Abstract
Differences in gene expression may play a major role in speciation and phenotypic diversity. We examined genome-wide differences in transcription factor (TF) binding in several humans and a single chimpanzee using chromatin immunoprecipitation followed by sequencing (ChIP-Seq). The binding sites of RNA Polymerase II (PolII) and a key regulator of immune responses, NFκB (p65), were mapped in ten lymphoblastoid cell lines and 25% and 7.5% of the respective binding regions were found to differ between individuals. Binding differences were frequently associated with SNPs and genomic structural variants (SVs) and were often correlated with differences in gene expression, suggesting functional consequences of binding variation. Furthermore, comparing PolII binding between human and chimpanzee suggests extensive divergence in TF binding. Our results indicate that many differences in individuals and species occur at the level of TF binding and provide insight into the genetic events responsible for these differences.
Text
Differences in gene expression have been observed in a variety of species (1–3). However, the extent to which TF binding differences occur both within individuals and closely related species and the global relationship between TF binding and genetic variation are largely unexplored (4). We used ChIP-Seq to map NFκB and PolII binding sites in ten humans: five are of European ancestry (including a parent-offspring trio), two of eastern Asian ancestry, and three of Nigerian ancestry (Table S1); nine of these have been analyzed by the HapMap (5) and the 1000 Genomes (http://1000genomes.org) Project, and one represents an individual for which high resolution SV maps are available (6, 7). All individuals but one were females; in pair-wise comparisons modest differences in TF binding were observed between the male and nine females; our analyses thus combined results from all ten. For comparison we also analyzed PolII-binding in a female chimpanzee.
We used stringent criteria to identify binding peaks (8), and clustered them into discrete “binding regions” (BRs) (9), yielding a total of 15,522 and 19,061 BRs for NFκB and PolII, respectively. Within BRs, most peaks were similar in position and magnitude among individuals (Fig. S1A). However, significant differences in binding were observed (Fig. S1A) and the Spearman correlation coefficients among replicates of different individuals (median values 0.79 and 0.90 for NFκB and PolII, respectively) were less than that of biological replicates of a given individual (median values 0.90 and 0.95, respectively; Fig. S2A, Table S2). 7.5% and 25% of the NFκB and PolII BRs, respectively, differed significantly between two individuals (ANOVA-test (9), Bonferroni-adjusted P-value <0.05; SOM and Fig. S3C), and many variable BRs exhibited >2 fold magnitude differences in binding (Fig. S3D). Variable BRs for both NFκB and PolII were often coassociated (P<1e-4; permutation test; Fig. 1D, Fig. S4), a correlation that is particularly strong for BRs <10kb apart (Fig. S4A). Variable NFκB and PolII regions were also often coassociated (P=2.80E-25, Kolmogorov-Smirnov, Table S3; Fig. S4A), even though the NFκB and PolII data are from TNFα-treated and untreated cells, respectively. These results suggest adjacent binding sites and BRs may influence one another, perhaps through cooperative binding or interactions with other proteins.
Fig. 1.
Effect of SNPs on NFκB and PolII binding. (A) Signal tracks of a NFκB motif and a TATA-box demonstrate effects of B-SNPs on TF binding, with correlations in the expected direction (i.e., with “correct trend”). (B) Fold enrichments for cumulative SNP-differences affecting BRs and for single SNPs affecting motifs, in pair-wise comparisons between individuals relative to the overall frequency of binding differences for NFκB (7.5%) and PolII (25%). (C) B-SNPs affecting motifs frequently lead to binding differences with “correct trend”. *P<0.001, based on randomization tests involving 10,000 permutations, i.e. permutation tests). (D) BRs adjacent to differentially bound BRs are enriched for binding variation.
For both NFκB and PolII, BRs within 1 kb of transcription start sites (TSSs) of RefSeq genes showed less variability (6% and 25%, respectively) than intergenic peaks (8% and 28%) (P<1e-4; permutation-test); TSS BRs also revealed stronger ChIP-Seq signals (1.2 and 2.3 fold, respectively), with many exceptions (Fig. S5). The majority of binding regions (>70%) were occupied in two or more individuals, which argues against cell line artifacts (see Fig. S3B). The signal intensity for 40% and 53% of the BRs absent (i.e., “lost”) in one individual was similar to background for NFκB and PolII (9), respectively, suggesting complete absence of binding in these cases, rather than “threshold effects”.
BRs differing in TF occupancy among individuals often involve loci of potentially high interest. These include the RPS26, BLK, SP140, and ZNF804A genes for PolII, which have been associated with type 1 diabetes, systemic lupus erythematosus, chronic lymphatic leukemia, and schizophrenia, respectively, and ORMDL3, PTGER4, and LOC253039 for NFκB, associated with asthma, Crohn’s disease, and rheumatoid arthritis (see SOM). Genes with variability in PolII binding showed a slight enrichment with immunity and defense functional gene categories (P-value=0.045, Benjamini-Hochberg multiple testing correction) among target genes (9).
We examined the genetic contribution to binding variation using SNPs from the 1000 Genomes Project. Individual SNPs in NFκB and PolII BRs frequently affected binding (Fig. 1A, Fig. S6A), and the number of SNPs in BRs correlated with the frequency of significant binding differences (Fig. 1B). SNPs altering the NFκB DNA binding motif had a strong effect, elevating the frequency of significant binding differences by 2.4 fold. ~90% of the binding differences followed the expected trend in which better matches to the consensus yielded higher binding signals (P<1e-3; see Fig. 1C, Table S4, Fig. S6B). We call SNPs that putatively affect binding B-SNPs for Binding-SNPs.
We also searched for other associated DNA motifs, such as the Stat1 motif (previously associated with NFκB-binding (10)), TATA-box, CAAT-box, and GC-box (11) and also performed de novo searches for enriched DNA motifs in BRs (9), which revealed BR enrichments for the NFκB-motif and the GC-box, along with additional motifs (Fig. S7). We assessed the effect of genetic variation on each of the motifs. SNPs in the Stat1 motif markedly elevated the frequency of significant NFκB binding differences (1.3-fold enrichment; P<1e-3, permutation-test; Fig. 1B), and 71% of the alterations in the Stat1 motif changed NFκB binding in the expected direction; i.e. improved Stat1 motif sequences increased NFκB binding (P<1e-3; see Fig. 1C, Table S4, Fig. S6B). For PolII, SNPs in the CAAT-box had a strong affect on binding (1.6 fold; P<1e-3), with 63% of cases displaying the correct trend, whereas SNPs in the TATA-box and GC-box had modest effects (1.5 fold and 1.3 fold, with 51% and 52% exhibiting the correct trend). The significant covariance in the Stat1 motif with NFκB binding differences and the NFY CAAT-box with PolII binding suggests a functional interaction of Stat1 with NFκB and NFY and PolII, respectively; the latter has been documented previously (12). We call this novel approach to examine coassociation of motifs with variable binding regions the Allele Binding Cooperativity test or “ABC test”.
We next analyzed the effect of SVs, >1kb genomic segments displaying copy-number variants (CNVs) or balanced inversions (6, 7, 13, 14). We probed high-density microarrays to identify CNVs in seven individuals ((9) Table S5) and combined these with CNVs from another survey (14). CNVs significantly elevated the frequency of BR differences between individuals by 5.1- and 2.0-fold for NFκB and PolII, respectively (P<1e-4, permutation-test; Fig. 2AB, Fig. S8, Table S6). Furthermore, the effect followed the correct trend in 90% and 80% of the respective NFκB and PolII cases (Fig. 2C); deletions reduced binding signals, whereas duplications elevated them. A combined set of high-resolution SVs identified by paired-end mapping (6, 13) also exhibited enrichment in binding differences for deletions intersecting with NFκB and PolII BRs (3.2 fold and 1.7-fold, respectively (P<1e-4, permutation-test)). Importantly, we found a 2.8-fold significant enrichment for inversions on NFκB BRs (P<1e-4, permutation-test), and a slight, non-significant enrichment for inversions on PolII BRs (Fig. 2B), suggesting that inversions may affect binding. We called SVs associated with binding “Binding-SVs” (B-SVs).
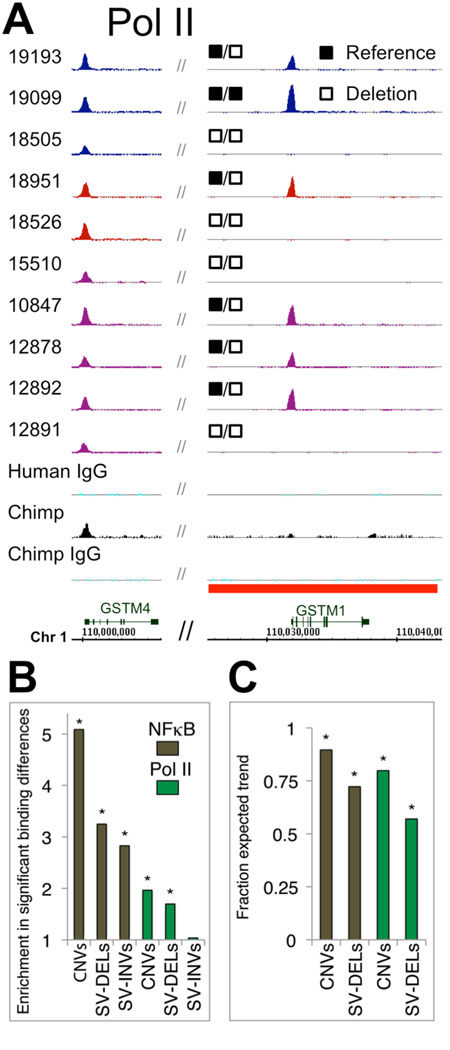
Fig. 2.
Effect of SVs on TF binding. (A) Example of a deletion affecting PolII binding. This example also shows a comparison of PolII occupancy in humans and a chimpanzee. A subset of individuals shares the chimpanzee binding phenotype. (B) Effect-sizes for microarray-based CNVs, SV-DELs (deletions identified by paired-end mapping), and SV-INVs (inversions detected by paired-end mapping). (C) Binding differences in regions displaying CNVs and SV-DELs frequently follow the “correct trend” in pair-wise comparisons between individuals. *P<0.01, based on permutation tests.
The total fraction of significant binding differences coinciding with genetic variations was 35% for NFκB and 26% for PolII (Table S7, Fig. S6C). 34% of the NFκB BRs intersect with SNP-differences between corresponding regions in different individuals (1% intersect with a known TF motif with SNPs falling both in the NFκB or the STAT1 motif; Table S8) and 3% with SVs (note: some SNPs coincide with SVs). Thus, genetic differences affecting the BR can be assigned to many, but not to the majority of, binding differences. Possible reasons for the remaining BR variation include trans-effects, epigenetic variation, as well as B-SNPs and B-SVs that were not ascertained. Some of the binding differences could be related to the different ages of the individuals.
We examined the effect of binding variation on gene expression by generation of deep RNA-Seq data from each cell line (9) and comparison with binding data (Fig. 3A, Fig. S9A). A significant correlation was observed (Spearman correlation coefficients of 0.475 and 0.461 for NFκB and PolII, respectively) (Fig. 3B, Fig. S9B, Table S9), suggesting an influence of binding differences on mRNA abundance. Examples of correlated genes include UGT2B17, GSTM1, and ZNF804A, encoding glucuronic acid and glutathione transferases and a gene linked to schizophrenia (see SOM). However, a number of BR differences were not associated with differences in gene expression and presumably compensatory (e.g. feedback) mechanisms influence the expression in these cases. We also examined the effect of B-SNPs with differences in both binding and gene expression and found that both NFκB and PolII binding and expression differences correlated with the presence of B-SNPs, including those in the NFκB- and Stat1-motif (for NFκB) and CAAT-, GC-, and TATA-box (for PolII) (Spearman: 0.48–0.82; Fig. 3C, Table S9). Copy number differences (i.e., B-SVs) also correlated with gene expression, albeit the correlation was not as strong as that of binding with gene expression (Table S10), indicating a more direct role for genetic variation on TF binding than on gene expression.
Fig. 3.
Correlation and effect sizes of TF binding and gene expression. (A) Example showing a correlation of binding and expression. This figure also shows a transgression event, in which the daughter displays a strong increase in binding relative to the parents. Continuous signal tracks shown in Fig. S10C. (B) Regions with binding variation correlate with differences in expression. Dark blue dots: PolII BRs displaying significant differences in binding in pair-wise comparisons between individuals; light blue dots: other BRs. The black lines demarcate data points that either fall two standard deviations outside the binding ratio or gene expression distributions. Indicated counts represent data points falling into the four corners for each data set. (C) Strong correlation between binding and gene expression at BRs in which a B-SNP intersects with the PolII specific CAAT-box. (D) Breakdown of segregation events in the trio showing the extent of BRs with candidate transgression events.
The observation that SNPs and SVs are frequently associated with binding differences suggests a crucial role of cis elements in the genetics of TF binding. We thus analyzed the segregation pattern of BR occupancy in the parent-offspring trio, and observed potential Mendelian segregation in >90% of BRs (Fig. S10A), although this was difficult to determine with certainty as not all alleles relevant to TF binding have been ascertained in the parents. Interestingly, 947 and 732 BRs were occupied by NFκB and PolII, respectively, in the child but not in the parents, indicative of transgression, in which a binding event was evident only in the offspring; Fig. 3AD, Fig. S10B, Tables S11, S12, S13).
We also examined whether some BRs are specific to certain populations. Although the number of individuals analyzed is small, the NFκB data revealed a total of 14 BRs that were specifically occupied or unoccupied in the African or Asian individuals (Table S14). For PolII, the chimpanzee data was used to infer gains and losses relative to the likely ancestral state of binding, and a total of 68 population specific occupancies (gains and losses) were identified in the three population groups (see Table S14). Overall, we found relatively few population-specific events, ~0.1% to ~0.4%, suggesting that most alleles affecting TF binding are shared among different populations.
Since humans and chimpanzees exhibit 5–10% differences in gene expression (15), we also examined divergence of TF binding among primates by analyzing PolII binding in a single chimpanzee. 15,418 (81%) of human BRs with corresponding syntenic regions in the chimpanzee genome were analyzed. The majority of PolII BRs were occupied both in humans and chimp (Fig. S11A). However, 32% of the BRs exhibited significant differences in binding (corrected P-value <0.05; e.g. Fig. 2A, Fig. 4A), a figure higher than that for human PolII variation (25%). Genes near regions uniquely occupied in the chimp were enriched in (i) nucleoside, nucleotide and nucleic acid metabolism; (ii) steroid metabolism (P-values: 3.60E-05 and 4.16E-04, respectively). Furthermore, BRs uniquely occupied in humans were significantly enriched in protein modification and mRNA transcription (Fischer Exact test (9), Benjamini-Hochberg P-values: 2.22E-89 and 9.08E-139, respectively; Table S15).
Fig. 4.
Comparison of PolII binding in humans and a chimpanzee. (A) Signal tracks for a peak found only in the chimpanzee. All ten individuals shown in Fig. S11B. B) Pie charts displaying occupancy by PolII of genomic regions where the chimp and human genomes are in synteny.
As in humans, relative differences identified in the chimpanzee were higher in intergenic BRs relative to BRs within 1 kb of a TSS: 33% of the syntenic intergenic PolII BRs differed significantly from the human samples, compared to 31% near TSSs (P<1e-4; permutation test). Consequently, human BRs near TSSs were generally more likely to be scored as “occupied” in chimpanzee (81%) than intergenic BRs (46%; Fig 4B). Furthermore, human BRs with strong binding signal (i.e., many mapped reads) are more frequently occupied in the chimpanzee than those with weaker signals (Fig. S11C), indicating either divergence of the weaker sites or signals that fell below the threshold at the low signal sites. Finally, we observed a general correlation between polymorphism and divergence in binding: i.e., variable BRs in humans displayed on average more divergence from chimpanzee BRs (in terms of fold-change in normalized read-counts) than non-variable BRs (Spearman 0.68; P=3.9e-07; see Fig. S11D).
Overall our data demonstrate extensive contributions of genetic variations on TF binding, many of which are expected to be functional through their affect on gene expression. Overall, the differences observed here (7.5% and 25% for NFκB and PolII, respectively, for humans; 32% for human/chimpanzee) greatly exceed estimates for sequence variation in coding sequences (estimated as 0.025% for humans (16) and 0.71% for human/chimpanzee (17)), suggesting a strong role for binding variation in human diversity. Extending mapping of B-SNPs and B-SVs for additional transcription factors will likely further inform on the genetic underpinnings of phenotypic diversity in humans.
References
- 1.Stranger BE, et al. Science. 2007 Feb 9;315:848. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rockman MV, Kruglyak L. Nat Rev Genet. 2006 Nov;7:862. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- 3.Skelly DA, Ronald J, Akey JM. Annu Rev Genomics Hum Genet. 2009;10:313. doi: 10.1146/annurev-genom-082908-150121. [DOI] [PubMed] [Google Scholar]
- 4.Borneman AR, et al. Science. 2007 Aug 10;317:815. doi: 10.1126/science.1140748. [DOI] [PubMed] [Google Scholar]
- 5.Frazer KA, et al. Nature. 2007 Oct 18;449:851. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korbel JO, et al. Science. 2007 Oct 19;318:420. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuzun E, et al. Nat Genet. 2005 Jul;37:727. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 8.Rozowsky J, et al. Nat Biotechnol. 2009 Jan;27:66. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Materials and Methods and supporting data are available at Science online.
- 10.Kramer OH, et al. Genes & Development. 2006;20:473. doi: 10.1101/gad.364306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. Nucleic Acids Res. 2004 Jan 1;32:D91. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faniello MC, et al. J Biol Chem. 1999 Mar 19;274:7623. doi: 10.1074/jbc.274.12.7623. [DOI] [PubMed] [Google Scholar]
- 13.Kidd JM, et al. Nature. 2008 May 1;453:56. [Google Scholar]
- 14.McCarroll SA, et al. Nat Genet. 2008 Oct;40:1166. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 15.Creely H, Khaitovich P. Prog Brain Res. 2006;158:295. doi: 10.1016/S0079-6123(06)58014-8. [DOI] [PubMed] [Google Scholar]
- 16.Levy S, et al. PLoS Biol. 2007 Sep 4;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe H, et al. Nature. 2004 May 27;429:382. [Google Scholar]
- 18.We thank the 1000 Genomes Project for early data access. This research was funded by grants from the NIH (MS, SW and MG), and by funding from the EMBL (JK), March of Dimes Foundation Grant (AU) and the NIH MSTP TG T32GM07205 (MK). MK was a Howard Hughes Medical Institute Medical Research Training Fellow. Data sets are available at GEO: GSE19486. MS is on the Scientific Advisory Board and a founder for both Affymetrix, Inc. and Metagenomix, Inc.