sequence in human and mouse genomes. ▼, potential Ets-1 binding site. (B) DNA sequence alignments of SBS1 and SBS2. Bold and underlined sequences indicate the location of SBSs. (C) ChIP assays for Smad3 were performed with chromatin from TEC and MEF cells treated with TGF-β1. Precipitated DNA was amplified with oligonucleotides spanning regions of SBSs. Total inputs are indicated.
miR-192 Is an Important Mediator of TGF-β Signaling in Renal Fibrosis In Vitro
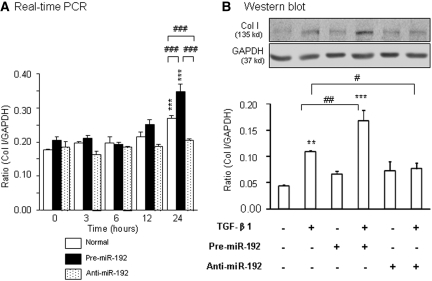
The functional importance of miR-192 in TGF-β–induced fibrosis was determined in vitro by transiently transfecting the miR-192 mimic (premiR-192) or miR-192 inhibitor (anti–miR-192), respectively, into normal rat TECs (NRK52E). As shown in Figure 7, TGF-β1–induced expression of collagen I at both mRNA and protein levels was significantly enhanced by overexpression of the premiR-192 but inhibited by the anti–miR-192 treatment.
Figure 7.
Overexpression of miR-192 promotes but inhibition of miR-192 blocks TGF-β1–induced collagen I mRNA and protein expression in rat TECs. (A and B) Real-time PCR (A) and Western blot (B) analysis. Real-time PCR and Western blot analyses show that TECs transfected with pre–miR-192 enhance TGF-β1-induced (2 ng/ml) collagen I mRNA and protein expression, which is abrogated by transfection with the anti–miR-192 inhibitor. Each bar represents the mean ± SEM for at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 versus either the control or time 0; #P < 0.05, ##P < 0.01, ###P < 0.001 as indicated.
Discussion
In this study, we found that miR-192 was highly upregulated in the diseased kidney with progressive renal fibrosis in a mouse model of UUO and in a rat model of remnant kidney disease. TGF-β1 was able to upregulate the miR-192 in rat TECs (NRK52E) in a time- and dosage-dependent manner. miR-192 was a mediator of TGF-β–induced renal fibrosis as evidenced by the findings that overexpression of miR-192 mimics enhanced TGF-β1–induced tubular collagen I expression, whereas inhibition of miR-192 by overexpressing anti–miR-192 blocked tubular collagen I expression in response to TGF-β1. Taken together, our results supported the findings of Kato et al.8 that miR-192 is upregulated by TGF-β1 and contributes to diabetic glomerulosclerosis. More important, this study, for the first time, identified the signaling mechanisms by which TGF-β signals through Smad3, not Smad2, to regulate positively miR-192 expression during renal fibrosis in vivo and in vitro.
In addition to the suggestion by Kato et al.8 that miR-192 is induced indirectly by TGF-β via ETS protein, we demonstrate in this study that Smad signaling is required for TGF-β–induced miR-192 expression during renal fibrosis, which is negatively regulated by Smad7. We found that TGF-β1 was able to induce miR-192 expression in TECs as early as 1 hour, which was tightly associated with a rapid activation of TGF-β/Smad2/3 as early as 5 minutes with the peak at 30 minutes to 1 hour.18 Furthermore, we found that blockade of the de novo protein synthesis by addition of cycloheximide did not alter TGF-β1–induced miR-192 expression in TECs, suggesting that TGF-β1 induces tubular miR-192 expression directly and does not require the new protein synthesis. Thus, activation of TGF-β/Smad signaling may directly regulate miR-192 expression in TECs, because miR-192 expression was tightly associated with activation of TGF-β/Smad2/3 in the fibrotic kidneys of UUO and remnant kidney disease.10,11 Inactivation of Smad2/3 by overexpressing Smad7 inhibits miR-192 expression and renal fibrosis in vivo in a rat remnant model of kidney disease11 and in vitro in TGF-β1–stimulated TECs. Furthermore, deletion of Smad7 greatly enhanced TGF-β/Smad signaling (Smad2/3),10 which promoted further miR-192 expression associated with progressive renal fibrosis in the UUO kidney. All of these findings supported an essential role for the Smad signaling pathway in regulating miR-192 expression and renal fibrosis. TGF-β1 may regulate the expression of miR-192 positively by Smad2/3 but negatively by Smad7. Inhibition of miR-192 expression may be a mechanism by which overexpression of Smad7 blocks but deletion of Smad7 promotes renal fibrosis in vivo and in vitro.10–13,19–21
Most important, we also found that TGF-β–induced tubular miR-192 expression was mediated by Smad3, not Smad2, because knockdown of Smad3, not Smad2, blocked TGF-β1–induced tubular miR-192 expression. This observation was further confirmed in Smad2 or Smad3 KO MEF cells and in vivo in the UUO kidney induced in Smad3 KO mice and conditional Smad2 KO mice. The finding of Smad3, not Smad2, to regulate miR-192 expression in response to TGF-β1 may be associated with the ability of Smad3 to bind DNA sequences directly through its MH1 domain, whereas Smad2 has no DNA-binding sequences and activates transcription indirectly by binding to transcriptional co-activators or repressors.22,23 Additional evidence supporting the Smad3-dependent miR-192 expression comes from the findings that Smad3-binding elements were found in the 5′ end of miR-192. Furthermore, by using ChIP assays, we successfully demonstrated that an antibody against Smad3 could successfully immunoprecipitate the DNA fragments containing the potential Smad3-binding sites, supporting that Smad3 could physically interact with the promoter region of miR-192. Thus, miR-192 expression mediated by Smad3, not Smad2, may account for our recent findings that Smad3, not Smad2, mediates renal and cardiovascular fibrosis in vivo and in vitro under diabetic and hypertensive conditions13,14; however, no enhancement of the ChIP signals upon TGF-β treatment was observed, suggesting that other factors may possibly influence Smad3 binding to the miR-192 promoter during TGF-β–induced miR-192 transcription. Because our recent study showed that p53 promotes renal injury in acute aristolochic acid nephropathy24 and miR-192 can be transcriptionally induced by p5325–27 in colon cancer cells, regulation of miR-192 transcription during renal injury may not be limited by TGF-β/Smad3 signaling alone; therefore, further investigation into the functional importance of miR-192 in transcriptional regulation of fibrosis in response to TGF-β/Smad3 signaling is needed.
Predicted and validated target genes for miR-192 have been published in several studies.8,26–28 During diabetic nephropathy, miR-192 has been shown to target both ZEB1 and ZEB2 in human TECs and only ZEB2 in mouse MCs.8,9 Both ZEB1 and ZEB2 are known to repress the E-cadherin transcription but to activate the vimentin expression during EMT.29 In addition, miR-192 has been demonstrated to contribute to the upregulation of collagen type I α2 in mouse MCs and isolated glomeruli from diabetic mouse.8 In human TECs, miR-192 seems to associate with maintenance of the E-cadherin expression9; therefore, miR-192 may be functionally important in kidney diseases. Normally, miR-192 is preferentially expressed in kidney as compared with miRNA expression profiles in different organs.6–8 Within rat kidneys, miR-192 is expressed at least 20-fold higher in the renal cortex than in the medulla,6 suggesting that miR-192 may be important for normal renal physiology, such as the regulation of the molecular machinery used for sodium transport in renal epithelial cells.30 This study provides the additional evidence that increased miR-192 expression is associated with renal fibrosis in normal rat TECs and in rodent models of either obstructive or hypertensive nephropathy.
Although our findings from this study were consistent with previous studies that miR-192 expression is induced in MCs by TGF-β1 and is upregulated in diabetic mouse kidney8 and in hypertensive nephropathy and IgA nephropathy,31,32 they were inconsistent with the recent findings reported by Krupa et al.9 that miR-192 expression was reduced by TGF-β1 in human TECs (HK-2) and downregulated in human diabetic nephropathy. Although these discrepancies are yet unexplained, the differences in the disease nature and conditions, the cell types with or without modifications, and the time and dosages of TGF-β1 used should be considered. In this study, we used a low dosage of TGF-β1 (2 ng/ml) to induce miR-192 expression significantly at 1 hour in normal rat TECs (NRK52E). In contrast, Krupa et al.9 used a high dosage of TGF-β1 (10 ng/ml) to reduce miR-192 at 96 hours in modified human HK-2 TECs. This late response may not be a direct effect of TGF-β1. It is possible that miR-192 may also be regulated by other factors under high concentrations of TGF-β1 or diabetic conditions. In addition, we found that there were two SBSs in the promoter of rodent miR-192 but only one SBS in human miR-192 promoter. This difference may also explain the discrepancy of miR-192 expression in diabetic nephropathy between humans and rodents. Nevertheless, in the study by Krupa et al.,9 the functional importance of miR-192 in regulating the key feature of fibrosis such as collagen matrix expression has not been investigated, which prevents further discussion into the discrepancy among these studies.
In summary, we demonstrated that miR-192 is upregulated in the fibrotic kidney associated with activation of TGF-β/Smad signaling. TGF-β1 regulates miR-192 expression positively by Smad3, not Smad2, and negatively by a Smad7-dependent mechanism. Thus, results from this study demonstrate a potential importance of miR-192 as a downstream mediator of TGF-β/Smad3 in renal fibrosis.
Concise Methods
Cell Culture
The NRK52E normal rat TEC line was obtained from American Type Culture Collection and grown in DMEM/LG containing 0.5% FBS with human TGF-β1 (R&D Systems, Minneapolis, MN) at concentrations of 0.00, 0.25, 0.50, 1.00, 2.00, and 4.00 ng/ml in the presence or absence of a neutralizing TGF-β antibody (10 μg/ml; R&D Systems) for periods of 0, 1, 3, 6, 12, and 24 hours for miR-192 detection.
To investigate the negative regulating role of Smad7 in TGF-β–induced miR-192 expression, we used a stable doxycycline (Dox)-regulated Smad7-expressing NRK52E cell line18 and induced Smad7 transgene expression by addition of Dox at an optimal concentration (2 μg/ml) for 24 hours. To further dissect the specific role of Smad2 and Smad3 in TGF-β–induced miR-192 expression, we generated and characterized stable cell lines with Smad2 and Smad3 gene knockdown in NRK52E as described previously.14 To examine whether de novo protein synthesis was required for miR-192 expression, we pretreated cells with 20 μM cycloheximide (Sigma, St. Louis, MO) for 30 minutes before TGF-β1 stimulations and cultured them for periods as described previously.
Transient Transfection of Cells with microRNAs
NRK52E cells were reverse-transfected with premiR-192 or anti–miR-192 (Ambion, Austin, TX) at a final concentration of 30 nM by using siPORT NeoFX transfection reagent (Ambion) in six-well plates. The control experiments were transfected with scramble RNA ODN (30 nM/ml; Ambion). Then the cells were stimulated with human TGF-β1 (R&D Systems) at 2 ng/ml for 3, 6, 12, and 24 hours in serum-free medium following our protocols.33
RNA Extraction and Quantitative Reverse Transcription–PCR Analysis
Total RNA was isolated from the cultured cells and kidney tissues by Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Template cDNA was prepared using reverse transcriptase, and miR-192 expression was quantified by real-time PCR by TaqMan MicroRNA Assay (Applied Biosystems, Foster City, CA) with small nuclear RNA U6 as an endogenous control for normalization according to the manufacturer's instructions. Gene expression profiles were normalized to U6 snRNA and calculated using the ΔΔCt (2−ΔΔCt) method.
Real-time reverse transcription–PCR of the fibrotic markers and Smad2 mRNA was performed as described previously.13 Ratios for mRNA/glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA were calculated using the ΔΔCt method for each sample and expressed as means ± SEM.
miRNA Expression Analyses
Total RNA (350 ng) was reverse-transcribed using TaqMan MicroRNA RT kit and Multiplex RT rodent primer pool (Applied Biosystems) according to the manufacturer's instructions. cDNAs were added to master mix (Applied Biosystems) and applied to miRNA TaqMan low-density array rodent panels 2.0 (Applied Biosystems) for the simultaneous quantification of 375 miRNAs according to the manufacturer's instructions. miRNA TaqMan assays were performed using 7900 HT Fast Real-Time PCR System (Applied Biosystems). Relative quantification was performed using ΔΔCt method, and the data were normalized with U6 and RNU48 (Applied Biosystems) as endogenous controls. Data were analyzed with Real-Time StatMiner (Integromics, Madrid, Spain). Significance was considered at P < 0.05.
ChIP Analysis
ChIP was performed by transcription factor ChIP kit according to the manufacturer's instructions. In brief, cells were cross-linked with 1% formaldehyde for 10 minutes at 37°C, quenched with glycine, and then sonicated using a Bioruptor (Diagenode, Liège, Belgium) to generate 300- to 600-bp DNA fragments. Immunoprecipitation was performed with the antibody against Smad3 (Upstate), and IgG was used as a control. Precipitated DNAs were detected by PCR using specific primers: SBS1 5′-AGCCAAGTGTCTCTCCGTGT-3′ and 5′-GGTGGCAGGAGGTGTGTACT-3′, SBS2 5′-CACTGTCCCAACATGCTCAC-3′ and 5′-GGTAGCAGGGAGAAGCAATG-3′, and SBS3 5′-CTTGGAAGGGATGAGAGTGC-3′ and 5′-GGCCAGCACCTAGAACTCAG-3′, The reaction mix consisted of 1.5 μl of template, 5.0 μl of 2× Bio-Rad iQ SYBR Green supermix, 0.3 μl of forward primer, 0.3 μl of reverse primer, and 2.9 μl of ddH2O. qPCR was performed as described already for quantitative real-time PCR.
Obstructive Kidney Disease Model
A UUO kidney disease model was induced in eight Smad3 WT/KO mice,34 Smad7 WT/KO mice,35 Smad2 floxed mice,36 and conditional Smad2 KO mice by left ureteral ligation as described previously.37 Conditional Smad2 KO mice were generated in Smad2 floxed/floxed mice by an ultrasound-microbubble–mediated Cre recombination technique as described previously13 and were characterized at the DNA level by PCR, mRNA level by real-time PCR, and protein expression levels by Western blot and immunohistochemistry with the anti-Smad2 antibody (Chemicon, Temecula, CA). Mice were killed on day 7 after the ligation. In addition, eight normal WT and eight normal transgenic mice were used as age-matched controls. The experimental procedures were approved by Animal Experimental Ethnic Committee of the Chinese University of Hong Kong.
Animal Model of Remnant Kidney Disease Treated with Smad7 Gene Transfer
A rat remnant kidney disease model was induced in male Sprague-Dawley rats by 5/6 subtotal nephrectomy as described previously.11 Immediately after the nephrectomy, groups of six animals received a ultrasound-microbubble–mediated, Dox-inducible Smad7 or empty vectors as described previously.11 All procedures were performed in accordance with institutional guidelines for animal care.
Western Blot Analysis
Western blot analysis was performed as described previously,14 with primary antibodies against collagen I (Southern Tech, Birmingham, AL) and GAPDH (Chemicon). After being incubated with an IRDyeTM800-conjugated secondary antibody (Rockland Immunochemicals, Gilbertsville, PA), the membranes were analyzed using LiCor/Odyssey infrared image system (LI-COR Biosciences, Lincoln, NE). Signal intensities from each Western blot were quantified by ImageJ software (National Institutes of Health, Bethesda, MD) and normalized to GAPDH levels.
Statistical Analysis
Each experiment was repeated at least three times throughout the study. Data were expressed as means ± SEM and analyzed using one-way ANOVA with Newman-Keuls comparison program from GraphPad Prism 5.0 (GraphPad Software, San Diego, CA).
Disclosures
None.
Acknowledgments
This work was supported by grants from the Research Grant Council of the Hong Kong SAR, China (GRF 768207 and 767508 and CUHK5/CRF/09 to H.Y.L., and 763908 and 764109 to A.C.C.).
We thank Dr. Erwin P. Bottinger for the Smad3 WT and KO MEF cell lines, Dr. Wansheng Wang for assistance in characterizing conditional Smad2 KO mice, and Dr. Chen-Chun Hou for preparing rat remnant kidney tissues.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
REFERENCES
- 1.Liu Y: Renal fibrosis: New insights into the pathogenesis and therapeutics. Kidney Int 69: 213–217, 2006 [DOI] [PubMed] [Google Scholar]
- 2.Bottinger EP, Bitzer M: TGF-beta signaling in renal disease. J Am Soc Nephrol 13: 2600–2610, 2002 [DOI] [PubMed] [Google Scholar]
- 3.Schnaper HW, Hayashida T, Poncelet AC: It's a Smad world: Regulation of TGF-beta signaling in the kidney. J Am Soc Nephrol 13: 1126–1128, 2002 [DOI] [PubMed] [Google Scholar]
- 4.Wang W, Koka V, Lan HY: Transforming growth factor-beta and Smad signalling in kidney diseases. Nephrology (Carlton) 10: 48–56, 2005 [DOI] [PubMed] [Google Scholar]
- 5.Zavadil J, Narasimhan M, Blumenberg M, Schneider RJ: Transforming growth factor-beta and microRNA:mRNA regulatory networks in epithelial plasticity. Cells Tissues Organs 185: 157–161, 2007 [DOI] [PubMed] [Google Scholar]
- 6.Tian Z, Greene AS, Pietrusz JL, Matus IR, Liang M: MicroRNA-target pairs in the rat kidney identified by microRNA microarray, proteomic, and bioinformatic analysis. Genome Res 18: 404–411, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun Y, Koo S, White N, Peralta E, Esau C, Dean NM, Perera RJ: Development of a micro-array to detect human and mouse microRNAs and characterization of expression in human organs. Nucleic Acids Res 32: e188, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R: MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci U S A 104: 3432–3437, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krupa A, Jenkins R, Luo DD, Lewis A, Phillips A, Fraser D: Loss of microRNA-192 promotes fibrogenesis in diabetic nephropathy. J Am Soc Nephrol 21: 438–447, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung AC, Huang XR, Zhou L, Heuchel R, Lai KN, Lan HY: Disruption of the Smad7 gene promotes renal fibrosis and inflammation in unilateral ureteral obstruction (UUO) in mice. Nephrol Dial Transplant 24: 1443–1454, 2009 [DOI] [PubMed] [Google Scholar]
- 11.Hou CC, Wang W, Huang XR, Fu P, Chen TH, Sheikh-Hamad D, Lan HY: Ultrasound-microbubble-mediated gene transfer of inducible Smad7 blocks transforming growth factor-beta signaling and fibrosis in rat remnant kidney. Am J Pathol 166: 761–771, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato M, Muragaki Y, Saika S, Roberts AB, Ooshima A: Targeted disruption of TGF-beta1/Smad3 signaling protects against renal tubulointerstitial fibrosis induced by unilateral ureteral obstruction. J Clin Invest 112: 1486–1494, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung AC, Zhang H, Kong YZ, Tan JJ, Huang XR, Kopp JB, Lan HY: Advanced glycation end-products induce tubular CTGF via TGF-beta-independent Smad3 signaling. J Am Soc Nephrol 21: 249–260, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang F, Chung AC, Huang XR, Lan HY: Angiotensin II induces connective tissue growth factor and collagen I expression via transforming growth factor-beta-dependent and -independent Smad pathways: The role of Smad3. Hypertension 54: 877–884, 2009 [DOI] [PubMed] [Google Scholar]
- 15.Zhou L, Fu P, Huang XR, Liu F, Chung AC, Lai KN, Lan HY: Mechanism of chronic aristolochic acid nephropathy: Role of Smad3. Am J Physiol Renal Physiol 298: F1006–F1017, 2010 [DOI] [PubMed] [Google Scholar]
- 16.Piek E, Ju WJ, Heyer J, Escalante-Alcalde D, Stewart CL, Weinstein M, Deng C, Kucherlapati R, Bottinger EP, Roberts AB: Functional characterization of transforming growth factor beta signaling in Smad2- and Smad3-deficient fibroblasts. J Biol Chem 276: 19945–19953, 2001 [DOI] [PubMed] [Google Scholar]
- 17.Loots GG, Ovcharenko I: rVISTA 2.0: Evolutionary analysis of transcription factor binding sites. Nucleic Acids Res 32: W217–W221, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li JH, Zhu HJ, Huang XR, Lai KN, Johnson RJ, Lan HY: Smad7 inhibits fibrotic effect of TGF-beta on renal tubular epithelial cells by blocking Smad2 activation. J Am Soc Nephrol 13: 1464–1472, 2002 [DOI] [PubMed] [Google Scholar]
- 19.Li JH, Huang XR, Zhu HJ, Johnson R, Lan HY: Role of TGF-beta signaling in extracellular matrix production under high glucose conditions. Kidney Int 63: 2010–2019, 2003 [DOI] [PubMed] [Google Scholar]
- 20.Fukasawa H, Yamamoto T, Togawa A, Ohashi N, Fujigaki Y, Oda T, Uchida C, Kitagawa K, Hattori T, Suzuki S, Kitagawa M, Hishida A: Down-regulation of Smad7 expression by ubiquitin-dependent degradation contributes to renal fibrosis in obstructive nephropathy in mice. Proc Natl Acad Sci U S A 101: 8687–8692, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang XR, Chung AC, Wang XJ, Lai KN, Lan HY: Mice overexpressing latent TGF-beta1 are protected against renal fibrosis in obstructive kidney disease. Am J Physiol Renal Physiol 295: F118–F127, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yagi K, Goto D, Hamamoto T, Takenoshita S, Kato M, Miyazono K: Alternatively spliced variant of Smad2 lacking exon 3: Comparison with wild-type Smad2 and Smad3. J Biol Chem 274: 703–709, 1999 [DOI] [PubMed] [Google Scholar]
- 23.Moustakas A, Heldin CH: The regulation of TGFbeta signal transduction. Development 136: 3699–3714, 2009 [DOI] [PubMed] [Google Scholar]
- 24.Zhou L, Fu P, Huang XR, Liu F, Lai KN, Lan HY: Activation of p53 promotes renal injury in acute aristolochic acid nephropathy. J Am Soc Nephrol 21: 31–41, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braun CJ, Zhang X, Savelyeva I, Wolff S, Moll UM, Schepeler T, Orntoft TF, Andersen CL, Dobbelstein M: p53-Responsive microRNAs 192 and 215 are capable of inducing cell cycle arrest. Cancer Res 68: 10094–10104, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Georges SA, Biery MC, Kim SY, Schelter JM, Guo J, Chang AN, Jackson AL, Carleton MO, Linsley PS, Cleary MA, Chau BN: Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res 68: 10105–10112, 2008 [DOI] [PubMed] [Google Scholar]
- 27.Song B, Wang Y, Kudo K, Gavin EJ, Xi Y, Ju J: miR-192 regulates dihydrofolate reductase and cellular proliferation through the p53-microRNA circuit. Clin Cancer Res 14: 8080–8086, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weber F, Teresi RE, Broelsch CE, Frilling A, Eng C: A limited set of human microRNA is deregulated in follicular thyroid carcinoma. J Clin Endocrinol Metab 91: 3584–3591, 2006 [DOI] [PubMed] [Google Scholar]
- 29.Miska EA: MicroRNAs: Keeping cells in formation. Nat Cell Biol 10: 501–502, 2008 [DOI] [PubMed] [Google Scholar]
- 30.Liang M, Liu Y, Mladinov D, Cowley AW, Jr, Trivedi H, Fang Y, Xu X, Ding X, Tian Z: MicroRNA: A new frontier in kidney and blood pressure research. Am J Physiol Renal Physiol 297: F553–F558, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang G, Kwan BC, Lai FM, Choi PC, Chow KM, Li PK, Szeto CC: Intrarenal expression of microRNAs in patients with IgA nephropathy. Lab Invest 90: 98–103, 2010 [DOI] [PubMed] [Google Scholar]
- 32.Wang G, Kwan BC, Lai FM, Choi PC, Chow KM, Li PK, Szeto CC: Intrarenal expression of miRNAs in patients with hypertensive nephrosclerosis. Am J Hypertens 23: 78–84, 2010 [DOI] [PubMed] [Google Scholar]
- 33.Li JH, Huang XR, Zhu HJ, Oldfield M, Cooper M, Truong LD, Johnson RJ, Lan HY: Advanced glycation end products activate Smad signaling via TGF-beta-dependent and independent mechanisms: Implications for diabetic renal and vascular disease. FASEB J 18: 176–178, 2004 [DOI] [PubMed] [Google Scholar]
- 34.Yang X, Letterio JJ, Lechleider RJ, Chen L, Hayman R, Gu H, Roberts AB, Deng C: Targeted disruption of SMAD3 results in impaired mucosal immunity and diminished T cell responsiveness to TGF-beta. EMBO J 18: 1280–1291, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li R, Rosendahl A, Brodin G, Cheng AM, Ahgren A, Sundquist C, Kulkarni S, Pawson T, Heldin CH, Heuchel RL: Deletion of exon I of SMAD7 in mice results in altered B cell responses. J Immunol 176: 6777–6784, 2006 [DOI] [PubMed] [Google Scholar]
- 36.Ju W, Ogawa A, Heyer J, Nierhof D, Yu L, Kucherlapati R, Shafritz DA, Bottinger EP: Deletion of Smad2 in mouse liver reveals novel functions in hepatocyte growth and differentiation. Mol Cell Biol 26: 654–667, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lan HY, Mu W, Tomita N, Huang XR, Li JH, Zhu HJ, Morishita R, Johnson RJ: Inhibition of renal fibrosis by gene transfer of inducible Smad7 using ultrasound-microbubble system in rat UUO model. J Am Soc Nephrol 14: 1535–1548, 2003 [DOI] [PubMed] [Google Scholar]