Abstract
The post-translational methylation of α-amino groups was first discovered over 30 years ago on the bacterial ribosomal proteins L16 and L331–2, but almost nothing is known about the function or enzymology of this modification. Several other bacterial and eukaryotic proteins have since been shown to be α-N-methylated3–10. However, the Ran guanine nucleotide-exchange factor, RCC1, is the only protein for which any biological function of α-N-methylation has been identified3, 11. Methylation-defective mutants of RCC1 have reduced affinity for DNA and cause mitotic defects3, 11, but further characterization of this modification has been hindered by ignorance of the responsible methyltransferase. All fungal and animal N-terminally methylated proteins contain a unique N-terminal motif, Met-(Ala/Pro/Ser)-Pro-Lys, indicating they may be targets of the same, unknown enzyme3,12. The initiating Met is cleaved, and the exposed α-amino group is mono-, di-, or trimethylated. Here we report the discovery of the first α-N-methyltransferase, which we named N-terminal RCC1 methyltransferase (NRMT). Substrate docking and mutational analysis of RCC1 defined the NRMT recognition sequence and enabled the identification of numerous new methylation targets, including SET/TAF-I/PHAPII and the retinoblastoma protein, RB. Knockdown of NRMT recapitulates the multi-spindle phenotype seen with methylation-defective RCC1 mutants3, demonstrating the importance of alpha-N-methylation for normal bipolar spindle formation and chromosome segregation.
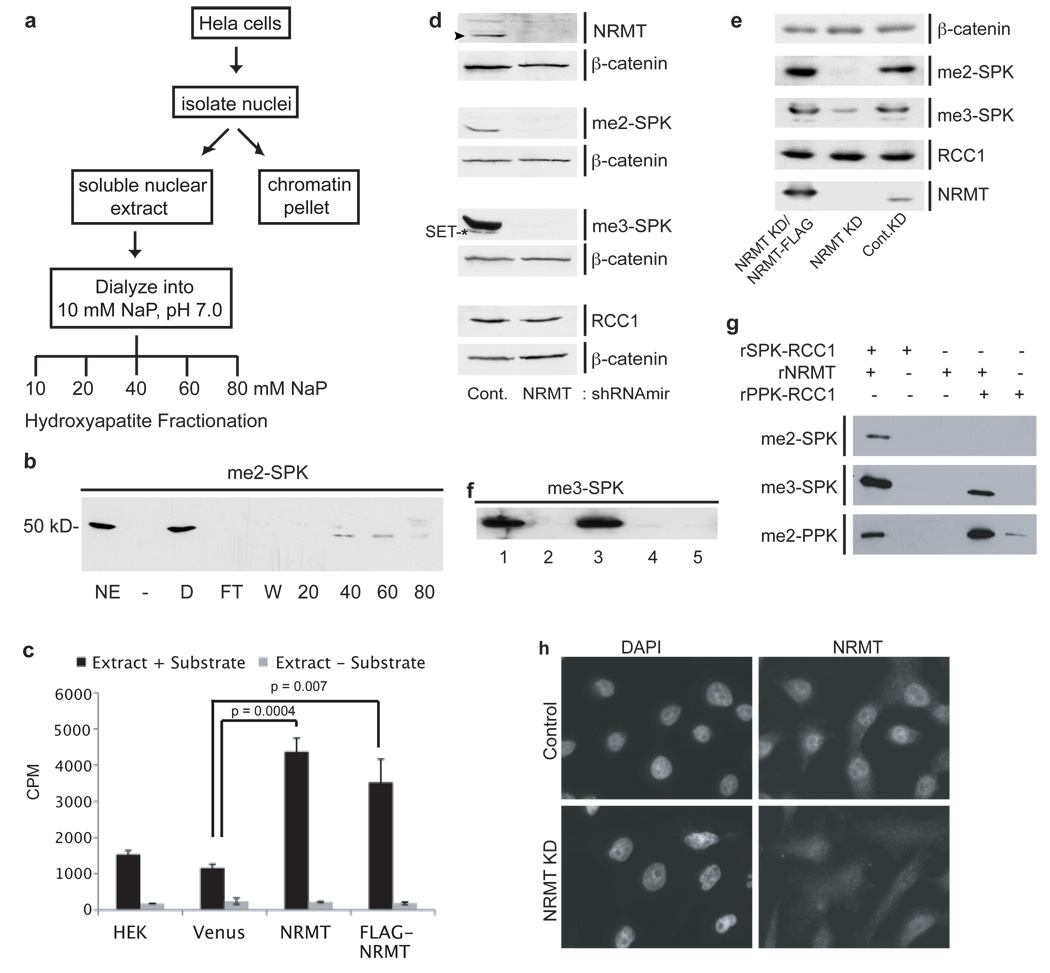
To purify the RCC1 N-terminal methyltransferase, soluble HeLa nuclear extract was fractionated over hydroxyapatite13,14, 15 (Fig. 1a). Step-elutions were performed with increasing sodium phosphate. Fractions were assayed for activity by immunoblotting methyltransferase assays with anti-me2-SPK or by ELISA assay (Supplementary Fig. S3a). RCC1 methylation activity eluted in the 40 – 60 mM fractions (Fig. 1b). The 40 mM fraction was analyzed by mass spectrometry (MS). Among peptides for >100 genes, 2 were detected and manually confirmed for an uncharacterized methyltransferase, METTL11a/C9orf32/Ad-00316 (Gene ID: 28989). METTL11a (now renamed NRMT) encodes a 25 kDa protein in the methyltransferase 11 family, most members of which methylate metabolites or other small molecules. NRMT lacks a SET domain but possesses a Rossman-like α/β fold. According to GeneNote and Oncomine, it is ubiquitously expressed in normal tissue and robustly over-expressed in gastrointestinal cancers. It has been conserved throughout eukaryotic evolution (Supplementary Fig. S1), but next to nothing is known about the function of NRMT orthologues in any model organism.
Figure 1. Mettl11a is the α-N-RCC1 methyltransferase (NRMT).
a, Purification scheme for RCC1 N-terminal methyltransferase. b, In vitro methylation assay (NE–nuclear extract, D–dialyzed nuclear extract, FT–flow thru, W-wash) showing activity elutes in the 40 and 60 mM NaP fractions. c, ELISA assay showing NRMT over-expression increases RCC1 α-N methylation. Data were analyzed by two-tailed independent t tests. n=3–5 independent reactions per condition. Error bars represent +/− 1 s.d. d, Lentiviral knockdown of NRMT in 293LT cells significantly decreases NRMT (arrowhead), di- and trimethylated RCC1, and levels of another methylated protein, later shown to be SET(*), as compared to control cells. β-catenin was the loading control. e, Expression of murine NRMT-FLAG rescues RCC1 methylation levels in NRMT knockdown cells. f, In vitro methylation assays showing immunoprecipitated FLAG-NRMT methylates RCC1-His6. (1-IP input, 2-Substrate only, 3-FLAG-NRMT Elution, 4-FLAG-NRMT elution without substrate, 5-Control IP elution). g, His6-NRMT di-and trimethylates SPK-RCC1-His6 and dimethylates PPK-RCC1-His6. h, Immunofluorescence of endogenous NRMT in HeLa cells +/− NRMT siRNA.
To determine whether NRMT is the authentic RCC1 α-N-methyltransferase, we over-expressed it in HEK 293LT cells and tested nuclear extracts for RCC1 methylation activity by ELISA. Over-expression of NRMT increased α-N-methylation 3-fold as compared to a pK-YFP transfected control (Fig. 1c). Similar results were obtained using N-terminally tagged FLAG-NRMT (Fig. 1c). FLAG-NRMT immunoprecipitated from 293LT cells and eluted with FLAG peptide also methylated recombinant RCC1-His6 in vitro (Fig. 1f). The methylation was verified to be on the N-terminal Ser by Fourier transform mass spectrometry (FTMS) (Supplementary Fig. S2a). This method was used because standard approaches cannot readily distinguish between trimethylation and acetylation. Depleting NRMT in 293LT cells, using lentivirus, significantly decreased methylation of endogenous RCC1, while not affecting overall RCC1 level (Fig. 1d). Similar results were obtained by depleting NRMT in HeLa cells using short interfering RNAs (siRNAs) (Supplementary Fig. S3b). Rabbit polyclonal antibodies generated against a unique C-terminal NRMT peptide confirmed effective knockdown of NRMT levels by the lentivirus and siRNAs (Fig. 1d and Supplementary Fig. S3b). Control virus had no effect as compared to untransfected 293LT cells (Supplementary Fig. S3c). Importantly, RCC1 methylation was rescued by expression of murine NRMT-FLAG, which is not targeted by the human shRNA (Fig. 1e), confirming that off-target effects of the RNAi were not responsible for the loss of methylation. Together, these data conclusively prove that NRMT is the predominant α-N-methyltransferase for RCC1.
Rabbit polyclonal antibody (anti-me2-PPK) against a methylated peptide corresponding to mouse RCC1 also detected methylation by NRMT of RCC1 possessing a Pro2 residue (Fig. 1g). PPK-RCC1 is present in all mammalian species except humans and chimpanzees. RCC1 methylation activity was originally found in the nuclear extract of HeLa cells3 and immunofluorescence of endogenous NRMT, or imaging of a NRMT-GFP fusion protein, showed the enzyme is predominantly nuclear (Fig. 1h and Supplementary Figs. S3d, e). siRNA against NRMT abolished the nuclear staining, confirming this localization pattern (Fig. 1h). Together, these data prove that NRMT is the authentic α-N-RCC1 methyltransferase and that it can recognize variants of the consensus sequence from different species.
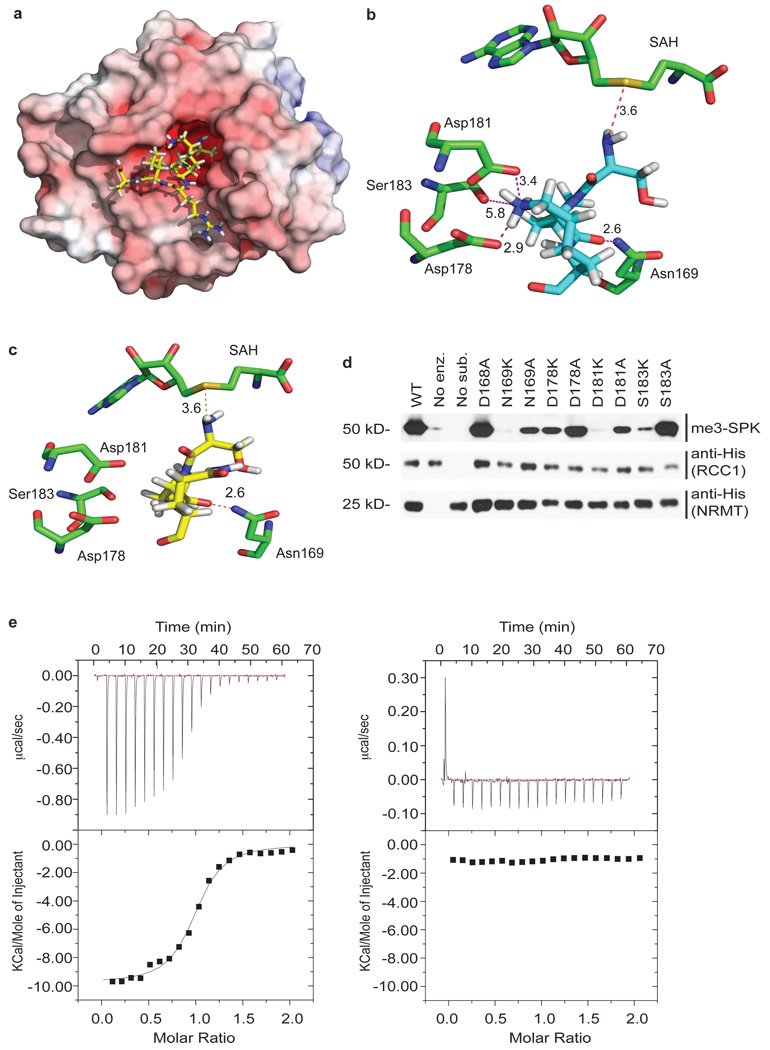
The crystal structure of NRMT in complex with S-adenosyl-homocysteine (SAH) was solved to 1.75 Å resolution by the Structural Genomics Consortium (PDB entry 2EX4, at http://www.pdb.org/pdb/explore/explore.do?structureId=2EX4). A large cavity opposite the SAH binding site could accommodate N-terminal peptides (Fig. 2a) and contains an arrangement of aromatic residues similar to those in chromo domains. We used ICM-PRO to model an RCC1 N-terminal peptide (Ser-Pro-Lys-Arg-Ile-Ala) in the putative active site (Fig. 2a). In this model, only the first 3 residues (Ser-Pro-Lys) interact with NRMT. The optimum conformation positions the substrate α-NH2 close to the SAH, in the correct orientation for methyl transfer, within 3.6Å of the sulfur atom. The peptide Lys4 side chain forms hydrogen bonds and electrostatic interactions with acidic residues at the lip of the active site. A stabilizing effect on Lys4 results mainly from Asp178 and Asp181 (2.9Å, and 3.4Å from the ε-NH2, respectively) plus a weaker effect of Ser183 (Fig. 2b). A similar structural motif was reported previously to coordinate lysine residues17. Other peptides, with Pro2 or Ala2, adopt the same conformation (data not shown); however, substitution of Lys4 by Gln prevented interactions with the basic residues at the lip of the active site (Fig. 2c).
Figure 2. Substrate recognition motif of NRMT.
a, NRMT structure with distribution of electrostatic potential. Electrostatic potential was calculated using APBS Tools Plug in23 with default settings, visualized with PyMol program (DeLano Scientific LLC) and colored from red (−1 kT) to blue (+1 kT). SPK-RCC1 hexapeptide (SPKRIA) is modeled into the active site. b, Schematic view of the interactions between the RCC1 substrate peptide (light blue) and the conserved residues of NRMT. Interactions between Lys4 of the RCC1 peptide and Asp178, Asp181, and Ser183 of NRMT are shown. All distances are in Å. c, Modeling of RCC1 mutant hexapeptide (yellow), in which Lys4 was replaced by Gln, into NRMT active site. d, Mutational analysis of NRMT residues predicted to be involved in SPK-RCC1 recognition. e, The affinity of the enzyme was measured using isothermal titration calorimetry. Wild-type 12-residue RCC1 N-terminal peptide showed exothermic binding to NRMT (ΔH= −9.8kcal/mol) with a Kd 70µM (left panel). A mutant peptide in which Gln replaced Lys4 showed no detectable binding (right panel).
To test the requirement for Lys4 in substrate binding to NRMT, we measured its affinity, by isothermal titration calorimetry, for wild-type RCC1 N-terminal peptide and a mutant peptide in which Gln replaced Lys4. Wild-type peptide showed exothermic binding to NRMT (ΔH= −9.8kcal/mol, Kd 70µM), while RCC1(Gln4) did not bind detectably (Fig. 2e). Lys4 is, therefore, an essential determinant of NRMT substrate specificity. In vitro methylation assays confirmed that only the wild-type peptide can be methylated by recombinant NRMT (Supplementary Fig. S3f). A second key interaction involves H-bonding between the Pro3 carbonyl and the Asn169 amido group (Fig. 2b). NRMT(Lys169) had no activity (Fig. 2d), while mutation of Asp168, which does not interact with the peptide substrate, had no effect on methylation (Fig. 2d). Mutating residues (Asp178, Asp181) at the lip of the active site to Ala decreased enzyme activity, which was further decreased by reverse-charge mutagenesis to Lys (Fig. 2d). Mutating Ser183 to Lys also decreased activity (Fig. 2d). Together, these data strongly support the model for substrate binding predicted from the structure and docking analysis.
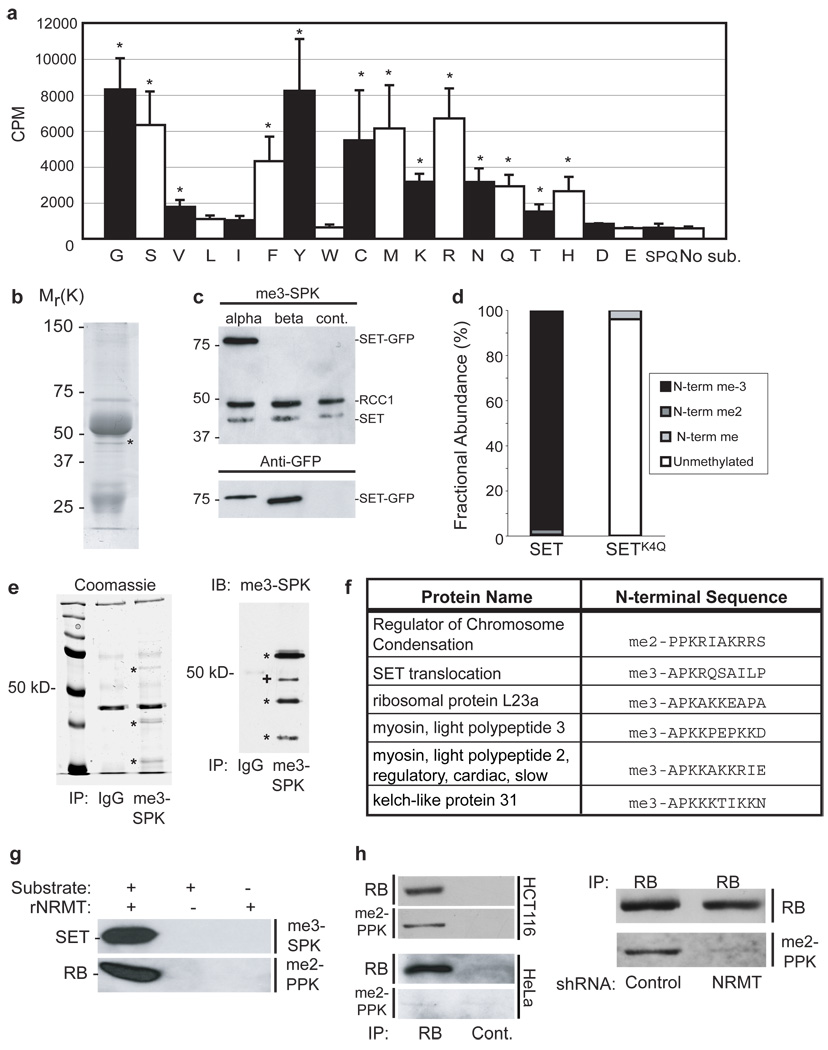
We found previously that α-N-methylation occurred on RCC1 containing Ser2, Pro2 or Ala23. To extend this analysis, we mutated the second residue to each of the other 17 amino acids, using a system in which Factor X cleavage provides efficient exposure of this residue. Testing the cleaved proteins for methylation revealed the enzyme to be promiscuous. Only acidic residues, some hydrophobic residues, and Trp gave no detectable methylation (Fig. 3a). Downstream of the x-Pro-Lys motif, mutagenesis had variable effects, but did not abolish methylation by NRMT (data not shown). We conclude that the x-Pro-Lys motif is important for recognition by NRMT, but that substrate specificity is likely also controlled by the efficiency with which the initiating Met is cleaved.
Figure 3. NRMT methylates many targets, including SETα and RB.
a, His6-NRMT can methylate RCC1-His6 with any amino acid in the second position except Leu, Ile, Trp, Asp, and Glu. Data were compared to the unmethylatable SPQ mutant by two-tailed independent t tests. * indicates P<0.01. n=3 independent reactions per mutant. Error bars: +/− 1 s.d. b, Coomassie stain of ~40 kD protein immunoprecipitated by anti-me3-SPK (*). c, SETα-GFP is recognized by the anti-me3-SPK antibody. d, MS analysis showing SETα-FLAG is stoichiometrically N-terminally trimethylated in HeLa cells. Mutation of SETα Lys4 to Gln abolishes methylation. e, Immunoprecipitations with anti-me3-SPK from mouse spleen (shown) and heart lysate produce at least 3 specific bands visible by Coomassie staining (*). These bands (*), and one additional (+), are recognized by immunoblotting with anti-me3-SPK and correspond to the sizes of the identified mouse proteins (kelch-like 31=70kD, RCC1=45 kD, SET=33kD, and myosin light chain or ribosomal protein L23a ~20 kD). f, Table of N-terminal sequence and methylation status of identified targets. g, NRMT-His6 can methylate in vitro the N-termini of SET and RB fused to GFP. h, Endogenous RB from HCT116 cells, but not HeLa cells, is recognized by the anti-me2-PPK antibody (left panel) and depletion of NRMT in HCT116 cells substantially reduces RB α-N methylation (right panel).
Using Met-(Ala/Ser/Pro)-Pro-Lys, we searched Genbank for candidate substrates of NRMT. More than 35 annotated genes contain this N-terminal motif (Supplementary Table 1). To screen tissues for α-N-methylated proteins, we immunoblotted mouse tissue lysates with our me3-SPK and me2-PPK antibodies. In addition to RCC1, the immunoblots picked up >10 other proteins (Supplementary Fig. S4a). All tissues contained RCC1 N-methylation activity (Supplementary Fig. S4b). To validate the Genbank search, we asked if any proteins detected by the antibodies corresponded to predicted substrates. Proteins were precipitated from HeLa cell lysates using anti-me3-SPK and separated by SDS-PAGE. Besides RCC1 and antibody chains, one additional band was visible at a size also detected by immunoblotting (Fig. 3b). This band was identified by MS as SET/TAF-I/PHAP-II, a predicted substrate for NRMT (Supplementary Table 1). SET has two splice variants, α and β. Only SET α begins with the NRMT consensus. When C-terminally tagged SETα-GFP and SETβ-GFP were expressed in HeLa cells, only SETα was recognized by anti-me3-SPK (Fig. 3c). In addition, SETα-FLAG was expressed in Hela cells, immunoprecipitated, and analyzed by MS. The protein was 96.5% trimethylated on its N-terminal Ser (Fig. 3d and Supplementary Fig. S2b). Similar to RCC13, SETα-FLAG with a Gln4 mutation was mostly unmodified, with only 4% mono-methylation, further confirming the importance of Lys4 (Fig. 3d). Finally, a band corresponding to methylated SET was reduced in cells expressing shRNA or siRNA against NRMT (Fig. 1d and Supplementary Fig. S3b).
We next used the anti-me3-SPK antibody, cross-linked to Protein A/G agarose, to immunoprecipitate proteins from mouse spleen and cardiac lysates. Multiple methylated bands were detected (Fig. 3e), and six N-terminally methylated proteins were identified by MS (Fig. 3f). Two, RCC1 and SET, confirm the validity of the approach (Supplementary Fig. S6). The other four - kelch-like protein 3118; (Supplementary Fig. S4c), ribosomal protein L23a, myosin light chain 2, and myosin light chain 3 (Supplementary Figs. S7 and S8) – confirm the predictive power of the identified motif, and expand the verified NRMT substrates.
Among the predicted substrates was the tumor suppressor protein RB. Given the importance of this protein in cell cycle control, we asked whether it too is a bona fide NRMT substrate. First, recombinant NRMT was able to methylate the N-terminal tails of both SETα and RB in vitro (Fig 3g). Second, endogenous RB immunoprecipitated from the colon cancer line HCT116, which has high levels of functional RB19, was recognized by anti-me2-PPK (Fig. 3h). Third, when NRMT was depleted from HCT116 cells, RB methylation was substantially reduced (Fig. 3h and Supplementary Fig. S3g). Surprisingly, RB from HeLa cells, which is inactivated by the E7 oncoprotein20, 21, was not methylated (Fig. 3h), even though HeLa cells express NRMT and contain methylated RCC1 and SET.
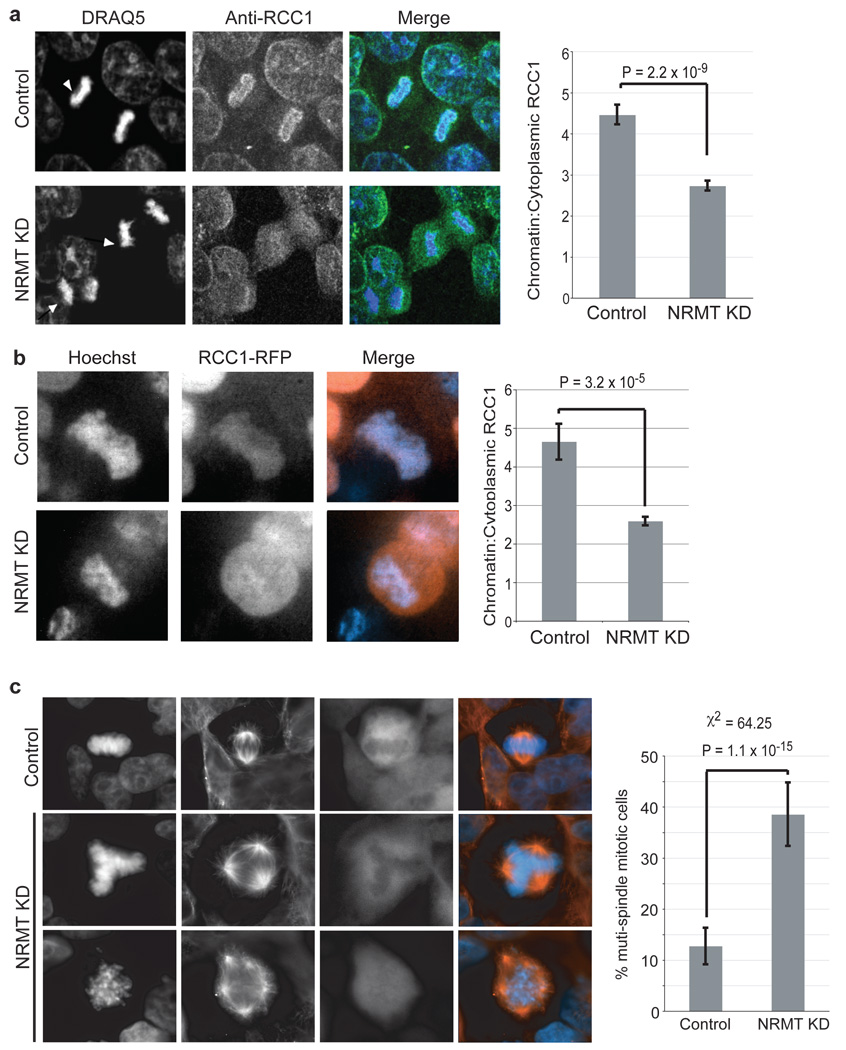
Non-methylatable mutants of RCC1 are defective in chromatin association, and their expression in a wild-type background produces supernumerary spindle poles and mis-segregation of mitotic chromosomes, most likely due to the disruption of the Ran gradient3, 11. To test directly the importance of RCC1 α-N-methylation for its function, we used stably silenced NRMT expression in 293LT cells (Fig. 1d), and examined the distribution of RCC1 during mitosis. RCC1 was significantly more diffuse in mitotic cells lacking NRMT than in control cells (Fig. 4a). The mean chromatin/cytoplasm intensity ratio of RCC1 immunofluorescence was ~2-fold greater in the control cells (Fig. 4a). This redistribution could also be observed in living 293LT cells depleted of NRMT and expressing RCC1–RFP (Fig. 4b).
Figure 4. Silencing NRMT reduces RCC1 association with chromatin, and increases frequency of mitotic defects.
a, Lentiviral silencing of NRMT decreases RCC1 association with chromatin in mitotic (arrowheads) 293LT cells. Chromatin to cytoplasmic ratio of endogenous RCC1 is 2x higher in control, compared to NRMT-depleted mitotic cells. Data analyzed by two-tailed independent t tests. n=50 mitotic cells for each condition. Error bars: +/− 1 s.e.m. b, Lentiviral silencing of NRMT decreases the association of RCC1-RFP with chromatin in live 293LT cells. Data were analyzed by two-tailed independent t tests. n=50 mitotic cells for each condition. Error bars: +/− 1 s.e.m. c, Lentiviral silencing of NRMT increases the frequency of supernumerary spindles to 3x higher than that of control cells. Data were analyzed by a χ2 test. n=109 mitotic cells for each condition. Error bars: +/− 1 s.d.
Strikingly, NRMT-depleted 293LT cells in mitosis exhibited >3x more supernumerary spindles than the control (Fig. 4c). These data, together with our previous studies3, argue that one essential role for methylation RCC1 is to stabilize chromatin association, and that this association is necessary for proper mitotic division. However, neither silencing of NRMT nor mutation of the methylation motif significantly altered RB nuclear localization or intranuclear dynamics in HCT116 cells (Supplementary Fig. S9), indicating that the function of the α-N-methylation is not solely to stabilize chromatin associations, but may have a more general role in the regulation of electrostatic interactions.
This study identifies the long-sought eukaryotic α-N-methyltransferase as a conserved member of a superfamily of non-SET domain enzymes and verifies 6 new protein targets. It is likely that NRMT orthologues throughout the Eukarya possess the same specificity and function, because recent screens have detected N-terminal dimethylation of the Arabidopsis and yeast L12 proteins, which possess the same NRMT consensus motif (PPK) as is found in almost all other eukaryotes5, 22. Further analysis of L12 and other target proteins will likely reveal multiple functions for α-N-methylation.
METHODS SUMMARY
In vitro methylation assays
Substrate proteins were expressed with C-terminal His6 tags, and assayed in 50mM Tris, 50mM K Acetate, pH 8.0, with 100 µM SAM as the methyl donor. All reactions were incubated 1 h at 30°C and analyzed for methylation by immunoblot. For ELISA assays, substrates and enzyme were mixed with 0.55 µCi 3H-SAM. Reactions were incubated as above then transferred to Protein A-coated ELISA plates pretreated with anti-me2-SPK. After 1 h, the reaction was removed and wells were treated with 1% SDS for 30 min. This solution was then quantified for incorporated 3H-methyl by scintillation counting.
Screening mutant NRMT substrates
Efficient removal of the initiating Met is dependent on the identity of the second residue. To avoid this problem, we created fusions from residues 2–10 of RCC1 with His6-tag plus Factor X cleavage site at the N-terminus, and C-terminal GFP. Cleavage with Factor X exposes the second residue of the RCC1 N-terminus. XPK-EGFP substrate proteins were expressed in E.coli and purified on Ni-NTA beads, then cleaved using Factor X.
LS-MS/MS identification of novel NRMT substrates
On-bead endoproteinase Glu-C and Asp-N digestions were used to identify the immunoprecipitated proteins. The retained N-termini were then eluted from the beads and analyzed for methylation (Supplementary Fig. S5).
Supplementary Material
Acknowledgments
This work was supported by research grants from the National Institutes of Health to I.G.M., D.F.M, and W.M. C.S.T. was the recipient of a Post-doctoral fellowship from the National Institutes of Health.
Footnotes
Full Methods and associated references are available in the online version of the paper at www.nature.com/nature.
Author Contributions C.S.T. purified NRMT and performed the experiments characterizing its function and targets and wrote the paper with I.G.M. J.J.P. performed the ITC experiments and the RCC1 mutagenesis, and the docking experiments with the aid of M.S. T.M.S. and J.L.B. performed the mass spectrometry with J.S. D.F.H directed the mass spectrometry. W.M. coordinated the experiments of J.P. I.G.M directed the biochemical and cell biological studies.
References
- 1.Brosius J, Chen R. The primary structure of protein L16 located at the peptidyltransferase center of Escherichia coli ribosomes. FEBS Lett. 1976;68:105–109. doi: 10.1016/0014-5793(76)80415-2. [DOI] [PubMed] [Google Scholar]
- 2.Wittmann-Liebold B, Pannenbecker R. Primary structure of protein L33 from the large subunit of the Escherichia coli ribosome. FEBS Lett. 1976;68:115–118. doi: 10.1016/0014-5793(76)80417-6. [DOI] [PubMed] [Google Scholar]
- 3.Chen T, et al. N-terminal alpha-methylation of RCC1 is necessary for stable chromatin association and normal mitosis. Nat Cell Biol. 2007;9:596–603. doi: 10.1038/ncb1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meng F, et al. Molecular-level description of proteins from saccharomyces cerevisiae using quadrupole FT hybrid mass spectrometry for top down proteomics. Anal Chem. 2004;76:2852–2858. doi: 10.1021/ac0354903. [DOI] [PubMed] [Google Scholar]
- 5.Sadaie M, Shinmyozu K, Nakayama J. A conserved SET domain methyltransferase, Set11, modifies ribosomal protein Rpl12 in fission yeast. J Biol Chem. 2008;283:7185–7195. doi: 10.1074/jbc.M709429200. [DOI] [PubMed] [Google Scholar]
- 6.Pettigrew GW, Smith M. Novel N-terminal protein blocking group identified as dimethylproline. Nature. 1977;265:661–662. doi: 10.1038/265661a0. [DOI] [PubMed] [Google Scholar]
- 7.Martinage A, Briand G, Van Dorsselaer A, Turner CH, Sautiere P. Primary structure of histone H2B from gonads of the starfish Asterias rubens. Identification of an N-dimethylproline residue at the amino-terminal. Eur J Biochem. 1985;147:351–359. doi: 10.1111/j.1432-1033.1985.tb08757.x. [DOI] [PubMed] [Google Scholar]
- 8.Nomoto M, Hayashi H, Iwai K. Tetrahymena histone H2B. Complete amino acid sequence. J Biochem. 1982;91:897–904. doi: 10.1093/oxfordjournals.jbchem.a133778. [DOI] [PubMed] [Google Scholar]
- 9.Grimm R, et al. Postimport methylation of the small subunit of ribulose-1,5-bisphosphate carboxylase in chloroplasts. FEBS Lett. 1997;408:350–354. doi: 10.1016/s0014-5793(97)00462-6. [DOI] [PubMed] [Google Scholar]
- 10.Henry GD, et al. The occurrence of alpha-N-trimethylalanine as the N-terminal amino acid of some myosin light chains. FEBS Lett. 1982;144:11–15. doi: 10.1016/0014-5793(82)80558-9. [DOI] [PubMed] [Google Scholar]
- 11.Hao Y, Macara IG. Regulation of chromatin binding by a conformational switch in the tail of the Ran exchange factor RCC1. J Cell Biol. 2008;182:827–836. doi: 10.1083/jcb.200803110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stock A, Clarke S, Clarke C, Stock J. N-terminal methylation of proteins: structure, function and specificity. FEBS Lett. 1987;220:8–14. doi: 10.1016/0014-5793(87)80866-9. [DOI] [PubMed] [Google Scholar]
- 13.Bloom KS, Anderson JN. Fractionation and characterization of chromosomal proteins by the hydroxyapatite dissociation method. J Biol Chem. 1978;253:4446–4450. [PubMed] [Google Scholar]
- 14.Simon RH, Felsenfeld G. A new procedure for purifying histone pairs H2A + H2B and H3 + H4 from chromatin using hydroxylapatite. Nucleic Acids Res. 1979;6:689–696. doi: 10.1093/nar/6.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duncan EM, et al. Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell. 2008;135:284–294. doi: 10.1016/j.cell.2008.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu RM, et al. Gene expression profiling in the human hypothalamus-pituitary-adrenal axis and full-length cDNA cloning. Proc Natl Acad Sci U S A. 2000;97:9543–9548. doi: 10.1073/pnas.160270997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheerer P, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 18.Yu W, et al. A novel human BTB-kelch protein KLHL31, strongly expressed in muscle and heart, inhibits transcriptional activities of TRE and SRE. Mol Cells. 2008;26:443–453. [PubMed] [Google Scholar]
- 19.Yamamoto H, et al. Paradoxical increase in retinoblastoma protein in colorectal carcinomas may protect cells from apoptosis. Clin Cancer Res. 1999;5:1805–1815. [PubMed] [Google Scholar]
- 20.Boyer SN, Wazer DE, Band V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996;56:4620–4624. [PubMed] [Google Scholar]
- 21.Dyson N, Howley PM, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 22.Carroll AJ, Heazlewood JL, Ito J, Millar AH. Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post-translational modification. Mol Cell Proteomics. 2008;7:347–369. doi: 10.1074/mcp.M700052-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.