Abstract
Insulin-like growth factor binding protein 7 (IGFBP7) functions mostly independent of the IGF signaling pathway and acts as a tumor suppressor in multiple cancers, but roles of IGFBP7 genetic variants in cancer remains unknown. In a hospital-based study of 1,065 patients with squamous cell carcinoma of head and neck (SCCHN) and 1,112 cancer-free controls of non-Hispanic whites, we investigated associations between two putatively functional IGFBP7 promoter single nucleotide polymorphisms (SNPs) (−702G>C, rs11573014 and −418G>A, rs4075349) and SCCHN risk. A significantly lower SCCHN risk was observed in those subjects carrying −418AG (adjusted OR=0.82, 95% CI=0.67–0.99) and −418AG+AA (adjusted OR=0.82, 95% CI=0.69–0.99) genotypes than those carrying the −418GG genotype, but not for the −702G>C SNP. However, those subjects carrying two common homozygous genotypes of these two SNPs (−418GG and −702GG) had an increased risk (adjusted OR=1.21, 95% CI=1.00-0.1.46) than did those carrying variant genotypes (−418AG+AA and −702CG+CC). This increased risk was more evident in subgroups of never smokers and subjects with oral cancer. Further functional analysis showed that the IGFBP7 −418A allele had significantly higher promoter and DNA-protein binding activities than did the G allele, suggesting a tumor suppressor role of this allelic change in the SCCHN etiology. We conclude that the functional variant −418 G>C in the IGFBP7 promoter is associated with reduced risk of SCCHN, likely by enhancing the IGFBP7 promoter and DNA-protein binding activities. Larger studies are needed to validate our findings.
Keywords: IGFBP7, case-control study, tumor suppressor gene, head and neck cancer, promoter polymorphism
Introduction
Squamous cell carcinoma of the head and neck (SCCHN) is a heterogeneous cancer originated from the oral cavity, oropharynx, hypopharynx and larynx, which accounts for over 90% of all types of head and neck cancer [1]. In 2009, there were estimated 48,010 new cases and 11,260 deaths of SCCHN in the United States [2]. While the history of tobacco and alcohol use remains the most well-recognized risk factors for SCCHN, increasing evidence suggests that genetic variations in genes involved in carcinogen metabolism, cell cycle regulation, DNA repair, apoptosis and other cellular processes play important roles in the etiology of SCCHN [3,4].
The IGFs (insulin-like growth factors) signaling pathway regulates cell growth, differentiation and apoptosis through a system consisting of IGF-I, IGF-II, IGF receptors (IGF-IR and IGF-IIR), insulin-like growth factor binding proteins (IGFBPs) and IGFBP proteases [5,6]. To date, there are at least six reported and well-characterized IGFBPs (IGFBP-1 to -6), all containing highly conserved cysteine-rich N-terminal and C-terminal domains and a variable mid-region [7]. These six IGFBPs have been characterized by their high binding affinities to IGFs and found to be able to either positively or negatively regulate the IGFs signaling pathway, depending on different cell types [7,8]. Recently, several new cysteine-rich proteins have been identified, named as IGFBP-related proteins (IGFBP-rPs) that share structural similarities with conventional IGFBPs in the N-terminal domain but bind to IGFs with a much lower affinity [7]. IGFBP-rP1 (also called IGFBP7, MAC25, TAF, or PSF), the first identified member of IGFBP-rPs, has been reported to function mainly in an IGF-independent manner, because of its weak binding affinity to IGFs [9].
Indeed, studies have shown that IGFBP7 is involved in multiple pathways. For example, IGFBP7 binds to insulin with a high and specific affinity [9], and it is regulated through the proteolytic cleavage by a membrane-bound serine proteinase matriptase [10]. In addition, DNA methylation, retinoid acid, and transforming growth factor beta 1 (TGF-β1) have all been reported to be regulators of the IGFBP7 transcription [11–13]. The expression level of IGFBP7 has also been reported to be associated with tumor development; and in many cancer types, such as cancers of the breast, prostate, lung, colon and rectum, and thyroid, IGFBP7 was reported to act as a tumor suppressor through the regulation of cell proliferation, cell adhesion, apoptosis, cellular senescence and angiogenesis [14–23]. Paradoxically, several studies suggested an oncogenic role of IGFBP7 in glioma, because IGFBP7 was shown to induce cell growth, migration and angiogenesis of glioma cancer cells [12,24]. These studies suggest that IGFBP7 may play different roles in different cancers.
Human IGFBP7 is a 30 kDa protein encoded by the IGFBP7 gene that is located on chromosome 4q12–13 with five exons. Although IGFBP7 is believed to play important roles in the development of multiple cancers, no well-designed association studies have been conducted to assess the roles of genetic polymorphisms of the IGFBP7 gene in cancer risk. It has been hypothesized that the genetic variations in the promoter-regulatory region might affect the gene expression level and, therefore, modifying the cancer susceptibility. Thus, we conducted a hospital-based case-control study of 1,065 SCCHN patients and 1,112 cancer-free controls of non-Hispanic whites to evaluate the association between the promoter polymorphisms of IGFBP7 and SCCHN risk and performed additional experiments to unravel the underlying molecular mechanisms.
Materials and methods
Study subjects
The recruitment of study subjects for the present study has been previously described [25,26]. Briefly, we initially recruited 1,111 newly diagnosed, untreated SCCHN (i.e., cancers of oral cavity, oropharynx, hypopharynx, and larynx) patients and 1,130 cancer-free controls at The University of Texas M. D. Anderson Cancer Center between October 1999 and October 2007. The SCCHN patients and cancer-free controls were all non-Hispanic whites and frequency matched by age (± 5 years) and sex. Sixteen patients with second primary tumors, primary tumors of nasopharynx or sinonasal tract, or any histopathologic diagnoses other than SCCHN were excluded. The self-reported, cancer-free controls were recruited from those outpatient clinics visitors at M. D. Anderson Cancer Center, who agreed to participate in the study and were not genetically related to the patients already enrolled in this study. All potential study subjects were first surveyed by using a short questionnaire to determine their willingness to participate in this study. In the face-to-face interview by a trained interviewer using a structured questionnaire, the subjects provided information about demographics and risk factors, including age, sex, ethnicity, history of tobacco and alcohol consumption from each individual after having signed a written consent. Among all eligible subjects, the response rate for SCCHN patients and cancer-free controls were approximately 93% and 85%, respectively. Among these SCCHN cases and controls, 30 cases and 18 controls either failed to be genotyped or showed inconsistent genotyping results in repeated genotyping and thus were excluded from the analysis. Therefore, our final analysis included 1,065 SCCHN patients and 1,112 cancer-free controls. Each subject donated one-time 30-mL venous blood to be used for biomarker assays, including DNA extraction for genotyping. The research protocol was approved by the M.D. Anderson Institutional Review Board.
Polymorphism selection and senotyping of IGFBP7
As of April, 2010, the IGFBP7 gene reportedly had 951 variants or single nucleotide polymorphisms (SNP) (http://www.ncbi.nlm.nih.gov/projects/SNP/), most of which are extremely rare, nonfunctional, or not validated by additional genotyping in the HapMap database (http://hapmap.ncbi.nlm.nih.gov/). In the NCBI SNP database, there are only four common (MAF- minor allele frequency ≥ 0.05) potentially functional promoter SNPs (rs4074555, rs11573018, rs4075349 and rs11573014). However, the first two SNPs (rs4074555 and rs11573018) failed to be genotyped in all currently available genotyping platforms, including polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP), SNPlex, and Taqman, nor these two SNPs were validated by the HapMap Project. Although rs4074555 and rs11573018 were genotyped in the NIH Polymorphism Discovery Resource (NIHPDR) 90 individual screening subset (http://www.ncbi.nlm.nih.gov/) with MAF of 0.193 and 0.119, respectively, these two initial reported SNPs may have to be re-examined in the population of non-Hispanic Caucasian due to the possibly ethnic difference in the allele/genotype frequencies. Therefore, we genotyped the other two SNPs (−702G>C, rs11573014 and −418G>A, rs4075349) by the SNPlex assay in the DNA Core Facility at MD Anderson Cancer Center, according to the protocol of manufacturer (Applied Biosystems, Foster City, CA). The data output from the SNPlex assays were analyzed in the GeneMapper software (Applied Biosystems) to determine the genotypes. The samples failed to be genotyped in the SNPlex assays were also re-evaluated with the PCR-RFLP method. Approximately 10% of the samples were randomly selected and repeated with RFLP, and the error rate was less than 1%. One case with inconsistent genotype results was excluded from the final analysis.
Construction of reporter plasmids
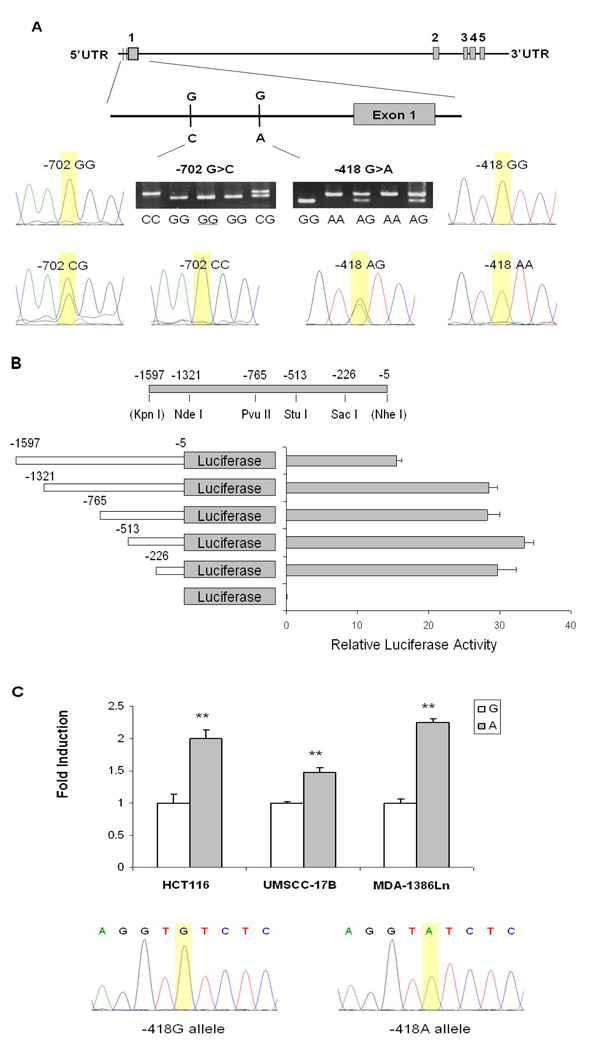
We first amplified a 1,593-bp IGFBP7 promoter with PCR (from −1597 to −5 relative to the transcription start site) from human head and neck carcinoma cell line MDA-1386Ln (obtained from Dr. Jeffrey N. Myers at M. D. Anderson Cancer center), which has a GG homozygous genotype at −418 position, with the primers (forward) 5’-AAGGTACCAGTGAGCCAAAATCTCACCACT-3’ and (reverse) 5’-AAGCTAGCGCGCGAGTGAGCCGTGTC-3’. The underlined sequences represented the KpnI and NheI restriction enzyme sites in forward and reverse primers, respectively. The amplified PCR products were then cloned into the basic-pGL3 firefly luciferase vector (Promega, Madison, WI) at the KpnI and NheI restriction sites. Different fragments of the IGFBP7 promoter were generated with a series of deletions of KpnI plus either NdeI, PvuII, StuI, or SacI (New England Biolabs, Beverly, MA), on the cloned IGFBP7-PGL3 containing the 1,593-bp promoter, followed by the Klenow enzyme (Sigma –Aldrich, St. Louis, MO) treatment and re-ligation. The corresponding A allele containing IGFBP7-PGL3 was generated with a site-directed mutagenesis kit, according to the manufacturer’s protocol (Agilent Technologies Stratagene, La Jolla, CA). The −418G and −418A constructs were sequenced to confirm the orientation and integrity of each insert (Figure 1).
Figure 1.
The IGFBP7gene structure, its promoter reporter gene constructs, and luciferase assays. A, Genomic structure and locations of two selected promoter SNPs, alone with their genotyping assays with sequencing confirmation; B, Identification of the functional region of the IGFBP7 promoter by a series of sequential deletions. C, Luciferase assay for the two IGFBP7 promoter constructs with −513 to −5 sequences relative to the transcriptional starting site, and containing G and A alleles at −418, respectively. The −418G and −418A constructs were sequenced to confirm the orientation and integrity. **denotes P < 0.01 for the difference between alleles G and A.
Transient transfection and luciferase reporter gene assay
The human colon cancer cell line HCT116 (a gift from Dr. Bert Vogelstein of John Hopkins University School of Medicine) and head and neck carcinoma cell line MDA-1386Ln were cultured in 1X Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (Sigma-Aldrich, MO), and human head and neck carcinoma cell line UMSCC-17B (a gift from Dr. Reuben Lotan at M. D. Anderson Cancer Center) was cultured in DMEM/F12 medium with 10% fetal bovine serum at 37 °C in 5% CO2. A total of 1×105 of each cell line were plated onto each well of the 24-well plates and transiently transfectd with 1.0 µg of −418G or −418A reporter constructs using FuGENE HD transfection reagent (Roche Applied Science, IN). The 50 ng of p-TK renilla luciferase (pRL-TK) (Promega) was co-transfected as an internal control. After 48 hr of transfection, the luciferase activities of both firefly and renilla luciferase were quantified by a Dual-Luciferase Reporter Assay System (Promega), and the ratios of these two activities were obtained as relative luciferase activity, according to the manufacturer’s instructions (Monolight™ 3010 Luminometer, BD Biosciences, San Jose, CA). The fold induction of relative luciferase activity was measured by setting reporter gene activity of construct with the G allele as 1. The experiment was performed in triplicates, and the mean and standard error of fold induction were calculated and tested by Student t test in Stata 10.0 (StataCorp LP, College Station, TX).
Nuclear extract preparation and electrophoretic mobility shift assay (EMSA)
The nuclear extracts from UMSCC-17B cells were prepared according to the method of Andrews and Faller [27]. Complementary single-stranded oligonucleotides (5’-GTAATGAGCACTCAGGTGTCTCAGGCCCA -3’ for the G allele and 5’-GTAATGAGCACTCAGGTATCTCAGGCCCA -3’ for the A allele) were biotin-labeled using the 3’-end biotin labeling kit (Thermo Scientific, Rockford, IL) and re-annealed to perform the DNA binding assays using the LightShift Chemiluminescent EMSA kit (Thermo Scientific, Rockford, IL). The competition was performed with a 50-fold excess of unlabeled oligonucleotides. Supershift experiments were performed with anti-GATA2 antibody (Abcam Inc., Cambridge, MA) or nonspecific rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA).
Statistical analysis
The chi-square tests were used to test the differences in the distributions of categorical variables, including age groups, sex, tobacco and alcohol use, and genotypes of IGFBP7 polymorphisms between the cases and controls. Crude and adjusted (for age, sex, smoking and drinking status) odds ratios (ORs) and 95% confidence intervals (CIs) were obtained from unconditional univariate and multivariable logistic regression analyses to evaluate the associations between IGFBP7 polymorphisms and SCCHN risk, which were also stratified by the subgroups of age, sex, tobacco use, alcohol use, and tumor sites. Homogeneity of ORs between different strata and trend tests were evaluated using a Cochrane-Mantel-Haenszel test as previously described [28], followed by analyses of gene-environment interactions, which were evaluated by the P value for the interaction term in multivariate logistic regression models with adjustment for age, sex, smoking and drinking status. Haplotypes for the IGFBP7 polymorphisms were constructed according to the PHASE program [29,30], and Pearson’s chi-square test was used to test for the difference in haplotypes distributions between cases and controls. All tests were two-sided, and P < 0.05 was considered significant. All data were analyzed with SAS statistical software program (SAS/STAT version 9.1.3; SAS Institute Inc., Cary, NC).
Results
Characteristics of the study population
In our final analysis, 1,065 SCCHN cases and 1,112 controls of non-Hispanic whites were adequately matched by age and sex (P = 0.643 and 0.605, respectively). The mean age ± standard deviation was 57.2 ± 11.2 years (range 18–90) for cases and 56.7 ± 11.0 (range 20–87) for controls. The median age for both cases and controls was 57 years, and there were more males than females in both cases (75% vs 25%) and controls (76% vs 24%). Comparing with controls, the cases were more likely to be smokers and drinkers (P < 0.001 for both), which were further adjusted for in the subsequent multivariate logistic regression analyses. The likely human papillomavirus (HPV)-associated oropharyngeal cancers accounted for approximately half of the cases (50.4%) in the study population (Table I).
Table I.
Demographic characteristics of SCCHN cases and cancer-free controls of a non-Hispanic population
| Variables | Cases No. (%) |
Controls No. (%) |
P valuea |
|---|---|---|---|
| All subjects | 1,065 (100.0) | 1,112 (100.0) | |
| Age group (y) | |||
| Range | 18–90 | 20–87 | 0.643 |
| Mean | 57.2 ±11.2 | 56.7±11.0 | |
| ≤ 57 (median) | 567 (53.2) | 581 (52.2) | |
| >57 (median) | 498 (46.8) | 531 (47.8) | |
| Sex | 0.605 | ||
| Female | 262 (24.6) | 263 (23.7) | |
| Males | 803 (75.4) | 849 (76.4) | |
| Smoking Status | <0.001 | ||
| Never | 298 (30.0) | 544 (48.9) | |
| Former | 363 (34.1) | 406 (36.5) | |
| Current | 404 (37.9) | 162 (14.6) | |
| Alcohol Status | <0.001 | ||
| Never | 296 (27.8) | 488 (43.9) | |
| Former | 229 (21.5) | 180 (16.2) | |
| Current | 540 (50.7) | 444 (39.9) | |
| Tumor Sites | |||
| Oral Cavity | 318 (29.9) | ||
| Oropharynx | 537 (50.4) | ||
| Hypopharynx | 42 ( 3.9) | ||
| Larynx | 168 (15.8) |
Two-sided χ2 test.
Association between IGFBP7 polymorphisms and risk of SCCHN
The IGFBP7 gene structure and locations of the two SNPs (−702G>C, rs11573014 and −418G>A, rs4075349) as well as their genotyping assays with sequencing confirmation are displayed in Figure 1A. The distribution of −702G>C genotypes in control subjects agreed with the Hardy-Weinberg equilibrium (HWE) (P = 0.156), but the distribution of −418G>A genotypes in control subjects deviated from HWE (P = 0.045), suggesting potential biases may exist. When the −418GG genotype was used as the reference, a reduced risk of SCCHN was significantly associated with the AG genotype (adjusted OR, 0.82; 95% CI, 0.67–0.99) but not the −418AA genotype (adjusted OR, 0.85; 95%CI, 0.65–1.11) (likely due to fewer subjects in this subgroup) after the adjustment of age, sex, smoking and drinking statues. In the dominant model, the combined −418AG+AA genotypes were also associated with significantly reduced risk of SCCHN (adjusted OR, 0.82; 95% CI, 0.69–0.99). However, no significantly altered risk was observed for the variant −702CG and CC genotypes, when the −702GG genotype was used as the reference (Table II).
Table II.
Association between genotypes of IGFBP7 polymorphisms and risk of SCCHN in a non-Hispanic white population
| SNP | Cases No. (%) |
Controls No. (%) |
P value |
Crude OR (95% CI) |
Adjusted OR (95% CI)a |
|---|---|---|---|---|---|
| All subjects | 1,065 | 1,112 | |||
| Genotypes | |||||
| −418G>A (rs4075349) | 0.136b | ||||
| GG | 395 (37.1) | 369 (33.2) | 1.00 | 1.00 | |
| AG | 506 (47.5) | 571 (51.4) | 0.83 (0.69–1.00) | 0.82 (0.67–0.99) | |
| AA | 164 (15.4) | 172 (15.5) | 0.89 (0.69–1.15) | 0.85 (0.65–1.11) | |
| AG+AA | 670 (62.9) | 743 (66.8) | 0.056c | 0.84 (0.71–1.01) | 0.82 (0.69–0.99) |
| A allele frequency | 0.392 | 0.411 | 0.212d | ||
| − 702G>C (rs11573014) | 0.172b | ||||
| GG | 794 (74.6) | 791 (71.1) | 1.00 | 1.00 | |
| CG | 251 (23.6) | 301 (27.1) | 0.83 (0.68–1.01) | 0.83 (0.68–1.02) | |
| CC | 20 ( 1.9) | 20 ( 1.8) | 1.00 (0.53–1.87) | 0.91 (0.47–1.74) | |
| CG+CC | 271 (25.5) | 321 (28.9) | 0.073c | 0.84 (0.70–1.02) | 0.83 (0.68–1.02) |
| C allele frequency | 0.137 | 0.153 | 0.146d | ||
| No. of favorable genotypes in the combined genotypesf |
0.081e | ||||
| 0 (−418GG and −702GG) | 379 (36.13) | 356 (32.4) | 1.00 | 1.00 | |
| 1 (−418AG+AA and −702GG) | 415 (39.6) | 435 (39.6) | 0.90 (0.74–1.09) | 0.89 (0.73–1.10) | |
| 2 (−418AG+AA and −702CG+CC) | 255 (24.3) | 308 (28.0) | 0.78 (0.62–0.97) | 0.76 (0.60–0.96) | |
| P value for trend | 0.026 | 0.071 | |||
| ≥1 Other genotypes | 686 (64.4) | 804 (68.0) | 0.078‖ | 1.00 | 1.00 |
| 0 (−418GG and −702GG) | 379 (35.6) | 356 (32.0) | 1.17 (0.98–1.40) | 1.21 (1.00–1.46) |
Adjusted for age, sex, smoking and alcohol status in the logistic regression model.
Two-sided χ2 test for the distribution of three genotypes.
Two-sided χ2 test for distribution of combined genotypes.
Two-sided χ2 test for differences in allele distributions between cases and controls.
Two-sided χ2 test for distribution of all combined genotypes.
The number of favorable genotypes used for the calculation were −418AG+AA, and −702CG+CC: 0 = carried common homozygous −418GG+ −702GG, 1 = only carried one variant genotype of −418AG+AA, and 2 = both of the variants genotypes (−418AG+AA and −702CG+CC).
The combined analysis of the two SNPs substantially improved the risk estimates, in which a reduced risk of SCCHN was observed in those who carried both of the variant genotypes (OR, 0.76 and 95% CI, 0.60–0.96 for −418AG+AA and −702CG+CC) in a variant-genotype dose-response manner (P for trend = 0.026 for crude ORs and 0.071 for adjusted ORs) (Table II). In other words, an increased risk of SCCHN was observed in those subjects carrying the common homozygous genotypes of both SNPs (−418GG and −702GG) (adjusted OR, 1.21, 95% CI, 1.00-0.1.46) than those carrying variant genotypes (−418AG+AA and −702CG+CC) (Table II).
To further evaluate the association between the combined variant genotypes of IGFBP7 −418G>A and −702G>C SNPs and SCCHN risk, we conducted a stratified analysis by subgroups of age, sex, smoking and drinking status, and tumor sites. As shown in Table III, an increased risk of SCCHN associated with the common homozygous genotypes of both SNPs (−418GG and −702GG) was more evident in the subgroups of never smokers (adjusted OR, 1.36; 95% CI, 1.01–1.83) and patients with cancer of the oral cavity (adjusted OR, 1.35; 95% CI, 1.02–1.77), compared with those having any of the variant (other) genotypes (Table III). However, there was no evident of an interaction between the combined genotypes and the variables used for the stratification (data not shown).
Table III.
Stratification analysis of the association with SCCHN risk by the combined IGFBP7genotypes (− 418G>A and −702G>C)
| Variables | Cases (n=1,065) | Controls (n=1,112) | Crude OR (95% CI) Other genotypes (reference) vs GG/GG |
Adjusted OR (95% CI)a Other genotypes (reference) vs. GG/GG |
||
|---|---|---|---|---|---|---|
| Other genotypesb n (%) |
GG/GG (−418/−702) n (%) |
Other genotypesb n (%) |
GG/GG (−418/−702) n (%) |
|||
| All subjects | 686 (64.4) | 379 (35.6) | 804 (68.0) | 356 (32.0) | 1.17 (0.98–1.40) | 1.21 (1.00–1.46) |
| Age (median) | ||||||
| ≤ 57 | 361 (63.7) | 206 (36.3) | 389 (67.0) | 192 (33.0) | 1.16 (0.91–1.47) | 1.16 (0.90–1.50) |
| > 57 | 325 (65.3) | 173 (34.7) | 367 (69.1) | 164 (30.9) | 1.19 (0.92–1.55) | 1.26 (0.95–1.66) |
| Sex | ||||||
| Female | 158 (60.3) | 104 (39.7) | 179 (68.1) | 84 (31.9) | 1.40 (0.98–2.01) | 1.38 (0.94–2.03) |
| Male | 528 (65.8) | 275 (34.2) | 577 (68.0) | 272 (30.0) | 1.11 (0.90–1.36) | 1.15 (0.93–1.43) |
| Smoking Status | ||||||
| Never | 181 (60.7) | 117 (39.3) | 367 (67.5) | 177 (32.5) | 1.34 (1.00–1.80) | 1.36 (1.01–1.83) |
| Former | 247 (68.0) | 116 (32.0) | 280 (69.0) | 126 (31.0) | 1.04 (0.77–1.42) | 1.06 (0.78–1.50) |
| Current | 258 (63.9) | 146 (36.1) | 109 (67.3) | 53 (32.7) | 1.16 (0.79–1.71) | 1.11 (0.74–1.66) |
| Drinking Status | ||||||
| Never | 185 (62.5) | 111 (37.5) | 325 (66.6) | 163 (33.4) | 1.20 (0.89–1.62) | 1.21 (0.90–1.65) |
| Former | 143 (62.5) | 86 (37.5) | 120 (66.7) | 60 (33.3) | 1.20 (0.80–1.81) | 1.22 (0.80–1.87) |
| Current | 358 (66.3) | 182 (33.7) | 311 (70.1) | 133 (29.9) | 1.19 (0.91–1.56) | 1.14 (0.86–1.52) |
| Tumor Site | ||||||
| Oral Cavity | 193 (60.7) | 125 (39.3) | 756 (68.0) | 356 (32.0) | 1.38 (1.06–1.78) | 1.35 (1.02–1.77) |
| Oropharynx | 354 (65.9) | 183 (34.1) | 756 (68.0) | 356 (32.0) | 1.10 (0.88–1.37) | 1.16 (0.93–1.45) |
| Larynx/Hypopharynx | 139 (66.2) | 71 (33.8) | 756 (68.0) | 356 (32.0) | 1.09 (0.79–1.48) | 1.07 (0.75–1.52) |
Adjusted for age, sex, smoking and alcohol status in the logistic regression model.
those who carried at least one variant genotype (−418AG+AA or −702CG+CC).
G to A allelic change at −418 location of the IGFBP7 promoter resulted in the increased promoter activity
We used a series of 5’ deletion methods to determine the regions of the promoter required for the IGFBP7 gene expression. As shown in Figure 1B, we found that the reporter gene construct containing sequences from −513 to −5 relative to the transcriptional starting site of IGFBP7 had the highest luciferase activity. The luciferase activity was not increased in longer fragments but was dramatically decreased when the region of −1597 to −1321 was included. Therefore, we believed that the region from −513 to −5 was the functional promoter region, which is consistent with our finding that only the −418G>A SNP was individually associated with SCCHN risk. We then used this −513 to −5 region to determine the allele-specific effect of −418G>A on the IGFBP7 promoter activity. Compared with the −418G, the IGFBP7 promoter containing −418A showed a significantly lower reporter gene expression in two human head and neck carcinoma cell lines MDA-1386Ln and UMSCC-17B and a human colon cancer cell line HCT116 (P < 0.01 for all) (Figure 1C), suggesting that the G to A allelic change at the IGFBP7 −418 site resulted in increased promoter activity in a non-tissue specific manner.
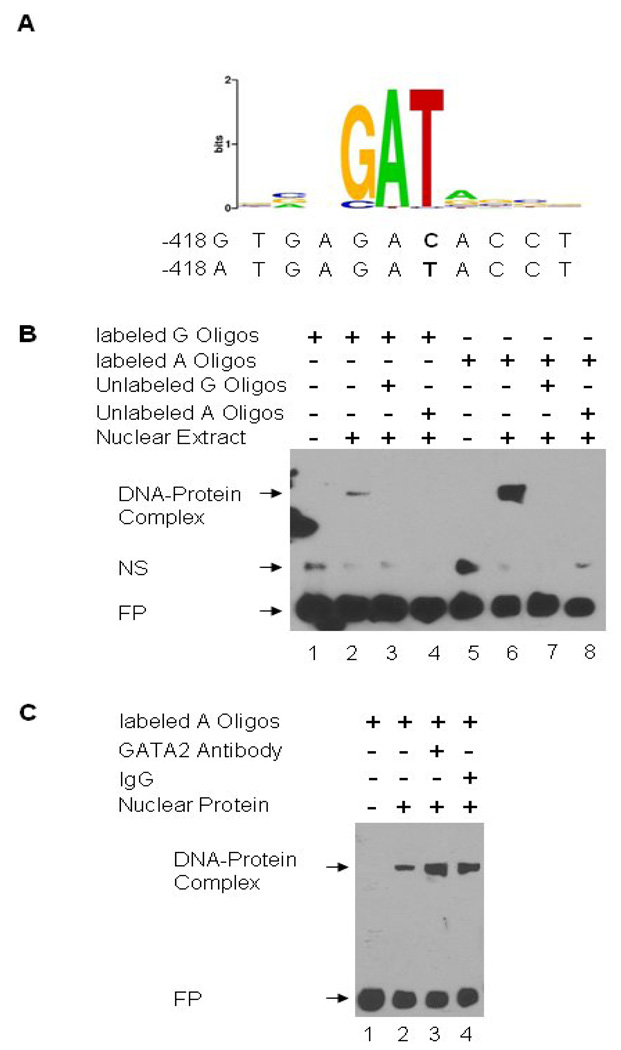
To further determine the underlying mechanism for the differential regulation of the IGFBP7 promoter activity at −418G>A, we used TFSearch (http://www.cbrc.jp/research/db/TFSEARCH.html) and transcription element search software (TESS) (http://www.cbil.upenn.edu/cgi-bin/tess/tess) to predict any potential transcription binding site change at −418G>A. We found that the GATA-binding protein 2 (GATA-2) was predicted by both sites to be able to potentially bind to G and A alleles differentially. The predicted GATA-2 binding motif from TESS is shown in Figure 2A. Based on this prediction, we therefore preformed an electrophoretic mobility-shift assay (EMSA) to test whether GATA-2 differentially binds to the G and A alleles at −418 site of IGFBP7. As shown in Figure 2B, the oligonucleotide containing A allele had a significantly stronger binding to the nuclear protein extract from UMSCC-17B cells than that with the G allele, and the excess of unlabeled oligonucleotides with the G or A allele competed for this binding activity. This result is consistent with that from the luciferase reporter gene assay. However, GATA-2 antibody did not result in the supershift (Figure 2B), indicating that, contrary to the prediction by TFSearch and TESS, GATA-2 may not be the transcriptional factor that binds to the promoter of IGFBP7 at −418 location. Therefore, more intensive molecular characterization of this binding site around the −418 location is warranted.
Figure 2.
Electrophoretic mobility shift assay (EMSA) for detection of DNA-protein binding at −418 of the IGFBP7 promoter. A, Prediction of GATA-2 binding site at the IGFBP7 promoter −418 position by TESS (http://www.cbil.upenn.edu/cgi-bin/tess/tess), the letter shown in bold representing the position of −418G>A; B, EMSA with biotin-labeled −418 G (lanes 1 to 4) or A probes (lanes 5 to 8) and UMSCC-17B nuclear extract; C, Supershift assay. GATA-2 antibody and normal rabbit IgG were used to determine the binding specificity. NS, non-specific binding; FP, free probe.
Discussion
In this hospital-based case-control study of 1,065 SCCHN patients and 1,112 cancer-free controls in non-Hispanic whites, we investigated whether the two common, validated IGFBP7 promoter polymorphisms are associated with SCCHN risk. We found that IGFBP7 −418AG and AG+AA genotypes were significantly associated with reduced risk of SCCHN. This effect was further enhanced when the −418AG+AA variant genotypes were combined with the −702CG+CC variant genotypes, compared with the combined −418GG and −702 GG genotypes. In other words, those who carried the combined −418GG and −702 GG genotypes had a statistically significantly increased SCCHN risk, compared with those who carried any of the variant genotypes (i.e., −418AG, −418AA, −702CG and −702CC). Further stratified analysis showed that this effect was more profound in the subgroup of women (borderline significant), never smokers and those who had oral cavity cancer. Additional functional analyses showed that the IGFBP7 promoter harboring the −418A allele had both significantly higher luciferase activity and DNA-protein binding activity, suggesting that the change of the IGFBP7 G allele to A allele may play a tumor suppressor role in the etiology of SCCHN. However, it remains unknown what transcription factors may bind to this polymorphic site in the promoter. To the best of our knowledge, this is the first epidemiological study to have investigated the roles of IGFBP7 polymorphisms on susceptibility to cancer and the related molecular mechanisms.
Although we could not find relevant data from published studies to compare with our findings, our relatively large study size appeared to have an adequate statistical power to detect the effects of IGFBP7 variant genotypes on SCCHN risk. The −702G>C (rs11573014) was only genotyped in the NIHPDR 90 subset with C allele frequency of 0.112 and CG and GG (but no CC) genotype frequencies of 22.4% and 77.6%, respectively. These frequency data are similar to those (0.153 for C allele and 27.1% and 71.1% for CG and GG genotype frequencies) in our 1,112 controls (Table 2). The −418G>A (rs4075349) was the only one genotyped in the HapMap database (http://hapmap.ncbi.nlm.nih.gov/- dbSNP b126) for 226 Utah residents with Northern and Western European ancestry from the CEPH collection with T/A allele frequency of 0.407 (compared to 0.411 for our 1,112 controls) and CC/GG, CT/GA and TT/AA genotype frequencies of 32.7%, 53.1% and 14.2% (compared to 33.2%, 51.4% and 15.5% for our 1,112 controls), respectively. These data suggest that any genotype errors in our controls are unlikely to have an effect on our findings. However, the corresponding allele and genotype frequencies were 0.667 and 11.9%, 42.9% and 45.2% for 168 Han Chinese in Beijing (CHB) and 0.688 and 8.9%, 44.6% and 46.4% for 224 Africans of Yoruba in Ibadan, Nigeria (http://hapmap.ncbi.nlm.nih.gov/ - dbSNP b126), suggesting some ethnic difference in both allele and genotype frequencies of this −418G>A SNP.
We noticed that the distribution of IGFBP7 −418G>A genotypes deviated from HWE in control subjects (P<0.05). Such HWE deviation may be caused by violation of its assumptions [31], in which one of the possible explanations is genotyping error, and the other is selection bias. We believe that genotyping error is unlikely to cause the deviation from HWE, because we did not observe any significant discrepancy in the results of both SNPlex and PCR-RFLP assays for approximately 10% of repeated genotyping samples, and the genotypes were also confirmed by direct sequencing. In addition, the genotyping frequencies of −418G>A three genotypes were close to those reported in Hapmap, suggesting the distribution of these genotypes may have been selected for, through evolution. Another possible reason for the violation of HWE is that the controls were recruited from cancer-free, outpatient clinics visitors at MD Anderson Cancer Center; therefore, they may not fully represent the general population, an inherent limitation of the hospital-based case-control study. Therefore, our results may not be generalizable to the general population. Nevertheless, our finding is biologically plausible as evidenced by a series of functional analyses of the impact of variant alleles on the IGFBP7 regulation. Furthermore, our relatively large study size appeared to have an adequate statistical power to detect the effects of IGFBP7 variant genotypes on SCCHN risk. Our finding of an increased risk associated with genotypes other the combined GG/GG genotypes in the subgroup of never smokers is consistent with the notion that susceptible individuals are more sensitive to expose to low doses of exposure. However, this subgroup finding often suffers from reduced statistical power or simply due to chances. Therefore, such a finding needs to be validated in future larger studies.
IGFBP7 is encoded by the IGFBP7 gene and has been shown to interact with multiple proteins, playing important roles in the development of multiple cancers, mostly independent of the IGF signaling. For example, IGFBP7, along with other tumor suppressors, including insulin-degrading enzyme (IDE) and phosphatase and tensin homolog (PTEN), can antagonize insulin and protect another tumor suppressor gene, retinoblastoma protein (RB), from inactivation, therefore helping suppress cancer development [32]. A recent study has also showed that the activated BRAF oncogene (BRAF600E) induces melanoma cell senescence and apoptosis through regulation of the secreted IGFBP7 protein [33]. Moreover, experiments both in human cell lines and mouse models support a potential role of IGFBP7 in the treatment of melanoma and colorectal cancer [34]. In lung cancer, over-expression of IGFBP7 reduces the anchorage-independent growth, activates caspase-3, and induces apoptosis in lung cancer cells [17]. Taken together, these studies and others have demonstrated that IGFBP7 acts as a tumor suppressor gene in most cancer types, such as cancers of the breast, prostate, colon and rectum, melanoma and lung, through the induction of senescence and apoptosis, or the inhibition of proliferation in cancer cells [17,18,21,33,35].
In the current study we demonstrated that IGFBP7 −418A had a higher promoter activity than −418G and that IGFBP7 −418A variant genotypes were associated with reduced risk of SCCHN, supporting a tumor suppressor role of IGFBP7 in SCCHN, a finding consistent with that of previously published studies. In an early study for the characterization of the murine mac25 gene promoter, Sp1 and methylation were found to regulate the expression of IGFBP7 [36]. More recently, the transcriptional factor AP-1 was identified to be able to bind to the promoter region of IGFBP7, participating in regulation of the IGFBP7 expression by BRAF600E in melanoma cells [33]. In addition, several studies showed that IGFBP7 expression was regulated by some epigenetic mechanisms. For example, hypermethylation of CpG islands in the IGFBP7 gene silenced the IGFBP7 expression, and demethylation resulted in the activation of IGFBP7 [37–39]. In our present study, we found that the higher promoter activity driven by the −418A allele of IGFBP7 may be regulated by the direct DNA-protein binding, but subsequent experiments ruled out GATA-2 as the potential transcriptional factor that could differentially bind to −418A and −418G as predicted by bioinformatics tools.
In summary, our study indicated that individuals carrying the IGFBP7 −418A variant genotypes may have a reduced risk of SCCHN. This is because the IGFBP7 promoter with −418A has a higher promoter activity than −418G, which may result in different expression levels of IGFBP7. Therefore, the change of IGFBP7 −418G to −418G may play a tumor suppressor role in SCCHN. However, the exact underlying molecular mechanisms for the roles of the IGFBP7 −418 G>A polymorphism and IGFBP7 expression in the etiology of SCCHN need to be further unraveled by more rigorous mechanistic studies. Although our study population is relatively large, an uncontrolled selection bias may have existed due to the nature of hospital-based case-control study. Considering this limitation, other large, population-based, preferably prospective studies are needed to further validate our findings.
Acknowledgments
We thank Margaret Lung and Kathryn L Tipton. for their assistance in recruiting the subjects and gathering the questionnaire information; Zhensheng Liu, Sheng Wei, Yawei Qiao, Jianzhong He, Kejing Xu and Min Zhao for laboratory assistance.
Grant support
This work was partly supported by the National Institute of Health grants R01 CA131274 (Q. Wei), R01 ES011740 (Q. Wei), P50 CA097007 (Scott Lippman), and P30 CA016672 (The University of Texas M. D. Anderson Cancer Center) and a cancer prevention fellowship supported by R25T CA057730 (to Robert M. Chamberlain and Shine Chang).
Abbreviations
- CI
confidence interval
- IGFBP7
insulin-like growth factor binding protein 7
- MAF
minor allele frequency
- OR
odds ratio
- PCR
polymerase chain reaction
- SCCHN
Squamous cell carcinoma of the head and neck
- SNP
single nucleotide polymorphism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Forastiere A, Koch W, Trotti A, Sidransky D. Head and neck cancer. N Engl J Med. 2001;345:1890–1900. doi: 10.1056/NEJMra001375. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009 doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 3.Ho T, Wei Q, Sturgis EM. Epidemiology of carcinogen metabolism genes and risk of squamous cell carcinoma of the head and neck. Head Neck. 2007;29:682–699. doi: 10.1002/hed.20570. [DOI] [PubMed] [Google Scholar]
- 4.Neumann AS, Sturgis EM, Wei Q. Nucleotide excision repair as a marker for susceptibility to tobacco-related cancers: a review of molecular epidemiological studies. Mol Carcinog. 2005;42:65–92. doi: 10.1002/mc.20069. [DOI] [PubMed] [Google Scholar]
- 5.Kim JJ, Accili D. Signalling through IGF-I and insulin receptors: where is the specificity? Growth Horm IGF Res. 2002;12:84–90. doi: 10.1054/ghir.2002.0265. [DOI] [PubMed] [Google Scholar]
- 6.Valentinis B, Baserga R. IGF-I receptor signalling in transformation and differentiation. Mol Pathol. 2001;54:133–137. doi: 10.1136/mp.54.3.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwa V, Oh Y, Rosenfeld RG. The insulin-like growth factor-binding protein (IGFBP) superfamily. Endocr Rev. 1999;20:761–787. doi: 10.1210/edrv.20.6.0382. [DOI] [PubMed] [Google Scholar]
- 8.Firth SM, Baxter RC. Cellular actions of the insulin-like growth factor binding proteins. Endocr Rev. 2002;23:824–854. doi: 10.1210/er.2001-0033. [DOI] [PubMed] [Google Scholar]
- 9.Oh Y, Nagalla SR, Yamanaka Y, Kim HS, Wilson E, Rosenfeld RG. Synthesis and characterization of insulin-like growth factor-binding protein (IGFBP)-7. Recombinant human mac25 protein specifically binds IGF-I and -II. J Biol Chem. 1996;271:30322–30325. doi: 10.1074/jbc.271.48.30322. [DOI] [PubMed] [Google Scholar]
- 10.Ahmed S, Jin X, Yagi M, Yasuda C, Sato Y, Higashi S, Lin CY, Dickson RB, Miyazaki K. Identification of membrane-bound serine proteinase matriptase as processing enzyme of insulin-like growth factor binding protein-related protein-1 (IGFBP-rP1/angiomodulin/mac25) FEBS J. 2006;273:615–627. doi: 10.1111/j.1742-4658.2005.05094.x. [DOI] [PubMed] [Google Scholar]
- 11.Komatsu S, Okazaki Y, Tateno M, Kawai J, Konno H, Kusakabe M, Yoshiki A, Muramatsu M, Held WA, Hayashizaki Y. Methylation and downregulated expression of mac25/insulin-like growth factor binding protein-7 is associated with liver tumorigenesis in SV40T/t antigen transgenic mice, screened by restriction landmark genomic scanning for methylation (RLGS-M) Biochem Biophys Res Commun. 2000;267:109–117. doi: 10.1006/bbrc.1999.1937. [DOI] [PubMed] [Google Scholar]
- 12.Pen A, Moreno MJ, Durocher Y, Deb-Rinker P, Stanimirovic DB. Glioblastoma-secreted factors induce IGFBP7 and angiogenesis by modulating Smad-2-dependent TGF-beta signaling. Oncogene. 2008;27:6834–6844. doi: 10.1038/onc.2008.287. [DOI] [PubMed] [Google Scholar]
- 13.Swisshelm K, Ryan K, Tsuchiya K, Sager R. Enhanced expression of an insulin growth factor-like binding protein (mac25) in senescent human mammary epithelial cells and induced expression with retinoic acid. Proc Natl Acad Sci U S A. 1995;92:4472–4476. doi: 10.1073/pnas.92.10.4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akaogi K, Okabe Y, Sato J, Nagashima Y, Yasumitsu H, Sugahara K, Miyazaki K. Specific accumulation of tumor-derived adhesion factor in tumor blood vessels and in capillary tube-like structures of cultured vascular endothelial cells. Proc Natl Acad Sci U S A. 1996;93:8384–8389. doi: 10.1073/pnas.93.16.8384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burger AM, Leyland-Jones B, Banerjee K, Spyropoulos DD, Seth AK. Essential roles of IGFBP-3 and IGFBP-rP1 in breast cancer. Eur J Cancer. 2005;41:1515–1527. doi: 10.1016/j.ejca.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 16.Burger AM, Zhang X, Li H, Ostrowski JL, Beatty B, Venanzoni M, Papas T, Seth A. Down-regulation of T1A12/mac25, a novel insulin-like growth factor binding protein related gene, is associated with disease progression in breast carcinomas. Oncogene. 1998;16:2459–2467. doi: 10.1038/sj.onc.1201772. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y, Pacyna-Gengelbach M, Ye F, Knosel T, Lund P, Deutschmann N, Schluns K, Kotb WF, Sers C, Yasumoto H, Usui T, Petersen I. Insulin-like growth factor binding protein-related protein 1 (IGFBP-rP1) has potential tumour-suppressive activity in human lung cancer. J Pathol. 2007;211:431–438. doi: 10.1002/path.2132. [DOI] [PubMed] [Google Scholar]
- 18.Ma Y, Lu B, Ruan W, Wang H, Lin J, Hu H, Deng H, Huang Q, Lai M. Tumor suppressor gene insulin-like growth factor binding protein-related protein 1 (IGFBP-rP1) induces senescence-like growth arrest in colorectal cancer cells. Exp Mol Pathol. 2008;85:141–145. doi: 10.1016/j.yexmp.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 19.Ruan W, Xu E, Xu F, Ma Y, Deng H, Huang Q, Lv B, Hu H, Lin J, Cui J, Di M, Dong J, Lai M. IGFBP7 plays a potential tumor suppressor role in colorectal carcinogenesis. Cancer Biol Ther. 2007;6:354–359. doi: 10.4161/cbt.6.3.3702. [DOI] [PubMed] [Google Scholar]
- 20.Sato Y, Chen Z, Miyazaki K. Strong suppression of tumor growth by insulin-like growth factor-binding protein-related protein 1/tumor-derived cell adhesion factor/mac25. Cancer Sci. 2007;98:1055–1063. doi: 10.1111/j.1349-7006.2007.00502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sprenger CC, Damon SE, Hwa V, Rosenfeld RG, Plymate SR. Insulin-like growth factor binding protein-related protein 1 (IGFBP-rP1) is a potential tumor suppressor protein for prostate cancer. Cancer Res. 1999;59:2370–2375. [PubMed] [Google Scholar]
- 22.Vizioli MG, Sensi M, Miranda C, Cleris L, Formelli F, Anania MC, Pierotti MA, Greco A. IGFBP7: an oncosuppressor gene in thyroid carcinogenesis. Oncogene. 2010 May 3; doi: 10.1038/onc.2010.136. advance online publication, doi:10.1038/onc2010.136 (2010) [DOI] [PubMed] [Google Scholar]
- 23.Wilson HM, Birnbaum RS, Poot M, Quinn LS, Swisshelm K. Insulin-like growth factor binding protein-related protein 1 inhibits proliferation of MCF-7 breast cancer cells via a senescence-like mechanism. Cell Growth Differ. 2002;13:205–213. [PubMed] [Google Scholar]
- 24.Jiang W, Xiang C, Cazacu S, Brodie C, Mikkelsen T. Insulin-like growth factor binding protein 7 mediates glioma cell growth and migration. Neoplasia. 2008;10:1335–1342. doi: 10.1593/neo.08694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu J, Hu Z, Wei S, Wang LE, Liu Z, El-Naggar AK, Sturgis EM, Wei Q. A novel functional variant (−842G>C) in the PIN1 promoter contributes to decreased risk of squamous cell carcinoma of the head and neck by diminishing the promoter activity. Carcinogenesis. 2009;30:1717–1721. doi: 10.1093/carcin/bgp171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niu J, Huang YJ, Wang LE, Sturgis EM, Wei Q. Genetic polymorphisms in the PTPN13 gene and risk of squamous cell carcinoma of head and neck. Carcinogenesis. 2009;30:2053–2058. doi: 10.1093/carcin/bgp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andrews NC, Faller DV. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong Y, Miao X, Zhang X, Ding F, Luo A, Guo Y, Tan W, Liu Z, Lin D. The role of P53 and MDM2 polymorphisms in the risk of esophageal squamous cell carcinoma. Cancer Res. 2005;65:9582–9587. doi: 10.1158/0008-5472.CAN-05-1460. [DOI] [PubMed] [Google Scholar]
- 29.Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trikalinos TA, Salanti G, Khoury MJ, Ioannidis JP. Impact of violations and deviations in Hardy-Weinberg equilibrium on postulated gene-disease associations. Am J Epidemiol. 2006;163:300–309. doi: 10.1093/aje/kwj046. [DOI] [PubMed] [Google Scholar]
- 32.Radulescu RT. One for all and all for one: RB defends the cell while IDE, PTEN and IGFBP-7 antagonize insulin and IGFs to protect RB. Med Hypotheses. 2007;69:1018–1020. doi: 10.1016/j.mehy.2007.02.042. [DOI] [PubMed] [Google Scholar]
- 33.Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132:363–374. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wajapeyee N, Kapoor V, Mahalingam M, Green MR. Efficacy of IGFBP7 for treatment of metastatic melanoma and other cancers in mouse models and human cell lines. Mol Cancer Ther. 2009;8:3009–3014. doi: 10.1158/1535-7163.MCT-09-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tennant MK, Vessella RL, Sprenger CC, Sikes RA, Hwa V, Baker LD, Plymate SR. Insulin-like growth factor binding protein-related protein 1 (IGFBP-rP1/mac 25) is reduced in human prostate cancer and is inversely related to tumor volume and proliferation index in Lucap 23.12 xenografts. Prostate. 2003;56:115–122. doi: 10.1002/pros.10223. [DOI] [PubMed] [Google Scholar]
- 36.Kanemitsu N, Kato MV, Miki T, Komatsu S, Okazaki Y, Hayashizaki Y, Sakai T. Characterization of the promoter of the murine mac25 gene. Biochem Biophys Res Commun. 2000;279:251–257. doi: 10.1006/bbrc.2000.3944. [DOI] [PubMed] [Google Scholar]
- 37.Lin J, Lai M, Huang Q, Ma Y, Cui J, Ruan W. Methylation patterns of IGFBP7 in colon cancer cell lines are associated with levels of gene expression. J Pathol. 2007;212:83–90. doi: 10.1002/path.2144. [DOI] [PubMed] [Google Scholar]
- 38.Lin J, Lai M, Huang Q, Ruan W, Ma Y, Cui J. Reactivation of IGFBP7 by DNA demethylation inhibits human colon cancer cell growth in vitro. Cancer Biol Ther. 2008;7:1896–1900. doi: 10.4161/cbt.7.12.6937. [DOI] [PubMed] [Google Scholar]
- 39.Ye F, Chen Y, Knosel T, Schluns K, Pacyna-Gengelbach M, Deutschmann N, Lai M, Petersen I. Decreased expression of insulin-like growth factor binding protein 7 in human colorectal carcinoma is related to DNA methylation. J Cancer Res Clin Oncol. 2007;133:305–314. doi: 10.1007/s00432-006-0171-z. [DOI] [PubMed] [Google Scholar]