Summary
Ubiquitin (Ub) provides the recognition and specificity required to deliver proteins to the eukaryotic proteasome for destruction. Prokaryotic ubiquitin-like protein (Pup) is functionally analogous to Ub in Mycobacterium tuberculosis (Mtb) as it dooms proteins to the Mtb proteasome. Studies suggest that Pup and Ub do not share similar mechanisms of activation and conjugation to target proteins. Dop (deamidase of Pup; Mtb Rv2112c/MT2172) deamidates the carboxyl-terminal glutamine of Pup to glutamate, preparing it for ligation to target proteins by proteasome accessory factor A (PafA). While studies have shed light on the conjugation of Pup to proteins, it was not known if Pup could be removed from substrates in a manner analogous to the deconjugation of Ub from eukaryotic proteins. Here, we show that Mycobacteria have a “depupylase” activity provided by Dop. The discovery of a depupylase strengthens the parallels between the Pup and Ub tagging systems of prokaryotes and eukaryotes, respectively.
Introduction
Proteasomes are compartmentalized proteases found in all eukaryotes and archaea, and bacteria of the class Actinomycetes [reviewed in (Cerda-Maira and Darwin, 2009)] and Nitrospira (De Mot, 2007). In the eukaryotic proteasome pathway, ubiquitin (Ub) provides the recognition and specificity required to target proteins to the proteasome for proteolysis [reviewed in (Pickart and Cohen, 2004)]. Ub is activated in a multi-step process prior to conjugation to lysine (Lys) residues on protein substrates [reviewed in (Kerscher et al., 2006)]. Additional Ub molecules can be added to Ub attached to a target substrate, and it is these polyubiquitin chains that are recognized by receptor proteins associated with the proteasome. At the proteasome, polyubiquitin chains are usually removed by deubiquitinating enzymes (DUBs) prior to substrate degradation [reviewed in (Finley, 2009)]. DUB activity not only allows Ub recycling, but also can control or prevent the degradation of specific substrates by the proteasome [reviewed in (Komander et al., 2009; Reyes-Turcu et al., 2009)].
Unlike eukaryotes, proteasome-bearing mycobacteria do not have a Ub system to target proteins for proteolysis but instead use the small protein Pup (Burns et al., 2009; Pearce et al., 2008). Pup is an intrinsically disordered protein with no sequence or structural homology to Ub (Chen et al., 2009; Liao et al., 2009; Sutter et al., 2009). Although the end-point for both the Ub and Pup degradation systems is a proteasome, the enzymology of ubiquitylation and pupylation appear completely different. For one, despite the presence of a di-Gly motif near the C-terminus of Pup, Pup conjugates to substrate lysines via a glutamate (Glu), not glycine. However, Pup is synthesized with a C-terminal glutamine (Gln), which is deamidated by Dop prior to substrate ligation by PafA (Striebel et al., 2009). Neither Dop nor PafA is similar to Ub-activating enzymes, and bioinformatic analysis suggests these enzymes are structurally similar to the carboxylate-amine/ammonia ligase super family of glutamine synthetases (Iyer et al., 2008). Several members of this family catalyze the ligation of amine groups with glutamate γ-carboxylates, resulting in an amide linkage.
Although studies have shed light on Pup activation and conjugation to target proteins, little was known about how pupylated substrates are processed upon delivery to the proteasome until recently (Striebel et al., 2010). One might predict that a method to recycle Pup for subsequent ligations would be energetically favorable and efficient. In recent work, however, Striebel and co-workers describe the in vitro reconstitution of Mpa- and proteasome-dependent degradation of pupylated substrates, and show that Pup is not removed, but can be degraded following targeting of substrates to the proteasome. Here, we describe a “depupylase” (DPUP) activity that removes Pup from two proteasomal substrates in vitro. Our data also suggest that at least one substrate is depupylated in an Mpa-dependent manner in vivo. Therefore, the DPUP may function similarly to DUBs in eukaryotes to prevent or promote proteasomal degradation of certain proteins in Mycobacteria.
Identification of a depupylase activity
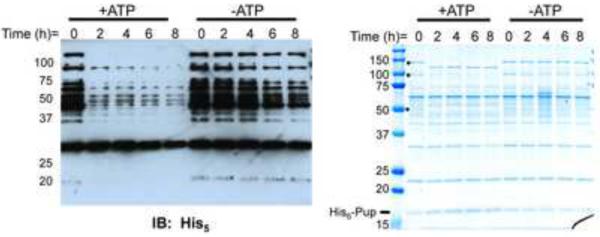
In previous studies, pupylated proteins (herein called the “pupylome”) were purified using His6-tagged Pup from both Mtb and Mycobacterium smegmatis (Msm) (Festa et al., 2010; Watrous et al., 2010). Upon in vitro analysis of the purified pupylome from Msm, we observed an apparent adenosine triphosphate (ATP)-dependent decay of numerous proteins as evidenced by the loss of the His6-Pup signal (Fig. 1, left). We hypothesized that, despite the inability of mass spectrometry to identify proteasome subunits in the pupylome (Festa et al., 2010; Watrous et al., 2010), enough proteasomes and Mycobacterial proteasome-associated ATPase (Mpa) co-purified with the pupylome and were responsible for its decay. To determine if the total protein level of the pupylome changed over time, we analyzed the reaction by Coomassie brilliant blue (CBB) staining (Fig. 1, right). There was little change in the amount of total protein over time, suggesting that the proteins were not being completely destroyed, as would be expected for proteasome-dependent degradation. Interestingly, it appeared as if at least one protein dropped in molecular weight (MW) while several others disappeared. Although it is possible that many of the proteins observed in the Coomassie stained gel are not true targets of pupylation but rather co-purify with pupylated proteins, the loss of Pup signal along with the apparent lack of total protein degradation suggested that the pupylome co-purified with an enzyme capable of removing Pup from proteins.
Fig. 1.
The pupylome is unstable in the presence of ATP. Stability of the pupylome in the presence and absence of ATP was analyzed by anti-His5 immunoblotting (left) and CBB staining (right) of 12% SDS-PAGE gels. Black dots indicate protein bands that change in the presence of ATP over time.
Mtb dop mutant lysates cannot depupylate proteasome substrates
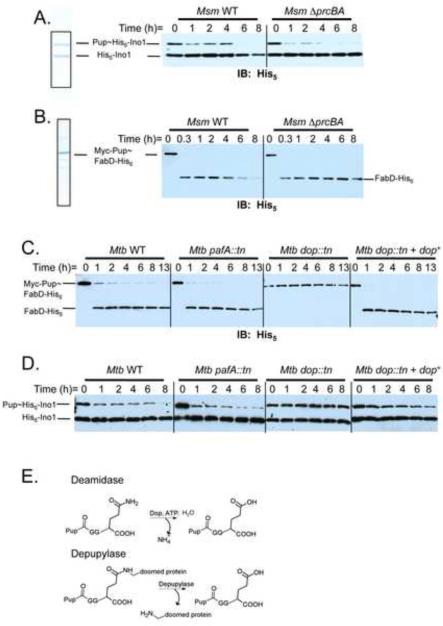
To investigate the instability of pupylated proteins further, we purified previously characterized pupylated proteasome substrates Msm Pup~His6-Ino1 (inositol 1-phosphate synthetase) (Fig. 2A) (Burns et al., 2009; Festa et al., 2010) and Mtb Myc-Pup~FabD-His6 (malonyl coA-acyl carrier protein) (Fig. 2B) (Pearce et al., 2006; Pearce et al., 2008) and monitored their stability in mycobacterial cell lysates. Pup~His6-Ino1 co-purifies with unpupylated His6-Ino1; because mycobacterial Ino1 forms tetramers (Norman et al., 2002), we presume that not every subunit is pupylated, resulting in the co-purification of pupylated and unpupylated Ino1. Upon addition of wild type (WT) Msm lysates to the Pup~His6-Ino1/His6-Ino1 mixture, we observed the disappearance of both Pup~His6-Ino1 as well as the unpupylated His6-Ino1 (Fig. 2A, left). In contrast, Pup~His6-Ino1, but not its unpupylated counterpart, nearly disappeared from lysates of proteasome-deleted Msm (Fig. 2A, right). These data indicate that the disappearance of Pup~His6-Ino1 does not require the proteasome, consistent with the presence of a DPUP activity in the lysates. It also appeared that proteasome activity was responsible for the reduction of His6-Ino1 over time, although the degradation was slow (Fig. 2A). Both Pup~His6-Ino1 and His6-Ino1 were stable in E. coli lysates (Fig. S1A), showing the reactions are mycobacteria-specific. The stability of another proteasome substrate, Myc-Pup~FabD-His6, was also examined (Pearce et al., 2006; Pearce et al., 2008). We used a tandem affinity purification protocol to purify Mtb Myc-Pup~FabD-His6 and similar to the Ino1 result, incubation of Myc-Pup~FabD-His6 in WT Msm lysates resulted in the formation of depupylated FabD-His6 followed by its degradation (Fig. 2B, left). This depupylated intermediate formed but appeared stable in proteasome mutant lysates (Fig. 2B, right), again showing that depupylation, but not degradation, could occur in the absence of proteasome activity.
Fig. 2.
Depupylation of substrates in mycobacterial lysates. (A) Pup~His6-Ino1 and His6-Ino1 were purified by Ni-NTA affinity chromatography as previously described (left) (Burns et al., 2009). The Pup~His6-Ino1/His6-Ino1 mixture and (B) Myc-Pup~FabD-His6 were added to Msm WT and Msm ΔprcBA lysates and stability was monitored by removing aliquots at the indicated times and adding sample buffer to stop the reaction. Samples were analyzed by 12% SDS-PAGE followed by immunoblotting with antibodies to His5. (C)–(D) Same assay in (B) but using Mtb WT and proteasome-associated mutant lysates. The dop mutation was complemented in single copy. (E) Deamidase reaction catalyzed by Dop (top) resembles the putative depupylase reaction (bottom). “tn” indicates the transposon ΦMycoMarT7. All strains are described in Table S1. See also figure S1.
We next investigated the stability of pupylated substrates in lysates of proteasome-associated mutants of Mtb (Table S1). Proteasome-inhibited or -depleted Mtb does not grow well in vitro (Darwin et al., 2003; Gandotra et al., 2007) and therefore could not be analyzed in this study. Myc-Pup~FabD-His6 was depupylated in lysates of WT and pafA (Pup ligase mutant) strains of Mtb (Fig. 2C). Surprisingly, we observed no depupylation of Myc-Pup~FabD-His6 in the dop lysate (Fig. 2C, third panel). A similar pattern was observed for the Pup~His6-Ino1 substrate (Fig. 2D). Depupylation was fully restored upon the genetic complementation of the dop mutation (Fig. 2C, D, far right). Complete degradation of FabD-His6 or His6-Ino1 was not observed, suggesting that either Mtb proteasome activity is particularly slow or inefficient in vitro. Nonetheless, these results demonstrated that Dop participates in the deconjugation of Pup from at least two targets of the proteasome.
The Dop-catalyzed deamidation reaction of Pup chemically resembles a putative depupylation reaction (Fig. 2E) and requires ATP binding but not hydrolysis (Striebel et al., 2009). An abundant protein species of the appropriate molecular weight for Dop was apparent in the pupylome reactions at all time points (Fig. 1A, right). To determine if this protein could be Dop, we performed immunoblot analysis with antibodies to Dop on the pupylome sample in Fig. 1A. Dop was present in the pupylome at its predicted molecular weight, suggesting that it interacts robustly, but not covalently, with one or more proteins in the pupylome (Fig. S1B). This result also likely explains why Dop was identified by mass spectrometry in the Mtb pupylome (Festa et al., 2010).
Recombinant Dop removes Pup from two mycobacterial proteasome substrates
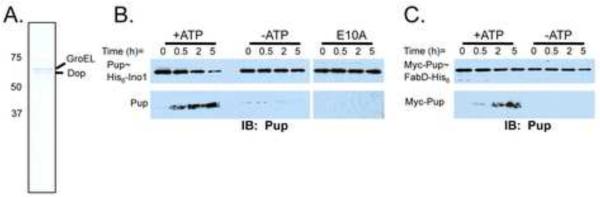
To test the DPUP activity of Dop in vitro, we purified His6-tagged Mtb Dop (MtDop) from a proteasome deletion mutant strain of Msm (ΔprcBA) (Burns et al., 2009) to ensure no contamination with proteasomes (Fig. 3A). Recombinant MtDop-His6 was highly insoluble when over-produced in E. coli (data not shown). Purification from Msm was done as described elsewhere (Striebel et al., 2009). We noticed that soluble Dop co-purified with GroEL, a chaperonin, after size exclusion chromatography, which may explain why Dop is insoluble when over-produced in E. coli. Consistent with this observation, a previous report showed that GroEL co-purified with Dop, Mpa and PafA when mycobacterial lysates were put over a Pup column to identify Pup-binding proteins (Striebel et al., 2009). Although GroEL is a common “contaminant” of protein preparations, it is possible that GroEL either maintains proper Dop folding, or itself participates in the Pup-proteasome system. Thus, all Dop assays were done in the presence of co-purified GroEL.
Fig. 3.
Recombinant Mtb Dop has depupylase activity. (A) CBB staining of His6-tagged MtDop purified from ΔprcBAMsm. (B) Depupylation of Pup~His6-Ino1/His6-Ino1 was monitored by removing aliquots at the indicated times and adding sample buffer to stop the reaction. Samples were analyzed by immunoblotting with antibodies to Pup. The same substrate was analyzed with mtDopE10A purified from ΔprcBAMsm. ATP was added to the reaction. (C) Another substrate, Myc-Pup~FabD-His6, was also assayed for depupylation by MtDop-His6. Free Pup was detected with the more sensitive chemiluminescent “Femto” reagent (see Supplemental Experimental Procedures). See also figure S2.
MtDop-His6 deamidated the C-terminal glutamine of Pup to glutamate (Fig. S2A), which was consistent with the previously reported activity of Dop (Striebel et al., 2009). To determine if MtDop-His6 had depupylase activity, we monitored the stability of Msm Pup~His6-Ino1 in the presence of Dop. When MtDop-His6 was added to the Pup~His6-Ino1/His6-Ino1 mixture, we observed the disappearance of Pup~His6-Ino1 (Fig. 3B, left) accompanied by the formation of free His6-Pup (Fig. 3B). It was previously shown that a mutation in the putative ATP binding site of Dop (DopE10A) was unable to complement the pupylation deficiency of a dop Msm mutant, demonstrating a role for this amino acid in deamidation (Imkamp et al., 2009). Therefore, we introduced this mutation into mtDop-His6 to further test Mtb Dop, and not a contaminant, as the DPUP. MtDopE10A failed to depupylate Pup~His6-Ino1 in vitro (Fig. 3B, right). Additionally, Dop converted Myc-Pup~FabD-His6 into FabD-His6 and Myc-Pup (Fig. 3C).
Similar to the deamidase reaction catalyzed by Dop (Striebel et al., 2009), the DPUP reaction was ATP-dependent, but ATP was not hydrolyzed during the course of the reaction; both adenosine-5'-[(α,β)-methyleno]triphosphate (ApCPP) and adenosine 5′-(β,γ-imido) triphosphate (AMP-PNP), non-hydrolyzable ATP analogues, could substitute for ATP (Fig. S2B). Additionally, MtDop-His6 was unable to hydrolyze linear Pup-substrate fusions (Fig. S2C), suggesting Dop is a strict isopeptidase.
Role of Dop in proteasomal degradation in vitro
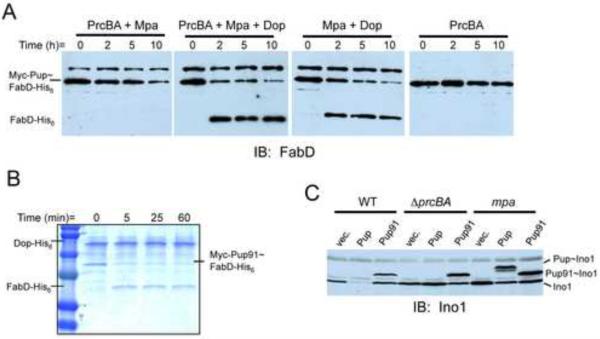
In an elegant study, Striebel and co-workers reported the degradation of pupylated substrates by Mpa and proteasomes in vitro (Striebel et al., 2010). Dop was not required for reconstitution and Pup was degraded during the course of the reaction. To determine whether the addition of Dop could enhance degradation by first depupylating the substrate, we reconstituted degradation with Mtb proteasomes and Mpa using our model substrate, Myc-Pup~FabD-His6 (Fig. 4A, left panel). Interestingly, the addition of recombinant Dop did not enhance degradation but appeared to rescue FabD-His6 from proteasomal degradation (Fig. 4A, second panel). These results suggest that depupylation by Dop is not required for Mpa/proteasome-dependent degradation of pupylated substrates in vitro, consistent with the Striebel and co-workers report.
Fig. 4.
In vitro and in vivo functional analysis of Dop and depupylation. (A) Reconstitution of proteasome-Mpa degradation of the substrate Myc-Pup~FabD-His6 showed that Dop could rescue FabD from degradation. Substrate stability was monitored by removing aliquots at the indicated times and adding sample buffer to stop the reaction. Samples were analyzed by SDS-PAGE followed by immunoblotting with polyclonal antibodies to Mtb FabD-His6. The cross-reactive band is recombinant Mpa. (B) In a reaction similar to that in Fig 3, Myc-Pup91~FabD-His6 was tested as a substrate for MtDop-His6 and analyzed by CBB staining. (C) Analysis of Ino1 in Msm ectopically over expressing his6-pup91 or his6-pup. Free Ino1 accumulates in degradation defective Msm (ΔprcBA or mpa) or in Msm expressing his6-pup91. Pup~Ino1 only accumulated in the mpa mutant.
Mpa-dependent depupylation of the proteasome substrate Ino1 in vivo
Previous studies demonstrated that the C-terminal half of Pup is necessary and sufficient to attach to proteins (Burns et al., 2010). To determine if Dop could remove truncated Pup from target proteins, we over-produced Myc-Pup91~FabD-His6, which has 29 amino acids deleted from the N-terminus of Pup (“Pup91”), and tested it as a substrate for Dop. Dop was able to deconjugate Pup91 from FabD, suggesting that the N-terminus of Pup is not necessarily required for depupylation in vitro (Fig. 4B).
Although the N-terminal half of Pup is dispensable for the conjugation and deconjugation of target proteins, it is required for proteasomal degradation in vivo and in vitro (Burns et al., 2010; Striebel et al., 2010). It was previously shown that Mpa engages the N-terminus of Pup to unfold a substrate, delivering it to the proteasome for degradation (Striebel et al., 2010). N-terminal truncations of Pup disrupt this activity, preventing Mpa-dependent proteasomal degradation. Consistent with these data, we showed that the over expression of Pup91 in both WT and proteasome-deleted Msm (ΔprcBA) results in an accumulation of both Ino1 and Pup91~Ino1 (Burns et al., 2010) (Fig. 4C). We thus concluded that the N-terminal half of Pup was required to initiate degradation of Pup~Ino1 in vivo.
If Pup~Ino1 were a direct substrate of the proteasome in vivo, we would expect to observe the accumulation full-length Pup~Ino1 in a proteasome mutant. However, the over production of full-length Pup in the ΔprcBA strain did not result in the accumulation of Pup~Ino1; instead only free Ino1 accumulated (Burns et al., 2010) (Fig. 4C). We hypothesized that Pup~Ino1, but not Pup91~Ino1, was depupylated in the ΔprcBA strain, and that the N-terminus of Pup was not only needed for Ino1 degradation, but was also required for the depupylation of Pup~Ino1 in vivo. Because the N-terminus of Pup is recognized by Mpa for substrate unfolding and there appears to be a defect in the removal of Pup91 from Ino1 in Msm, we hypothesized that the engagement of Mpa with the N-terminus of Pup facilitates the depupylation of Pup~Ino1 in vivo. To test this, we analyzed Ino1 in an mpa mutant of Msm overproducing Pup91 or Pup. Similar to WT and proteasome-deleted Msm, Pup91~Ino1 accumulated in the mpa mutant (Fig. 4C). In contrast to what was observed with the WT and proteasome-deleted Msm, Pup~Ino1 accumulated in the mpa mutant (Fig. 4C), suggesting that Pup is not removed from Ino1 in this strain due to the absence of Mpa. Preliminary data suggest that Mpa does not enhance depupylation in vitro (data not shown); however, this was not surprising because Dop could remove a truncated form of Pup from a substrate in vitro under the conditions tested (Fig. 4B). The requirement of Mpa for depupylation in vivo but not in vitro may be due to the presence of co-factors that regulate Dop activity in the intact cells, a hypothesis under active investigation.
Discussion
In this study we show that pupylation, like many post-translational modifications eukaryotes, is reversible. We demonstrated that Dop, an enzyme previously identified as the deamidase of Pup, appears to remove Pup from at least two proteins, but can also depupylate many proteins in vitro (Fig. 1), suggesting it is a general DPUP. It remains to be determined if this activity facilitates or rescues proteins (or both) from proteasomal degradation.
Dop appears to have two functions in organisms where Pup terminates in glutamine: a deamidase activity to convert glutamine to glutamate, which ultimately conjugates to target proteins; and a DPUP activity to remove Pup from the target proteins. The deamidase and DPUP reactions catalyzed by Dop require similar chemistries (Fig. 2E): both reactions involve the hydrolysis of an amide bond at the C-terminus of Pup. Without structural information, it is unclear if both reactions occur at the same active site of Dop, however, we hypothesize that this is highly probable due to the similarity of the proposed catalyzed reactions. Additionally, because Dop acts as both a deamidase and DPUP, our data support the observation that the Pup~target isopeptide bond is between the side chain carboxylate of glutamate (Sutter et al., 2010).
It remains to be determined if DPUP and proteasome activities are coupled in vivo, and if the mechanisms controlling depupylation are substrate dependent. Previous work strongly implicates a single Pup ligase in Mtb for all pupylation (Pearce et al., 2008). Data presented here suggest that there may be a single DPUP for all pupylated substrates, which could be hundreds of proteins (Festa et al., 2010; Watrous et al., 2010), thus novel mechanisms may be involved to impart specificity on Pup attachment and removal from its substrates.
Interestingly, we showed that Mpa, which structurally resembles the eukaryotic p97/valosin-containing protein ATPase complex (Darwin et al., 2005), appears to facilitate depupylation of Ino1 in vivo (Fig. 4). p97 is an ATPase associated with a number of cellular processes, including membrane fusion, endoplasmic reticulum-associated degradation, and proteasomal degradation (Brunger and DeLaBarre, 2003; Wang et al., 2004; Ye et al., 2001). It is able to interact with Ub conjugation machinery and either promote or inhibit ubiquitylation, thus influencing the stability of proteins. It was recently shown that p97 forms a complex with the COP9 signalosome (CSN) and the isopeptidase and deubiquitinase activities of CSN regulate the amount of ubiquitylated substrates bound to p97/VCP (Cayli et al., 2009). Perhaps functionally similar to CSN association with p97/VCP, our preliminary in vivo data suggest that Dop, possibly via other factors, interacts with Mpa to regulate the pupylation status of certain substrates. Additional experiments are underway to test this hypothesis.
Dop was proposed to have a function beyond deamidation because of its presence in bacterial species in which Pup terminates in Glu, presumably rendering deamidation unnecessary (Striebel et al., 2009). Our studies may have now answered the perplexing question as to why Dop is conserved in these bacteria. Important functions of Dop in these organisms may include the removal of Pup from substrates to facilitate delivery into the proteasome, or to rescue proteins from a doomed fate. In addition, like Ub, Pup might be recycled in Mtb, a hypothesis supported by recent data (Cerda-Maira et al., 2010). We presume that depupylation, like deubiquitination, is a highly regulated process. As with Ub and its related modifiers, the removal of Pup may be tightly controlled, and determine not only the stability of a target protein, but also its localization or activity.
Despite the functional similarities between the Pup and Ub systems, recent work has highlighted the biochemical differences between the conjugation pathways, as well as structural differences between Ub and Pup [reviewed in (Burns and Darwin, 2010)]. Our studies show that bacteria have not only evolved to covalently attach small protein modifiers to other proteins, but have also developed a system to reverse this process. Taken together, it is apparent that prokaryotes and eukaryotes have developed distinct but parallel mechanisms to regulate protein stability by proteasomes.
Experimental Procedures
Bacterial strains and culture conditions
Bacterial strains used in this study are listed in Table S1. Details for culture conditions are described in the Supplemental Experimental Procedures.
Plasmids, primers and antibodies
Plasmids and primers used in this study are listed in Table S1. All primers were purchased from Invitrogen. Phusion DNA polymerase (for all PCR) and restriction enzymes were purchased from New England Biolabs. Details for plasmid constructions are described in the Supplemental Experimental Procedures.
For polyclonal antibody production, Mtb Dop-His6 was purified from E. coli. Dop-His6 was purified under denaturing conditions as described in the QIAexpressionist manual, separated by SDS-PAGE and used for the immunization of rabbits at Covance (Denver, PA). Monoclonal Penta-His antibodies were from Qiagen and Pup and Ino1 polyclonal rabbit antibodies are described elsewhere (Festa et al., 2010; Pearce et al., 2008). Horseradish peroxidase (HRP) coupled anti-rabbit secondary antibodies were used according to manufacturer's instructions (GE/Amersham). Detection of HRP was performed using either SuperSignal West Pico or West Femto Chemiluminescent Substrate (ThermoScientific).
Pupylome instability assay
The Msm pupylome was purified as previously described (Burns et al., 2009). 20 μg of the pupylome was incubated with 5 mM ATP, 5 mM MgCl2, and 1 mM DTT in 50 mM Tris-HCl, pH 8 in a final volume of 150 ml. At indicated times, samples were withdrawn and added to SDS loading buffer. Samples were analyzed by SDS-PAGE and/or transferred to nitrocellulose membrane and blocked with 3% BSA. Detection with horseradish peroxidase was performed using either SuperSignal West Pico or West Femto Chemiluminescent substrate (ThermoScientific).
Dop-His6, Pup~His6-Ino1, Myc-Pup~FabD-His6 and Myc-Pup91~FabD-His6 purification and DPUP and proteasome reconstitution assays
Purification of recombinant protein from Msm and details for reconstitution assays are described in the Supplemental Experimental Procedures.
In vivo substrate accumulation assays
Experiments were performed as previously described (Burns et al., 2010).
Highlights.
Deconjugation of a bacterial post-translational modifier.
Dop catalyzes the removal of Pup from two proteasome substrates.
Dop is active in the presence or absence of proteasomes.
Supplementary Material
Acknowledgments
We are grateful to A. Darwin, D. Finley, T. Huang, and K. Walters for critical review of drafts of this manuscript. We thank J. Raper and R. Thomson for use of their FPLC instrumentation and Superdex200 column. We thank C. Barry for gifts of the proteasome mutant Msm and the polyG vector, and G. DeMartino and R. Hampton for helpful conversations. This work was supported by NIH R01 grants HL92774 awarded to K.H.D., AI070285 awarded to H.L., and AI 30036, 37856, and 36973 awarded to W.R.B. K.H.D is supported by a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brunger AT, DeLaBarre B. NSF and p97/VCP: similar at first, different at last. FEBS Lett. 2003;555:126–133. doi: 10.1016/s0014-5793(03)01107-4. [DOI] [PubMed] [Google Scholar]
- Burns KE, Darwin KH. Pupylation versus ubiquitylation: tagging for proteasome-dependent degradation. Cell Microbiol. 2010;12:424–431. doi: 10.1111/j.1462-5822.2010.01447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KE, Liu WT, Boshoff HI, Dorrestein PC, Barry CE., 3rd Proteasomal protein degradation in Mycobacteria is dependent upon a prokaryotic ubiquitin-like protein. J Biol Chem. 2009;284:3069–3075. doi: 10.1074/jbc.M808032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KE, Pearce MJ, Darwin KH. Prokaryotic ubiquitin-like protein provides a two-part degron to Mycobacterium proteasome substrates. J Bacteriol. 2010;192:2933–2935. doi: 10.1128/JB.01639-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayli S, Klug J, Chapiro J, Frohlich S, Krasteva G, Orel L, Meihardt A. COP9 Signalosome Interacts ATP-dependently with p91/Valosin-containg Protein (VCP) and Controls the Ubiquitination Status of Proteins Bound to p97/VCP. Journal of Biological Chemistry. 2009;284:34944–34953. doi: 10.1074/jbc.M109.037952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerda-Maira F, Darwin KH. The Mycobacterium tuberculosis proteasome: more than just a barrel-shaped protease. Microbes Infect. 2009;11:1150–1155. doi: 10.1016/j.micinf.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerda-Maira F, Pearce MJ, Fuortes M, Bishai WR, Hubbard SR, Darwin KH. Molecular Analysis of the Prokaryotic Ubiquitin-Like Protein (Pup) Conjugation Pathway in Mycobacterium tuberculosis. Mol Microbiol. 2010 doi: 10.1111/j.1365-2958.2010.07276.x. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Solomon WC, Kang Y, Cerda-Maira F, Darwin KH, Walters KJ. Prokaryotic ubiquitin-like protein pup is intrinsically disordered. J Mol Biol. 2009;392:208–217. doi: 10.1016/j.jmb.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin KH, Ehrt S, Weich N, Gutierrez-Ramos J-C, Nathan CF. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302:1963–1966. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- Darwin KH, Lin G, Chen Z, Li H, Nathan CF. Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol Microbiol. 2005;55:561–571. doi: 10.1111/j.1365-2958.2004.04403.x. [DOI] [PubMed] [Google Scholar]
- De Mot R. Actinomycete-like proteasomes in a Gram-negative bacterium. Trends Microbiol. 2007;15:335–338. doi: 10.1016/j.tim.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Festa RA, McAllister F, Pearce MJ, Mintseris J, Burns KE, Gygi SP, Darwin KH. Prokaryotic ubiquitin-like protein (Pup) proteome of Mycobacterium tuberculosis. PLoS One. 2010;5:e8589. doi: 10.1371/journal.pone.0008589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandotra S, Schnappinger D, Monteleone M, Hillen W, Ehrt S. In vivo gene silencing identifies the Mycobacterium tuberculosis proteasome as essential for the bacteria to persist in mice. Nat Med. 2007;13:1515–1520. doi: 10.1038/nm1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imkamp F, Rosenberger T, Striebel F, Keller PM, Amstutz B, Sander P, Weber-Ban E. Deletion of dop in Mycobacterium smegmatis abolishes pupylation of protein substrates in vivo. Mol Microbiol. 2009;75:744–754. doi: 10.1111/j.1365-2958.2009.07013.x. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Burroughs AM, Aravind L. Unraveling the biochemistry and provenance of pupylation: a prokaryotic analog of ubiquitination. Biol Direct. 2008;3:45. doi: 10.1186/1745-6150-3-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- Liao S, Shang Q, Zhang X, Zhang J, Xu C, Tu X. Pup, a prokaryotic ubiquitin-like protein, is an intrinsically disordered protein. Biochem J. 2009;422:207–215. doi: 10.1042/BJ20090738. [DOI] [PubMed] [Google Scholar]
- Norman RA, McAlister MS, Murray-Rust J, Movahedzadeh F, Stoker NG, McDonald NQ. Crystal structure of inositol 1-phosphate synthase from Mycobacterium tuberculosis, a key enzyme in phosphatidylinositol synthesis. Structure. 2002;10:393–402. doi: 10.1016/s0969-2126(02)00718-9. [DOI] [PubMed] [Google Scholar]
- Pearce MJ, Arora P, Festa RA, Butler-Wu SM, Gokhale RS, Darwin KH. Identification of substrates of the Mycobacterium tuberculosis proteasome. EMBO J. 2006;25:5423–5432. doi: 10.1038/sj.emboj.7601405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322:1104–1107. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striebel F, Hunkeler M, Summer H, Weber-Ban E. The mycobacterial Mpa-proteasome unfolds and degrades pupylated substrates by engaging Pup's N-terminus. EMBO J. 2010;29:1262–1271. doi: 10.1038/emboj.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striebel F, Imkamp F, Sutter M, Steiner M, Mamedov A, Weber-Ban E. Bacterial ubiquitin-like modifier Pup is deamidated and conjugated to substrates by distinct but homologous enzymes. Nat Struct Mol Biol. 2009;16:647–651. doi: 10.1038/nsmb.1597. [DOI] [PubMed] [Google Scholar]
- Sutter M, Damberger FF, Imkamp F, Allain FH, Weber-Ban E. Prokaryotic ubiquitin-like protein (Pup) is coupled to substrates via the side chain of its C-terminal glutamate. J Am Chem Soc. 2010;132:5610–5612. doi: 10.1021/ja910546x. [DOI] [PubMed] [Google Scholar]
- Sutter M, Striebel F, Damberger FF, Allain FH, Weber-Ban E. A distinct structural region of the prokaryotic ubiquitin-like protein (Pup) is recognized by the N-terminal domain of the proteasomal ATPase Mpa. FEBS Lett. 2009;583:3151–3157. doi: 10.1016/j.febslet.2009.09.020. [DOI] [PubMed] [Google Scholar]
- Wang Q, Song C, Li CC. Molecular perspectives on p97-VCP: progress in understanding its structure and diverse biological functions. J Struct Biol. 2004;146:44–57. doi: 10.1016/j.jsb.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Watrous J, Burns K, Liu WT, Patel A, Hook V, Bafna V, Barry CE, 3rd, Bark S, Dorrestein PC. Expansion of the mycobacterial “PUPylome”. Mol Biosyst. 2010;6:376–385. doi: 10.1039/b916104j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.