Abstract
Grid technologies are appealing to deal with the challenges raised by computational neurosciences and support multi-centric brain studies. However, core grids middleware hardly cope with the complex neuroimaging data representation and multi-layer data federation needs. Moreover, legacy neuroscience environments need to be preserved and cannot be simply superseded by grid services. This paper describes the NeuroLOG platform design and implementation, shedding light on its Data Management Layer. It addresses the integration of brain image files, associated relational metadata and neuroscience semantic data in a heterogeneous distributed environment, integrating legacy data managers through a mediation layer.
Keywords: Distributed Data Management, Relational Data, Semantic Data
1. Federating multi-centric neuroimaging data resources
The clinical world has witnessed the generalization of radiology data acquisition in digital format, the increasing importance of imaging techniques in healthcare (prognosis, diagnosis, planning, guided intervention), and more than two decades of digital image analysis techniques capability improvement. In particular, neurosciences are highly depending on brain imaging modalities (primarily MR and PET) and computerized image analysis techniques. Computational neuroscience has emerged as a discipline of its own, demonstrating the power of computing techniques to analyze neurological data sets and study the brain functions. The informatics technologies exploited by neuroscientists have evolved with the state of the art in computer science. Recently, an increasing effort has been invested in grid technologies for neurosciences to extend the computing infrastructures deployed within each brain imaging center and to face the challenges raised by modern multi-patients statistical studies or biomodeling activities. This momentum is testified through the emergence of targeted projects such as NeuroGrid [13], BIRN [5] or neuGrid [2], accompanied by the development and distribution of large-scale neuroimage resources sharing infrastructures such as NeuroBase [3] or ADNI [1].
To support multi-centric neuroscience studies, the NeuroLOG project1 similarly aims at integrating neurological resources into a collaborative platform that federates local neuroscience resources published by several centers, bridging legacy informatics environments deployed over these centers and leveraging grid technologies. The grand objective is the provision of an unprecedented scale platform dedicated to neurosciences addressing both the needs for individual resources control and easy exchanges of resources between contributing institutes. A particularly important milestone in the completion of the NeuroLOG road-map is the Data Management Layer (DML) architecture and design. It is based on the integration of cutting-edge technologies in the area of data federation, distributed databases and knowledge representation/exploitation, as well as the development of new techniques for ensuring data coherence, completeness and quality. Given the sensitive nature of the data manipulated, an important aspect of the DML is its security architecture. However, it could not be described in details in this contribution. The interested reader can refer to [12].
The NeuroLOG prototype platform currently integrates data from 5 different centers. The major challenges addressed by the middleware design are: (i) the integration of pre-existing and independently managed data repositories, including image files and relational data, with heterogeneous data schemas; (ii) the provision of a coherent federated view of all data available; (iii) the representation of cross-health enterprise clinical data sets used in multi-centric studies; (iv) the capture, representation and exploitation of domain-specific knowledge; and (v) the autonomous operation of the federated centers through a weak coupling of the data repositories.
Some of the targeted objectives are difficult to fulfill simultaneously (e.g. preserving databases coherence and ensuring centers autonomy) and trade-offs have to be found. The rest of this document describes the NeuroLOG DML designed to integrate all these concerns. The corner-stone of the DML is a data schema federating existing ones, later on referred to as Federated Schema (FS). The FS design was based on a sound semantics capture methodology. It is encompassing both domain-specific relational information and technical information needed to associate radiology images with metadata as well as ensuring the overall platform coherence. Center-specific databases are dynamically mapped to the FS. Dedicated ontologies were designed for the needs of the FS specification. They are further on exploited to enrich the data repositories with semantic data and infer new domain knowledge.
2. Data Management Layer overview
The NeuroLOG DML integrates and delivers to the users multiple types of data. Raw radiology data is stored as image files. It is complemented by relational metadata. In the NeuroLOG platform, each neuroscience center contributes by exposing part of its image files and associated metadata. The middleware provides secure file transfer across-centers through encrypted communication channels and a federated RDBM query engine for accessing distributed metadata. In addition the middleware translates part of the available metadata into a semantic representation which allows for advanced querying. The semantic representation is grounded on a federation-wide common ontology specified with OWL-Lite. The semantic annotations are encoded as RDF triplets and stored in a centralized repository. To provide this complete panel of functionality, the DML integrates multiple technologies developed in the domains of data grids, Semantic Web and distributed RDBM.
2.1. Related work
Many different tools have been developed to achieve grid data integration, although to the best of our knowledge none of them encompasses all facets addressed by the DML. The Canadian Brain Imaging Research Network (CBRAIN) and the neuGrid EU project exploit LORIS [19], a centralized database system dedicated to brain data. TRENCADIS [6] is a distributed environment to share DICOM objects over multiple administrative domains. It uses simplified ontologies for federation, taking advantage of the structured DICOM representation. The caBIG cancer Biomedical Informatics Grid [8] similarly relies on a common vocabulary specification (VCDE). The BirnLex task force [5] develops elaborated ontologies but their exploitation is limited to formalize the domain terminology. The DML is a distributed system with dynamic data mediation capability, exploiting ontologies and the Conceptual Resource Search Engine (CORESE) semantic data manipulation engine [9] for new knowledge inference.
For data files management, grid file catalogs such as the gLite LFC (LHC File Catalog) [20] provide a unified hierarchical view on files distributed over distributed storage resources. The catalog is complemented by a file transfer interface. For instance, the gLite middleware complies with the Storage Resource Management (SRM) interface. The DICOM standard protocol has been integrated in several grid middleware services such as the gLite Medical Data Manager [18] and GLOBUS Medicus [11]. The DML provides an upper layer which indexes both LFC files and local site files that are not managed through SRM. Compliance with DICOM was not required however, since data sources deployed in neuroscience centers are typically non-DICOM data servers.
The ARDA metadata catalog project (AMGA) interface [16] was developed to provide a grid credential-compliant secure and homogeneous interface to various relational RDBM. It has been used for structured medical metadata storage, such as DICOM-Structured Reports [7]. The DML is based on a JDBC interface instead, and integrates the DataFederator [10] mediation and multi-databases unification layers that are not available with AMGA. The OGSA-DAI data integration layer [14] could have been considered as an alternative, although it does not provide the same level of distributed relational queries optimization as DataFederator.
2.2. Data repositories
As illustrated in figure 1, from the end-users point of view, the DML appears as three data repositories providing a unified virtual view of the data fragments distributed over the participating sites: a relational metadata repository, a file catalog and a semantic repository. The complete DML is structured according to the metadata repository. Data files are indexed through this catalog and raw semantic data is extracted out of its content.
Figure 1.
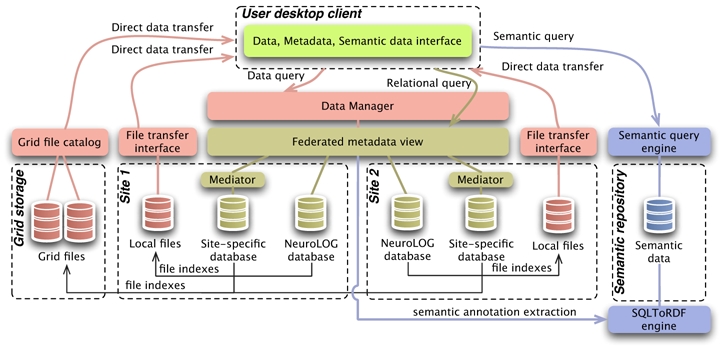
Data Management Layer overview
No virtual file hierarchy tree is exposed to the users. The different directory structures and file name conventions adopted by the participating sites would make a file hierarchy difficult to comprehend and navigate. Instead, metadata is easily browsable and refers to the relevant image files. In addition, files stored on the EGEE grid infrastructure can similarly be indexed from the DML metadata and transparently be integrated.
The semantic repository is centralized due to the current limitations of the tooling available to manipulate semantic annotations. It is composed of several ontologies (see section 3.1), associated rules, and RDF annotations compliant with the ontologies taxonomy. The annotations are primarily extracted from relational metadata and later on completed by platform usage information.
An important aspect of the DML is to preserve the legacy metadata repositories of each participating site. Metadata repositories contain rich clinical information which has been either automatically extracted from DICOM image headers or manually filled-in by neuroscientists. The installation of the NeuroLOG middleware does not interfere with the legacy environments which continue to be managed and accessible locally through site-specific tools. Instead, the DML includes a mediation layer that adapts to the site-specific data structures, preserving the complete autonomy of the sites and the backward compatibility with years of efforts dedicated to the development of the local environments. The integration of the radiology image files is simple through file indexing as described above. The most challenging aspect is the integration of the relational databases deployed over the centers.
Given that the DML makes no assumption on the data management policies implemented nor has any control over the relational entities managed within each site, it has a read-only access to these site-specific databases. To hold middleware-managed entities and the administration metadata needed for distributed operations such as access control information or data files indexing, an additional NeuroLOG database, structured according to the FS, is deployed at each site. The relational mediation layer of the DML performs a mapping of site-specific entities to the FS. Thus both the site-specific database and the NeuroLOG database share the same external view and browse interface.
2.3. Technical implementation
As the rest of the NeuroLOG middleware, the DML is architected as a set of collaborative Web Services implemented in java and hosted in Apache Tomcat servers deployed at each participating site. The site-specific relational mediation layers are topped with a JDBC-compliant distributed relational query engine. Local file servers are topped with a data management layer capable of identifying the location of any file from its identifier (making use of the distributed relational engine) and delivering data files directly to the client over an HTTPS channel. Grid files are also recognized and transferred directly to the client using the GridFTP protocol. The semantic repository is a simple set of RDF files produced by the METAMorphoses translation tool [21]. The CORESE semantic query engine, topped with a Web Service interface, is used for querying semantic information.
3. Data Management Layer design
3.1. Federated Schema
Federating heterogeneous data from several independent databases raises the issue of referring to common semantics. Our common semantics are defined by means of an ontology, called OntoNeurolog. The design of this ontology is an important part of the project and should be seen as a major deliverable, which can be used in other similar projects.
OntoNeurolog reuses and extends the OntoNeurobase ontology [22]. Both were designed using the same methodological framework [22], based on: (1) the use of a foundational ontology called DOLCE (Descriptive Ontology for Linguistic and Cognitive Engineering) [17], that provides the philosophical principles underlying the overall modeling, (2) the use of a number of core ontologies, that provide generic, basic and minimal concepts and relations in specific domains such as Artefacts, Participant roles, Information and Discourse acts, (3) the use of OntoSpec [15] as an initial semi-formal representation. Two implementations were manually derived from this initial highly expressive representation: an OWL-Lite implementation and a relational schema.
The design of the ontology was envisaged as an iterative process, with three development stages, delivering three successive versions. The first focused on data sets (i.e. images) and the entities involved in their generation and use (Subjects that they concern, Studies and Examinations in the context of which they were obtained, Centers that produced them, etc.). The second puts emphasis on representing other facets of the subjects’ state, as explored using neuropsychological instruments and behavioral scales [4]. It also goes into further details about MR acquisition protocols and MR sequences. The third is still being developed and will focus on Region of interest (ROI) and ROI annotations, and on the annotation of processing tools shared as Web Services.
The relational FS is derived from the ontology. As a relational model, it can only partially implement it. In particular, complex class hierarchies, as well as properties of relationships are partial. Nevertheless, the reference to the ontology is useful to associate precise semantics to the databases’ entities denoted by table names and column names, and facilitate their use as a pivot to integrate heterogeneous data. Furthermore, the joint integration of the relational FS and the semantic repository into the DML ensures (i) compatibility with the legacy metadata stores federated, (ii) performance of relational query languages, and (iii) advanced data search capability exploiting the complete expressivity power of a semantic representation and the associated query language.
3.2. Mediation layer
Although NeuroLOG sites host databases with heterogeneous engines and schemas, they do share common concerns: they deal with the same entities such as brain images and studies. In order to come up with a consistent federated view, each site aligns its own legacy database onto the FS, hence allowing the DML to handle multi-centric queries. The alignment is performed by mapping site-specific columns and tables to their equivalent in the FS (no mapping is needed for the NeuroLOG databases that already adopts the FS). However, the semantics of an entity in a site-specific database may be slightly different from the semantics of this entity in the NeuroLOG ontology. Depending on the way entities were designed, the mapping may be a rather straight process, or it may require some choices in order to preserve the original semantics as much as possible as illustrated in Figure 2. For instance, a straight mapping (four first fields of the Sutdy entity) transforms local IDs (primary keys) into global IDs to avoid ID overlaps; another one maps values from a domain or an enumerated type to a system-wide domain of values, e.g.: {true, false} to {1, 2}. In a more complex case (fifth field of the Study entity), each line of a multiple-valued column of the source table (centers) has to be converted into several lines of the target table (Center), while generating relations between the Study and the Center in an intermediate relation table.
Figure 2.
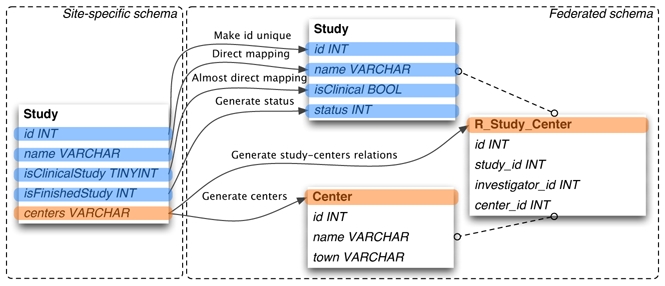
Mappings between site-specific and common schemas
Complex mappings raise questions about how trustful the mapped data is, with regards to the semantics of the source data. Typically, mappings may (i) narrow down a concept, that is possibly loose information, e.g. map {left, right, converted left, ambidextrous} to {left, right}; (ii) conversely, broaden a concept; and (iii) come up with relations that do not exist in the site-specific database. Such issues challenge the global coherence of the federated data, and should be answered in collaboration with site-specific schema experts. In case mappings raise unacceptable inconsistencies or approximations, either (i) the site-specific schema is not sufficient to express the required semantics, a schema change should be considered (if this is acceptable for the site) or (ii) the ontology lacks some semantics details and must be changed to reflect this.
3.3. Ensuring coherence of distributed metadata
The NeuroLOG metadata mediation layer provides a federated view of entities distributed over several sites. The coherence of the metadata system may be challenged due to (i) relations between entities stored in different databases (cross-site entities referencing), and (ii) physical objects represented by multiple instances in different databases (multiple instances of an entity).
Cross-site entities referencing
Entities are represented in a relational schema as table lines, indexed by a local identifier (the primary key). Within an RDBM, references to other entities are secondary (foreign) keys whose coherence is guaranteed within the same database. In the federated view though, referred entities may be stored in any of the two databases of each site, and on any site. Therefore a local identifier is mapped into a global system-wide identifier as the triplet <site>:<database>:<local_ID> by the DataFederator site-specific mapper. Cross-site references typically occur for DataSets resulting from the processing of parent DataSets from different sites, or multi-centric studies relating to Subjects and DataSets from different sites.
Multiple instances of an entity
Site-specific databases are meant to store entities such as Subjects, Studies or clinical Centers. Given that they are managed independently on each site, there is no guarantee that some physical entity will not appear in several databases. In addition, these instances may not necessarily be coherent. Some multiple instances occurrence may be a rare event (e.g. a subject may be scanned on purpose in different centers but this represents a minority of the acquisitions), while other occurence may be rather common (e.g. a multi-centric study involves several sites, each one holding an instance of the study). Also some multiple instances occurrence may be critical (e.g. a multi-centric study must clearly be identified), while other occurence may not be critical (e.g. multiple definition of a clinical center may not be harmful). Multiple instances of an entity need to be identified manually when they occur, a master/slave tagging is then applied: a single instance is tagged as master while all others are slaves. Through its specific mapping, a site only reports its proper entities and its master entities while ignoring slaves. This ensures that a single instance of each entity appears in the federated view.
DML coherence
The distributed metadata management is supported to a large extent by the DataFederator tool as the mappings are used to (i) provide the federated view, (ii) reference entities across sites, and (iii) manage multiple instances (master/slave). However, the overall coherence of the federated view is not guaranteed and it is challenged by sites autonomous behavior. For instance, an entity might be removed by the local RDBM independently to potential cross-site references. This enforces the DML to restore coherence through a curation process. Consequently, a curation procedure is proposed to periodically detect and solve inconsistencies. For instance, when a master is deleted, one of the slaves is promoted as new master. The principle of multiple instances management can be applied to the replication of some critical entities whose uncontrolled update would strongly impact the federation. An assessment of the most critical entities is done and cross-referenced critical entities are replicated locally to the reference site as a slave. This improves both reliability and performance of the DML.
3.4. Semantic repository
The rationale for building and exploiting a semantic repository is that the semantics of the shared information stored in the different NeuroLOG databases cannot be fully represented in relational databases. As far as classes are concerned, such knowledge can easily be represented using ontologies. The semantic repository relates the entities and relationships represented in the databases to the corresponding classes and properties of the ontology, using RDF triplets. It is supposed to allow “intelligent” querying, that explicitly uses the knowledge embedded in the ontology, such as the taxonomy of classes, the associated axioms, or the characteristics of the object properties (e.g. transitivity). The NeuroLOG client desktop application allows users to query the semantic repository through the CORESE semantic search engine. While end-users submit predefined SPARQL queries, expert users can edit and submit their own queries.
Semantic data is extracted from the metadata, thanks to the METAmorphoses tool (SQLToRDF engine in Figure 1). It exploits a specific schema mapping language specifying the mapping between some parts of the database schema and the ontology. It queries the FS schema through the JDBC interface and generates an RDF document.
3.5. Distributed files
Data files stored on the NeuroLOG platform and the EGEE grid infrastructure are federated through the index held in the federated relational view. A federation-wide unique identifier is associated to each file (the gLite Grid Unique IDentifiers are reused for grid files and local identifiers prefixed with a unique site name are assigned to local files). The file identifier is associated to relevant entities (DataSet, etc) through the FS. File discovery is performed by metadata browsing. Once file identifiers have been selected by the client, data file queries are sent to the data management service, as illustrated in Figure 1. If needed, this service exposes the data files on an HTTPS server. This is usually the case for legacy neuroimaging centers which do not expose their data to the Internet. Conversely, grid files are already exposed through a standard SRM interface implementing various transfer protocols. File transfers are performed directly between the storage resource and the client, using one of the supported protocols (HTTP/HTTPS file transfer, GridFTP or Web Service optimized streaming - MTOM).
The data file manager thus implements an extended file index that covers both grid and local files. In addition, a strict and fine-grained file access control policy is implemented to restrict file transfers to authorized clients. This policy, described in [12], ensures that each contributing site preserves full control over its data files, while enabling across-sites file exchanges.
4. System deployment and usage
4.1. Prototype infrastructure configuration
The NeuroLOG platform is currently deployed on five participating sites, namely: ASCLEPIOS (Nice), GIN (Grenoble), IRISA (Rennes), IFR49 (Paris) and I3S (Nice). Each site provides either data or processing tools (or both), depending on their specific research material, and proposes a secured access to its registered users. Each site deploys a NeuroLOG site server consisting of a web application container (Apache Tomcat) hosting the NeuroLOG middleware (including all DML core services), and a DataFederator query server that maps the site-specific database to the FS. Site servers are the entry point for Java desktop clients to securely connect to and interact with the whole federation.
4.2. Shared data
Three main clinical applications are considered: Multiple Sclerosis, Stroke and Alzheimer. Presently, the five distributed databases contain essentially structural MR images (T1-weighted, T2-weighted, Flair and Diffusion), brief information about subjects (sex, age, patient or healthy volunteer) and the corresponding name of the study in which the subject was involved. For each MR image, the modality (MR Dataset), the type (e.g. reconstructed or non reconstructed), the nature (e.g. T1-weighted or Diffusion) and the explore entity (e.g. Anatomical or Functional) are available. Because of their importance for patient retrieval and collecting, information about the neuropsychological and behavioral scores such as MMSE (Mini Mental State Examination) or CDR-SoB (Clinical Dementia Rating Scale Sum of Boxes) will be added soon.
4.3. Typical use case
DataFederator together with the NeuroLOG middleware provide a consistent relational federated view of the multiple sites databases. The NeuroLOG client application provides users with the ability to query data from the federated view, and ultimately collect data sets of interest. Querying data is designed as browsing through a tree of metadata where branches are parent-child entity relations defined by the ontology, typically relations between Subject, Study and Dataset, and leaves are data sets. At each step the query may be refined using search criteria matching entity properties of the ontology.
A typical example of using the architecture is illustrated in figure 3. From left to right, all clinical studies are first searched for and three of them are selected in the result panel (bottom left). Second, male subjects, involved in the selected studies, between 40 and 55 years old are searched for. 14 patients are retrieved (4 from ASCLEPIOS, 5 from IRISA, 5 from IFR49) out of which 4 subjects are selected. Third, data sets acquired for those subjects whithin those studies are queried: all MR reconstructed datasets, which explored anatomy and are T1-Weighted are searched for. 12 data sets are retrieved from ASCLEPIOS (8) and IRISA (4). The identified data sets can then be added to the user cart (not shown on this figure), and the corresponding images downloaded to the user’s computer for visualization with the client image viewer or local storage.
Figure 3.
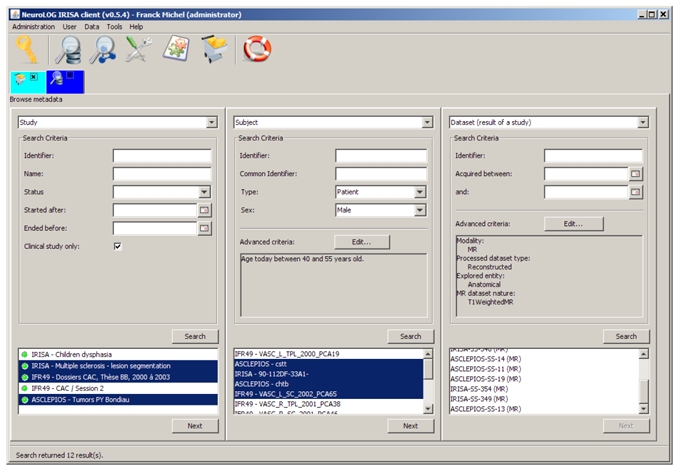
Navigating federated metadata to retrieve data sets
Additionally, users have the ability to view detailed data of any entity retrieved from queries in different ways: (i) either in tabular view (bottom-right window in figure 4) which allows to sort entites according to any criteria and change the selection as desired to go on with the next step of the browsing; (ii) or in a tree view (top-left window on figure 4) which root is the selected entity; descendent nodes reflect the relations between this entity and other entities as defined in the ontology.
Figure 4.
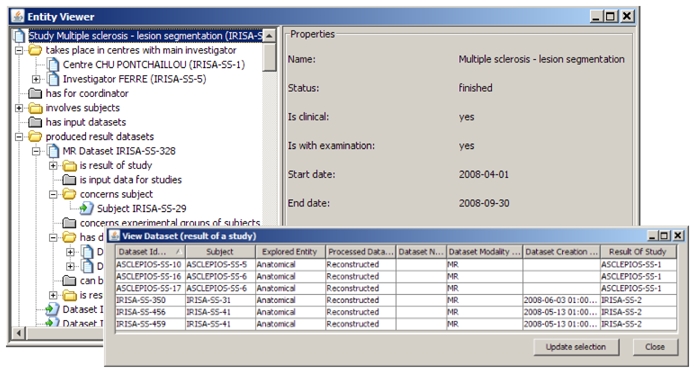
Different ways of viewing entities
4.4. System scalability
The NeuroLOG middleware is intended to be executed on high-reliability academic or professional networks. It is admitted that the connectivity between participating sites is hardly interrupted. Entities cross-site referencing (discussed in section 3.3) implies that some queries can only be resolved at the global system level. In particular, some join operations will be treated at the DataFederator engine level (rather than by the sites RDBM). Given the high-reliability assumption, referencing entities across sites is considered reasonable for all non-critical relations.
In a longer term, especially in the perspective of extending the system to less reliable networks (e.g. small radiology centers using ADSL connections), reliability can be reinforced by caching cross-referenced entities. However, providing a reliable metadata distribution manager in a low reliability environment is out of the scope of the NeuroLOG project as it would require modifications of the DataFederator tool to handle caches internally. Another issue to be addressed with the proposed solution is the behavior of DataFederator in case of site failure (server unreachable, application error): currently, a single site failure in a federated request will cause DataFederator to report an error and return no result at all. Although such failures are assumed to be rare, this behavior would be damageable and it is preferable to report all entities that could be collected in a best-effort way, while informing the querier that the result may be incomplete. In this perspective, we plan to implement a procedure to automatically detect site failures and run an automatic reconfiguration excluding the unreachable sites.
5. Conclusions and perspectives
This paper describes the NeuroLOG Data Management Layer, which provides a federated view of distributed neuroimaging resources (images, associated metadata and semantic data) avoiding interferences with the legacy environments. The approach adopted is grounded on a sound ontology design methodology from which the cross-sites Federated Schema is derived. The integration of a new site requires more than middleware services deployment. A manual process of local data representation analysis and relational schema mapping design has to be completed. The benefit of this work is a semantically tightly integrated data federation over data sources provisioned by loosely coupled, autonomous sites. These properties are critical, in a domain where only data sets acquired following rigorous protocols and properly documented can be actually exploited.
In addition to the data federation, image processing tools and image analysis procedures management capabilities (workflows) are currently being integrated into the NeuroLOG environment.
Acknowledgments
This work is funded by French National Agency for Research, NeuroLOG project, under contract number ANR-06-TLOG-024. The NeuroLOG platforms owes a lot to our clinical colleagues providing the databases exploited (Pr Gilles Edan and Dr Jean-Christophe Ferré in Rennes, Pr Jean-François Lebas in Grenoble, clinicians of the neuroradiology and neuropsychology departments of the Pitié Salpétrière hospital). We are grateful for the support provided by the CORESE development team (Dr Olivier Corby, Dr Catherine Faron-Zucker).
Footnotes
NeuroLOG: http://neurolog.polytech.unice.fr
References
- 1.ADNI (Alzheimer’s Disease Neuroimaging Initiative) http://www.loni.ucla.edu/ADNI.
- 2.Anjum A, Bloodsworth P, Habib I, Lansdale T, McClatchey R, Mehmood Y. HealthGrid’09. IOS Press; Jun, 2009. Reusable Services from the neuGRID Project for Grid-Based Health Applications; pp. 283–288. [PubMed] [Google Scholar]
- 3.Barillot C, Benali H, Dojat M, Gaignard A, Gibaud B, Kinkingnéhun S, Matsumoto J-P, Pélégrini-Issac M, Simon E, Temal L. HealthGrid’06. Spain: Jun, 2006. Federating Distributed and Heterogeneous Information Sources in Neuroimaging: The NeuroBase Project; pp. 3–13. [PubMed] [Google Scholar]
- 4.Batrancourt B, Dojat M, Gibaud B, Kassel G. A core ontology of instruments used for neurological, behavioural and cognitive assessments. 6th International Conference on Formal Ontology in Information Systems (FOIS); Toronto, Canada. May 2010. [Google Scholar]
- 5.BIRN (Biomedical Imaging Research Network) http://www.nbirn.net.
- 6.Blanquer Espert I, Hernández García V, Meseguer Anastásio J, Segrelles Quilis JD. Content-based organisation of virtual repositories of DICOM objects. Future Generation Computer Systems (FGCS) 2009 Jun;25(6):627–637. [Google Scholar]
- 7.Blanquer Espert I, Hernández García V, Salavert J, Segrelles Quilis JD. Using Grid-Enabled Distributed Metadata Database to Index DICOM-SR. HealthGrid’09. 2009 Jun;:117–126. [PubMed] [Google Scholar]
- 8.CaBIG (National Cancer Institute) http://cabig.cancer.gov.
- 9.Corby O, Dieng-Kuntz R, Faron-Zucker C. Prestigious Applications of Intelligent Systems, ECAI. Vol. 6. Valencia; Spain: 2004. Querying the semantic web with CORESE search engine; pp. 705–709. [Google Scholar]
- 10.Data Federator. http://www.sap.com/solutions/sapbusinessobjects/large/information-management/data-integration.
- 11.Erberich S, Silverstein J, Chervenak AL, Schuler R, Nelson M, Kesselman C. Globus MEDICUS - Federation of DICOM Medical Imaging Devices into Healthcare Grids. Studies in Health Technology and Informatics. 2007;126:269–278. [PubMed] [Google Scholar]
- 12.Gaignard A, Montagnat J. HealthGrid. IOS Press; Jun, 2009. A distributed security policy for neuroradiological data sharing; p. 262. [PubMed] [Google Scholar]
- 13.Geddes J, Mackay C, Lloyd S, Simpson A, Power D, Russell D, Katzarova M, Rossor M, Fox N, Fletcher J, Hill D, McLeish K, Hajnal J, Lawrie S, Job D, McIntosh A, Wardlaw J, Sandercock P, Palmer J, Perry D, Procter R, Ure J, Bath P, Watson G. The Challenges of Developing a Collaborative Data and Compute Grid for Neurosciences. 19th IEEE International Symposium on Computer-Based Medical Systems; Salt Lake City, Utah. June 2006; IEEE Computer Society; pp. 81–86. [Google Scholar]
- 14.Grant A, Antonioletti M, Hume AC, Krause A, Dobrzelecki B, Jackson MJ, Parsons M, Atkinson MP, Theocharopoulos E. OGSA-DAI: Middleware for data integration: Selected applications. ESCIENCE ’08: Proceedings of the 2008 Fourth IEEE International Conference on eScience; Washington, DC, USA. IEEE Computer Society; 2008. p. 343. [Google Scholar]
- 15.Kassel G. CoRR. 2005. Integration of the dolce top-level ontology into the ontospec methodology. abs/cs/0510050. [Google Scholar]
- 16.Koblitz B, Santos N, Pose V. The amga metadata service. Journal of Grid Computing. 2008 Mar;6(1):61–76. [Google Scholar]
- 17.Masolo C, Borgo S, Gangemi A, Guarino N, Oltramari A. Ontology library. WonderWeb Deliverable. 2003;D18 [Google Scholar]
- 18.Montagnat J, Frohner Á, Jouvenot D, Pera C, Kunszt P, Koblitz B, Santos N, Loomis C, Texier R, Lingrand D, Guio P, Brito Da Rocha R, Sobreira de Almeida A, Farkas Z. A Secure Grid Medical Data Manager Interfaced to the gLite Middleware. Journal of Grid Computing. 2008 Mar;6(1):45–59. [Google Scholar]
- 19.Redolfi A, McClatchey R, Anjum A, Zijdenbos A, Manset D, Barkhof F, Spenger C, Legré Y, Wahlund LO, Barattieri C, Frisoni G. Grid infrastructures for computational neuroscience: the neuGRID example. Future Neurology. 2009;4(6):703–722. [Google Scholar]
- 20.Stewart GA, Cameron D, Cowan GA, McCance G. Storage and data management in EGEE. ACSW ’07: Proceedings of the fifth Australasian symposium on ACSW frontiers; Darlinghurst, Australia. Australian Computer Society, Inc; 2007. pp. 69–77. [Google Scholar]
- 21.Svihla M, Jelínek I. Benchmarking rdf production tools. In: Wagner R, Revell N, Pernul G, editors. DEXA, volume 4653 of Lecture Notes in Computer Science. Springer; 2007. pp. 700–709. [Google Scholar]
- 22.Temal L, Dojat M, Kassel G, Gibaud B. Towards an ontology for sharing medical images and regions of interest in neuroimaging. J of Biomedical Informatics. 2008;41(5):766–778. doi: 10.1016/j.jbi.2008.03.002. [DOI] [PubMed] [Google Scholar]


