Abstract
Proteasomes, the primary mediators of ubiquitin-protein conjugate degradation, are regulated through complex and poorly understood mechanisms. Here we show that Usp14, a proteasome-associated deubiquitinating enzyme, can inhibit the degradation of ubiquitin-protein conjugates, in vivo and in vitro. A catalytically inactive variant of Usp14 has reduced inhibitory activity, suggesting that inhibition is mediated by trimming of the ubiquitin chain on the substrate. A high-throughput screen identified a selective small-molecule inhibitor of the deubiquitinating activity of human Usp14. Treatment of cultured cells with this compound enhanced degradation of several proteasome substrates that have been implicated in neurodegenerative disease. Usp14 inhibition accelerated the degradation of oxidized proteins and enhanced resistance to oxidative stress. Enhancement of proteasome activity through inhibition of Usp14 may offer a strategy to reduce the levels of aberrant proteins in cells under proteotoxic stress.
The proteasome is essential for life in eukaryotes and regulates many aspects of cell physiology1,2. Most of its substrates are targeted to the proteasome via ubiquitination. The proteasome holoenzyme is composed of a 19-subunit regulatory particle (known as the RP, 19S complex, or PA700) and a 28-subunit core particle (known as the CP, or 20S complex). Substrate first binds the RP, and is then actively translocated to the CP, where it is degraded. The mechanisms regulating proteasome activity remain poorly understood, but involve numerous proteins that reversibly associate with it. Some bind the RP and deliver ubiquitin-conjugates to the proteasome, while others open the axial channel into the CP. A third class of associated proteins, composed of ubiquitin ligases and deubiquitinating enzymes (DUBs), modifies proteasome-bound ubiquitin chains. Ubiquitin chains vary in their linkage type and length, and longer variants interact more strongly with the proteasome3. The extension and disassembly of chains at the proteasome may alter substrate degradation rates by changing substrate affinity for the proteasome.
Mammalian proteasomes are associated with three DUBs: Rpn11, Uch37, and Usp14 (refs 4–22). Uch37 and Usp14 associate reversibly with the proteasome, whereas Rpn11 is a stoichiometric subunit1. These enzymes reside on the RP and remove ubiquitin from the substrate prior to substrate degradation. The release of ubiquitin spares it from degradation, minimizing fluctuations in ubiquitin pools. The activity of Rpn11 on the substrate’s ubiquitin chain is thought to be delayed until the proteasome is committed to degrading the substrate4,5. Rpn11 then cuts at the base of a ubiquitin chain, freeing substrate5. Thus, removal of the ubiquitin chain by Rpn11 can promote substrate translocation into the CP to be hydrolyzed4,5. However, deubiquitination prior to commitment might inhibit substrate degradation, since ubiquitin targets the protein for degradation6.
In contrast to Rpn11, Usp14 and Uch37 can attack ubiquitin chains independently of commitment to substrate degradation. Uch37, and perhaps Usp14, disassemble the chain from its substrate-distal tip6,15,16, thus shortening chains rather than removing them en bloc. Little is known about such “chain-trimming” reactions6–8. One model is that chain trimming increases the ability of proteasomes to discriminate between long and short multiubiquitin chains6. Here we show that a small-molecule inhibitor of deubiquitination by Usp14 stimulates protein degradation in vitro and in vivo. These findings reveal that in vivo proteasome function is limited by Usp14-dependent chain-trimming, implying that otherwise competent substrates of the proteasome can be rejected when chain trimming is faster than competing steps leading to substrate degradation.
Usp14 inhibits the proteasome in vitro
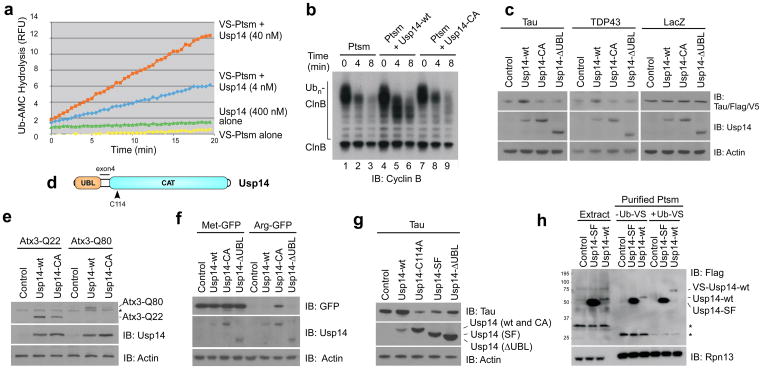
We have previously shown that Ubp6, the yeast ortholog of Usp14, is a potent inhibitor of the proteasome16. To test whether this is true of Usp14 from humans, we first developed a purification procedure that results in proteasomes lacking detectable Usp14 (modified from ref 23). Such proteasomes retain high levels of ubiquitin-AMC (Ub-AMC) hydrolyzing activity (data not shown), which is presumably Uch37-dependent (Supplementary Fig. 1). This activity can be inhibited irreversibly using ubiquitin-vinylsulfone (Ub-VS)24, which forms an adduct with the active site Cys in DUBs of the thiol protease class. When such “VS-proteasomes” were reconstituted with recombinant Usp14 (Supplementary Fig. 2), Ub-AMC hydrolyzing activity was increased 800-fold over that of isolated Usp14 (Fig. 1a). Thus, the deubiquitinating activity of Usp14 is activated by proteasomes (see also refs 10,11,15,18,22). Using the Ub-AMC assay, the affinity of Usp14 for the proteasome was found to be 4 nM (Supplementary Fig. 3).
Figure 1. Usp14 is an inhibitor of the proteasome.
a, Ub-AMC hydrolysis assay of Usp14 activity in the presence or absence of Ub-VS treated human proteasome (VS-proteasome; 1 nM). RFU, relative fluorescence units. Ptsm, 26S proteasome.
b, In vitro degradation assay with polyubiquitinated cyclin B (Ubn-ClnB), human proteasome (4 nM), and wild-type Usp14 (Usp14-wt) or mutant Usp14-CA (60 nM). Samples in b, c, and e-h analyzed by SDS-PAGE/immunoblotting (IB).
c, Plasmids expressing Tau, TDP-43Flag, or LacZV5 were cotransfected into usp14−/− MEFs with variants of Usp14Flag as indicated. Samples collected 2 days post-transfection. Actin, load control.
d, Diagram of human Usp14, showing ubiquitin-like (UBL) and catalytic (CAT) domains. C114, active site cysteine. Splice variant Usp14-SF is produced from an mRNA lacking exon 4 (ref 12).
e, Flag-tagged Ataxin3-Q22 or -Q80 was coexpressed with Usp14 variants in usp14−/− MEFs and detected with anti-Flag antibodies.
f, Arg-GFP or control Met-GFP coexpressed with Usp14 variants in usp14−/− MEFs.
g, As c except in HEK293 cells.
h, Usp14-SF associates with but is not activated by proteasomes. Each variant of Usp14Flag was expressed in HEK293T cells containing tagged hRpn11, and proteasomes were affinity purified. Where indicated, Ub-VS was incubated with lysate prior to proteasome purification. Extract samples represent 5% of total. Asterisks, nonspecific signals. Proteasome subunit Rpn13, load control. Control samples, empty vector. Equal cell numbers were used for each lane.
Proteasomes reconstituted with a saturating amount of Usp14 were challenged with a model proteasome substrate, ubiquitinated cyclin B (Ub-cyclin B). Like Ubp6, Usp14 inhibited the degradation of Ub-cyclin B (Fig. 1b). An active site mutant of Usp14, Usp14-C114A (hereafter Usp14-CA), showed little inhibitory effect, pointing to chain trimming as the basis for inhibition. Indeed, extensive trimming of ubiquitin from cyclin B, as indicated by the shift of Ub-cyclin B bands to a lower molecular mass, was evident in the presence of Usp14 but not Usp14-CA (Fig. 1b). Apparently, complete deubiquitination of cyclin B is not required to suppress degradation, because Usp14 rapidly removed only a portion of the ubiquitin from cyclin B, even upon longer incubation (Fig. 1b). The strong dependence of chain trimming on Usp14 was unexpected because active Uch37 was present in these proteasomes (Supplementary Fig. 1). When Ub-AMC is used as a substrate, Uch37 activity predominates over that of Usp14 (refs 7,8), but a true ubiquitin-protein conjugate, Ub-cyclin B, shows the inverse effect. The lack of significant inhibition of degradation seen with Usp14-CA was also surprising because Ubp6 exhibits a noncatalytic mechanism of proteasome inhibition16. As described below, a noncatalytic effect is seen with Usp14, though it is not well visualized in this assay.
Usp14 inhibits protein turnover in cells
To verify that Usp14 inhibits the proteasome in cells, we expressed Usp14 variants in usp14−/− murine embryonic fibroblasts (MEFs), together with proteasome substrates. As substrates we initially chose two proteins that are critical in neurodegenerative diseases, Tau and TDP-43 (refs 25,26). Tau is thought to undergo proteasomal degradation27,28. Tau and TDP-43 showed increased levels when co-expressed with wild-type Usp14 in usp14−/− MEFs, suggesting proteasome inhibition by Usp14 (Fig. 1c). No effect was seen at the mRNA level (Supplementary Fig. 4). As seen in vitro, Usp14-CA showed little or no activity in the assay. Thus, the ability of the proteasome to degrade Tau and TDP-43 in MEFs appears to be suppressed by trimming of ubiquitin from these substrates. The N-terminal ubiquitin-like (UBL) domain of Usp14 (Fig. 1d) is its principal proteasome-binding site15, and accordingly its deletion abrogated the Usp14 effect (Fig. 1c). The effects on Tau levels reflected accelerated degradation: Tau disappeared more slowly from cells expressing Usp14 in a chase experiment (Supplementary Fig. 5). Usp14 did not affect the levels of two proteins that are thought to be stable, LacZ and actin (Fig. 1c). In MEFs and other cell types, wild-type Usp14 was usually expressed at lower levels than Usp14-CA (Fig. 1), suggesting that Usp14 may be subject to autoregulation.
Another protein linked to neurodegeneration and thought to be a substrate of the proteasome is ataxin-3 (Atx3)29. Poly-Q expanded forms of Atx3 give rise to spinocerebellar ataxia 3. Both wild-type (Q22) and expanded forms of Atx3 (Q80) were stabilized by expression of Usp14 in usp14−/− MEFs (Fig. 1e). Expression of wild-type Usp14 engendered stronger accumulation of Atx3 than expression of Usp14-CA. However, in contrast to Tau and TDP-43, the stabilizing effect of catalytically inactive Usp14 was substantial for Atx3-Q22 and Atx3-Q80. Stabilization of Atx3 by Usp14-CA presumably represents the same noncatalytic effect as described for Ubp6 in S. cerevisiae16. Thus the noncatalytic inhibitory effect is apparently conserved in evolution. A noncatalytic effect was also observed for a model substrate of the proteasome30,31, Arg-GFP (Fig. 1f). Wild-type Usp14 was ineffective in Arg-GFP stabilization in comparison to Usp14-CA (Fig. 1f), the noncatalytic effect being dominant. Met-GFP, a stable protein, was unaffected by Usp14. It will be interesting to determine what substrate features underlie the differing sensitivities of these substrates to catalytic and noncatalytic inhibition of degradation.
The effect of Usp14 on Tau degradation was confirmed in HEK293 cells. As in MEFs, Usp14 overexpression stabilized Tau (Fig. 1g). Results obtained with Usp14 mutants differed somewhat from those obtained using MEFs, as expected, given that HEK293 cells express endogenous Usp14; the expression of Usp14-CA in usp14−/− MEFs had no effect on Tau, whereas in HEK293 cells the Usp14-CA mutant produced accelerated Tau degradation (Fig. 1g). This result presumably reflects displacement of endogenous Usp14 from the proteasome. As expected, deletion of the UBL domain attenuated the dominant negative effect (Fig. 1g). In contrast to Usp14-ΔUBL, the short form (SF) of Usp14, expressed from a developmentally regulated18 mRNA that lacks a 33-codon junctional exon (exon 4) between the UBL domain and the catalytic domain12,18 (Fig. 1d), did exhibit a dominant negative effect (Fig. 1g). This result suggested that Usp14-SF might bind proteasomes and counter the action of full-length Usp14. Thus, Usp14-SF may be an endogenous inhibitor of Usp14 activity. Consistent with this possibility, Usp14-SF binds proteasomes, but is not activated enzymatically by proteasome binding, as shown by its inability to react with Ub-VS (Fig. 1h). Usp14-SF also appears to lack noncatalytic proteasome-inhibitory capacity, because its expression in usp14−/− MEFs did not stabilize Arg-GFP (Supplementary Fig. 6).
A selective small-molecule inhibitor of Usp14
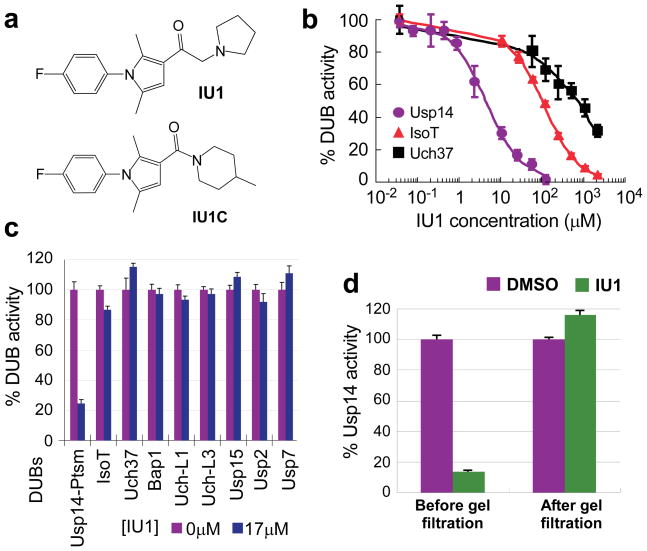
The results above suggested that chain trimming at the proteasome antagonizes degradation of multiple substrates. Therefore, a small-molecule inhibitor of Usp14 might enhance proteasome activity. We screened 63,052 compounds for the ability to inhibit Usp14, using VS-proteasomes reconstituted with Usp14 and assayed with Ub-AMC, and identified 215 as true Usp14 inhibitors (details in Supplementary Methods, Supplementary Table, and Supplementary Fig. 7). When the hits were counterscreened against a panel of DUBs, only three showed selectivity for Usp14. We proceeded with more detailed studies of the strongest hit, 1-[1-(4-fluorophenyl)-2,5-dimethylpyrrol-3-yl]-2-pyrrolidin-1-ylethanone, referred to below as IU1 (Fig. 2a). Its structure is suggestive of an active-site-directed thiol protease inhibitor. The IC50 of IU1 for Usp14 is 4–5 μM (Fig. 2b; Supplementary Fig. 8). IU1 failed to significantly inhibit eight DUBs of human origin (Figs. 2b, 2c; Supplementary Figs. 9, 10), as well as Ub-AMC hydrolysis by proteasomes lacking Usp14, which is attributable to Uch37 (Supplementary Fig. 8). We also identified a compound that is closely related to IU1 but does not inhibit Usp14 (IU1C; Fig. 2a; Supplementary Fig. 11), and used this as a specificity control in subsequent assays. In the absence of proteasomes, Usp14 was insensitive to IU1 (Supplementary Fig. 8), suggesting that IU1 binds specifically to the activated form of Usp14. IU1 could potentially inhibit Usp14 by preventing its docking on the proteasome, but direct tests of this scenario proved negative (Supplementary Fig. 12). Usp14 inhibition was rapidly established upon addition of IU1 and rapidly reversed upon its removal (Fig. 2d and Supplementary Fig. 13).
Figure 2. IU1 inhibits human Usp14 specifically and reversibly.
a, Chemical structures of IU1 and IU1C. Analytical data shown in Supplementary Fig. 16.
b, IC50 determination for IU1 inhibition of Ub-AMC hydrolysis by proteasome-bound Usp14 (4.7 ± 0.7 μM), IsoT (100 ± 0.4μM), and Uch37 (0.7 ± 0.3 mM).
c, Ub-AMC (1 μM) hydrolysis assays showing specificity of IU1 for Usp14.
d, Reversibility of Usp14 inhibition. 60 nM Usp14 and 5 nM human proteasome were treated with vehicle (DMSO) or 100 μM IU1 for 2 hr. After spin gel-filtration, proteins were assayed for Ub-AMC hydrolysis. All values are presented as mean ± s.d. (n=3).
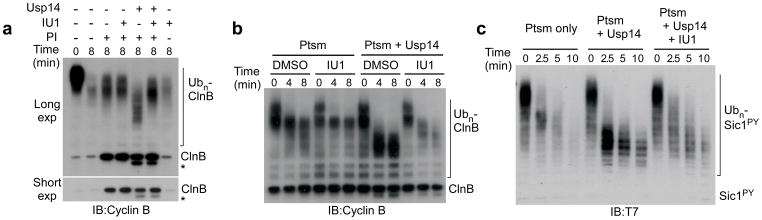
We employed Ub-cyclin B to test whether IU1 could inhibit the trimming of ubiquitin chains by the proteasome. To separate chain trimming from substrate degradation, these assays were done in the presence of proteasome inhibitors4. When proteasomes lacking Usp14 were tested, IU1 had no effect on ubiquitin chain release (Fig. 3a), which is likely Rpn11-dependent4. Chain trimming was strongly enhanced by Usp14, as apparent from the increased electrophoretic mobility of Ub-cyclin B species. Addition of IU1 to the assay reversed this effect (Fig. 3a; see also Supplementary Fig. 14).
Figure 3. IU1 inhibits chain trimming and stimulates substrate degradation in vitro.
a, Chain trimming assays. Samples contained 4 nM proteasome, and Usp14 was added at 15-fold molar excess over proteasome. IU1 was added at 50 μM and proteasome inhibitors (PI) at 5 μM (PS-341, epoxomicin). Asterisk, cyclin B species derived from residual thrombin from Usp14 preparation16. All panels, SDS-PAGE/immunoblot analysis.
b, In vitro Ubn-ClnB degradation assay (IU1 at 34 μM).
c, In vitro degradation assay with polyubiquitinated Sic1PY, human proteasome (5 nM), and Usp14-wt (75 nM) in the absence or presence of IU1 (75 μM).
We next tested whether IU1 could enhance degradation. Proteasomal degradation of Ub-cyclin B was indeed dramatically stimulated by IU1 (Fig. 3b). Proteasomes lacking Usp14 were insensitive to IU1 (Fig. 3b), and cyclin B degradation was unaffected by IU1 when proteasomes were reconstituted with Usp14-CA (Supplementary Fig. 15). Thus, suppression of chain trimming by IU1 may account for its enhancement of degradation. IU1 also promoted degradation of Sic1, a CDK inhibitor from S. cerevisiae (Fig. 3c). These assays employed a modified form of Sic1 in which the PY element signals ubiquitination32. The Ub-cyclin B used in these assays carries mixed ubiquitin chains linked via K48, K63, and K11 residues33. In contrast, ubiquitin chains formed on Ub-Sic1PY are homogenously linked via K6332. Chains of different topologies may therefore be susceptible to Usp14-dependent regulation.
A Usp14 inhibitor accelerates proteolysis in cells
To determine whether IU1 is cell permeable, it was added to cultures and cell-associated IU1 was quantified by LC-MS or UV absorption. When added at 50 μM, IU1 reached an apparent intracellular concentration of ~13 μM within 1 hour, which remained constant over the course of the experiment (Supplementary Figs. 17–19). Effects of IU1 on the viability of MEFs only became apparent at 250μM (Supplementary Figs. 20, 21). Moreover, IU1 did not noticeably induce apoptosis (Supplementary Fig. 22). When cell proliferation was measured in real time, slight inhibition became apparent at 120 μM (Supplementary Fig. 21). In the case of both cell viability and proliferation assays, usp14−/− MEFs were no less sensitive than wild-type, indicating that IU1 toxicity at high concentrations was independent of Usp14 inhibition.
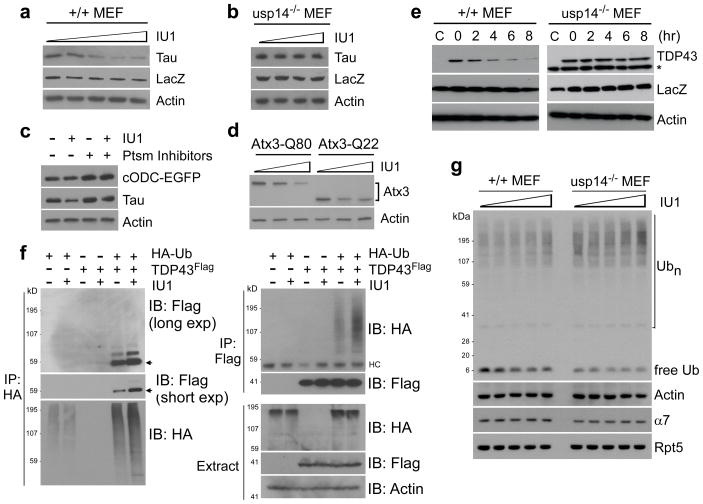
To determine whether IU1 could enhance proteasome function in cells, we expressed Tau in MEFs treated with sub-cytotoxic doses of IU1. IU1 induced dose-dependent reduction in Tau levels, with a strong effect seen at 50 μM (Fig. 4a; Supplementary Fig. 23). Thus, IU1 treatment affected Tau similarly to Usp14-CA (Fig. 1c), consistent with active site inhibition. No effect was seen on Tau mRNA (Supplementary Fig. 24). When usp14−/− MEFs were treated with IU1, no effect on Tau was observed, indicating that IU1 enhances Tau degradation through inhibiting Usp14 (Fig. 4b). Based on these and previous experiments (Fig. 2c), nonspecific inhibition of other DUBs by IU1 does not affect proteasome function at this dose. The effect of IU1 on Tau degradation was independent of autophagy (Supplementary Fig. 25). Several other proteins implicated in proteotoxic mechanisms–TDP-43, ataxin-3, and glial fibrillary acidic protein (GFAP)–were similarly depleted from MEFs by IU1 (Figs. 4d, 4e; Supplementary Fig. 26). The effectiveness of IU1 in neurons, where proteotoxic mechanisms are commonly observed, has not been examined.
Figure 4. IU1 enhances proteasomal degradation in vivo.
All panels show SDS-PAGE/immunoblot data.
a, 36 hours after cotransfecting wild-type MEFs with plasmids expressing Tau and LacZV5, cells were incubated with 0, 25, 50, 75, or 100 μM of IU1 for 6 hr. LacZ, transfection control. Actin, loading control.
b, As in a except that MEFs were usp14−/− and IU1 was at 0, 10, 50, or 100 μM.
c, Tau and Ub-independent proteasome substrate cODC-EGFP were coexpressed in wild-type MEFs and incubated with 50 μM IU1 for 6hr. Proteasome inhibitors were MG132 (30 μM) and PS-341 (10μM).
d, As b except with Atx3-Q80 and Atx3-Q22.
e, TDP-43Flag was cotransfected with a LacZ-expressing plasmid into either wild-type or usp14−/− MEFs, then treated with IU1 (75 μM) for the time indicated. Asterisk, nonspecific signal.
f, HA-tagged Ub and/or Flag-tagged TDP-43 were transiently overexpressed in wild-type MEFs with 50μM IU1 incubation for 6 hr. Proteasome inhibitors (20 μM MG132, 10 μM PS-341) were added 4 hr before lysis. Lysates were subjected to immunoprecipitation with anti-HA or anti-Flag. Arrows indicate likely ubiquitinated TDP-43 species. HC, heavy chain.
g, Wild-type MEF and usp14−/− MEF cells were treated with IU1 (0, 25, 50, 75, or 100 μM) for 6 hr, followed by analysis for ubiquitin, actin, CP subunit α7, and RP subunit Rpt5.
IU1 enhanced the extent of ubiquitin modification of TDP-43 in cells, perhaps accounting for its accelerated degradation (Fig. 4f). In contrast, little change was seen in bulk cellular ubiquitin conjugates (Fig. 4g; Supplementary Fig. 27). Free ubiquitin was reduced following IU1 addition, and, as the dose of IU1 increased, the level of free ubiquitin in wild-type MEFs approached that of untreated usp14−/− MEFs (Fig. 4g). Previous work showed that Usp14 and Ubp6 assist in maintaining of cellular ubiquitin pools by suppressing proteasomal degradation of ubiquitin11,13,14,17,34,35. The conjugated rather than free form of ubiquitin is most subject to degradation36. By separating ubiquitin from its conjugative target, Usp14 antagonizes this pathway of ubiquitin degradation.
Enhanced protein degradation in cells treated with IU1 could result from increased synthesis of proteasomes; however, no significant changes in proteasome composition were seen after IU1 treatment (Supplementary Fig. 28). Usp14 is known to affect gating of the proteasome21, but this does not appear to be critical in the mode of action of IU1 (unpublished data). The detailed similarities observed between the effects of mutational inactivation of Usp14’s catalytic site and IU1 treatment, as well as the observation that Usp14 is required for IU1 to affect protein degradation, provide strong evidence for the importance of chain trimming by Usp14. In addition, IU1 had little or no effect on the in vivo degradation of a ubiquitin-independent substrate37 of the proteasome, cODC-EGFP (Fig. 4c). Similar results were obtained in vitro with antizyme-promoted ODC degradation (data not shown). The effects of IU1 are likely restricted to ubiquitin-dependent proteasome substrates, based on its mode of action, but further characterization is required to establish this. Finally, Arg-GFP levels were constant upon treatment with IU1, when assayed in cells expressing Usp14-CA, suggesting that IU1 does not influence Usp14’s noncatalytic inhibitory effect (Supplementary Fig. 26).
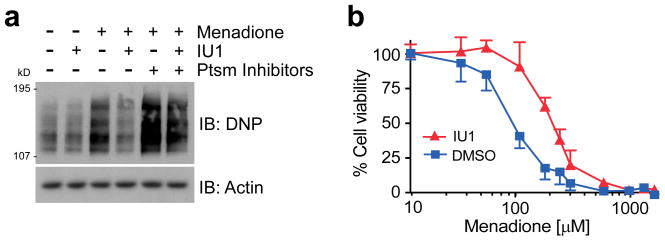
Oxidized proteins form a class of misfolded proteasome substrates that increase with age and are apparently toxic when they accumulate38,39. We induced protein oxidation by treating cells with menadione, and visualized oxidized species via their carbonyl groups. IU1 treatment strongly reduced the accumulation of oxidized proteins (Fig. 5a). When proteasome inhibitor was added, the effect of IU1 was attenuated, suggesting that IU1 does not prevent the oxidation reaction itself. IU1 treatment reduced menadione toxicity substantially in HEK293 cells (Fig. 5b), strongly supporting the hypothesis that proteins are critical targets of oxidative damage. IU1 also reduced the toxicity of an unrelated oxidizing agent, hydrogen peroxide (data not shown). IU1C, the IU1 variant that is inactive against Usp14, failed to reduce menadione cytotoxicity (Supplementary Fig. 29). In summary, these experiments suggest that IU1 can promote cell survival during proteotoxic stress.
Figure 5. IU1 alleviates cytotoxicity induced by oxidative stress.
a, HEK293 cells were preincubated with IU1 (75 μM) or proteasome inhibitors (20 μM MG132, 10 μM PS-341) for 4 hr, then treated with menadione (300 μM) for 60 min. Lysates were treated with DNPH and immunoblotted with anti-DNP antibody to assay oxidized proteins.
b, Cell survival under oxidative stress measured using the MTT assay. HEK293 cells were pretreated with 50 μM IU1 for 2 hr. Menadione was added, followed by 4-hr incubation. IU1 effects comparable to those of panels a and b were obtained in wild-type but not usp14−/− MEFs (data not shown). Values are represented as mean ± s.d. (n=3).
Ubiquitin chain trimming antagonizes proteasome function
We report here that a small molecule selected for its capacity to inhibit the proteasome-associated deubiquitinating enzyme Usp14 strongly enhances substrate degradation by the proteasome in cells. This is the first evidence that chain trimming by Usp14 or its yeast ortholog Ubp6 inhibits proteasome activity through deubiquitination. The trimming of substrate-bound ubiquitin chains on the proteasome appears to govern the degradation rates of many ubiquitin-protein conjugates. Under normal growth conditions, the proteasome may be subject to tonic inhibition through this mechanism.
The scope of proteasome inhibition by chain trimming is not limited to substrates bearing only one or a few ubiquitin groups6, as shown by studies of cyclin B and Sic1PY degradation. Also, suppression of degradation by chain trimming does not appear to be restricted to proteins that are not proper substrates of the proteasome. Chain trimming may be a more general, though not universal, mechanism for regulating protein turnover rates, suppressing the degradation of some substrates but not others. Further work will be required to determine what fraction of proteasome substrates can be regulated through this pathway, and what distinguishing features they share.
We also report that Usp14 can inhibit proteasome function noncatalytically, as previously observed for its yeast ortholog Ubp6 (ref 16). The capacity of Usp14 to inhibit proteasomes via two distinct pathways has important implications. Briefly, the noncatalytic effect, in slowing substrate degradation at the proteasome, may allow individual substrates to be docked at the proteasome for a longer time, thus enhancing the effectiveness of chain trimming by allowing ubiquitin chains to be trimmed more extensively. However, the two modes of proteasome inhibition are not obligatorily coupled, because proteasome substrates exhibited differing relative sensitivities to catalytic and noncatalytic inhibition by Usp14 (Fig. 1c–f). It will be important to identify the mechanistic basis of these differences.
Both Usp14 and Uch37 exhibit chain-trimming activity, but the effectiveness of IU1 in reducing chain trimming and stimulating proteasome activity suggests that the redundancy between these two proteasome-bound enzymes may be less than expected. Uch37 does not readily substitute for Usp14. Proteasomes are associated with multiple ubiquitin receptors, and the relative susceptibility of substrates to chain trimming by Usp14 and Uch37 may depend on which receptors are engaged with a given substrate and the positioning of these receptors with respect to Usp14 and Uch37. For example, Uch37 binds proteasomes via a ubiquitin receptor, Rpn13 (refs 24,40–42). Whether chain trimming by Uch37 can suppress proteasome activity as powerfully as Usp14 will require further study.
Although eukaryotic cells require proteasome function, proteasome inhibitors have proven highly effective in the treatment of multiple myeloma43, and may have additional applications44. In other contexts, however, enhancement of proteasome activity might be beneficial45,46. For example, enhanced proteasome function could have applications in disorders that result from partial loss of function mutations in ubiquitin pathway components47. More generally, many diseases, including major neurodegenerative diseases, are thought to be caused by the accumulation of misfolded proteins48–50. Misfolding, which renders these proteins toxic, can also mark them as substrates of the ubiquitin-proteasome and autophagy pathways50. Enhanced proteasome function, as induced by inhibitors of chain trimming, could therefore potentially be used to eliminate such toxic proteins more effectively.
METHODS SUMMARY
For expression in mammalian cells we employed full-length Usp14 (Usp14-wt) and its splice variant lacking exon 4 (Usp14-SF) subcloned into pcDNA3.1 (Invitrogen) as previously described19. The Usp14-C114A and Usp14-ΔUBL constructs were generated in the same vector by PCR-mediated mutagenesis. Human proteasomes were affinity-purified on a large scale from a stable HEK293 cell line harboring HTBH-tagged hRpn11 (a kind gift from L. Huang). 10 μl of Usp14 was dispensed into 384-well low volume plates in duplicate using a Wellmate dispenser. 33.3 nl of compound from the library was transferred into the wells using a Seiko pin transfer robotic system, followed by preincubation for ~30 min. To initiate the reaction, 10 μl of VS-proteasome plus Ub-AMC mixture was added to each well. The sources of compound libraries for screening were as follows: Maybridge, Asinex, ActiMol TimTec, ChemBridge, ChemDiv, Enamine, and MMV1. Primary hits were defined by ‘robust’ Z-score analysis (Supplementary Fig. 7). To obtain dose-response curves, curve fitting was performed by the four parameter logistic model or the three parameter fixed bottom model using SigmaPlot 9.0 according to guidelines from NIH Chemical Genomics Center. The gene trap allele, usp14rrk114 (ref 19), is referred to here as usp14−/−. For additional details see Supplementary Information.
Supplementary Material
Acknowledgments
We thank K. Gordon, J.Y. Suk, and N. Bays for advice and assistance, and members of the Finley lab for comments on the manuscript. We thank everyone at the ICCB facility at Harvard Medical School, where the HT screen was carried out. We also thank N. Hathaway for ubiquitinated cyclin B, L. Huang for the tagged proteasome cell line, R. Baker for anti-Usp14 antibody, G. DeMartino for anti-Uch37 antibody, K. Wilkinson and K. Walters for DUB enzymes, as well as C. Seong, M. Kim, S.M. Lim, and D. Waterman for assistance in some experiments. For plasmids we thank K. Walters, M. Sowa, W. Harper, V. Lee, F. Baralle, H. Paulson, Y. T. Kwon, M. Masucci, M.-K. Kwak, P. Coffino, and C. Kahana. This work was supported by grants from the National Institutes of Health (DK082906 to D.F., GM65592 to D.F., GM66492 to R.W.K., and NS047533 to S.M.W.); the Harvard Technology Development Accelerator Fund (D.F.); Merck & Co. (D.F. and R.W.K.); and Johnson & Johnson (D.F. and R.W.K.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Author contributions. B.-H.L. carried out screening and most in vitro studies, and M.J.L. chemical analysis and most cell-based assays. R.W.K. and D.F. were responsible for overall design and oversight of the project. S.P., S.E., and N.D. provided skilled assistance in proteasome biochemistry and assays. D.-C.O, C.G., and S.P.G. designed and carried out chemistry studies. P.-C.C, S.M.W., and J.H. provided key reagents and intellectual input. Many authors contributed to preparation of the manuscript.
References
- 1.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schrader EK, Harstad KG, Matouschek A. Targeting proteins for degradation. Nat Chem Biol. 2009;5:815–822. doi: 10.1038/nchembio.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verma R, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 5.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 6.Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- 7.Koulich E, Li X, DeMartino GN. Relative structural and functional roles of multiple deubiquitylating proteins associated with mammalian 26S proteasome. Mol Biol Cell. 2008;19:1072–1082. doi: 10.1091/mbc.E07-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobson AD, et al. The lysine 48 and lysine 63 ubiquitin conjugates are processed differently by the 26S proteasome. J Biol Chem. 2009;284:35485–35494. doi: 10.1074/jbc.M109.052928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verma R, et al. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borodovsky A, et al. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 2001;20:5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leggett DS, et al. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 12.Wilson SM, et al. Synaptic defects in ataxia mice result from a mutation in Usp14, encoding a ubiquitin-specific protease. Nat Genet. 2002;32:420–405. doi: 10.1038/ng1006. [DOI] [PubMed] [Google Scholar]
- 13.Chernova TA, et al. Pleiotropic effects of Ubp6 loss on drug sensitivities and yeast prion are due to depletion of the free ubiquitin pool. J Biol Chem. 2003;278:52102–52115. doi: 10.1074/jbc.M310283200. [DOI] [PubMed] [Google Scholar]
- 14.Anderson C, et al. Loss of Usp14 results in reduced levels of ubiquitin in ataxia mice. J Neurochem. 2005;95:724–731. doi: 10.1111/j.1471-4159.2005.03409.x. [DOI] [PubMed] [Google Scholar]
- 15.Hu M, et al. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme Usp14. EMBO J. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanna J, et al. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 17.Hanna J, Meides A, Zhang DP, Finley D. A ubiquitin stress response induces altered proteasome composition. Cell. 2007;129:747–759. doi: 10.1016/j.cell.2007.03.042. [DOI] [PubMed] [Google Scholar]
- 18.Crimmins S, et al. Transgenic rescue of ataxia mice with neuronal-specific expression of ubiquitin-specific protease 14. J Neurosci. 2006;26:11423–11431. doi: 10.1523/JNEUROSCI.3600-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crimmins S, et al. Transgenic rescue of ataxia mice reveals a male-specific sterility defect. Dev Biol. 2009;325:33–42. doi: 10.1016/j.ydbio.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen PC, et al. The proteasome-associated deubiquitinating enzyme Usp14 is essential for the maintenance of synaptic ubiquitin levels and the development of neuromuscular junctions. J Neurosci. 2009;29:10909–10919. doi: 10.1523/JNEUROSCI.2635-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peth A, Besche HC, Goldberg AL. Ubiquitinated proteins activate the proteasome by binding to Usp14/Upb6, which cause 20S gate opening. Mol Cell. 2009;36:794–804. doi: 10.1016/j.molcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Catic A, et al. Screen for ISG15-crossreactive deubiquitinases. PLoS One. 2007;2:e679. doi: 10.1371/journal.pone.0000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, et al. Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry. 2007;46:3553–3565. doi: 10.1021/bi061994u. [DOI] [PubMed] [Google Scholar]
- 24.Yao T, et al. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat Cell Biol. 2006;8:994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]
- 25.Spires-Jones TL, Stoothoff WH, de Calignon A, Jones PB, Hyman BT. Tau pathophysiology in neurodegeneration: a tangled issue. Trends Neurosci. 2009;32:150–159. doi: 10.1016/j.tins.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 26.Kwong LK, Uryu K, Trojanowski JQ, Lee VM. TDP-43 proteinopathies: neurodegenerative protein misfolding diseases without amyloidosis. Neurosignals. 2008;16:41–51. doi: 10.1159/000109758. [DOI] [PubMed] [Google Scholar]
- 27.David DC, et al. Proteasomal degradation of tau protein. J Neurochem. 2002;83:176–178. doi: 10.1046/j.1471-4159.2002.01137.x. [DOI] [PubMed] [Google Scholar]
- 28.Petrucelli L, et al. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum Mol Genet. 2004;13:703–714. doi: 10.1093/hmg/ddh083. [DOI] [PubMed] [Google Scholar]
- 29.Todi SV, et al. Cellular turnover of the polyglutamine disease protein ataxin-3 is regulated by its catalytic activity. J Biol Chem. 2007;282:29348–29358. doi: 10.1074/jbc.M704126200. [DOI] [PubMed] [Google Scholar]
- 30.Varshavsky A, Turner G, Du F, Xie Y. The ubiquitin system and the N-end rule pathway. Biol Chem. 2000;381:779–789. doi: 10.1515/BC.2000.101. [DOI] [PubMed] [Google Scholar]
- 31.Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat Biotechnol. 2000;18:538–543. doi: 10.1038/75406. [DOI] [PubMed] [Google Scholar]
- 32.Saeki Y, Isono E, Toh-E A. Preparation of ubiquitinated substrates by the PY motif-insertion method for monitoring proteasome activity. Meth Enzymol. 2005;399:215–227. doi: 10.1016/S0076-6879(05)99014-9. [DOI] [PubMed] [Google Scholar]
- 33.Kirkpatrick DS, et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 34.Amerik AY, Li SJ, Hochstrasser M. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol Chem. 2000;381:981–992. doi: 10.1515/BC.2000.121. [DOI] [PubMed] [Google Scholar]
- 35.Hanna J, Leggett DS, Finley D. Ubiquitin depletion as a key mediator of toxicity by translational inhibitors. Mol Cell Biol. 2003;23:9251–9261. doi: 10.1128/MCB.23.24.9251-9261.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shabek N, Herman-Bachinsky Y, Ciechanover A. Ubiquitin degradation with its substrate, or as a monomer in a ubiquitination-independent mode, provides clues to proteasome regulation. Proc Natl Acad Sci USA. 2009;106:11907–11912. doi: 10.1073/pnas.0905746106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoyt MA, Zhang M, Coffino P. Probing the ubiquitin/proteasome system with ornithine decarboxylase, a ubiquitin-independent substrate. Meth Enzymol. 2005;398:399–413. doi: 10.1016/S0076-6879(05)98033-6. [DOI] [PubMed] [Google Scholar]
- 38.Stadtman ER. Protein oxidation and aging. Free Radic Res. 2006;40:1250–1258. doi: 10.1080/10715760600918142. [DOI] [PubMed] [Google Scholar]
- 39.Ahmed EK, Picot CR, Bulteau AL, Friguet B. Protein oxidative modifications and replicative senescence of WI-38 human embryonic fibroblasts. Ann NY Acad Sci. 2007;1119:88–96. doi: 10.1196/annals.1404.020. [DOI] [PubMed] [Google Scholar]
- 40.Hamazaki J, et al. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 2006;25:4524–4536. doi: 10.1038/sj.emboj.7601338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiu XB, et al. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 2006;25:5742–5753. doi: 10.1038/sj.emboj.7601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Husnjak K, et al. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chauhan D, Bianchi G, Anderson KC. Targeting the UPS as therapy in multiple myeloma. BMC Biochem. 2008;9(Suppl 1):S1. doi: 10.1186/1471-2091-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muchamuel T, et al. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat Med. 2009;15:781–787. doi: 10.1038/nm.1978. [DOI] [PubMed] [Google Scholar]
- 45.Chondrogianni N, et al. Overexpression of proteasome β5 subunit increases the amount of assembled proteasome and confers ameliorated response to oxidative stress and higher survival rates. J Biol Chem. 2005;280:11840–11850. doi: 10.1074/jbc.M413007200. [DOI] [PubMed] [Google Scholar]
- 46.Tonoki A, et al. Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol Cell Biol. 2009;29:1095–1106. doi: 10.1128/MCB.01227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lehman NL. The ubiquitin proteasome system in neuropathology. Acta Neuropathol. 2009;118:329–347. doi: 10.1007/s00401-009-0560-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hinault MP, Ben-Zvi A, Goloubinoff P. Chaperones and proteases: cellular fold-controlling factors of proteins in neurodegenerative diseases and aging. J Mol Neurosci. 2006;30:249–265. doi: 10.1385/JMN:30:3:249. [DOI] [PubMed] [Google Scholar]
- 49.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 50.Goldberg AL. Protein degradation and protection against misfolded and damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.