Abstract
Myc oncoproteins promote continuous cell growth, in part by controlling the transcription of key cell cycle regulators. Here, we report that c-Myc regulates the expression of Aurora A and B kinases (Aurka and Aurkb), and that Aurka and Aurkb transcripts and protein levels are highly elevated in Myc-driven B-cell lymphomas in both mice and humans. The induction of Aurka by Myc is transcriptional and is directly mediated via E-boxes, whereas Aurkb is regulated indirectly. Blocking Aurka/b kinase activity with a selective Aurora kinase inhibitor triggers transient mitotic arrest, polyploidization, and apoptosis of Myc-induced lymphomas. These phenotypes are selectively bypassed by a kinase inhibitor-resistant Aurkb mutant, demonstrating that Aurkb is the primary therapeutic target in the context of Myc. Importantly, apoptosis provoked by Aurk inhibition was p53 independent, suggesting that Aurka/Aurkb inhibitors will show efficacy in treating primary or relapsed malignancies having Myc involvement and/or loss of p53 function.
Introduction
Myc oncogenes (c-Myc, N-Myc, and L-Myc) are overexpressed in approximately 70% of tumors, by virtue of chromosomal amplifications or translocations, or through mutations in pathways that normally restrict Myc expression.1 Myc oncoproteins function as basic/helix-loop-helix/leucine zipper transcription factors that normally coordinate cell growth (mass) and metabolism with cell division.2,3 Accordingly, when overexpressed Myc oncoproteins accelerate cell growth and block terminal differentiation and withdrawal from the cell cycle, they also promote cell migration and tumor cell metastasis and trigger tumor angiogenesis.4–9
c-Myc (hereafter Myc) accelerates transit through the G1 and S phases of the cell cycle by inducing the transcription of cyclin D2 and Cdk4, and the cyclin-D2-Cdk4 complex then activates cyclin E-Cdk2 complexes by sequestering the universal Cdk inhibitor p27Kip1.10,11 Further, Myc activates the transcription of E2f1, a transcription factor that induces cyclin E.12 Activated cyclin E-Cdk2 complexes then phosphorylate p27Kip1 on threonine-187, a modification recognized by the F-box protein Skp2 component of the SCFSkp2 complex that ubiquitylates p27Kip1 and targets it for degradation by the proteasome.13 In addition, Myc induces the expression of both Skp2 and its allosteric regulator Cks1 to increase levels of the SCFSkp2 complex, and this response is also essential for suppressing p27Kip1.14 Finally, the reductions in p27kip1 levels triggered by Myc, coupled with its induction of cyclin B1, augment the activity of Cdk1-complexes that drive cells into and through mitosis.15,16
The Aurora kinases A, B, and C (Aurka, Aurkb, and Aurkc) are highly conserved serine/threonine kinases that play essential and distinct roles in mitosis.17 Specifically, Aurka is required for the assembly of the mitotic spindle, where it accumulates on centrosomes at the spindle poles during prophase until metaphase. Accordingly, loss of Aurka triggers early embryonic lethality in mice.18 By contrast, Aurkb is required for mitotic progression and cytokinesis and is localized, along with inner centromeric protein and survivin, at centromeres and the spindle midzone during the metaphase to anaphase transition.17,19
Both Aurka and Aurkb have been implicated in tumorigenesis; and as kinases, they are attractive therapeutic targets. For example, AURKA is amplified in a variety of human cancers, and increased levels of Aurkb have been reported in aggressive malignancies.19 Furthermore, both Aurka and Aurkb augment Ras-induced transformation.20,21 However, Aurka also appears to harness tumorigenesis, as Aurka heterozygosity increases the rate of tumor development; and, in the absence of p53, Aurka inhibition can provide a growth advantage.18,22
Recently, Aurka was shown to stabilize N-Myc protein in neuroblastoma, by directing a K48 to K63/K11 switch in its ubiquitylation by the E3 ubiquitin ligase Fbw7.23 Here we report a new link between Aurora kinases and Myc, where both Aurka and Aurkb are up-regulated by Myc ex vivo and in vivo, and where these kinases are required for the maintenance of Myc-driven B-cell lymphoma.
Methods
Mice and tumor analysis
All animal experiments were performed in accordance with regional/local animal ethics committee approvals from Reg Obb (Regierung von Oberbayern). Eμ-Myc transgenic mice (C57B/6)24 and lymphoma-transplanted C57/B6 syngeneic mice (Harlan Laboratories) were observed for signs of morbidity and tumor development. Tumors were harvested after death of mice, fixed in formalin or snap-frozen in liquid nitrogen, and processed for histology and analysis of RNA and protein. To obtain wild-type and precancerous Eμ-Myc B cells, mice were killed and their bone marrow and spleens were harvested. Red cell lysis was performed using an ammonium chloride/potassium bicarbonate solution. Cells were then resuspended, incubated with B220-MicroBeads, and enriched by magnetic cell sorting for B cells, according to the manufacturer's instructions (Miltenyi Biotec), and used for further analyses.
With institutional review board approval from St Jude Children's Research Hospital, and after informed consent according to the Declaration of Helsinki, tumors from Burkitt lymphoma (BL) patients were banked. RNA and protein were extracted from these tumors. As a control, pooled peripheral blood mononuclear cells from healthy donors were enriched using CD19-MicroBeads, according to the manufacturer's instructions (Miltenyi Biotec), and RNA and protein was prepared.
Plasmids and antibodies
The Aurka and Aurkb cDNAs in pEGFP-C1 were kindly provided by Erich Nigg (Martinsried, Germany). Details of their cloning into the pBabe-Puro expression vector and their mutagenesis (QuikChange Kit; Stratagene) are available on request. To generate Aurka- and Aurkb-promoter reporter plasmids, a 1533-bp sequence upstream of the ATG site of Aurka, and a 2037-bp sequence upstream of the ATG site of Aurkb, were polymerase chain reaction (PCR)-amplified from mouse genomic DNA using primers shown in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article), digested and cloned into pGL3 (Promega).
Information regarding all of the antibodies used in our studies is provided in supplemental Table 1.
Cell culture
P493-6 cells were kindly provided by G. Bornkamm (Munich, Germany) and were cultured and treated with tetracycline (Sigma-Aldrich) as previously described.25 Mouse Eμ-Myc B lymphoma cells were established from single-cell suspensions of tumors arising in Eμ-Myc mice by serial passage in culture. Lymphoma lines that harbor wild-type or mutant p53 were used in this study. Eμ-Myc lymphoma cells or NIH-3T3 cells (DSMZ) were infected with retrovirus (pBABE-puro, pBABE-Aurka, pBABE-Aurkb, MSCV-Myc-ERTAM-IRES-puro, MSCV-c-Myc-IRES-GFP [gift of C. Schmitt, Berlin, Germany] or vector control MSCV-IRES-GFP) and puromycin-treated (8 μg/mL) or green fluorescent protein (GFP)-sorted by flow cytometry to more than 98% GFP+ cells. Eμ-Myc lymphoma cells were infected with lentivirus generated using the commercially available Mission-shRNA expression system (Sigma-Aldrich) for knockdown of murine Aurka and Aurkb using specific shRNAs according to the manufacturer's instructions. Transient transfections of NIH-3T3 cells were performed using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen).
Cell viability and apoptosis assays
The cell viability/proliferation was assessed using the CellTiter 96 NonRadioactive Cell Proliferation Assay according to the manufacturer's instructions (Promega). Briefly, 105 cells were plated into a 96-well plate and treated as indicated in the figure legends, and the optical density was determined (490 nm). To assess apoptosis, 5 × 105 cells were stained with propidium iodide (PI) and annexin V–fluorescein isothiocyanate (Annexin-V-Fluos Kit, Roche Applied Sciences). After incubation, cells were washed, resuspended in phosphate-buffered saline, and analyzed by flow cytometry. Apoptosis and cell cycle distribution were measured by lysing cells for 30 minutes at 37°C in Vindelöv solution (20mM Tris, 100mM NaCl, 50 μg/mL PI, 20 μg/mL RNAse, and 0.1% NP40 adjusted to pH 8.0) followed by analysis of DNA content in the FL2 channel (linear mode, cell cycle) or FL3 channel (logarithmic mode, apoptosis). Cells with one log less and above to diploid DNA content were considered apoptotic (sub-G1).
RNA preparation and analyses
Expression profiling was performed using previously generated Affymetrix data (Supplemental data). For reverse transcriptase (RT) quantitative PCR, RNA was prepared from cultured cells or tumors using the RNeasy kit (QIAGEN). cDNA was prepared from 1μg RNA using the iScript cDNA synthesis kit (Bio-Rad). Quantitative RT-PCR was performed using an iCycler machine (Bio-Rad) and the iTaq SYBR Green kit (Bio-Rad). Details on data analyses performed by comparing ΔΔCt values with a control sample set as 1 and primer sequences are given in supplemental Table 1.
Immunoblotting
Protein extracts (20 μg or 50 μg per lane) were electrophoretically separated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels, transferred to membranes (Protran, Schleicher & Schuell), and blotted with specific antibodies (supplemental Table 1). Ponceau Red staining of the nitrocellulose membranes after transfer also confirmed equal protein loading.
Aurka and Aurkb promoter Luciferase reporter assays
NIH-3T3 cells were transfected with pGL3-Aurka, pGL3-Aurkb, or pGL3 firefly luciferase reporter plasmids together with pRL-SV40 (Renilla luciferase). Luciferase activity was determined according to the manufacturer's instructions (Promega). The relative luciferase activity was determined by calculating the ratio of firefly to cotransfected Renilla luciferase activity.
ChIP analysis
Balb/c-3T3 mouse fibroblasts expressing mouse Myc fused to the estrogen receptor (Myc-ER) were cultured for 48 hours in 0.2% serum and then stimulated for 6 hours with or without 4-hydroxytamoxifen (4HT; 2μM). Chromatin from 3 × 107 cells/condition was cross-linked, isolated, sonicated, and reverse cross-linked.26 Myc protein was immunoprecipitated from sonicated chromatin using an anti-c-Myc rabbit polyclonal antibody (supplemental Table 1) and magnetic protein G beads (Active Motif). A 1:100 volume of chromatin was processed without antibody and used as total input control for quantitative PCR. Quantitative PCR was run on immunoprecipitated and control chromatin using primers designed to detect E-boxes in the mouse Aurka and Aurkb genes (supplemental Table 1). Control primers were designed to amplify chromatin that did not contain an E-box sequence. The mouse carbamoyl-phospate synthetase gene (Cad), an established direct Myc target gene, was used as a positive control for Myc ChIP.27 Percentage total chromatin immunoprecipitated was calculated using the equation % total = 2 Ct input − Ct ChIP × % input used for chromatin immunoprecipitation (ChIP).28
Histology and immunohistochemistry
Organ tissue samples were obtained from lymphoma-bearing mice. Slides of 5- to 6-μm sections cut from formalin-fixed, paraffin-embedded tissues were deparaffinized and stained with hematoxylin and eosin (Dako), dehydrated, and then covered with a coverslip. Cultured lymphoma cells were collected by cytospin and stained with Giemsa-May-Grünwald solution. For immunohistochemistry 2-μm sections were deparaffinized. Antigen retrieval was performed by pressure cooking in citrate buffer (pH 6) for 7 minutes. Primary antibodies (supplemental Table 1) were incubated overnight at 4°C. Antibody detection was performed using the Dako REAL detection kit (Dako) according to the manufacturer's protocol. Two independent operators analyzed (n = 3) high-power fields (40×) of n = 5 samples of BL or mantle cell lymphoma (MCL).
PET
Before administration of the radiotracer [18F]deoxyglucose (FDG; Department of Radiopharmacy, TU München, München, Germany), all mice were anesthetized with 4% isoflurane (Abbott GmbH) using a veterinary anesthesia System (Vetland Medical Sales and Services). During positron emission tomography (PET) imaging, the dose was reduced to 1.5% isoflurane. Dynamic imaging with [18F]FDG-PET was performed using a small animal PET system (MicroPET Focus 120, Siemens Preclinical Solutions); 3.7 to 7.4 MBq [18F]FDG/mouse was injected intravenously. Data were acquired for 60 minutes. All acquisitions were performed in list-mode format and histogrammed into a framed sinogram. The sinogram was reconstructed into a 128 × 128 × 95 voxel image using the filtered back-projection method with a cutoff at the Nyquist frequency. The voxel size equals 0.433 × 0.433 × 0.796 mm3. The data were normalized and corrected for random coincidences, dead time, and decay.
Statistical analysis
Statistical analyses were performed using the statistical functions of Excel 2007 (Microsoft) or GraphPad Prism 5 (GraphPad Software). Bars represent the mean plus or minus SD or SEM. All statistical analyses were t tests. Only P values less than .05 were considered statistically significant.
Results
Myc up-regulates the expression of Aurora kinases
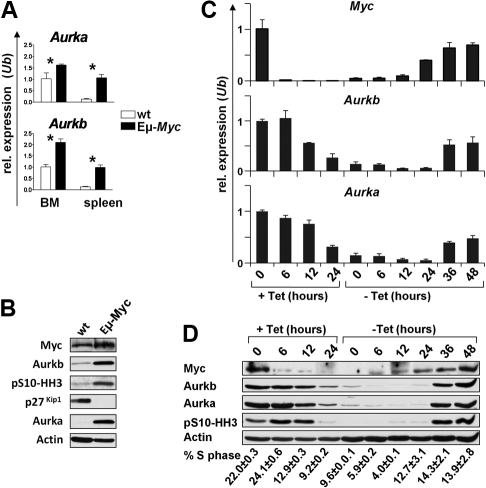
Myc induces the expression of a large cast of genes involved in cell cycle entry and transit, and many of these play roles in the G1-S phase transition.11,29 To assess this in an in vivo context, we analyzed their mRNA levels in splenic B220+ B cells of precancerous (4- to 6-week-old) Eμ-Myc transgenic mice, a model of human B-cell lymphoma,24 and compared these with levels expressed in splenic B220+ B cells of wild-type, nontransgenic littermates using expression profiling (supplemental Figure 1). Two of the mRNAs highly elevated in Eμ-Myc B cells encode Aurora kinase A (Aurka) and Aurora kinase B (Aurkb; supplemental Figure 1), which are both essential for progression and completion of mitosis.19 The up-regulation of Aurka and Aurkb mRNAs in Eμ-Myc B cells was confirmed by quantitative RT-PCR (Figure 1A). Furthermore, immunoblotting demonstrated that Aurka and Aurkb protein levels were also markedly elevated in premalignant Eμ-Myc B cells versus wild-type B cells (Figure 1B). Finally, in premalignant Eμ-Myc B cells, there were much higher levels of serine-10 phosphorylated histone H3 (pS10-HH3; Figure 1B), an established Aurkb substrate.30 Thus, Myc expression in vivo leads to marked increases in the level and activity of Aurora kinases.
Figure 1.
Myc up-regulates the expression of Aurka and Aurkb. (A) Quantitative SYBR Green real-time PCR analysis (quantitative RT-PCR) of Aurka and Aurkb transcript levels in bone marrow (BM) and splenic (spleen) B220+ B cells from 4-week-old wild-type (wt; white bars) or Eμ-Myc (black bars) littermate mice. Levels of mRNA are standardized to the expression of Ubiquitin (Ub). Bars represent the mean of 3 experiments ± SEM. *P < .05. (B) Immunoblot analysis of the indicated proteins in splenic B220+ B cells from 4-week-old wild-type (wt) or Eμ-Myc-transgenic littermate mice. (C) Quantitative RT-PCR analysis of c-Myc, Aurka, and Aurkb RNA expression in P493-6 human B cells that bear a tetracycline-regulated Myc gene. The cells were treated with tetracycline for the indicated time (+Tet), washed, and cultured in tetracycline-free medium (−Tet). Levels of mRNAs were standardized to the expression of Ubiquitin (Ub), which is not regulated by Myc. (D) Top panel: Immunoblot analysis of the indicated proteins from the same P493-6 cells as in panel C. The numbers below the immunoblots indicate the percentage of cells in S phase (PI staining) assessed by flow cytometry. Tet indicates tetracycline.
The up-regulation of Aurka and Aurkb levels and activity in premalignant Eμ-Myc B cells might simply reflect that a higher percentage of these B cells are cycling and transiting mitosis. To address whether Aurka and Aurkb were up-regulated in response to c-Myc, we used human P493-6 B cells whose proliferation can be driven by a tetracycline-off regulated c-Myc transgene.25 As expected, the removal of tetracycline resulted in marked increases in the levels of c-Myc mRNA and protein, and this was followed by marked increases in Aurka and Aurkb mRNA and protein (Figure 1C-D) and by marked increases in the levels of pS10-HH3 (Figure 1D), as well as restored proliferation. Thus, activation or overexpression of Myc results in marked increases in the levels and activity of Aurka and Aurkb both in vivo and ex vivo.
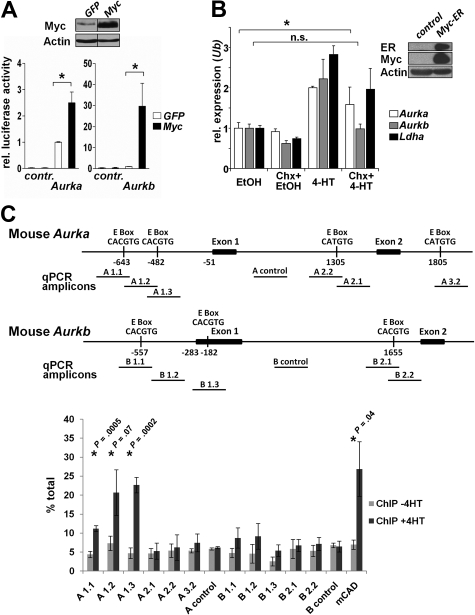
To investigate whether Aurka and Aurkb were transcriptionally induced by Myc, we initially assessed the regulation of their promoters by Myc. The regulatory regions of the murine and human AURKA and AURKB promoters contain Myc-responsive CACGTG, CACATG, CTCGTG, and CATGTG E-box sequences.31 We generated luciferase reporter-based plasmids by cloning the murine promoter sequences of Aurka and Aurkb into the pGL3 luciferase reporter plasmid. Pools of NIH-3T3 cells expressing Myc-GFP or GFP alone (Figure 2A top panel, > 98% GFP+ cells as assessed by flow cytometry) were then transfected with these reporter plasmids and luciferase activity determined after normalizing for transfection efficiency. Notably, Myc-overexpressing fibroblasts had 2- to 3-fold increases in Aurka promoter activity (P = .009) and approximately 30-fold increases in Aurkb promoter activity (P = .038) compared with GFP-expressing control cells (Figure 2A).
Figure 2.
Aurka is a direct Myc target gene, whereas Aurkb is indirectly up-regulated by Myc. (A) Top panel: Immunoblot analysis of NIH-3T3 cells infected with Myc or GFP control retrovirus. Vertical lines have been inserted to indicate a repositioned gel. Bottom panel: NIH-3T3 cells were transiently transfected with the indicated firefly luciferase promoter-reporter constructs and assessed for relative luciferase activity. Shown is the relative luciferase activity in Myc-expressing versus GFP-only expressing cells. Renilla luciferase plasmids were cotransfected to normalize for transfection efficiency. Bars represent the mean ± SEM of 3 independent experiments, each performed in duplicate. *P < .05. (B) Right panel: Immunoblot analysis of NIH-3T3 cells infected with retroviruses encoding MSCV-Myc-ER-IRES-Puro. Left panel: After puromycin selection, cells expressing Myc-ER were cultured in the presence or absence of 4HT and/or cycloheximide (Chx) for 4 hours to activate preexisting Myc-ER in the presence or absence of protein synthesis. Ethanol (EtOH) was the vehicle for 4HT. The cells were harvested and RNA prepared for quantitative RT-PCR in which primers directed against Aurka, Aurkb, Ldha, and Ubiquitin (Ub) were used. Levels of mRNA were standardized to the expression of Ub. Bars represent the mean ± SD from 3 experiments. The up-regulation of all genes is significant on Myc activation in the absence of Chx (P < .05). The induction of Aurka by Myc is significant in the presence of Chx. *P < .05. n.s. indicates not significant. (C) ChIP analysis for Myc binding to mouse Aurk genes in Balb/c-3T3 fibroblasts expressing Myc-ER. Top panel: Map of the murine Aurka and Aurkb gene regions analyzed and the location of the E-boxes and amplicons used to detect Myc binding by quantitative PCR after ChIP. Amplicons A1.1, A1.2, A1.3, A2.2, A.2.1, A3.2, and A control detect Aurka, whereas amplicons B1.1, B1.2, B1.3, B2.1, B2.2, and B control detect Aurkb genomic DNA. Chromatin was isolated from cells 6 hours after activation of Myc-ER (+4HT) or uninduced (−4HT) and immunoprecipitated with antibody against c-Myc. Quantitative PCR was carried out on immunoprecipitated chromatin and input chromatin and expressed as percentage total using the calculation % total = 2Ct input − Ct ChIP × % input used for ChIP.28 Shown is the mean of 3 experiments ± SD. *P values comparing −4HT and +4HT samples.
To assess whether Aurka and Aurkb were directly regulated by Myc, we infected NIH-3T3 fibroblasts with a retrovirus harboring Myc-ERTAM, a fusion protein where Myc can be activated by treatment with tamoxifen. If a gene is activated by tamoxifen in Myc-ER-expressing cells in the presence of the protein synthesis inhibitor cycloheximide, the gene might be a direct target induced by Myc. Interestingly, although the mouse Aurka promoter was weakly induced by Myc compared with Myc-mediated regulation of the Aurkb promoter, Aurka mRNA was induced similar to the established direct Myc target gene Lactate dehydrogenase A (Ldha),32 whereas the induction of Aurkb mRNA was dependent on de novo protein synthesis (Figure 2B).
We next tested whether Myc protein could bind to the endogenous promoter-regulatory regions of mouse and human Aurka and Aurkb genes by ChIP assays (Figure 2C; supplemental Figure 2). Chromatin was isolated from Balb/c-3T3 cells expressing Myc-ER that were cultured with and without tamoxifen and precipitated with an antibody against Myc. Chromatin from the mouse Aurka gene that contains 2 E-boxes located upstream of exon 1 (A1.1, A1.2, and A1.3) was specifically enriched after the activation of Myc-ER (+4HT). This suggests that Myc binds either or both of these E-boxes present in the mouse Aurka gene. By contrast, Myc was not enriched at the other E-box in Aurka or to any of the E-boxes in the Aurkb promoter-regulatory region. Collectively, these data suggest that Myc directly regulates the transcription of Aurka and indirectly regulates the transcription of Aurkb in mouse. Interestingly, although the human AURK genes contain E-boxes, they are not conserved with those found in mouse, and we did not detect an interaction of Myc with either human gene by ChIP assays (supplemental Figure 2). Collectively, these data indicate that the up-regulation of murine and human Aurka and Aurkb transcription by Myc has both direct and indirect components.
Aurka and Aurkb overexpression is a hallmark of Myc-induced B-cell lymphoma
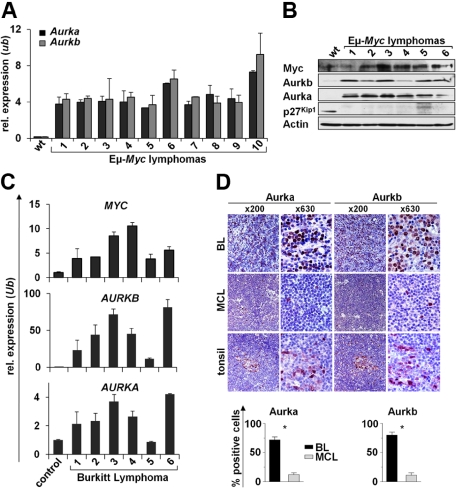
Given that Aurka and Aurkb expression was elevated in premalignant Eμ-Myc B cells, we reasoned that they should be highly expressed in the Myc-driven lymphomas that arise in Eμ-Myc transgenic mice and in human BL. Expression microarray analysis showed that several genes associated with mitosis were elevated in premalignant Eμ-Myc B cells compared with wild-type B cells and that this response was augmented in the lymphomas that arose in Eμ-Myc transgenics (supplemental Figure 1). Quantitative RT-PCR and Western blot analyses confirmed that Aurka and Aurkb mRNA and protein levels were highly elevated in all Eμ-Myc lymphomas relative to levels expressed in wild-type B cells (Figure 3A-B). Furthermore, increased levels of AURKA and AURKB mRNAs paralleled levels of elevated MYC transcripts in human pediatric BL samples assessed by quantitative RT-PCR. Indeed, all human BL expressed markedly higher levels of AURKB mRNA (10- to 80-fold higher) compared with normal control CD19+ B cells. Increases in AURKA expression were less striking but were still elevated relative to levels of AURKA expressed in normal CD19+ B cells (Figure 3C). High Aurk levels in BL compared with MCL were confirmed on protein level by immunohistochemistry (Figure 3D). Importantly, when assessing 5 primary BL samples by fluorescence in situ hybridization, we did not detect AURKA or AURKB gene amplification (supplemental Figure 3). Thus, overexpression of Aurka, and especially Aurkb, is a hallmark of Myc-driven B-cell lymphoma in mice and humans.
Figure 3.
Elevated Aurka and Aurkb are hallmarks of Myc-driven lymphoma. (A) Quantitative RT-PCR analysis of Aurka and Aurkb expression in 10 Eμ-Myc lymphomas versus splenic wild-type (wt) B220+ B cells. Levels of mRNA were normalized to the expression of Ubiquitin (Ub). (B) Immunoblot analysis of the indicated proteins in wild-type (wt) control splenic B220+ B cells vs Eμ-Myc lymphomas. (C) Quantitative RT-PCR analysis of Myc, Aurka, and Aurkb mRNA expression in 6 human BL samples compared with CD19+ control B cells. Levels of mRNA were normalized to the expression of Ubiquitin (Ub). (D) Immunohistochemical analysis of Aurka and Aurkb expression in BL, MCL, and control tissue (tonsils). Top panel: Representative samples at low- (original magnification ×200) and high-power (original magnification ×630) views. Image acquisition: Zeiss Axioplan 2 microscope; 63×/0.95 numeric aperture (NA) Plan-Neofluar air objective (high-power views), 20×/0.5 NA Plan-Neofluar air objective (low-power views); Zeiss Axiocam MRc 5 camera; Axiovision Rel 4.6 scanning software. Bottom panels: Percentage of cells that stain positive for Aurka and Aurkb. A grid ocular objective was used to count 400 cells over 3 high-power fields (original magnification ×40). Bars represent the mean percentage of positive cells from 5 BL and 5 MCL samples ± SD. *Significant difference (P < .001).
Aurora kinase function is required for the maintenance of Myc-driven lymphoma
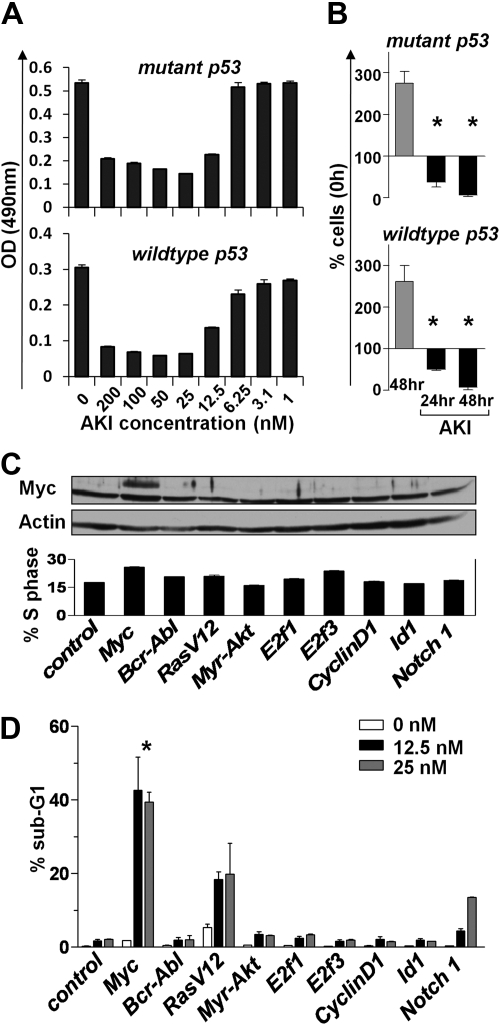
Myc-overexpressing cells are particularly sensitive to signals or agents that target the G2/M transition of the cell cycle.33,34 Thus, we hypothesized that Aurora kinase inhibitor (AKI) might compromise the maintenance of Myc-driven lymphomas. To test this notion, we assessed the consequences of inhibiting Aurora kinases in Eμ-Myc lymphomas that differ in their p53 status (wild-type and mutant p53). Interestingly, lymphoma lines of both p53 genotypes were highly susceptible to the pan-AKI AS703569 when cultured in vitro. Even at relatively low doses (25nM), AKI treatment completely inhibited lymphoma cell proliferation, and prolonged exposure resulted in a significant reduction in cell numbers (Figure 4A-B). To assess whether there is a selective sensitivity of Myc-expressing cells to Aurora kinase inhibition, we examined the effects of the AKI AS703569 on immortalized rodent Rat-1 fibroblasts that were stably infected with control virus or viruses encoding a variety of oncogenes,33 including Myc (Figure 4C top panel). As expected, Myc and several other oncogenes triggered increases of cells in S phase (Figure 4C bottom panel). Quite strikingly, Myc-overexpressing Rat-1 cells were significantly more sensitive to AKI treatment than cells overexpressing other oncogenes at AKI concentrations as low as 12.5nM (Figure 4D). Thus, the AKI AS703569 selectively induces cell death of Myc-overexpressing cells.
Figure 4.
Aurk inhibition blocks Myc-driven proliferation. (A) Primary Eμ-Myc lymphoma cells (having either wild-type or mutant p53) were cultured ex vivo until feeder-independent lines were established. These were treated for 24 hours with the indicated concentrations of the AKI AS703569. Effects on proliferation were assessed using an MTT assay. Shown is a representative experiment of 3 experiments performed. Bars represent the mean ± SD. (B) Proliferation of primary ex vivo cultured Eμ-Myc lymphoma cells treated for the indicated times with 25nM AS703569 (AKI). Control cells (gray bars) were left untreated for 48 hours. Bars represent the mean ± SEM percentage of cells compared with input cells set as 100% of 3 independent experiments. *Significant difference compared with control cells (P < .01). (C) Rat-1 fibroblasts were stably infected with the retroviruses expressing the indicated oncogenes and were then treated with the indicated doses of the AKI AS703569. Top panel: Immunoblot analysis of c-Myc levels. Actin was used as a loading control. Bottom panel: Percentage of Rat-1 cells in S phase assessed by PI staining for DNA content. (D) The antiproliferative effect of the AKI AS703569 in Rat-1 cells driven by the indicated oncogenes. Asynchronously growing cells were treated with AKI AS703569 at the indicated concentrations (white, black, and gray bars) for 48 hours and assessed for cell death by flow cytometric analysis of DNA content (PI). Bars represent the mean percentage of cells with a sub-G1 DNA content ± SD of 3 independent experiments. *P < .05 of Myc-infected cells compared with other cells.
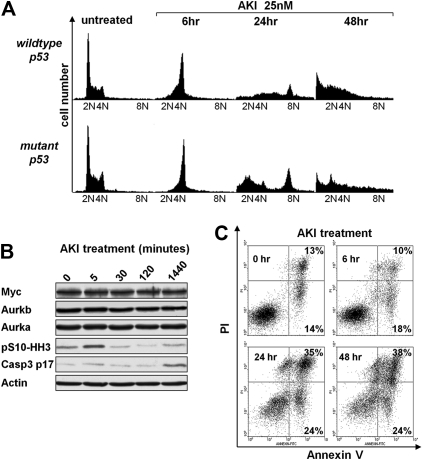
Previous studies have shown that Aurka or Aurkb inhibition leads to defects in mitosis and cytokinesis.17–19 Exposure of Eμ-Myc lymphoma cells to AS703569 resulted in a rapid and significant accumulation of cells in G2/M (Figure 5A), followed by apoptosis (sub-G1 cells [Figure 5A and supplemental Figure 4], and annexin-V+ cells [Figure 5C]) and/or to an increased number of cells with 8N DNA content (Figure 5A; supplemental Figure 4). Giemsa staining of these cells confirmed the accumulation of Eμ-Myc lymphoma cells at prometaphase or metaphase as early as 6 hours of treatment with AS703569, and this was followed by the appearance of polyploid cells (supplemental Figure 5A). Finally, as expected, AS703569 treatment of Eμ-Myc lymphoma cells led to marked and rapid decreases in the levels of pS10-HH3 and to the protracted appearance of cleaved caspase-3 (Figure 5B), consistent with the apoptotic effects of this agent on Eμ-Myc lymphoma (Figure 5C). When Eμ-Myc lymphoma cells were pretreated with the pan-caspase inhibitor Q-VD-OPH and then exposed to the AKI, AS703569 apoptosis was significantly reduced and significantly more cells acquired a tetraploid or octaploid DNA content (supplemental Figure 5B). Thus, Aurora kinase inhibition leads to defective mitosis and provokes apoptosis of Myc-driven lymphoma cells.
Figure 5.
Aurk inhibition triggers mitotic arrest and polyploidy and induces apoptosis of Eμ-Myc lymphoma cells irrespective of the p53 status. (A) Eμ-Myc lymphoma cells cultured ex vivo were treated for the indicated times with 25nM AS703569 (AKI) and assessed for DNA content by flow cytometric analysis of PI-stained cells. The PI-stained cells were analyzed using the FL2 channel in a linear scale. Shown are representative histograms. The quantification and statistical analyses of 3 independently performed experiments are provided in supplemental Figure 4. (B) Eμ-Myc lymphoma cells were treated for the indicated times with the AKI AS703569 (25nM). Expression of the indicated proteins was assessed using immunoblotting. (C) Untreated Eμ-Myc lymphoma cells or cells treated with 25nM AS703569 (AKI) for the indicated times were analyzed for their apoptotic index by staining with annexin V–PI. Cells in the top right quadrant represent late apoptotic/necrotic cells. Cells in the bottom right quadrant represent early apoptotic cells. The percentage of cells in these quadrants is given. Shown is one representative dot blot graph of 3 independently performed experiments.
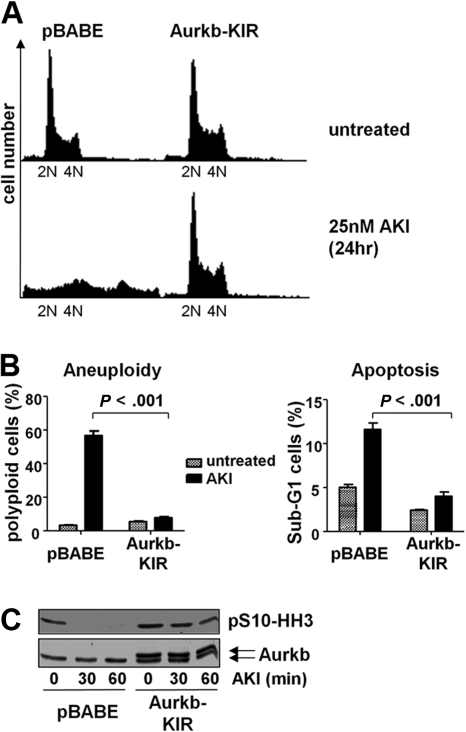
To confirm that AS703569 was indeed selective as an inhibitor of Aurka or Aurkb in Myc-driven lymphoma, we designed mutations in Aurka/b by computer-based methods that are predicted to be resistant to this inhibitor. These mutations were introduced into Aurka (G216V) or Aurkb (G160V) by site-directed mutagenesis, and these mutants were overexpressed in Eμ-Myc lymphoma cells using retroviruses harboring these kinase inhibitor-resistant (KIR) mutants (Figure 6C). Notably, Aurkb-KIR-expressing cells showed a significant resistance to AS703569 with regard to the occurrence of aneuploidy as well as apoptosis (Figure 6A-B), and the decrease of Aurkb activity as assessed by phosphorylation of S10-HH3 was greatly diminished (Figure 6C). This response was not manifest in Aurka-KIR-expressing lymphoma cells (data not shown). Thus, overexpression of Aurkb-KIR is sufficient to confer profound resistance to the inhibitor AS703569. Short-term experiments that applied shRNA-mediated knockdown further established that Aurkb is the primary target of AS703569, as knockdown of Aurkb induced the drug-induced phenotype. However, long-term knockdown of either Aurka or Aurkb was not compatible with long-term lymphoma cell growth (data not shown). We conclude that Aurkb is the central target for Aurora kinase inhibition in Myc-driven cells.
Figure 6.
Overexpression of an AKI-resistant mutant of Aurkb renders Myc-expressing lymphoma cells resistant to Aurk inhibition. (A) Eμ-Myc lymphoma cells were infected with control virus (pBABE) or pBABE-AurkbG160V (Aurkb-KIR) encoding retrovirus. The cells were then selected with puromycin and treated with 25nM AS703569 (AKI) for 24 hours. The representative histogram shows the DNA content assessed by PI staining. (B) Quantification of aneuploidy and apoptosis on AKI treatment in control cells and cells expressing of Aurk-KIR. Cells were treated with carrier only (untreated) or 25nM AS703569 (AKI) and subjected to DNA content analysis using PI staining and flow cytometry. The PI-stained cells were analyzed using the FL2 channel in a linear scale. Apoptosis measurements were based on the percentage of cells that carried less than diploid DNA content (Sub-G1) in the FL3 channel in a logarithmic scale. (C) Control cells (pBABE) or cells expressing mutant Aurkb (Aurkb-KIR) were treated for the indicated time with 25nM AS703569 (AKI) and assessed for Aurkb and phosphorylated S10-HH3 levels by immunoblotting. Note that Aurkb-KIR was Flag-tagged and migrates slightly slower than endogenous Aurkb.
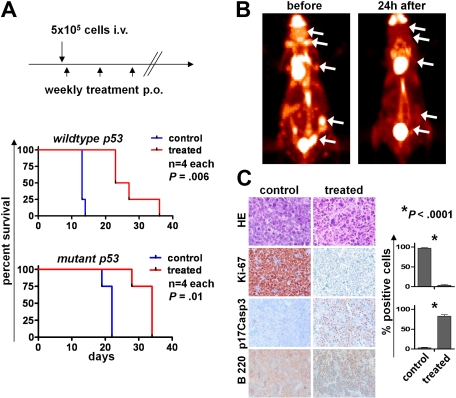
In syngeneic transplantations, Eμ-Myc lymphomas induce rapid disease. To evaluate whether inhibition of Aurora kinases compromised Myc-induced tumorigenesis in vivo, syngeneic C57/B6 mice were intravenously injected with 5 × 105 Eμ-Myc lymphoma cells, and after 24 hours they were then treated with vehicle or with AS703569 (60 mg/kg) once per week by oral gavage. Mice were then observed for signs of lymphoma onset. As expected, mice receiving vehicle alone developed rapid and disseminated lymphoma within 2 to 3 weeks (Figure 7A). Quite strikingly, tumorigenesis was markedly delayed in transplantation recipients receiving AS703569 treatment (Figure 7A; 2 independent experiments shown).
Figure 7.
Aurk activity is required for maintenance of Myc-driven lymphoma in vivo. (A) Top panel: Schedule of lymphoma cell injection and AS703569 treatment (60 mg/kg body weight, once weekly per oral gavage). Bottom panels: Survival curves. p53 wild-type or p53 mutant Eμ-Myc lymphoma cells were injected intravenously into syngeneic recipients, and mice were followed for lymphoma onset. The differences between the control and Aurk inhibitor treatment groups (treated) are statistically significant. (B) FDG-PET scans were performed before and 24 hours after a single oral treatment with Aurk inhibitor (AS703569, 60 mg/kg body weight) in a mouse with manifest lymphoma. (C) Histologic assessment of Aurk inhibition in manifest lymphomas. Top row: Hematoxylin and eosin (HE) staining shows massive necrosis after Aurk inhibition in treated versus control mice (lymph node). Immunohistochemistry for the indicated markers shows a reduction of proliferating cells as assessed by Ki-67 positivity (second row) and increased apoptotic cell death assessed by staining for cleaved (p17) caspase-3 (p17Casp3; third row). Image acquisition: Zeiss Axioplan 2 microscope; 20×/0.5 NA Plan-Neofluar air objective; Zeiss Axiocam MRc 5 camera; Axiovision Rel 4.6 scanning software; original magnification ×200. Right panel: Quantification of the percentage of Ki-67 and p17Casp3-positive cells in control lymphomas or lymphomas from mice treated 24 hours with a single dose of the Aurk inhibitor AS703569. Bars represent the mean percentage ± SEM of positive cells from lymph nodes from 3 different control or AS703569-treated mice.
To evaluate the efficacy of this Aurk inhibition in animals with established disease, untreated mice that had been injected with 5 × 105 Eμ-Myc lymphoma cells and that had developed clinically manifest lymphoma received FDG-PET imaging before and 24 hours after AS703569 treatment. Quite remarkably, one application of AS703569 was sufficient to significantly reduce the uptake of FDG to lymphoma sites (Figure 7B). Histologic examination revealed a highly significant decrease in cell proliferation assessed by Ki-67 staining and massive lymphoma cell death assessed by staining for the active form of caspase-3 (Figure 7C). Thus, Myc-driven lymphoma cells are highly susceptible to Aurk inhibition in vivo, and Aurk inhibition represents an effective treatment for established, highly aggressive disease.
Discussion
Myc controls, by both direct and indirect means, the expression of key regulators of cell cycle transit to coordinate cell growth and metabolism with cell division.2,3,29,35 This includes the regulation of cyclin D2, Cdk4, E2f1, and p27Kip1 during G1-S phase transition,10–12,14,15,36 and components of the prereplication complex, BRCA1 and cyclin E and A, during entry and progression through S phase.16,37–40 Relatively little is known about Myc functions at the G2-M transition and its regulation of mitosis and cytokinesis, other than that cyclin B1, Cdc2 (Cdk1), Bub1b (BubR1), and Mad2 appear to be direct Myc targets.16,41 Here we provide evidence that Myc controls 2 other essential regulators of mitosis and cytokinesis, Aurka and Aurkb.
Both Aurka and Aurkb mRNAs are elevated in response to Myc in human and mouse cells, and both genes harbor E-boxes in their promoter-regulatory regions. However, our studies suggest that only Aurka is directly regulated by Myc, and this is restricted to mouse cells, as Myc binds to the mouse Aurka promoter but does not bind to E-boxes present in the human AURKA gene under conditions where there is a robust induction of AURKA mRNA in response to Myc. Thus, Myc either binds to other regions in the human AURKA locus to mediate transcription or the up-regulation of both Aurka and Aurkb is Myc dependent but occurs through additional mechanisms of transcriptional control.19 Collectively, these findings indicate that Myc regulates murine and human Aurka and Aurkb gene expression in a direct or an indirect noncanonical manner to control cell cycle traverse into and through mitosis.
Entry into mitosis is triggered by the activated CyclinB1/Cdk1 complex (the M-phase-promoting factor), which is exquisitely controlled during the cell cycle.42 Cyclin B1 is a well-established Myc transcription target gene,16 and loss of c-Myc delays the G2-M transition. Further, high Myc levels prevent G2 arrest.43 We predict that the control of the G2/M transition by Myc includes its induction of Aurka because Aurka phosphorylates and activates the Cdc25B phosphatase at centrosomes that then trigger activation of the CyclinB/Cdk1 complexes.44 In this scenario, Myc's induction of Aurka would provide proper levels of Aurka that are required for centrosome maturation, centrosome separation, and spindle assembly.19
In normal cells, Myc's oncogenic effects are effectively harnessed by apoptotic checkpoints that appear to sense inappropriate rates of cell cycle progression. Specifically, Myc overexpression activates the p53 apoptotic program by activating the DNA damage response through ATM45 or via inducing the nucleolar tumor suppressor Arf that inactivates Mdm2.46 Both of these responses lead to the induction of a cast of p53 apoptotic target genes. Accordingly, loss-of-function mutations in these tumor suppressors are hallmarks of Myc-driven malignancies.7 Interestingly, there are also interactions between Aurka and p53, where Aurka directly phosphorylates p53 to augment p53 protein turnover and transcriptional activity.47 Surprisingly, in cells that have suffered p53 loss, inhibition of Aurka has been shown to provide a growth advantage.22 Our data challenge this notion, as at least in the context of Myc-driven B-cell lymphomas inhibition of Aurka/Aurkb leads to robust death irrespective of p53 status.
In Myc-expressing B cells, inhibition of Aurka/Aurkb activity or knockdown of their expression provokes defects in cytokinesis leading to endoreduplication, and triggers apoptosis. These findings somewhat differ from those of others who have shown that pharmacologic inhibition of Aurka/Aurkb leads to higher levels of endoreduplication than we observe.48 Rather, in the context of Myc overexpression the predominant outcome of canceling Aurka/Aurkb activity is apoptosis, suggesting that Aurk may suppress Myc-induced apoptosis in the G2-M phase of the cell cycle. Although we do not yet know the exact chain of events that sensitize Myc-expressing cells to apoptosis after Aurk inhibition, it is tempting to speculate that this involves bypass of mitotic checkpoints and/or that Aurkb inhibition also blocks the antiapoptotic function of Survivin.49 The fact that the kinase-inhibitor resistant form of Aurkb was sufficient to suppress the effects of a pan-Aurk inhibitor supports this notion. In addition, we also show that apoptosis in response to Aurk inhibition is at least somewhat selective for Myc-overexpressing cells, as there are clearly stronger effects of Aurka/Aurkb inhibition in cells whose proliferative programs are driven by Myc versus other oncogenes. It has to be considered, however, that Myc overexpression is often associated with particularly high levels of proliferation. Together with the work of others,33,50 our data point to a particular sensitivity of Myc-transformed cells to agents that trigger checkpoints at the G2-M transition.
Myc and Aurora kinases have heretofore been linked through their central functions at the G2-M transition, in the development of genomic instability, and through p53 activation and function. Our findings that Myc induces Aurka and up-regulates Aurkb and that this response is essential for maintenance of Myc-driven malignancies, even in fully established disease, strongly suggest that agents that inhibit these kinases are attractive therapeutics for rapidly dividing malignancies having Myc involvement.
Supplementary Material
Acknowledgments
The authors thank Sabine Stritzke, Michaela Wagner, Madlen Oelsner, and the staff of the Department of Nuclear Medicine, TU München, for expert technical assistance; the ZPF animal facility (TU München) and Umeå Transgene Core Facility for animal care; Erich Nigg (Max-Planck Institut, Martinsried, Germany) for providing Aurka and Aurkb expression plasmids; Georg Bornkamm (Helmholtz Zentrum München, Munich, Germany) for providing P493-6 cells; Drs Mihaela Onciu and John Sandlund (St Jude Children's Research Hospital, Memphis, TN) for providing BL samples; and EMD-Serono (Rockland, MA) for kindly providing AS703569.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB TRR 54, U.K.; and SFB 824, A.B), the Swedish Cancer Society, the Association of International Cancer Research, the Swedish Research Council, the Kempe Foundation and Umeå University (J.A.N.), the National Institutes of Health (grant CA076379; J.L.C.), and funds from the State of Florida to Scripps Florida (J.L.C.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: U.K., J.A.N., J.L.C., J.D., and C.P. designed the research; J.d.H., S.R., A.B., A.H., J.R.D., M.R., M.S., A.G., N.v.B., N.G., and M.K. performed experiments and analyzed the data; M.A.H. analyzed the microarray data; U.K., J.A.N., and J.L.C. wrote the paper; and all authors reviewed the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ulrich Keller, III Medical Department, Technische Universität München, Ismaninger Str 15, 81675 Munich, Germany; e-mail: ulrich.keller@lrz.tum.de.
References
- 1.Boxer LM, Dang CV. Translocations involving c-myc and c-myc function. Oncogene. 2001;20(40):5595–5610. doi: 10.1038/sj.onc.1204595. [DOI] [PubMed] [Google Scholar]
- 2.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19(1):1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernard S, Eilers M. Control of cell proliferation and growth by Myc proteins. Results Probl Cell Differ. 2006;42:329–342. doi: 10.1007/400_004. [DOI] [PubMed] [Google Scholar]
- 4.Baudino TA, McKay C, Pendeville-Samain H, et al. c-Myc is essential for vasculogenesis and angiogenesis during development and tumor progression. Genes Dev. 2002;16(19):2530–2543. doi: 10.1101/gad.1024602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pelengaris S, Khan M, Evan GI. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 2002;109(3):321–334. doi: 10.1016/s0092-8674(02)00738-9. [DOI] [PubMed] [Google Scholar]
- 6.Roussel MF, Cleveland JL, Shurtleff SA, Sherr CJ. Myc rescue of a mutant CSF-1 receptor impaired in mitogenic signalling. Nature. 1991;353(6342):361–363. doi: 10.1038/353361a0. [DOI] [PubMed] [Google Scholar]
- 7.Nilsson JA, Cleveland JL. Myc pathways provoking cell suicide and cancer. Oncogene. 2003;22(56):9007–9021. doi: 10.1038/sj.onc.1207261. [DOI] [PubMed] [Google Scholar]
- 8.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 9.Coppola JA, Cole MD. Constitutive c-myc oncogene expression blocks mouse erythroleukaemia cell differentiation but not commitment. Nature. 1986;320(6064):760–763. doi: 10.1038/320760a0. [DOI] [PubMed] [Google Scholar]
- 10.Bouchard C, Thieke K, Maier A, et al. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 1999;18(19):5321–5333. doi: 10.1093/emboj/18.19.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hermeking H, Rago C, Schuhmacher M, et al. Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci U S A. 2000;97(5):2229–2234. doi: 10.1073/pnas.050586197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leone G, DeGregori J, Sears R, Jakoi L, Nevins JR. Myc and Ras collaborate in inducing accumulation of active cyclin E/Cdk2 and E2F. Nature. 1997;387(6631):422–426. doi: 10.1038/387422a0. [DOI] [PubMed] [Google Scholar]
- 13.Bartek J, Lukas J. p27 destruction: Cks1 pulls the trigger. Nat Cell Biol. 2001;3(4):E95–E98. doi: 10.1038/35070160. [DOI] [PubMed] [Google Scholar]
- 14.Keller UB, Old JB, Dorsey FC, et al. Myc targets Cks1 to provoke the suppression of p27Kip1, proliferation and lymphomagenesis. EMBO J. 2007;26(10):2562–2574. doi: 10.1038/sj.emboj.7601691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vlach J, Hennecke S, Alevizopoulos K, Conti D, Amati B. Growth arrest by the cyclin-dependent kinase inhibitor p27Kip1 is abrogated by c-Myc. EMBO J. 1996;15(23):6595–6604. [PMC free article] [PubMed] [Google Scholar]
- 16.Menssen A, Hermeking H. Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc Natl Acad Sci U S A. 2002;99(9):6274–6279. doi: 10.1073/pnas.082005599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrews PD, Knatko E, Moore WJ, Swedlow JR. Mitotic mechanics: the auroras come into view. Curr Opin Cell Biol. 2003;15(6):672–683. doi: 10.1016/j.ceb.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 18.Lu LY, Wood JL, Ye L, et al. Aurora A is essential for early embryonic development and tumor suppression. J Biol Chem. 2008;283(46):31785–31790. doi: 10.1074/jbc.M805880200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vader G, Lens SM. The Aurora kinase family in cell division and cancer. Biochim Biophys Acta. 2008;1786(1):60–72. doi: 10.1016/j.bbcan.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Kanda A, Kawai H, Suto S, et al. Aurora-B/AIM-1 kinase activity is involved in Ras-mediated cell transformation. Oncogene. 2005;24(49):7266–7272. doi: 10.1038/sj.onc.1208884. [DOI] [PubMed] [Google Scholar]
- 21.Tatsuka M, Sato S, Kitajima S, et al. Overexpression of Aurora-A potentiates HRAS-mediated oncogenic transformation and is implicated in oral carcinogenesis. Oncogene. 2005;24(6):1122–1127. doi: 10.1038/sj.onc.1208293. [DOI] [PubMed] [Google Scholar]
- 22.Mao JH, Wu D, Perez-Losada J, et al. Crosstalk between Aurora-A and p53: frequent deletion or downregulation of Aurora-A in tumors from p53 null mice. Cancer Cell. 2007;11(2):161–173. doi: 10.1016/j.ccr.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otto T, Horn S, Brockmann M, et al. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell. 2009;15(1):67–78. doi: 10.1016/j.ccr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 24.Adams JM, Harris AW, Pinkert CA, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318(6046):533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 25.Pajic A, Spitkovsky D, Christoph B, et al. Cell cycle activation by c-myc in a Burkitt lymphoma model cell line. Int J Cancer. 2000;87(6):787–793. doi: 10.1002/1097-0215(20000915)87:6<787::aid-ijc4>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 26.Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1(2):729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyd KE, Farnham PJ. Myc versus USF: discrimination at the cad gene is determined by core promoter elements. Mol Cell Biol. 1997;17(5):2529–2537. doi: 10.1128/mcb.17.5.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001;15(16):2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amati B, Alevizopoulos K, Vlach J. Myc and the cell cycle. Front Biosci. 1998;3:250–268. doi: 10.2741/a239. [DOI] [PubMed] [Google Scholar]
- 30.Hirota T, Lipp JJ, Toh BH, Peters JM. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 2005;438(7071):1176–1180. doi: 10.1038/nature04254. [DOI] [PubMed] [Google Scholar]
- 31.Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251(4998):1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 32.Shim H, Dolde C, Lewis BC, et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A. 1997;94(13):6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goga A, Yang D, Tward AD, Morgan DO, Bishop JM. Inhibition of CDK1 as a potential therapy for tumors over-expressing MYC. Nat Med. 2007;13(7):820–827. doi: 10.1038/nm1606. [DOI] [PubMed] [Google Scholar]
- 34.Hoglund A, Nilsson LM, Forshell LP, Maclean KH, Nilsson JA. Myc sensitizes p53-deficient cancer cells to the DNA-damaging effects of the DNA methyltransferase inhibitor decitabine. Blood. 2009;113(18):4281–4288. doi: 10.1182/blood-2008-10-183475. [DOI] [PubMed] [Google Scholar]
- 35.Iritani BM, Eisenman RN. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc Natl Acad Sci U S A. 1999;96(23):13180–13185. doi: 10.1073/pnas.96.23.13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baudino TA, Maclean KH, Brennan J, et al. Myc-mediated proliferation and lymphomagenesis, but not apoptosis, are compromised by E2f1 loss. Mol Cell. 2003;11(4):905–914. doi: 10.1016/s1097-2765(03)00102-3. [DOI] [PubMed] [Google Scholar]
- 37.Grandori C, Gomez-Roman N, Felton-Edkins ZA, et al. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7(3):311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- 38.Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003;421(6920):290–294. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- 39.Robinson K, Asawachaicharn N, Galloway DA, Grandori C. c-Myc accelerates S-phase and requires WRN to avoid replication stress. PLoS ONE. 2009;4(6):e5951. doi: 10.1371/journal.pone.0005951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dominguez-Sola D, Ying CY, Grandori C, et al. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448(7152):445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 41.Menssen A, Epanchintsev A, Lodygin D, et al. c-MYC delays prometaphase by direct transactivation of MAD2 and BubR1: identification of mechanisms underlying c-MYC-induced DNA damage and chromosomal instability. Cell Cycle. 2007;6(3):339–352. doi: 10.4161/cc.6.3.3808. [DOI] [PubMed] [Google Scholar]
- 42.Minshull J, Blow JJ, Hunt T. Translation of cyclin mRNA is necessary for extracts of activated xenopus eggs to enter mitosis. Cell. 1989;56(6):947–956. doi: 10.1016/0092-8674(89)90628-4. [DOI] [PubMed] [Google Scholar]
- 43.Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8(10):1039–1048. [PubMed] [Google Scholar]
- 44.Lindqvist A, Kallstrom H, Lundgren A, Barsoum E, Rosenthal CK. Cdc25B cooperates with Cdc25A to induce mitosis but has a unique role in activating cyclin B1-Cdk1 at the centrosome. J Cell Biol. 2005;171(1):35–45. doi: 10.1083/jcb.200503066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reimann M, Loddenkemper C, Rudolph C, et al. The Myc-evoked DNA damage response accounts for treatment resistance in primary lymphomas in vivo. Blood. 2007;110(8):2996–3004. doi: 10.1182/blood-2007-02-075614. [DOI] [PubMed] [Google Scholar]
- 46.Zindy F, Eischen CM, Randle DH, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12(15):2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katayama H, Sasai K, Kawai H, et al. Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nat Genet. 2004;36(1):55–62. doi: 10.1038/ng1279. [DOI] [PubMed] [Google Scholar]
- 48.Yang J, Ikezoe T, Nishioka C, et al. AZD1152, a novel and selective aurora B kinase inhibitor, induces growth arrest, apoptosis, and sensitization for tubulin depolymerizing agent or topoisomerase II inhibitor in human acute leukemia cells in vitro and in vivo. Blood. 2007;110(6):2034–2040. doi: 10.1182/blood-2007-02-073700. [DOI] [PubMed] [Google Scholar]
- 49.Furuya M, Tsuji N, Kobayashi D, Watanabe N. Interaction between survivin and aurora-B kinase plays an important role in survivin-mediated up-regulation of human telomerase reverse transcriptase expression. Int J Oncol. 2009;34(4):1061–1068. doi: 10.3892/ijo_00000232. [DOI] [PubMed] [Google Scholar]
- 50.Li Q, Dang CV. c-Myc overexpression uncouples DNA replication from mitosis. Mol Cell Biol. 1999;19(8):5339–5351. doi: 10.1128/mcb.19.8.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.