SUMMARY
The neural crest is a multipotent stem cell-like population that is induced during gastrulation, but only acquires its characteristic morphology, migratory ability and gene expression profile after neurulation. This raises the intriguing possibility that precursors are actively maintained by epigenetic influences in a stem cell-like state. Accordingly, we report that dynamic histone modifications are critical for proper temporal control of neural crest gene expression in vivo. The histone demethylase, JumonjiD2A (JmjD2A/KDM4A), is expressed in the forming neural folds. Loss of JmjD2A function causes dramatic down-regulation of neural crest specifier genes analyzed by multiplex NanoString and in situ hybridization. Importantly, in vivo chromatin immunoprecipitation reveals direct stage-specific interactions of JmjD2A with regulatory regions of neural crest genes, and associated temporal modifications in methylation states of lysine residues directly affected by JmjD2A activity. Our findings show that chromatin modifications directly control neural crest genes in vertebrate embryos via modulating histone methylation.
INTRODUCTION
The neural crest (NC) is a multipotent stem cell-like population that migrates extensively and differentiates into diverse cell types in vertebrate embryos. In the chick embryo, neural crest induction initiates during gastrulation (Basch et al., 2006), leading to establishment of a precursor population at the border between presumptive neural and non-neural ectoderm. As the neural folds elevate (stage 7), these border cells begin to express premigratory neural crest markers. Finally, after neural tube closure (stages 8–10), these cells become distinct and emigrate from the central nervous system, migrating extensively and forming numerous derivatives (Crane and Trainor, 2006; Knecht and Bronner-Fraser, 2002).
A “hard-wired” gene regulatory network, formulated from data derived from several model vertebrates, is thought to mediate neural crest formation (Sauka-Spengler and Bronner-Fraser, 2008). Most notably, modules of transcription factors are proposed to function in sequential fashion to first specify the neural plate border and then the nascent neural crest. Finally, the process of neural crest differentiation involves the activity of NC specifier genes including SoxE transcription factors (Sox10 and Sox9), which regulate effector genes that imbue neural crest derivatives with characteristics of terminal differentiation (Sauka-Spengler and Bronner-Fraser, 2008). However, little is known about the hierarchical relationships, direct regulatory interactions, or connections between these different modules. The temporal gap between initial induction during gastrulation (Basch et al., 2006) and final appearance of the migratory, multipotent neural crest raises the fundamental question of what maintains neural crest cells in an undifferentiated, stem cell-like state in the intervening time.
The prolonged maintenance of stem cell properties is consistent with an important role for chromatin modifications in spatiotemporal control of the neural crest gene expression program. In vitro studies of embryonic stem cells suggest that these modification events may poise stem cell genes in a transcriptionally ready state via modifications of N-terminal histone tails. For example, histone methylation has been associated with both transcriptional activation and repression (Kouzarides, 2007). Whereas methylation of H3 lysines 4 and 36 (H3K4me2/me3 and H3K36me2/3) are enriched in transcriptionally active euchromatic regions, H3 lysines 9 and 27 (H3K9me3 and H3K27me3) are located in repressed sites (Nielsen et al., 2001; Shi et al., 2003) as well as associated with heterochromatin (Li et al., 2007; Nakayama et al., 2001; Peters et al., 2002). Histone demethylases such as members of the Jumonji family revert histone trimethylation (Tan et al., 2007). Unlike other demethylases, JmjD2/KDM4 proteins have been shown to remove both lysine 9 and 36 trimethyl marks (Couture et al., 2007).
An intriguing possibility is that such chromatin modifiers may play a critical role in the balance between differentiation and stem cell like behavior at the neural plate border and forming neural crest. However, little is known about the function of the Jumonji family or other epigenetic modifiers in vivo in the developing vertebrate embryo. As an accessible embryonic stem cell population, the neural crest offers an excellent experimental system to probe the role of chromatin modification during development in vivo.
RESULTS
Jumonji is expressed at key times/locations during neural crest development
The JmjD2/KDM4 subfamily consists of the JmjD2A-F genes, though JmjD2E and F appear to be pseudogenes (Katoh, 2004) and only the JmjD2A-D proteins possess histone demethylase activity (Cloos et al., 2006; Klose et al., 2006; Whetstine et al., 2006). By in silico analysis, only JmjD2A-C genes were detectable in the chick genome.
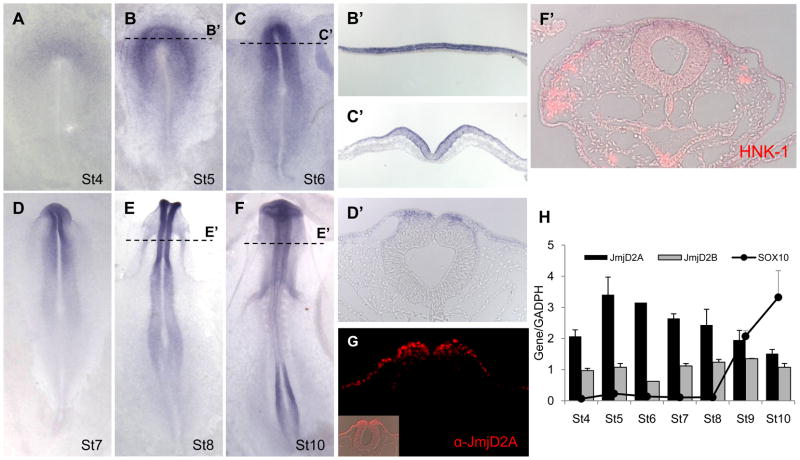
We examined the presence of all three transcripts by whole mount in situ hybridization and quantitative polymerase chain reaction (QPCR) from chick gastrulation through neural crest migration (Fig. 1, S1). Of these, JmjD2A demonstrated appropriate spatiotemporal expression consistent with playing a role in neural crest development (Fig. 1A–G). JmjD2A transcripts were observed in the neural plate, neural plate border and non-neural ectoderm (Fig. 1A–C). JmjD2A initially is expressed throughout the neural plate, partially overlapping with the neural plate border marker Pax7 (Basch et al., 2006), but resolves to the neural crest forming region by stage 5/6. As the neural folds elevate, JmjD2A was restricted to the dorsal neural folds, from which neural crest cells originate, as well as faintly expressed in non-neural ectoderm (Fig. 1D–E). Protein expression, as revealed by a JmjD2A specific antibody, paralleled that of the transcripts (Fig. 1G). After neural tube closure, only low levels of JmjD2A expression were observed on migrating HNK1+ neural crest cells (Fig. 1F). Parallel analysis by QPCR reveals the highest expression of JmjD2A at stage 5, decreasing from stage 8 onward; this is reciprocal to the onset of Sox10 expression, a key regulator of neural crest cell specification (Fig. 1H). In contrast to JmjD2A, JmjD2B was detected only by QPCR presenting an invariable patter of expressing (Fig. 1H), and JmjD2C was not detectable by either QPCR or in situ hybridization at the stages examined (Fig. S1). These data provide anatomical and temporal evidence suggesting that JmjD2A may be an important player in neural crest specification.
Figure 1. JmjD2A is expressed at the right time and location to play an important role in neural crest development.
(A–F) Expression pattern of JmjD2A in the early chick embryo by whole mount in situ hybridization at stages 4–10. Transverse sections (dotted lines) reveal specific expression in the neural plate at stage 5 (B′); in neural plate, neural plate border and non-neural ectoderm at stage 6 (C′); in dorsal neural tube at stage 8 (E′); and at low levels in migratory neural crest (evidenced by HNK-1 staining in red) at stage 10 (F′). (G) Immunohistochemistry using an anti-JmjD2A corroborates the protein expression at stage 8 in dorsal neural tube and non-neural ectoderm. (H) QPCR analyses show highest expression of JmjD2A (black bars) at stage 5, decreasing thereafter in a manner reciprocal to that of the neural crest specifier gene, Sox10 (solid line). JmjD2B (gray bars) present an invariable expression patter during the analyzed stages. Values represent the average of 3 samples run in triplicate ± SD. See also Figure S1.
Over-expression of JmjD2A in fibroblasts specifically depletes H3K9me3 and H3K36me3
To define the lysine specificity of chick JmjD2A, a vector containing the coding sequence of JmjD2A (JmjD2A-GFP) under the control β-actin promoter was transfected into chicken fibroblast cells (Fig. S2). The results show that cells over-expressing JmjD2A completely lack either H3K9me3 or H3K36me3 marks; and on the other hand, H3K27me3 methylation marks remain expressed. This activity is dependent on an intact JmjC domain, as amino-acid substitutions (ΔJmjD2A-GFP: G133A, G138A, G165A, G170A, S288N and T289B) in the catalytic domain (Chen et al., 2007) abrogate these effects. These results demonstrate that JmjD2A is specific for H3K9me3 and H3K36me3 substrates in these cells, consistent with previous reports.
Loss of JmjD2A leads to depletion of neural crest specifier genes
To examine the function of JmjD2A in neural crest specification, we tested its developmental effects on gene expression by a loss-of-function analysis using antisense morpholino oligonucleotides (MO) to specifically block translation of JmjD2A protein. We used two non-overlapping fluorescein-tagged MOs, one located just over the ATG codon and the second l38 nucleotides upstream of the start codon, with both yielding similar effects. MOs plus carrier DNA were injected on the left half of stage 4–5 embryos, such that the uninjected side served as an internal control. Immunoblotting confirmed a marked reduction of JmjD2A protein after morpholino knock-down on the injected side (IS) compared with the uninjected side (UIS) of the same embryos, as well as between the injected side of control-MO treated embryos (Fig. 3C). Furthermore, there was no significant change in either cell proliferation (P = 0.29, n = 5) or cell death (P = 0.32, n = 5), as assayed by TUNEL and phosphohistone 3 staining (Fig. S4B).
Figure 3. JmjD2A loss-of-function depletes Sox10 expression.
(A) Electroporation of fluorescent JmjD2A morpholino (JmjD2A-MO) into one side of the embryo (green staining in inset) causes a dramatic reduction in Sox10 expression in embryos examined at stage 8+ and 10, compared with the non-electroporated side or control morpholinated embryos (Control-MO). (B) Quantitation of the numbers of JmjD2A-MO treated embryos with either mild or strong phenotypes on the electroporated versus contralateral side compared with those observed in control-MO treated embryos. Asterisk indicates significant differences (P<0.01) by contingency table followed by chi-square test. Numbers represent individual embryos. (C) Western blots reveal specific reduction of JmjD2A protein expression on the JmjD2A-MO injected side (IS) compared with both the uninjected side (UIS) and the injected side of the Control-MO embryos. See also Figure S4.
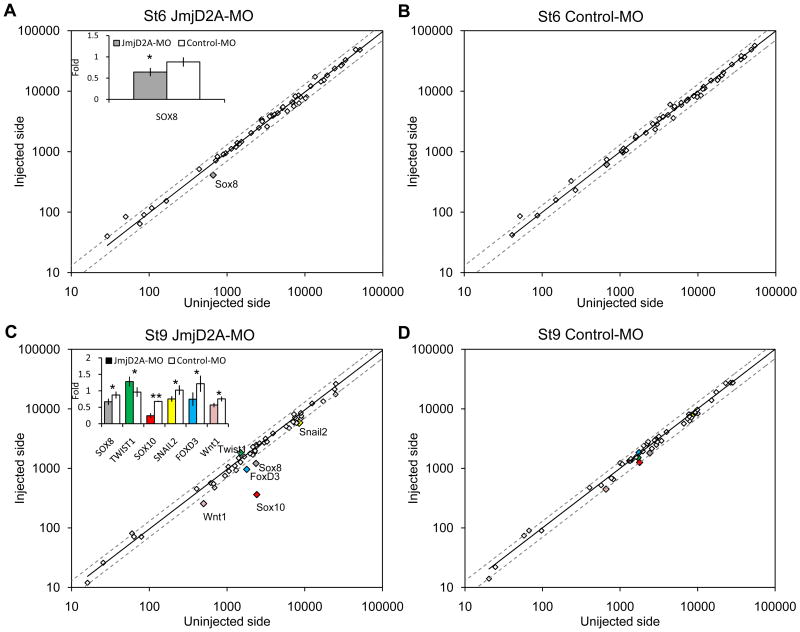
As a first step in assessing the effects of JmjD2A on a wide array of downstream genes, we used the NanoString nCounter technology (Fortina and Surrey, 2008) to perform a multiplex analysis to examine changes in transcript levels of 50 genes (Fig. 2, S3), including those expressed in the neural crest, neural plate, neural plate border, neural tube, ectodermal placodes and non-neural ectoderm, as well as genes involved in proliferation and apoptosis. Individual half embryos (injected versus non-injected side) were analyzed at st. 6 or st. 9 after treatment with JmjD2A- or control-MO (Fig. 2). No significant differences were observed at either stage between the two sides of control-MO treated embryos (Fig. 2B, C), with a less than 20% variation. As a consequence, we defined 25% (gray dotted line on figure 2) variation as our cut-off for statistical significance. At stage 6, the NanoString reads collected from four embryos showed that JmjD2A-MO caused a down-regulation of only the Sox8 (P=0.04) gene (inset in Fig. 2A). However, by stage 9, there was a significant reduction in gene expression for several neural crest specifier genes, with Sox10 (P=0.0002) most dramatically affected, as well as significant down-regulation of Sox8 (P=0.04), FoxD3 (P=0.04), Snail2 (P=0.02) and Wnt1 (P=0.01). Twist1 (P=0.03) was the only gene significantly up-regulated (inset in Fig. 2C) after JmjD2A knock-down. In contrast, there were no significant changes in the expression of several neural tube, ectodermal, neural plate and border genes, or in markers of proliferation and apoptosis.
Figure 2. NanoString nCounter quantification. JmjD2A loss-of-function affects neural crest specifier gene expression.
(A) At stage 6, the injected side of JmjD2A-MO treated embryos shows >25% reduction of Sox8 (gray) on the injected compared with the uninjected side. No significant changes were noted in other genes. (B) Control morpholino injected embryos had no significant differences between injected and uninjected side at stage 6. (C) At stage 9, the expression of several neural crest specifier genes, including Sox10 (red), FoxD3 (blue), Sox8 (gray) and Snail2 (yellow), as well as the signaling factor, Wnt1 (pink) were reduced more than 25% on the JmjD2A-MO injected side (IS) compared with the uninjected (UIS). In contrast, Twist1 (green) was upregulated on the morpholino-injected side. (D) Control morpholino injected embryos had no significant differences between injected and uninjected side at stage 9. Insets show fold expression differences (mean ± SD) between IS and UIS comparing JmjD2A-MO (n=4) and control-MO (n=3) treated embryos. Asterisks (* and **) indicate significant differences (P<0.05 and P<0.01, respectively) by t-Student test (B-D). Dotted line represents variation greater than 25% between IS and UIS. See also Figure S3.
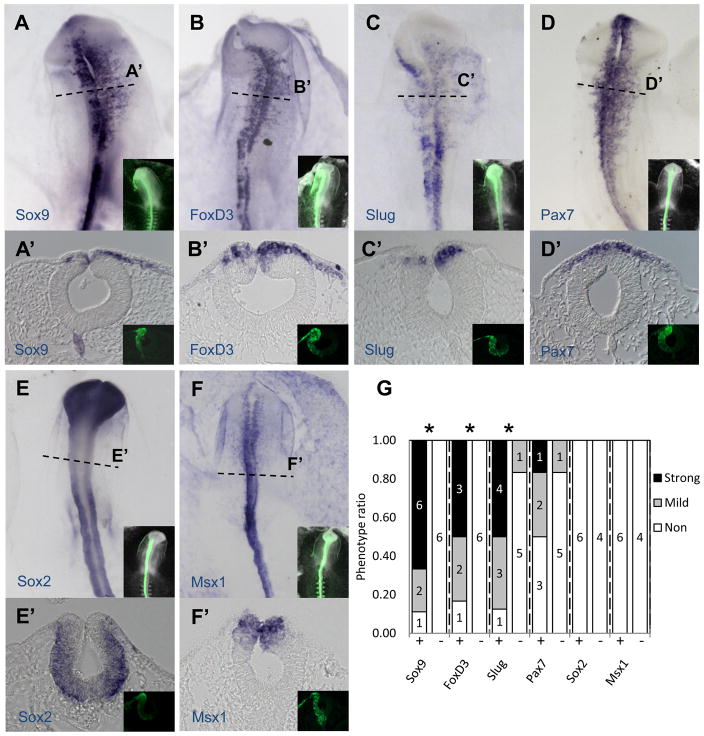
In order to corroborate the NanoString nCounter results and gain spatial information on expression on some of the genes affected by the JmjD2A-MO treatment, we perfomed in situ hybridization (Fig. 3A–B, 4). The results show that JmjD2A protein knock-down profoundly diminishes expression of neural crest specifier genes while having no effect on other genes in the neural tube. By categorizing embryos according to the severity of their phenotype as strong, mild or none (Fig. S4A), we observed significant reduction between morpholino-treated and control embryos in the expression of neural crest specifier genes Sox10 (P<0.01, n = 22), Sox9 (P<0.01, n = 9), FoxD3 (P<0.05, n = 6) and Snail2 (P<0.05, n = 8) in pre-migratory and migratory neural crest cells. On the other hand, we observed no significant effects on expression in the dorsal neural tube of Pax7, Msx1 and Sox2 (n = 6). As expected, control-MO treated embryos had no phenotype. These results are consistent with those observed in our NanoString experiments, with the addition of the neural crest specifier gene Sox9, for which we lacked a functional NanoString probe.
Figure 4. JmjD2A loss-of-function affects expression of neural crest specifier genes Sox9, FoxD3 and Snail2, but not dorsal neural tube markers.
As seen by in situ hybridization in whole mount (A–F) or transverse section (bottom of each panel) (A′–F′), electroporation of JmjD2A-MO causes a significant reduction in expression of Sox9, FoxD3 and Snail2, whereas no obvious effects were noted for Pax7, Sox2 and Msx1. Insets show the distribution of fluorescently labeled morpholino (green). Dotted lines indicate level of section. (G) Quantitation of the numbers of embryos showing a phenotype on the electroporated side of JmjD2A-MO (+) and Control-MO (−) treated embryos. Asterisk indicates P<0.05 versus control-MO by contingency table followed by chi-square. Numbers represent individual embryos. See also Figure S5.
To control for the specificity of our morpholino knock-down, rescue experiments were performed by co-electroporating the JmjD2A-MO, located upstream of the start codon, with a vector encoding full length JmjD2A protein (JmjD2A-H2RFP), plus nuclear RFP expressed as an independent protein due to the presence of an Internal Ribosome Entry Site (IRES) in the vector (Fig. 5). Co-electroporation of JmjD2A-MO plus JmjD2A-H2RFP resulted in a 25% reduction in the severity of the phenotype compared with co-electroporation of JmjD2A-MO plus empty vector (H2RFP), and was not significantly different from control-MO treated embryos. Importantly, the rescue effect was lost when the morpholino was co-electroporated with the mutated JmjD2A lacking catalytic activity (ΔJmjD2A-H2RFP) described above, restoring significant differences (P<0.01) from phenotypes observed in control-MO treated embryos.
Figure 5. Rescue experiment restored Sox10 expression.
(A) Electroporation of JmjD2A morpholino (JmjD2A-MO, in green) together with a vector containing the coding region of JmjD2A (JmjD2A-H2RFP, in red) rescues the depletion of Sox10 expression as assayed by ISH. In contrast, co-electroporation JmjD2A-MO plus an empty vector (H2RFP, in red) or catalytically-dead mutant of JmjD2A (ΔJmjD2A-H2RFP, in red) failed to rescue the loss-of-function phenotype. (B) Quantitation of the numbers of rescue experiment with either mild or strong phenotypes on the electroporated versus contralateral side compared with those observed in control-MO treated embryos. Asterisk indicates significant differences (P<0.01) by contingency table followed by chi-square test. Numbers represent individual embryos. See also Figure S2.
To look at long-term effect of the JmjD2A depletion, we examined the formation of ganglia in the head to which the neural crest contributes (trigeminal, geniculate, petrosal and nodose) formation (Fig. S5). The results reveal a severe defect in the cranial ganglia formation as assayed by β-neurotubulin (TuJ1) immunostaining at St. 16–17. This is particularly evident in the trigeminal ganglion, the largest of the cranial ganglia, which appears smaller and ill-formed on the injected side of JmjD2A-MO treated embryos compared with both the self uninjected side of the same embryo or with control-MO treated embryos.
Taken together, these results demonstrate that JmjD2A is required and remarkably specific for correct expression of several neural crest specifier genes, perturbation of which causes profound defects in neural crest derivatives. This highlights the importance of chromatin modifications during neural crest development.
H3K9me3 and H3K36me3 occupancy regulates neural crest specifier expression in vivo
In order to directly determine the importance of histone methylation in regulation of NC specifier gene expression, we performed chromatin immunoprecipitation (ChIP) with antibodies to H3K9me3 and H3K36me3 using dissected tissue from 30 embryos. The results reveal dynamic occupancy of sites in proximity to the transcriptional start site (TSS) of two important neural crest specifier genes, Sox10 and Snail2 (Fig. 6A).
Figure 6. Dynamic changes in H3K9me3, H3K36me3 and JmjD2A occupancy on neural crest specifier genes during chick development in vivo.
(A) Chromatin immunoprecipiation (ChIP) assay was used to assess the H3K9me3 and H3K36me3 occupancy around the transcriptional start site (TSS) of Sox10, Snail2 and β-actin. Three independent experiments were performed for each stage. The vertical axis represents % input (ChIP enriched/input) and horizontal axis the distance from the TSS in kilobases (Kb). Schematic diagrams represent primer location over the three analyzed genes and numbers represent approximate distance in Kb from the TSS. Cartoons schematize the two stages (stages 4–5 and 10–11) of analysis, with red indicating the region of tissue dissected and collected for ChIP. One representative sample is depicted per stage. The results show high occupancy of H3K9me3 at −0.5kb from the TSS of Sox10 and Snail2 genes at stage 3–4. This repressive mark appears to be eliminated at stage 10–11. In contrast, both Snail2 and β-actin genes shown high abundance of the active mark H3K36me3, mostly downstream of the TSS. (B) ChIP assays assessed binding of JmjD2A at -0.5Kb from the Sox10 and Snail2 TSS. The graphs show a mean ± SD of three independent experiments for each stage. At stage 3–4, JmjD2A significantly binds to both Sox10 (0.006% input) and Snail2 (0.007% input), when compared with two distant, control regions in their respective chromosome 1 (Chr1nc, 0.002% input) and 2 (Chr2nc, 0.003% input), as well as with β-actin locus (0.003% input). At stage 10–11, this binding was greatly reduced to 0.002 and 0.003% input on the Sox10 and Snail2 genes, respectively. Asterisk indicates P<0.05 versus respective negative control by paired t-student test. See also Figure S6.
Tissue was dissected from embryonic regions containing neural crest precursors from gastrula stage (stages 3–4) or from actively migrating stages (stages 10–11). Interestingly, we observed high and moderate occupancy of H3K9me3 at −0.5kb from the TSS of the both Sox10 and Snail2 at the late gastrula stage, respectively. This result is consistent with the possibility that this strong epigenetic repression mark inhibits transcription at this time point. Moreover, occupancy of H3K36m3, generally associated with active gene transcription, was high for the β-actin and Snail2, and low for Sox10. Taken together, the location and abundance of H3K9me3 and H3K36me3 clearly reflects the transcriptional state of a repressed Sox10, Snail2 poised for activation and active β-actin gene at late gastrula stages.
Interestingly, the occupancy of the “repressive” mark H3K9me3 near the Sox10 and Snail2 TSS was clearly reduced by times of active neural crest migration (stages 10–11) and approached levels similar to those observed for β-actin, consistent with high expression of these genes by this stage. On the other hand, occupancy on the H3K36me3 marks situated downstream of the TSS was still abundant for Snail2 and β-actin loci, but unchanged in Sox10. These findings suggest that the demethylation of H3K9me3 (repressive) marks plays an important role in regulating expression of neural crest specifier genes during chick development.
JmjD2A directly binds to regulatory regions of neural crest specifier genes in vivo
We next performed ChIP for JmjD2A as a function of developmental stages using dissected tissue from 30 embryos and analyzing by QPCR the occupancy around the TSS of Sox10, Snail2 and β-actin genes (Fig. 6B, S6). Interestingly, JmjD2A ChIP revealed stage dependent binding to a regulatory region located −0.5kb from the TSS of both Sox10 and Snail2 neural crest specifier genes. During gastrulation, there was 2.6 and 4 fold changes on the input enrichment near the Sox10 and Snail2 TSS region, respectively, relative to negative controls situated in ORF-free intergenic regions on the same chromosome, Chr1 (negative control for Sox10), Chr2 (negative control for Snail2). Moreover, no differences were observed when analyzing similar positions on the β-actin locus. In contrast to gastrulation, we failed to detect occupancy of JmjD2A around the TSS of the Sox10, Snail2 and β-actin genes during neural crest migration less than one day later. These data demonstrate that JmjD2A interacts in vivo in a time and location dependent manner on neural crest specifier genes, Sox10 and Snail2. Interestingly, the direct interaction of JmjD2A with neural crest regulatory regions correlates well with the histone modifications described in the figure 5, consistent with the idea that it derepresses those genes through H3K9me3 demethylation.
JmjD2A knock-down inhibits demethylation of H3K9me3 on the Sox10 promoter
To narrow the timing of JmjD2A action on neural crest genes, we performed ChIP with H3K9me3 and JmjD2A antibodies using dorsal neural tubes from embryos collected at stage 8 and 9 in order to explore when H3K9me3 is demethylated and how the timing correlates with binding of JmjD2A to the Sox10 promoter. The time period between stage 8 and 9 is key because it correlates with the first appearance of Sox10 transcripts in the neural crest by both in situ hybridization and quantitative PCR. The results reveal a fine time window of association between the JmjD2A and H3K9me3 occupancy on the Sox10 promoter (Fig. 7A, B). Interestingly, JmjD2A binds efficiently to −0.5kb on Sox10 genes at St. 8, but not at St. 9 by which time its association is not different from the negative control region (Chr1 nc). In a similar manner, the region −0.5Kb from the TSS of Sox10 gene was highly occupied by H3K9me3 at St. 8, but dropped precipitously at stage 9 to levels comparable to those observed on surrounding region (−1 and 0.5Kb from the TSS). These results reveal a rapid and dynamic process whereby JmjD2A is preloaded on the Sox10 locus and leaves upon demethylation, thus permitting activation of Sox10 transcription.
Figure 7. JmjD2A activity is responsible for derepression of Sox10 by demethylating H3K9me3.
ChIP was used to assay the occupancy of JmjD2A and H3K9me3 on the Sox10 gene from stage 8 and 9 wild type or morpholinated embyos. Two to three independent experiments were performed at each stage using 10 dorsal neural tubes per experiment. One representative sample is depicted per stage. The graphs show a mean ± SD. (A) JmjD2A occupancy at −0.5Kb from the TSS was high at stage 8 compared with a distant region in the same chromosome (Chr1nc). By stage 9, however, this binding was greatly reduced to values similar to those at Chr1nc. (B) Similarly, H3K9me3 occupancy at −0.5Kb from the TSS was high at stage 8 but reduced by stage 9, reaching values similar to surrounding positions (−1 and 0.5Kb from the TSS). (C) JmjD2A-MO completely abolished occupancy of JmjD2A at stage 8, reaching values similar those observed on the negative control Chr1nc. (D) In JmjD2A-MO treated embryos, H3K9me3 occupancy at stage 9 remained high on the areas surrounding the TSS when compared with Control-MO treated embryos.
Given the dynamic changes in both JmjD2A and H3K9me3 occupancy of the Sox10 promoter between stages 8 and 9, we examined the effects of JmjD2A-MO on Sox10 promoter occupancy at these key stages (Fig 7C, D). As predicted, the occupancy JmjD2A was completely abolished on the promoter in JmjD2A-MO compared with both control-MO and untreated embryos collected at stage 8. This is not surprising given the lack of JmjD2A protein in the JmjD2-MO embryos. On the other hand, H3K9me3 occupancy of the Sox10 promoter clearly increased in JmjD2A-MO treated embryos collected at stage 9, with highest levels at −0.5kb from the TSS. In contrast, control-MO embryos had similar levels at all positions analyzed, scoring values similar to those observed on untreated embryos at stage 9. As a further control, we use −0.5Kb from the TSS of β-actin gene, and observed no differences in H3K9me3 association between JmjD2A-MO and control-MO treated embryos (Fig. 7E). These results conclusively demonstrate that JmjD2A activity is responsible for demethylation of H3K9me3 at −0.5kb from the TSS of Sox10 gene which in turn derepresses expression of Sox10 transcripts.
DISCUSSION
Jumonji family members have been shown to function as powerful regulators of epigenetic events in cell culture (Nottke et al., 2009). However, little has been known about their roles and mechanism of action in the context of the whole organism. In the present work, we demonstrate that the Jumonji family member, JmjD2A, is critical for embryonic development, playing a specific role in allowing neural crest gene expression at the appropriate developmental time. This dramatic stage-dependent regulation of gene expression is made possible due to appropriate histone modifications. Thus, Jumonji dependent chromatin modification can be used to dynamically regulate rapidly changing gene regulatory networks such as the one underlying neural crest formation.
By specifically affecting the neural crest specifier module, our data place JmjD2A as a modulator that functions upstream of neural crest specifier genes. However, there is an intriguing delay between JmjD2A occupancy and reduction of H3K9me3 on the Sox10 locus, opening the possibility that other factors need to be recruited to the TSS. In this critical position, JmjD2A acts on neural crest precursors that are retained in a stem cell-like state but poised to initiate transcription upon removal of a repressive H3K9me3 mark. In this way, neural crest cells that are induced during gastrulation only initiate a bona fide neural crest program during neurulation.
Current thinking regarding the histone code involved in controlling differential methylation states suggests that H3K9me3 is linked to “silent” genes or pericentric heterocromatin (Peters et al., 2002), whereas H3K36me3 is linked to transcriptional elongation (Li et al., 2007) and suppression of inappropriate transcription (Carrozza et al., 2005; Keogh et al., 2005). Furthermore, H3K36me3 is generally restricted to the coding region downstream for the TSS, and is particularly enriched in exons. Our data suggest that the balance between bivalent JmjD2A histone targets, H3K9me3 and H3K36me3, reflecting repressive and activating marks respectively, may underlie the fine tuning that controls the timing of execution of a neural crest gene regulatory network program in vivo. However even when we observed high H3K36me3 occupancy at the 3′ region on actively transcribed genes (e.g. Snail2 and β-actin), we failed to observe a similar pattern on the Sox10 gene at stages of active transcription. An intriguing possibility in the case of Sox10 in neural crest cells is that the normal role of H3K36me3 is replaced by another epigenetic mark that allows it elongation, such as, H3K79me2 which also is associated with active transcription (Morillon et al., 2005; Schubeler et al., 2004).
A similar bivalent regulation between repressive (H3K27me3) and activating (H3K4me3) marks has been described in a large set of developmentally important genes in embryonic stem cells in vitro. Moreover, the activation of some of these genes correlates well with the demethylation of H3K27me3 (Azuara et al., 2006; Bernstein et al., 2006). These similarities raise the possibility that the flavor of the histone mark may regulate the degree of a cell’s multipotency. Genes that are repressed in multipotent cells contain distinct bivalent H3K9me3/H3K36me3 marks that render them poised for activation, and are distinguishable from those required for later differentiation genes. In fact, H3K9me3 and H3K36me3 marks can discriminate between genes that are expressed (H3K36me3) or stably repressed (H3K9me3). As a consequence, they may reflect the cell’s state and fate potential (Mikkelsen et al., 2007). To date, it remains unknown whether JmjD2A favors H3K9me3 or H3K36me3 sites in vivo. This opens the possibility that JmjD2A may form complexes with different proteins depending upon its local and changing intracellular environment. A similar scenario has been suggested previously for other demethylases (Metzger et al., 2005; Shi et al., 2003).
Our observation that JmjD2A is associated with key neural crest specifier genes gives an additional and dynamic level of complexity to the neural crest gene regulatory network (Sauka-Spengler and Bronner-Fraser, 2008), emphasizing the importance of chromatin modification in spatial and temporal control of gene expression during development Furthermore, these findings highlight the intriguing possibility that repressive histone marks influences may underlie the latency of the multipotent state of neural crest precursors between induction and overt morphological manifestation of the mature state.
EXPERIMENTAL PROCEDURES
Embryos
Fertilized chicken eggs were obtained from local commercial sources and incubated at 37° C to the desired stages according to the criteria of Hamburger and Hamilton. For in situ hybridization (ISH), embryos were fixed overnight in 4% paraformaldehyde (PFA) at 4°C, washed in PTW (PBS-DEPC treated containing 0.1% Tween) and dehydrated in a MeOH/PTW series at room temperature before being stored at −20°C in 100% MeOH.
RNA preparation and QPCR
RNA was prepared from individual embryos (n=3) using RNAqueous®-Micro isolation kit (Ambion) following manufacturers instruction. The obtained RNA was treated with DNaseI amplification grade (Invitrogen) and then reversed-transcribed to cDNA with SuperScript III (Invitrogen) using random hexamers. QPCR was performed using the 96-well plate ABI 7000 QPCR machine in a TaqMan assay (Applied Biosciences) with Sybrgreen Itaq Supermix with ROX (Bio-Rad), 150–450 nM of each primer, and 200–500 ng of cDNA in a 25 μl reaction volume. During the exponential phase of the QPCR reaction, a threshold cycle (CT) and baseline was set according to the protocols of Applied Biosystems. The results for different samples were then interpolated on a line created by running standard curves for each primer set and then normalized against the GADPH housekeeping gene. These calculations were performed according to the standard curve assay method which is detailed in the Applied Biosystems protocols. Three replicates of every sample and points of the standard curve were loaded.
Electroporation of antisense morpholinos and vectors
Two antisense morpholinos to JmjD2A were designed, one near the ATG codon (5′-TGAGGCTCTCCAGCTCCGAGGCCAT-3′) and the second located 38 nucleotides upstream of the start codon (5′-CGGTCAGCTCCGCGTCCCGCTTCTT-3′), with both yielding similar results. Injections of fluorescein-tagged morpholino (1 mM plus 0.3 mg/ml of plasmid DNA) and pCIG -H2RFP, -JmjD2A-H2RFP and/or -ΔJmjD2A-H2RFP (catalytically dead mutant carrying mutations on G133A, G138A, G165A, G170A, S288N and T289B) vectors were performed by air pressure using a glass micropipette targeted to the presumptive neural crest region at stages 4–5. Stages 4–5 electroporation was conducted on whole chick embryo explants placed ventral side up on filter paper rings. The morpholinos and vectors were injected on the left side of the embryo and platinum electrodes were placed vertically across the chick embryos and electroporated with five pulses of 7 V in 50 ms at 100 ms intervals. Embryos were cultured in 0.5 ml of albumen in tissue culture dishes to reach the desired stages. Embryos were then removed and, fixed overnight in 4% PFA at 4°C. Embryos were placed in PBS, viewed and photographed as whole-mounts using a fluorescence stereomicroscope to evidence electroporation efficiency. Embryos used for ISH were dehydrated in a MeOH/PTW series at room temperature before being stored at −20°C in 100% MeOH.
NanoString nCounter
Individual half of embryos treated with Control-MO and JmjD2A-MO were disaggregated in lysis buffer (Ambion) and stored at −80°C. The total RNA from the lysates were then allowed to hybridize with the capture and reporter probe and incubating overnight at 65°C according to nCounter™ Gene Expression Assay Manual. After the washes, the purified Target/Probe complexes were eluted off and immobilized in the cartridge for data collection carried out in the nCounter™ Digital Analyzer.
In situ hybridization (ISH)
Whole-mount chick ISH was performed as previously described (Kee 2001). Linealized plasmidic DNA templates used for digoxigenin-labeled RNA probes were JmjD2A, JmjD2B, JmjD2C, Sox10, Sox9, FoxD3, Snail2, Pax7, Msx1, and Sox2. Embryos were imaged as a whole-mount and after were transverse sectioned at 14–16 μm in a cryostat.
Cell death and proliferation
TUNEL labeling was performed using the TMR-In Situ Cell Death kit (Roche) and subsequently immunostained with the proliferation marker Phosphohistone 3 (rabbit anti-PH3, 1:500), and then detected using the secondary Alexa Fluor goat anti-rabbit 350 (1:1000). Cells were counted in the dorsal neural tube of both JmjD2A-MO and Control-MO, and expressed as a ratio between injected vs. unijected side.
Chromatin immunoprecipitation (ChIP)
Cells were extracted from presumptive neural plates and dorsal-rostral area (see cartoon in figure 4) from 30 embryos on three independent experiment at stages 3–4 and 10–11, respectively. The cells were then cross-linked and sonicated in an average size of 300–800 bp. Samples were split into five tubes for Input sample, mock control (rabbit anti-IgG) and target antibodies (anti-JmjD2A, anti-H3K9me3 and anti-H3K36me3, Abcam) bound to Protein A-magnetic beads (Invitrogen). The complexes were magnetically isolated, eluted and crosslink reversed. The DNA was purified, precipitated and finally used as a template for QPCR analyzes (see table 1S for list of primers). Every sample was load by three replicates each. The results were quantified using ΔΔCt method and calculated according to manufacturer’s instructions (Applied BioSystems). On the ChIPs performed on morpholinated treated embryos, 10 dorsal neural tube were dissected from JmjD2A-MO and Control-MO embryos at stages 8 and 9, and processed as above mentioned
Supplementary Material
Acknowledgments
We thank Dr. Scott Fraser for critical reading of the manuscript. This work was supported by USPHS grant HD037105.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFFERENCES
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Basch ML, Bronner-Fraser M, Garcia-Castro MI. Specification of the neural crest occurs during gastrulation and requires Pax7. Nature. 2006;441:218–222. doi: 10.1038/nature04684. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Chen Z, Zang J, Kappler J, Hong X, Crawford F, Wang Q, Lan F, Jiang C, Whetstine J, Dai S, et al. Structural basis of the recognition of a methylated histone tail by JMJD2A. Proc Natl Acad Sci U S A. 2007;104:10818–10823. doi: 10.1073/pnas.0704525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, Hansen KH, Helin K. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- Couture JF, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat Struct Mol Biol. 2007;14:689–695. doi: 10.1038/nsmb1273. [DOI] [PubMed] [Google Scholar]
- Crane JF, Trainor PA. Neural crest stem and progenitor cells. Annu Rev Cell Dev Biol. 2006;22:267–286. doi: 10.1146/annurev.cellbio.22.010305.103814. [DOI] [PubMed] [Google Scholar]
- Fortina P, Surrey S. Digital mRNA profiling. Nat Biotechnol. 2008;26:293–294. doi: 10.1038/nbt0308-293. [DOI] [PubMed] [Google Scholar]
- Katoh M. Identification and characterization of JMJD2 family genes in silico. Int J Oncol. 2004;24:1623–1628. [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- Knecht AK, Bronner-Fraser M. Induction of the neural crest: a multigene process. Nat Rev Genet. 2002;3:453–461. doi: 10.1038/nrg819. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillon A, Karabetsou N, Nair A, Mellor J. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol Cell. 2005;18:723–734. doi: 10.1016/j.molcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O’Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- Nottke A, Colaiacovo MP, Shi Y. Developmental roles of the histone lysine demethylases. Development. 2009;136:879–889. doi: 10.1242/dev.020966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, Mermoud JE, O’Carroll D, Pagani M, Schweizer D, Brockdorff N, Jenuwein T. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat Genet. 2002;30:77–80. doi: 10.1038/ng789. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O’Neill LP, Turner BM, Delrow J, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- Tan H, Wu S, Wang J, Zhao ZK. The JMJD2 members of histone demethylase revisited. Mol Biol Rep. 2007 doi: 10.1007/s11033-007-9121-3. [DOI] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.