Summary
PARP-1 is an abundant nuclear enzyme that regulates gene expression, although the underlying mechanisms are unclear. We examined the interplay between PARP-1, histone 3 lysine 4 trimethylation (H3K4me3), and linker histone H1 in the chromatin-dependent control of transcription. We show that PARP-1 is required for a series of molecular outcomes at the promoters of PARP-1 regulated genes, leading to a permissive chromatin environment that allows loading of the RNA Pol II machinery. PARP-1 does so by (1) preventing demethylation of H3K4me3 through the PARylation, inhibition, and exclusion of the histone demethylase KDM5B and (2) promoting the exclusion of H1 and the opening of promoter chromatin. Upon depletion of PARP-1, these outcomes do not occur efficiently. Interestingly, cellular signaling pathways can use the regulated depletion of PARP-1 to modulate these chromatin-related molecular outcomes. Collectively, our results help to elucidate the roles of PARP-1 in the regulation of chromatin structure and transcription.
Keywords: PARP-1, KDM5B, histone methylation, chromatin remodeling, transcription
Introduction
Chromatin, a repeating array of nucleosomes and nucleosome-binding proteins, plays key roles in the regulation of gene transcription by limiting (1) the loading of the RNA polymerase II (Pol II) machinery at gene promoters, (2) the initiation of transcription from transcription start sites (TSSs), and (3) the elongation of transcripts through the bodies of genes (Campos and Reinberg, 2009; Li et al., 2007). Several properties of chromatin contribute to its gene-regulating effects, including its composition (e.g., types and extent of histones modifications, repertoire of nucleosome-bound proteins) and structure (e.g., positioning and spacing of nucleosomes) (Campos and Reinberg, 2009; Li et al., 2007). Although great strides have been made, the molecular mechanisms by which specific chromatin-binding and histone-modifying proteins interact to alter chromatin structure and function to regulate transcription require further analysis.
Nucleosome-binding architectural proteins, such as poly(ADP-ribose) polymerase-1 (PARP-1) and the linker histone H1, promote structural alterations in chromatin and, as a consequence, modulate transcriptional responses (Happel and Doenecke, 2009; Kraus, 2008; Krishnakumar and Kraus, 2010; Tulin et al., 2003). Both PARP-1 and H1 can alter nucleosome spacing and promote the compaction of nucleosomal arrays (Happel and Doenecke, 2009; Kim et al., 2004; Wacker et al., 2007). Although PARP-1 and H1 elicit grossly similar alterations in chromatin structure in vitro (Kim et al., 2004; Wacker et al., 2007), their effects on chromatin structure, however, may be interpreted differently in vivo (Kraus, 2008). PARP-1 and H1 compete for binding to nucleosomes (Kim et al., 2004) and exhibit a reciprocal pattern of binding at actively transcribed promoters: H1 is depleted and PARP-1 is enriched (Krishnakumar et al., 2008). Both PARP-1 and H1 are widely distributed across the genome (Kim et al., 2004; Krishnakumar et al., 2008; Tulin and Spradling, 2003) and their depletion can promote large scale alterations in chromatin structure (Fan et al., 2005; Lu et al., 2009; Petesch and Lis, 2008; Tulin and Spradling, 2003), but their effects on transcription are limited to a subset of specific target genes (Fan et al., 2005; Frizzell et al., 2009; Lu et al., 2009).
Unlike H1, PARP-1 possesses an intrinsic enzymatic activity that catalyzes the polymerization of ADP-ribose units from donor NAD+ molecules on target proteins (D’Amours et al., 1999). Although PARP-1 is the major target for PARP-1-mediated poly(ADP-ribosyl)ation (PARylation) in vivo through an automodification reaction, a number of other targets have been also described, including core histones, H1, and a variety of nuclear proteins involved in gene regulation (D’Amours et al., 1999; Krishnakumar and Kraus, 2010). PARylation of protein targets by PARP-1 alters their function, typically in an inhibitory manner (Ju et al., 2006; Ju et al., 2004; Kraus, 2008; Krishnakumar and Kraus, 2010). For example, extensive PARylation of PARP-1 can inhibit its ability to bind nucleosomes (Kim et al., 2004; Wacker et al., 2007), while more modest PARylation of components of the TLE1 corepressor complex can promote its dissociation from target gene promoters (Ju et al., 2004). Although PARP-1 enzymatic activity has been shown to play a key role in the regulation of transcription in some contexts (Cohen-Armon et al., 2007; Ju et al., 2006; Ju et al., 2004; Krishnakumar and Kraus, 2010; Tulin and Spradling, 2003), in others it is dispensable (D’Amours et al., 1999; Hassa and Hottiger, 2002; Pavri et al., 2005). A clear consensus about the targets and role(s) of PARP-1 enzymatic activity during transcription has yet to emerge.
In addition to nucleosome-binding proteins, covalent posttranslational modifications of the amino-terminal tails of histone proteins, such as acetylation, phosphorylation, and methylation, can affect chromatin structure and function. These modifications can alter the charge of the histone tails and promote structural changes in nucleosomes, as well as create or destroy binding sites for chromatin-regulating proteins (Berger, 2007; Campos and Reinberg, 2009). Different histone modifications mark different functional regions of chromatin. For example, histone H3 lysine 4 trimethylation (H3K4me3) is enriched at TSSs and is positively correlated with gene expression (Ruthenburg et al., 2007; Santos-Rosa et al., 2002). This modification creates a binding site for structural modules (e.g., PHD fingers, chromodomains, and tudor domains) within a variety of proteins that regulate chromatin structure and transcription (Berger, 2007; Campos and Reinberg, 2009; Ruthenburg et al., 2007). Enzymes that remove the H3K4me3 mark, such as the KDM5 family of lysine-specific demethylases (e.g., KDM5B, a.k.a. JARID1B and PLU1), can reverse the actions of the proteins that bind the mark (Ruthenburg et al., 2007; Yamane et al., 2007).
Although recent studies have linked PARP-1 to the regulation of histone acetylation (Cohen-Armon et al., 2007), a clear understanding of how the chromatin-modulating effects of PARP-1 are coordinated with various covalent modifications to control transcriptional outcomes is lacking. In the present study, we have examined the mechanisms by which PARP-1 maintains an active chromatin environment at the promoters of target genes by exploring the sequence of events following depletion of PARP-1 that lead to transcriptional repression. We find that PARP-1 maintains H3K4me3 levels by PARylating and inhibiting the recruitment of the lysine-specific demethylase KDM5B. Collectively, our results elucidate a pathway by which the chromatin-modulating effects of PARP-1 are coordinated with a histone modification to regulate transcription.
Results
PARP-1 promotes the binding of RNA Pol II and components of the basal transcription machinery to the promoters of positively regulated target genes
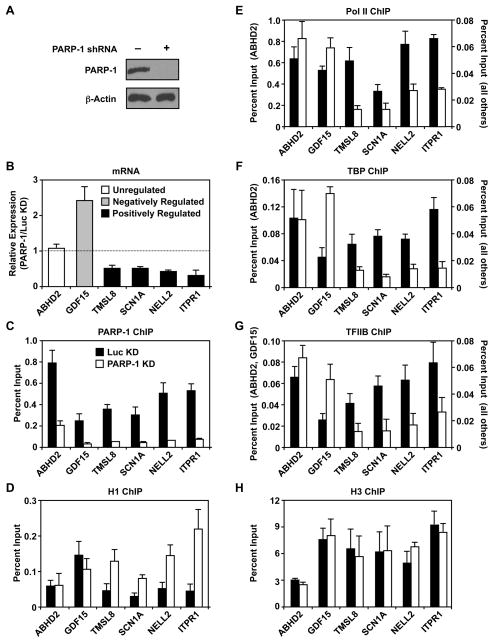
To better understand how PARP-1 modulates chromatin structure as a means of regulating gene expression, we conducted a series of molecular assays using MCF-7 human breast cancer cells. Given the growing interest in the role of PARP-1 in breast cancer biology (Frizzell and Kraus, 2009), as well as the considerable gene-specific and genomic data available regarding PARP-1 localization and function in MCF-7 cells (Krishnakumar and Kraus, 2010), this is an excellent model system to use. We used shRNA-mediated knockdown to explore PARP-1 function in these cells. As described previously (Frizzell et al., 2009; Krishnakumar et al., 2008), we see a robust knockdown of PARP-1 in this system compared to a control knockdown (luciferase; Luc) (Fig. 1A). We focused our studies on a set of previously characterized genes (i.e., TMSL8, SCN1A, NELL2, ITPR1; (Frizzell et al., 2009; Krishnakumar et al., 2008)) whose efficient expression requires PARP-1 (i.e., expression decreases by >50 percent upon PARP-1 knockdown; termed “positively regulated” genes in our shorthand; Fig. 1B). Genes in this set function in signaling, oncogenesis, and cell mobility (Table S1). For comparison, we also examined genes whose expression is inhibited by PARP-1 (i.e., expression increases >2-fold upon PARP-1 knockdown, such as GDF15; “negatively regulated”) or is unaffected by PARP-1 (e.g., ABHD2; “unregulated”) (Fig. 1B; Table S1).
Figure 1. PARP-1 promotes the binding of RNA Pol II and components of the basal transcription machinery to the promoters of positively regulated target genes.
(A) Western blot showing shRNA-mediated knockdown of PARP-1 with β-actin as an internal standard.
(B) Analysis of mRNA expression for six genes by RT-qPCR in MCF-7 cells with PARP-1 knockdown. The data are expressed relative to control (Luc) knockdown cells. Each bar represents the mean plus the SEM, n ≥ 3. The differences observed for all genes except ABHD2 are significant (Student’s t-test, p ≤ 0.05).
(C – H) ChIP-qPCR analyses of factor or histone levels at the promoters of the PARP-1 target genes from panel A in control (Luc) and PARP-1 knockdown (KD) cells. Each bar represents the mean plus the SEM, n ≥ 3. The differences observed for the positively regulated genes (i.e., TMSL8, SCN1A, NELL2, ITPR1) are significant for panels C – G. The differences observed for the negatively regulated gene (i.e., GDF15) are significant for panels C, F, and G (Student’s t-test, p ≤ 0.05).
(see Table S1)
In our initial studies, we examined the effects of PARP-1 depletion on the binding of the linker histone H1 and components of the Pol II transcription machinery at the promoters of these genes by using chromatin immunoprecipitation (ChIP) assays. As expected, knockdown of PARP-1 caused a dramatic reduction in PARP-1 ChIP signal for all genes tested (Fig. 1C). We have shown previously that H1 competes with PARP-1 for binding to promoter-proximal nucleosomes and represses gene transcription by Pol II (Kim et al., 2004; Krishnakumar et al., 2008). For the genes positively regulated by PARP-1, knockdown of PARP-1 caused an increase in H1 binding at the promoter (Fig. 1D) without a change in nucleosome density (as determined by histone H3 levels; Fig. 1H). This was accompanied by a significant reduction (>60%) in the promoter occupancy of TBP, TFIIB, and Pol II for all four genes in this group (Fig. 1, E – G). Opposite effects were observed for the negatively regulated gene GDF15 (i.e., the increase in gene expression upon PARP-1 knockdown was accompanied by an increase in the binding of TBP, TFIIB, and Pol II; Fig 1, E – G). As expected, ABHD2, a gene whose expression is not affected by PARP-1, showed no changes in these parameters upon PARP-1 knockdown (Fig 1, E – G). Taken together, these results indicate that PARP-1 modulates the occupancy of H1, TBP, TFIIB, and Pol II at target promoters to control gene expression.
PARP-1 maintains an open chromatin architecture at the TSSs of positively regulated target genes
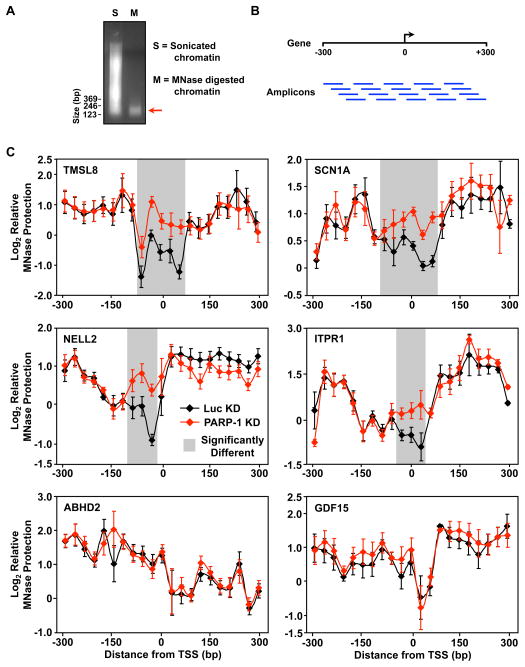
Previous studies have shown that PARP-1 can regulate chromatin structure, nucleosome compaction, and the accessibility of DNA in chromatin (Kim et al., 2004; Krishnakumar and Kraus, 2010; Wacker et al., 2007). Based on these results, as well as the data presented in Fig. 1, we hypothesized that PARP-1 might act to maintain an open chromatin architecture at the promoters of positively regulated target genes. To test this hypothesis, we examined the accessibility of promoter DNA surrounding the transcription start sites (TSSs; ca. −300 to +300 bp) to digestion by micrococcal nuclease (MNase). Mononucleosome-sized genomic DNA fragments (~160 bp; Fig. 2A) were released by MNase digestion from chromatin isolated from control and PARP-1 knockdown MCF-7 cells. We then used quantitative real-time PCR (qPCR) to tile through the promoter region with overlapping amplicons (21 amplicons for ~600 bp, ~30 bp overlap; Fig. 2B) to determine the extent of MNase digestion. As expected, for all genes we observed regions of increased MNase protection on either side of the TSS, likely due to −1 and +1 nucleosomes (Ozsolak et al., 2007; Schones et al., 2008) (Fig. 2C). In addition, we observed a dip in protection at or near the TSS, as described previously (Krishnakumar et al., 2008; Ozsolak et al., 2007; Schones et al., 2008). Knockdown of PARP-1 caused a significant increase in the MNase protection (i.e., decreased accessibility or a closed chromatin architecture) at or near the TSS for the positively regulated genes relative to the control knockdown (Fig. 2C; TMSL8, SCN1A, NELL2, ITPR1). This effect was not observed for GDF15 or ABHD2 (Fig. 2C). Together, the results from Figs. 1 and 2 are consistent with a model in which PARP-1 acts to maintain an open chromatin structure at the promoters of positively regulated target genes to allow loading of the transcriptional machinery and subsequent transcription by Pol II.
Figure 2. PARP-1 maintains an open chromatin architecture at the TSSs of positively regulated target genes.
(A) Agarose gel showing ethidium bromide-stained DNA from sonicated chromatin (S) or MNase digested chromatin (M) isolated from MCF-7 cells. Size markers are shown.
(B) Schematic of the qPCR amplicons used for the MNase protection assays. Overlapping amplicons were used to determine the enrichment of MNase digested DNA relative to sonicated genomic DNA at specific genomic locations spanning from −300 to +300 bp relative to the TSS for each gene indicated.
(C) MNase protection experiments in control (Luc) and PARP-1 knockdown (KD) MCF-7 cells. qPCR with overlapping amplicons (see Fig. 2B). Each point represents the mean ± the SEM, n ≥ 3. The shaded region indicates statistically significant differences between the Luc and PARP-1 knockdown conditions (Student’s T-test, p ≤ 0.05).
PARP-1 binding correlates with histone H3 lysine 4 trimethylation at many promoters across the genome
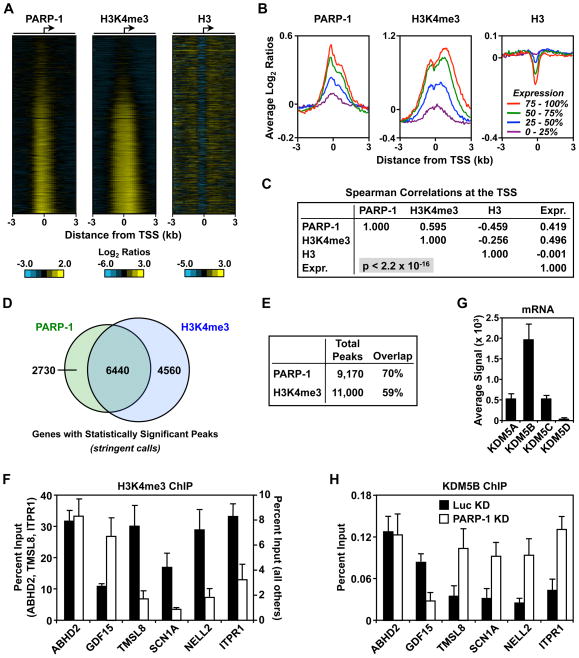
How might PARP-1 regulate promoter chromatin architecture? We considered the possibility of a mechanism involving histone modifications, which have been shown in many contexts to be correlated with specific chromatin states (Berger, 2007; Campos and Reinberg, 2009). We focused on histone H3 lysine 4 trimethylation (H3K4me3), which localizes to promoters and positively correlates with gene expression (Barski et al., 2007; Ruthenburg et al., 2007; Santos-Rosa et al., 2002). We used ChIP coupled with hybridization to RefSeq promoter arrays (i.e., ChIP-chip) to determine if the binding of PARP-1 to promoters might correlate with H3K4me3 at promoters across the genome. Indeed, the pattern of PARP-1 promoter localization that we have observed previously (Krishnakumar et al., 2008) was similar to the pattern of H3K4me3 in MCF-7 cells (Fig. 3, A and B). The levels of PARP-1 and H3K4me3 were positively correlated with each other (Spearman’s correlation coefficient 0.60, p < 2.2 × 10−16), and both were positively correlated with gene expression, as reported previously (Spearman’s correlation coefficients of 0.42 and 0.50, respectively, p < 2.2 × 10−16 for both) (Fig. 3, B and C) (Barski et al., 2007; Krishnakumar et al., 2008). We found statistically significant peaks of both PARP-1 and H3K4me3 at or near about one third of the ~23,500 TSSs on the array using stringent peak finding criteria (Fig. 3, D and E). The strong correlation of PARP-1 and H3K4me3 suggests a functional link between the two. We explored this possibility in the experiments noted below.
Figure 3. PARP-1 binding correlates with histone H3 lysine 4 trimethylation at promoters across the genome.
ChIP-chip analysis of PARP-1, H3K4me3, and H3 at ~23,500 TSSs (−3 kb to +3 kb).
(A) Heatmaps showing a visual representation of the ChIP-chip data.
(B) Average PARP-1, H3K4me3, and H3 ChIP-chip signals across promoters based on expression quartile (0 – 25% = lowest expression; 75 – 100% = highest expression).
(C) Multiple correlation analysis for PARP-1, H3K4me3, and H3 ChIP-chip signals at the TSS. The Spearman correlation coefficients and the p-value are shown.
(D and E) The total and overlapping peaks of PARP-1 and H3K4me3 are shown in a Venn diagram (panel D) and a table (panel E).
(F and H) ChIP-qPCR analysis of H3K4me3 and KDM5B levels at target gene promoters in control (Luc) and PARP-1 knockdown (KD) MCF-7 cells. Each bar represents the mean plus the SEM, n ≥ 3. The differences observed for GDF15, TMSL8, SCN1A, NELL2, ITPR1 are significant (Student’s t-test, p ≤ 0.05).
(G) KDM5B is the most highly expressed KDM5 isoform in MCF-7 cells. Average signal intensities for KDM5 isoforms from three different Affymetrix expression arrays. Each bar represents the mean plus the SEM.
PARP-1 prevents KDM5B-dependent demethylation of H3K4me3 at the promoters of positively regulated target genes
To determine if PARP-1 might play an active role in establishing or maintaining H3K4me3 at target gene promoters, we examined the effect of PARP-1 knockdown on H3K4me3 levels. For the positively regulated genes, H3K4me3 levels decreased by more than 65 percent upon PARP-1 knockdown (Fig. 3F). For the negatively regulated gene, GDF15, the opposite effect was observed, and for the unregulated gene, ABHD2, no effect was observed (Fig. 3F). These results correlate well with the effects of PARP-1 knockdown on Pol II, TBP, and TFIIB binding noted above (Fig. 1). Interestingly, TAF3 (a subunit of the TBP-containing complex TFIID) has been shown to bind to H3K4me3 (Vermeulen et al., 2007). Thus, the PARP-1-dependent reduction in H3K4me3 may reduce TBP binding, leading to a reduction in Pol II occupancy at the promoter.
We considered the possibility that PARP-1 might regulate promoter H3K4me3 levels by recruiting a histone methyltransferase or by blocking the recruitment of a histone demethylase. A number of methyltransferases in the TRX/MLL family can trimethylate H3K4 in mammalian cells (Berger, 2007), while the KDM5 demethylases (A – D) are the only enzymes known to remove the modification (Nottke et al., 2009; Ruthenburg et al., 2007). Of the four KDM5 isoforms, KDM5B (lysine demethylase 5B; a.k.a. PLU-1 or JARID1B) is the most highly expressed in MCF-7 cells (Fig. 3G). Thus, in our studies of PARP-1 function in MCF-7 cells, we focused on KDM5B. Knockdown of PARP-1 affected KDM5B binding at the promoters of the six genes tested in a predictable manner that precisely matched its effects on H3K4me3 levels (Fig. 3H). For example, PARP-1 knockdown increased KDM5B levels and decreased H3K4me3 levels at the promoters of the positively regulated genes, but not the negatively regulated or unregulated genes. These results suggest that PARP-1 prevents demethylation of H3K4me3 by KDM5B at the promoters of positively regulated target genes.
Antagonism of KDM5B-dependent H3K4me3 demethylation by PARP-1 facilitates the expression of positively regulated target genes
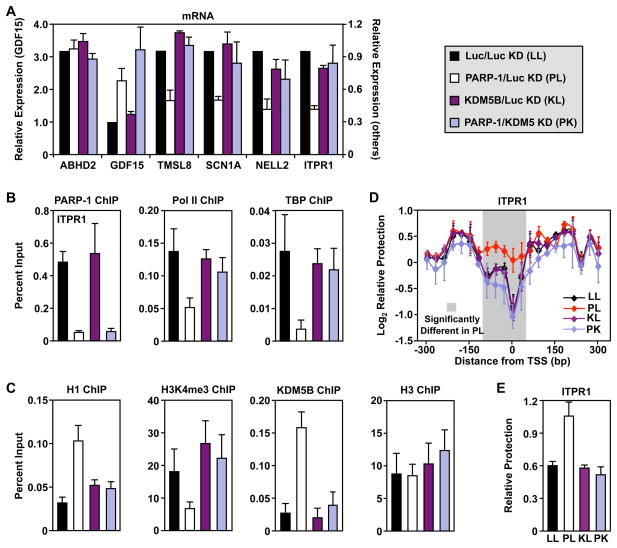
To further explore the role of PARP-1-dependent antagonism of KDM5B in target gene expression, we examined the effects of KDM5B knockdown using an shRNA sequence. According to our model, a key role of PARP-1 is to prevent KDM5B-dependent demethylation of H3K4me3; shRNA-mediated knockdown of PARP-1 allows demethylation of H3K4me3 by KDM5B and subsequent inhibition of transcription of the positively regulated genes. If this model is correct, then depletion of KDM5B should reverse the effects of PARP-1 knockdown. To test this model, we generated a set of cell lines with knockdown of PARP-1 + Luc (PL), KDM5B + Luc (KL), or PARP-1 + KDM5B (PK), matched with an appropriate control cell line expressing two shRNAs targeting luciferase (LL). We confirmed the knockdown of PARP-1 and KDM5B by Western blotting and RT-qPCR (Fig. S1, A and B). In addition, we showed that knockdown of KDM5B reduced its signal in ChIP assays (Fig. S3C). Although KDM5B knockdown alone had little effect on the expression of positively regulated target genes, it reversed the effect of PARP-1 knockdown on target gene mRNA levels (Fig. 4A). In contrast, for the negatively regulated gene, GDF15, knockdown of KDM5B did not reverse the effect of PARP-1 knockdown, suggesting a different mode of regulation (Fig. 4A). These results indicate that a loss of PARP-1 at the promoters of positively regulated target genes can be overcome by removing KDM5B, consistent with the hypothesis that these two proteins act antagonistically in the same pathway.
Figure 4. Antagonism of KDM5B-dependent H3K4me3 demethylation by PARP-1 promotes a transcriptionally permissive environment at the promoters of positively regulated target genes.
The experiments in this figure used MCF-7 cells with knockdown of PARP-1 + Luc (PL), KDM5B + Luc (KL), or PARP-1 + KDM5B (PK), matched with an appropriate control cell line expressing two shRNAs targeting luciferase (LL).
(A) Analysis of mRNA expression for six genes by RT-qPCR in PARP-1 and KDM5B knockdown MCF-7 cells. The data are expressed relative to LL cells. Each bar represents the mean plus the SEM, n ≥ 3. The differences observed for PL relative to LL are significant for all genes except ABHD2 (ANOVA, p ≤ 0.05).
(B and C) ChIP-qPCR analysis of factor, histone, and histone modification levels at the promoter of the PARP-1 target gene, ITPR1, in MCF-7 cells with PARP-1 and KDM5B knockdown, as indicated. Each bar represents the mean plus the SEM, n ≥ 3. The differences observed for PL relative to LL are significant for all ChIPs except H3 (ANOVA, p ≤ 0.05).
(D) MNase protection experiments in MCF-7 cells with PARP-1 and KDM5B knockdown, as indicated, for ITPR1. Each point represents the mean ± the SEM, n ≥ 3. The shaded region indicates statistically significant differences between the LL and PL cells (Student’s t-test, p ≤ 0.05).
(E) MNase protection data at the TSS from panel D, above. Each bar represents the mean plus the SEM, n ≥ 3. The differences observed for PL relative to LL are significant (ANOVA, p ≤ 0.05).
(see Fig. S1)
Next, to determine the consequences of PARP-1 antagonism of KDM5B, we assayed factor binding (Pol II, TBP, H1) and H3K4 trimethylation at the promoters of the PARP-1 target genes in the double knockdown cells. The results for the positively regulated gene, ITPR1, are shown in Figs. 4B and 4C for illustrative purposes, while the results for additional genes are shown in Fig. S1, D – I. As shown above, knockdown of PARP-1 decreased the binding of Pol II and TBP, as well as the levels of H3K4me3, at the promoters of the positively regulated genes (Fig. 4, B and C; Fig. S1, E, F, and H), while increasing the binding of H1 (Fig. 4C; Fig. S1G). As we observed with gene expression, knockdown of KDM5B had little effect on its own, but was able to reverse all of the effects of PARP-1 knockdown on the positively regulated genes (Pol II, TBP, H1, H3K4me3; Fig. 4, B and C; Fig. S1, D – I). These results indicate that removal of H3K4me3 is required for the binding of H1 and the eviction of Pol II from the promoters of positively regulated target genes. These data support a model in which antagonism of KDM5B binding and H3K4me3 demethylation by PARP-1 facilitates the expression of positively regulated target genes.
Antagonism of KDM5B-dependent H3K4me3 demethylation by PARP-1 maintains an open chromatin architecture at the TSSs of positively regulated target genes
In Fig. 2C, we showed that PARP-1 is required to maintain an open chromatin architecture at the TSSs of positively regulated genes. To determine if PARP-1’s antagonism of KDM5B and enhancement of H3K4 trimethylation levels plays a role in this process, we used the MNase protection assay described in Fig. 2 with PARP-1 and KDM5B double knockdown cells. As with the gene expression outcomes described above, if our model is correct, then depletion of KDM5B should reverse the effects of PARP-1 knockdown. As described previously (Fig. 2C), PARP-1 knockdown caused a significant increase in the MNase protection (i.e., decreased accessibility or a closed chromatin architecture) relative to the control knockdown near the TSSs of the positively regulated genes ITPR1 (Fig. 4, D and E) and SCN1A (Fig. S1J), but not for the unregulated gene ABHD2 (Fig. S1K). Although KDM5B knockdown alone had little effect on the promoter chromatin architecture of ITPR1 and SCN1A, it reversed the effect of PARP-1 knockdown on these genes (Fig. 4, D and E; Fig. S1J). These results indicate that PARP-1 maintains an open chromatin architecture at the TSSs of positively regulated genes by inhibiting KDM5B-mediated demethylation of H3K4me3.
PARP-1 catalytic activity is required to prevent the binding of KDM5B at the promoters of positively regulated target genes
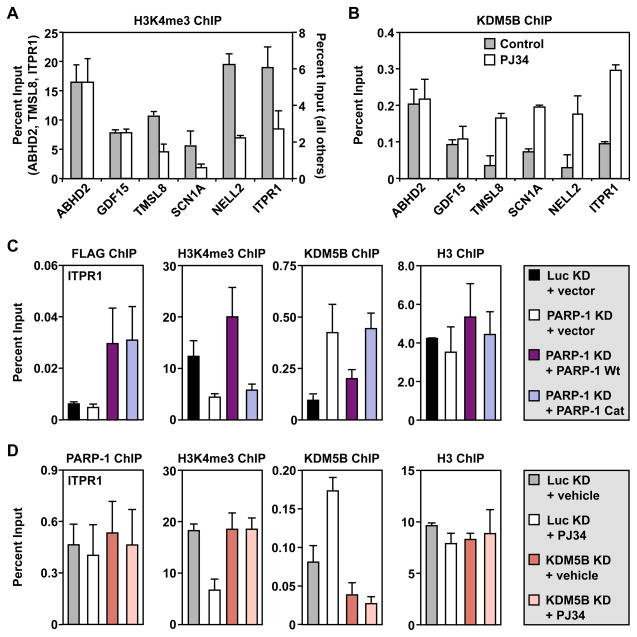
Previous studies from our lab and others have shown that PARP-1 catalytic activity is required for the PARP-1-dependent expression of some, but not all, target genes (D’Amours et al., 1999; Frizzell et al., 2009; Hassa and Hottiger, 2002; Ju et al., 2006; Krishnakumar and Kraus, 2010; Pavri et al., 2005; Tulin and Spradling, 2003). In some cases, PARP-1 catalytic activity may be required for one or more steps in the gene regulatory pathway, while the PARP-1 protein itself may be required for other steps. To test this possibility, we determined the effects of the PARP inhibitor, PJ34, on the same target genes and functional endpoints that we examined above. We have shown previously that PJ34 effectively inhibits PARP-1 enzymatic activity in MCF-7 cells (Frizzell et al., 2009). Interestingly, PJ34 had no effect on gene expression or the binding of PARP-1, H1, or Pol II at the promoters of the genes examined (Fig. S2, A – F). These results indicate that, unlike depletion of PARP-1 protein, inhibition of PARP-1 catalytic activity is not sufficient to inhibit the expression of the positively regulated genes. This suggests that the physical presence of the PARP-1 protein at the promoter is key for maintaining gene expression. However, PJ34 did reduce the levels of H3K4me3 and increase the levels of KDM5B at the same promoters (Fig. 5, A and B). Thus, of the various endpoints examined, PARP-1 catalytic activity is specifically directed at preventing demethylation of H3K4me3 by KDM5B.
Figure 5. The enzymatic activity of PARP-1 is required to antagonize the actions of KDM5B at target gene promoters.
(A and B) ChIP-qPCR analysis of H3K4me3 and KDM5B levels at target gene promoters in MCF-7 cells treated with or without 5 μM PJ34 for 1.5 hours before collection. Each bar represents the mean plus the SEM, n ≥ 3. The differences observed for TMSL8, SCN1A, NELL2, and ITPR1 are significant (Student’s t-test, p ≤ 0.05).
(C and D) ChIP-qPCR analysis of H3K4me3, KDM5B, H3, or ectopically expressed PARP-1 (FLAG) levels at the promoter of the PARP-1 target gene, ITPR1, in MCF-7 cells with or without: (1) PARP-1 knockdown (KD) or PARP-1 KD + ectopic expression of wild-type (Wt) or catalytically inactive (Cat) FLAG-tagged RNAi-resistant PARP-1 or (2) KDM5B KD or KDM5B KD + 5 μM PJ34 treatment for 1.5 hours before collection, as indicated. Each bar represents the mean plus the SEM, n ≥ 3. The differences observed for H3K4me3 and KDM5B are significant (ANOVA, p ≤ 0.05).
(see Fig. S2)
To explore this possibility in more detail, we determined if a catalytically inactive mutant of PARP-1 (E988K) could restore PARP-1 function in knockdown cells. We used previously described Luc or PARP-1 knockdown MCF-7 cells with or without ectopic expression of RNAi-resistant FLAG-tagged wild-type (Wt) or catalytically inactive PARP-1 (Cat) (Frizzell et al., 2009). The results for the positively regulated gene, ITPR1, are shown in Fig. 5C for illustrative purposes, while the results for additional genes are shown in Fig. S2, G – J. ChIP with FLAG antibody showed that both ectopically expressed Wt and Cat PARP-1 can bind to the promoter regions of target genes (Fig. 5C; Fig. S2J). Only Wt PARP-1, however, was able to inhibit KDM5B binding and restore H3K4me3 levels (Fig. 5C; Fig. S2, G and H). These results indicate that PARP-1 catalytic activity is required to prevent the binding of KDM5B at the promoters of target genes, in agreement with our results using PJ34. In this regard, knockdown of KDM5B reversed the effects of PJ34 treatment on H3K4me3 levels (Fig. 5D; Fig. S2, K and L), indicating that PJ34 (and PARP-1 catalytic activity) acts through a KDM5B-dependent pathway.
PARP-1 binds to, PARylates, and inhibits the histone demethylase activity of KDM5B
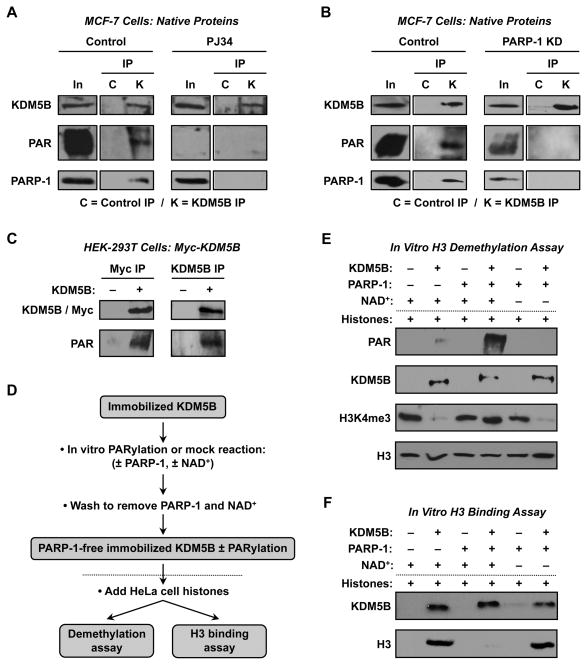
Based on the preceding results, we hypothesized that PARP-1 might interact with and poly(ADP-ribosyl)ate (PARylate) KDM5B in cells, thus preventing KDM5B from binding to promoters. To test this, we immunoprecipitated KDM5B from MCF-7 nuclear extracts and subjected the immunoprecipitated material to Western blotting using antibodies to KDM5B, PARP-1, and PAR. We found that native KDM5B immunoprecipitates native PARP-1 and is modified by PARylation (Fig 6, A and B). Treatment of the cells with PJ34 (Fig. 6A) and knockdown of PARP-1 (Fig. 6B) inhibited KDM5B PARylation, indicating that PARP-1 is the enzyme mediating the modification. The specificity of the PARylation of KDM5B was confirmed in HEK-293T cells with ectopic expression of Myc-KDM5B (Fig. 6C).
Figure 6. PARP-1 binds to, PARylates, and inhibits the histone demethylase activity of KDM5B.
(A and B) PARP-1 binds to and PARylates KDM5B in MCF-7 cells in the absence, but not the presence, of 5 μM PJ34 for 1.5 hours before collection (panel A), and in control knockdown (Luc), but not PARP-1 knockdown, cells (panel B). KDM5B was immunoprecipitated from nuclear extracts and the immunoprecipitated material (IP) was subjected to Western blotting using antibodies to KDM5B, PARP-1, and PAR. C, IP with non-specific control antibody; K, IP with KDM5B antibody; In, input.
(C) Myc-tagged KDM5B ectopically expressed in HEK-293T cells is PARylated. IP-Western blotting experiments with Myc IP/KDM5B Western and KDM5B IP/Myc Western as shown. The immunoprecipitated material was also subjected to Western blotting for PAR.
(D) Schematic diagram of the in vitro H3 demethylation and H3 binding assays shown in (E) and (F). Dotted line separating the PARylation and demethylation/binding reactions corresponds to the dotted lines in (E) and (F).
(E) NAD+-dependent PARylation of KDM5B by PARP-1 in vitro inhibits KDM5B histone demethylase activity. Reactions were set up as indicated using PARylated or unPARylated Myc-KDM5B immunoprecipitated from HEK-293T cells (or control IP material) and core histones from HeLa cells. The reactions were then subjected to Western blotting as indicated.
(F) NAD+-dependent PARylation of KDM5B by PARP-1 in vitro inhibits KDM5B histone H3 binding activity. Reactions were set up as in (E) with KDM5B immobilized on protein G resin. The bound material was washed and subjected to Western blotting as indicated. (see Fig. S3)
To determine the effect of PARylation on KDM5B histone demethylase activity, we set up an in vitro PARylation-coupled demethylation assay using recombinant KDM5B - either full-length Myc-KDM5B immunoprecipitated from transfected HEK-293T cells or an enzymatically amino-terminal fragment purified from bacteria, KDM5B-N. Immobilized KDM5B was subjected to PARylation (or mock PARylation) using recombinant PARP-1 and NAD+ under conditions described previously (Kim et al., 2004). The resin was then washed to remove the PARP-1 and NAD+, leaving immobilized PARP-1-free PARylated or unPARylated KDM5B (Fig. 6D). The immobilized KDM5B was used in H3 demethylation assays with native HeLa cell histones, which show readily detectable levels of H3K4me3 by Western blotting. As expected, the addition of Myc-KDM5B or KDM5B-N caused the demethylation of H3K4me3 (Fig. 6E; Fig. S3A). The demethylation was inhibited by the PARylation of KDM5B (Fig. 6E; Fig. S3A). Further examination using an in vitro histone H3 binding assay set up in parallel under the same conditions assay showed that this result is due, at least in part, to the inhibition of H3 binding by KDM5B (Fig. 6F; Fig. S3B). Taken together, these results indicate that PARP-1 binds to, PARylates, and inhibits the histone demethylase activity of KDM5B. These effects of PARP-1 on KDM5B are likely to underlie PARP-1’s ability to prevent the demethylation of H3K4me3 at the promoters of positively regulated target genes.
Removal of PARP-1 from promoters is a mechanism utilized by signaling pathways to negatively regulate gene expression
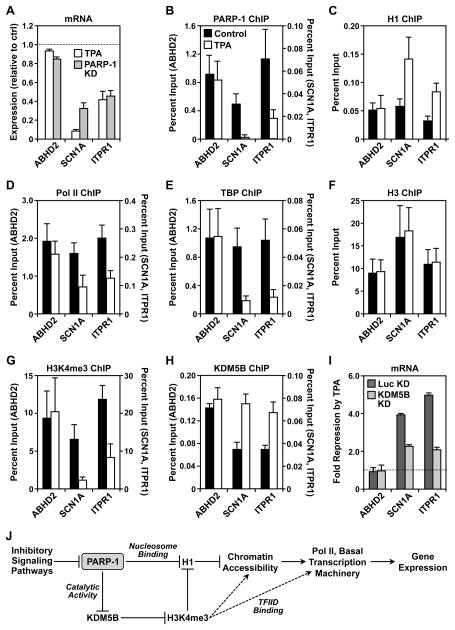
Having determined a specific role for PARP-1 in maintaining constitutive gene expression in MCF-7 cells under basal growth conditions, we next asked whether the PARP-1-dependent regulatory mechanisms which we uncovered might be applicable to signal-regulated transcription. MCF-7 cells initiate well characterized transcriptional responses to a wide variety of stimuli, including the phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA), a potent activator of the protein kinase C pathway. Many genes in MCF-7 cells are responsive to TPA treatment (e.g., (Cunliffe et al., 2003)), including SCN1A and ITPR1 (Fig. 6A). The expression of these two genes is inhibited by treatment with TPA (100 ng/ml TPA for 3 hours), an effect that is similar to PARP-1 knockdown (Fig. 7A). The expression of ABHD2, which we examined for comparison, was unaffected by TPA or PARP-1 knockdown (Fig, 7A).
Figure 7. Signaling in the PKC pathway promotes the removal of PARP-1 from promoters to negatively regulate gene expression.
(A) Analysis of mRNA expression for three genes by RT-qPCR in MCF-7 cells treated with 100 ng/ml TPA for 3 hours versus MCF-7 cells with PARP-1 knockdown. The data are expressed relative to untreated LL cells. Each bar represents the mean plus the SEM, n ≥ 3. The differences observed for all genes except ABHD2 are significant (Student’s T-test, p ≤ 0.05).
(B – H) ChIP-qPCR analysis of factor, histone, or histone modification levels at the promoters of three genes, as indicated, in MCF-7 cells with or without 100 ng/ml TPA treatment for 3 hours. Each bar represents the mean plus the SEM, n ≥ 3. The differences observed with TPA are significant for SCN1A and ITPR1 for panels C, D, E, G, and H (Student’s T-test, p ≤ 0.05).
(I) Analysis of mRNA expression for three genes by RT-qPCR in MCF-7 cells with KDM5B knockdown. The data are expressed relative to untreated LL cells. Each bar represents the mean plus the SEM, n ≥ 3. The differences observed for SCN1A and ITPR1 are significant (ANOVA, p ≤ 0.05).
(J) Model of PARP-1-dependent gene regulation derived from the data shown herein and the literature. Additional details are provided in the Discussion.
(see Fig. S4).
We considered the possibility that TPA-dependent signaling might inhibit the expression of SCN1A and ITPR1 by blocking the localization and/or function of PARP-1 at their promoters. Treatment with TPA promoted the release of PARP-1 from these promoters (Fig. 7B), perhaps as a direct target endpoint of the signaling pathway. Given that TPA promoted the release of PARP-1 from the SCN1A and ITPR1 promoters, we expected that TPA might promote the same molecular outcomes as PARP-1 knockdown at these promoters. In this regard, we observed an increase in H1 binding (Fig. 7C), a decrease in Pol II and TBP binding (Fig. 7, D and E), and a decrease in the levels of H3K4me3 (Fig. 7, F and G) upon TPA treatment. Thus, for these endpoints, TPA treatment and PARP-1 knockdown are functionally similar.
To determine if KDM5B might be involved in this pathway as well, we monitored H3K4me3 and KDM5B binding at the promoters in response to TPA treatment. As with PARP-1 knockdown, TPA treatment caused a decrease in H3K4me3 levels and an increase in KDM5B binding to the promoters (Fig. 7, G and H). To directly link KDM5B to these TPA-mediated effects, we examined the set of functional endpoints in MCF-7 cells subjected to knockdown of KDM5B. We found that knockdown of KDM5B inhibited TPA-mediated repression of SCN1A and ITPR1 (Fig. 7I). In addition, we found that knockdown of KDM5B inhibited TPA-mediated effects on Pol II, H1, and H3K4me3 (Fig. S4; compare the Luc KD + TPA versus the KDM5B KD + TPA conditions). Together, these results indicate that cellular signaling pathways, like those mediated by TPA, can regulate gene expression by abrogating PARP-1 binding at the promoter, which in turn allows H1 binding, KDM5B binding, and the removal of H3K4me3.
Discussion
In this study, we examined the functional interplay between PARP-1, H3K4 trimethylation, and the linker histone H1. We found that PARP-1 is required for a series of molecular outcomes at the promoters of genes whose expression is dependent on PARP-1 (“positively regulated” genes). Specifically, our experiments using shRNA-mediated knockdown show that PARP-1 establishes a permissive chromatin environment at the promoters of these genes by preventing the demethylation of H3K4me3 through the PARylation, inhibition, and exclusion of KDM5B, while promoting the exclusion of H1 and increased accessibility of the promoter DNA at the TSS. Upon depletion of PARP-1, these outcomes do not occur efficiently. The permissive chromatin environment is required for the efficient loading of the Pol II transcription machinery (e.g., Pol II, TBP, TFIIB) and subsequent transcription (Fig. 7). Finally, our results indicate that cellular signaling pathways use the regulated depletion of PARP-1 from promoters to inhibit gene expression.
PARP-1 prevents demethylation of H3K4me3 at the promoters of positively regulated genes by antagonizing KDM5B
PARP-1 has long been known to regulate chromatin structure, yet the underlying mechanisms and targets have not been clearly determined (Krishnakumar and Kraus, 2010). Our results indicate that PARP-1 regulates chromatin in at least two ways. First, it regulates the covalent posttranslational modification of chromatin by preventing demethylation of H3K4me3, a histone modification that is positively correlated with gene expression (Ruthenburg et al., 2007). Second, it regulates the composition of chromatin by promoting the exclusion of H1, a nucleosome-binding protein associated with gene repression (Happel and Doenecke, 2009).
With respect to the former, we have shown herein that PARP-1 promotes the exclusion of the H3K4me3 demethylase KDM5B from promoter chromatin (Fig. 3H) through a mechanism that requires PARP-1 catalytic activity (Fig. 5 and Fig. S2, G – M). As such, PARP-1 is able maintain the levels of H3K4me3 elevated at the promoters of positively regulated genes, which helps to maintain an active chromatin environment. The intimate interplay between PARP-1 and KDM5B is illustrated by our studies showing that (1) PARP-1 and H3K4me3 colocalize on many promoters across the genome (Fig. 3, A – E) and (2) the requirement for PARP-1 to maintain a variety of molecular outcomes at the promoters of positively regulated target genes (e.g., Pol II and TBP binding, H1 exclusion, transcription) is abrogated upon KDM5B knockdown (Fig. 4; Fig. S1, D – I). In addition, our data suggest that while H3K4me3 demethylation by KDM5B is necessary for reducing gene expression, it is not sufficient, and that PARP-1 protein must also be physically removed from the promoter to allow for H1 binding and subsequent changes in chromatin structure.
How does PARP-1 exclude KDM5B and prevent KDM5B-dependent H3K4me3 demethylation? Our results indicate that PARP-1 binds to and PARylates KDM5B (Figs. 6, A – C), which inhibits the H3K4me3 demethylase activity of KDM5B (Fig. 6E; Fig. S3A) and blocks its binding to promoter chromatin (Fig. 5, B and C). PARP-1-mediated PARylation has been shown to be an effective mechanism for inhibiting the activity of transcription-related proteins due to its ability to block protein-protein or protein-DNA interactions (Krishnakumar and Kraus, 2010). In this regard, we found that PARylation of KDM5B inhibits its ability to bind H3 (Fig. 6F; Fig. S3B). Previous studies have implicated PARP-1 enzymatic activity in the regulation of factor binding or exchange at target promoters (Ju et al., 2006; Ju et al., 2004). In this regard, PARP-1’s effects on KDM5B might be part of a broader process designed to establish a set of histone modifications permissive to transcription (i.e., regulating the binding of enzymes that elevate H3K4me3 and H3/H4 acetylation, or reduced H3K27me3).
What role might H3K4me3 play in the PARP-1-dependent transcription process? Previous studies have shown that this modification creates a binding site for structural modules within a variety of proteins that regulate chromatin structure and transcription (Ruthenburg et al., 2007), including the PHD finger of TAF3, a component of TFIID (Vermeulen et al., 2007). The binding of H3K4me3 by TAF3 has been shown to direct the binding of TFIID at promoters (Vermeulen et al., 2007). In this regard, we found that depletion of PARP-1, which reduces the levels of H3K4me3 at positively regulated promoters, also reduces the binding of TBP, another component of TFIID (Fig. 1F). These effects of PARP-1 on TBP binding are antagonized by KDM5B, and the requirement for PARP-1 to maintain TBP levels is abrogated upon knockdown of KDM5B (Fig. 4B; Fig. S1F). Thus, one aspect of PARP-1-dependent maintenance of H3K3 trimethylation may be the promotion of TFIID binding (Fig. 7J).
PARP-1 promotes the exclusion of H1 and the formation of open chromatin structures at the promoters of positively regulated genes
In addition to preventing H3K4me3 demethylation by promoting the exclusion of KDM5B, PARP-1 antagonizes the binding of H1 at the promoters of positively regulated target genes (Figs. 1D and 7J), likely through competition for overlapping binding sites on nucleosomes (Kim et al., 2004). PARP-1 catalytic activity is not required for this effect (Fig. S2C), consistent with the results of our revious biochemical assays showing that PARP-1 catalytic activity is not required for the displacement of H1 from nucleosomes (Kim et al., 2004). These results are also consistent with our genomic assays showing that PARP-1 and H1 exhibit a reciprocal pattern of binding at promoters across the genome (i.e., H1 is depleted where PARP-1 is enriched) (Krishnakumar et al., 2008). Our current results, however, suggest that additional mechanisms may also contribute to the PARP-1-dependent exclusion of H1 from promoter chromatin. For example, the effects of PARP-1 knockdown on H1 binding are abrogated upon KDM5B knockdown (Fig. 4C; Fig. S1G), suggesting a role for KDM5B. The actions of KDM5B in this PARP-1-dependent process may be through demethylation of H3K4me3 (Fig. 7), or perhaps demethylation of H1 or another chromatin- and transcription-related factor.
A key consequence of PARP-1-dependent antagonism of H1 binding is the maintenance of an open chromatin architecture at the TSSs of positively regulated genes. Previous studies have shown that H1 binding to nucleosomes can promote the formation of a closed chromatin architecture that is repressive to transcription (Happel and Doenecke, 2009). In such cases, removal of H1 is required for efficient transcription. In this regard, we found that PARP-1-dependent antagonism of H1 binding correlates with an open chromatin architecture (Fig. 2C) and the binding of the Pol II transcription machinery (Pol II, TBP, and TFIIB; Fig. 1, E, F and G). Our studies suggest a scenario in which PARP-1 acts upstream in an ordered series of events including the removal of H1, opening of the promoter chromatin architecture, and binding of the Pol II transcription machinery (Fig. 7J). This is consistent with models proposed in the literature (Campos and Reinberg, 2009; Li et al., 2007).
Requirement for PARP-1 catalytic activity at the promoters of positively regulated target genes
The role of PARP-1 enzymatic activity in transcriptional regulation has been studied in various gene contexts with different transcriptional activators (D’Amours et al., 1999; Frizzell et al., 2009; Krishnakumar and Kraus, 2010). In some cases, PARP-1 enzymatic activity was found to be required (e.g., with HES1 and Elk1) (Cohen-Armon et al., 2007; Ju et al., 2004), while in others it was not (e.g., NF-κB and retinoic acid receptor) (Hassa and Hottiger, 2002; Pavri et al., 2005). We found that PARP-1 enzymatic activity is not solely responsible for PARP-1-dependent effects on the transcription of positively regulated target genes (Fig. S2A), but it is required for some of the specific molecular outcomes that we tested. For example, PARP-1 enzymatic activity is required for the prevention of H3K4me3 demethylation through inhibition of KDM5B binding (Fig. 5; Fig. S2, G – M), while it is dispensable for the inhibition of H1, Pol II, and TBP binding (Supplemental Fig. S2, C, D and F).
Why might the increased KDM5B binding observed upon PARP-1 knockdown (Fig. 3H), which inhibits the transcription of positively regulated target genes (Fig. 1B), have a different effect than the increased KDM5B binding observed upon PARP-1 inhibition with PJ34 (Fig. 5B), which does not inhibit transcription of those genes (Supplemental Fig. S2A)? The answer is likely that with PARP-1 knockdown, the PARP-1 protein is removed, while with PJ34, it is not. Although inhibition of PARP-1 enzymatic activity might allow certain steps in the pathway involving KDM5B, the removal of the PARP-1 protein itself is required to allow the binding of H1 and subsequent steps (e.g., promoting a closed chromatin architecture, inhibiting Pol II occupancy, repressing transcription) (Fig. 7J). Although the specific molecular role(s) played by PARP-1 enzymatic activity in the transcription process have been difficult to sort out (Cohen-Armon et al., 2007; Frizzell et al., 2009; Hassa and Hottiger, 2002; Ju et al., 2006; Ju et al., 2004; Pavri et al., 2005), our results have helped to discern distinct molecular roles played by PARP-1 protein and PARP-1 enzymatic activity.
Cellular signaling pathways regulate PARP-1-dependent molecular outcomes at target gene promoters
PARP-1 has been implicated in a number of different signal-dependent gene regulatory pathways, including those mediated by activation of PKC, CaM kinase IIδ, estrogen signaling, retinoic acid signaling, or NF-κB-dependent proinflammatory signaling (Hassa and Hottiger, 2002; Ju et al., 2006; Ju et al., 2004; Krishnakumar and Kraus, 2010; Pavri et al., 2005). We found that signaling in the PKC pathway can inhibit the expression of a subset of genes positively regulated by PARP-1, including SCN1A and ITPR1 (Fig. 7A; Table S1). Our molecular analyses indicate that TPA promotes the loss of PARP-1 from the promoters of these genes (Fig. 7B). Like shRNA-mediated knockdown of PARP-1, the TPA-dependent reduction of PARP-1 binding at these promoters leads to an increase in the binding of H1 and KDM5B (Fig. 7, C and H), as well as a reduction in the binding of Pol II and TBP (Fig. 7, D and E), and the levels of H3K4me3 (Fig. 7G). Thus, modulation of PARP-1 binding to chromatin as an endpoint of cellular signaling pathways can provide a means of regulating gene expression programs important for cellular outcomes. These outcomes may play key roles in diseases such as breast cancer, a condition for which both PARP-1 and KDM5B have been implicated
Collectively, our results help to elucidate the mechanisms by which PARP-1 regulates chromatin structure and transcription. More broadly, our results help to clarify the molecular mechanisms by which specific chromatin-binding and histone-modifying proteins interact to alter chromatin structure and function to regulate gene transcription.
Experimental Procedures
Additional details about the experimental procedures can be found in the Supplemental Materials.
Cells lines and shRNA-mediated knockdown
MCF-7 cells were maintained and propagated as described previously (Frizzell et al., 2009). shRNA-mediated knockdown of PARP-1 and KDM5B using stable retrovirus-mediated gene transfer was performed as described previously (Frizzell et al., 2009). Control-matched cells expressing shRNAs targeting luciferase (Luc) were generated in parallel. Retrovirus infected cells were selected and maintained under appropriate drug conditions (0.5 μg/ml puromycin and 800 μg/ml G418). PARP-1 knockdown MCF-7 cells with ectopic re-expression of FLAG-tagged wild-type or catalytically inactive PARP-1 have been described previously (Frizzell et al., 2009)
mRNA expression analyses
Total RNA was isolated from cells at about 80% confluence using Trizol Reagent (Invitrogen), reverse transcribed, and subjected to real-time quantitative PCR (qPCR) using gene-specific primers. All target gene transcripts were normalized to the β-actin transcript.
Chromatin immunoprecipitation (ChIP) assays
ChIP-qPCR was performed as described previously (Krishnakumar et al., 2008). For ChIP assays with the KDM5B antibody, we included an additional crosslinking step with 10 mM dimethyl suberimidate (Pierce) for 10 min. prior to formaldehyde crosslinking.
Micrococcal nuclease (MNase) protection assays
Nuclei were prepared from formaldehyde-crosslinked cells and chromatin was isolated. The chromatin samples were divided into two aliquots, one of which was digested with MNase to yield mononucleosomes and the other was lightly sonicated. Both samples were then incubated at 65°C overnight to reverse the crosslinks. The DNA was cleared of protein and residual RNA by digestion with proteinase K and RNase H, respectively. The enrichment of MNase digested DNA relative to sonicated genomic DNA (“relative protection”) at specific genomic locations was determine by qPCR with overlapping amplicons.
Statistical tests for the gene-specific assays
Each gene-specific experiment (i.e., RT-qPCR, ChIP-qPCR, MNase-qPCR) was conducted a minimum of three times with independent biological replicates to ensure reproducibility. The significance of differences between experimental and control samples was determined using a Student’s t-test or ANOVA, as indicated in the figure legends.
ChIP-chip and genomic data analyses
ChIP for PARP-1, H3K4me3, and H3 coupled with hybridization to promoter microarrays was performed as described previously (Krishnakumar et al., 2008) using a custom RefSeq promoter array from NimbleGen (−3 kb to +3 kb relative to the TSS). Genomic data analyses using a 1 kb moving window with 250 bp steps were performed as previously described (Krishnakumar et al., 2008). The data can be accessed through the NCBI/GEO site using accession number GSE19619.
Immunoprecipitation of KDM5B from cells and detection of PARylation
Immunopreciptation of KDM5B from MCF-7 nuclear extracts was performed using an antibody to KDM5B versus a control rabbit IgG. The nuclear extract was incubated with 5 μl of either antibody and protein A-agarose beads for 16 hours at 4°C, followed by sequential washes with 100 mM, 150 mM, and 300 mM KCl. Immunoprecipitated proteins were subjected to Western blotting with antibodies to KDM5B, PAR, and PARP-1. Similar experiments were performed using HEK-293T cells with ectopic expression of full-length Myc-tagged KDM5B.
PARylation-coupled in vitro histone binding and demethylation assays
In vitro PARylation (or mock PARylation) reactions were set up using immobilized recombinant KDM5B (full-length Myc-tagged from mammalian cells or a 6xHis-tagged amino-terminal fragment from bacteria), recombinant 6xHis-PARP-1, and NAD+ as indicated in the figures. Subsequently, the immobilized KDM5B was washed to remove the PARP-1 and NAD+, leaving PARP-1-free PARylated or unPARylated KDM5B. H3 demethylation assays were set up using the immobilized KDM5B (or corresponding control resins from untransfected or uninduced cells). Histone binding assays were performed under similar conditions. The resin was then washed three times, boiled in SDS loading solution, and analyzed by Western blotting.
Supplementary Material
Acknowledgments
The authors would like to thank Gabor Bethlendy, Leo Iniguez, and Heidi Rosenbaum from Roche NimbleGen for promoter microarrays, reagents, and technical assistance with the ChIP-chip experiments; the students of the 2009 CSHL Eukaryotic Gene Expression course for assistance with the ChIP-chip experiments; Joyce Taylor-Papadimitriou and Degui Chen for KDM5B expression constructs, and John Lis, Raghuvir Viswanatha, and members of the Kraus and Lis labs for helpful discussions and comments on the manuscript. This work was supported by R01 DK069710 from the NIH/NIDDK to W.L.K. and a predoctoral fellowship from the American Heart Association to R.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- Cohen-Armon M, Visochek L, Rozensal D, Kalal A, Geistrikh I, Klein R, Bendetz-Nezer S, Yao Z, Seger R. DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: a link to histone acetylation. Mol Cell. 2007;25:297–308. doi: 10.1016/j.molcel.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Cunliffe HE, Ringner M, Bilke S, Walker RL, Cheung JM, Chen Y, Meltzer PS. The gene expression response of breast cancer to growth regulators: patterns and correlation with tumor expression profiles. Cancer Res. 2003;63:7158–7166. [PubMed] [Google Scholar]
- D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- Frizzell KM, Gamble MJ, Berrocal JG, Zhang T, Krishnakumar R, Cen Y, Sauve AA, Kraus WL. Global analysis of transcriptional regulation by poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in MCF-7 human breast cancer cells. J Biol Chem. 2009 doi: 10.1074/jbc.M109.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frizzell KM, Kraus WL. PARP inhibitors and the treatment of breast cancer: beyond BRCA1/2? Breast Can Res. 2009;11:111. doi: 10.1186/bcr2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happel N, Doenecke D. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene. 2009;431:1–12. doi: 10.1016/j.gene.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Hassa PO, Hottiger MO. The functional role of poly(ADP-ribose)polymerase 1 as novel coactivator of NF-kappaB in inflammatory disorders. Cell Mol Life Sci. 2002;59:1534–1553. doi: 10.1007/s00018-002-8527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- Ju BG, Solum D, Song EJ, Lee KJ, Rose DW, Glass CK, Rosenfeld MG. Activating the PARP-1 sensor component of the groucho/TLE1 corepressor complex mediates a CaMKinase IIdelta-dependent neurogenic gene activation pathway. Cell. 2004;119:815–829. doi: 10.1016/j.cell.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol. 2008;20:294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Molecular Cell. 2010;39 doi: 10.1016/j.molcel.2010.06.017. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lu X, Wontakal SN, Emelyanov AV, Morcillo P, Konev AY, Fyodorov DV, Skoultchi AI. Linker histone H1 is essential for Drosophila development, the establishment of pericentric heterochromatin, and a normal polytene chromosome structure. Genes Dev. 2009;23:452–465. doi: 10.1101/gad.1749309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottke A, Colaiacovo MP, Shi Y. Developmental roles of the histone lysine demethylases. Development. 2009;136:879–889. doi: 10.1242/dev.020966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Song JS, Liu XS, Fisher DE. High-throughput mapping of the chromatin structure of human promoters. Nat Biotechnol. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- Pavri R, Lewis B, Kim TK, Dilworth FJ, Erdjument-Bromage H, Tempst P, de Murcia G, Evans R, Chambon P, Reinberg D. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol Cell. 2005;18:83–96. doi: 10.1016/j.molcel.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin A, Chinenov Y, Spradling A. Regulation of chromatin structure and gene activity by poly(ADP-ribose) polymerases. Curr Top Dev Biol. 2003;56:55–83. doi: 10.1016/s0070-2153(03)01007-x. [DOI] [PubMed] [Google Scholar]
- Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Wacker DA, Ruhl DD, Balagamwala EH, Hope KM, Zhang T, Kraus WL. The DNA binding and catalytic domains of poly(ADP-ribose) polymerase 1 cooperate in the regulation of chromatin structure and transcription. Mol Cell Biol. 2007;27:7475–7485. doi: 10.1128/MCB.01314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P, Zhang Y. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell. 2007;25:801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.