Summary
Since its discovery a decade ago, RNA interference (RNAi) has been developed not only into powerful experimental tools but also into promising novel therapeutics. In contrast to conventional antiepileptic drugs that target specific proteins such as ion channels or receptors, RNAi –based therapeutics exploit an endogenous regulatory mechanism of gene expression and thereby are poised to prevent or reverse pathogenetic mechanisms involved in seizure development. Therapeutic RNAi has been widely explored for dominant targets involved in neurodegenerative diseases; however, their use for epilepsy therapy has received less attention. This review will discuss potential RNAi-based targets that are of interest for epilepsy therapy, including adenosine kinase (ADK), the key negative regulator of the brain’s endogenous anticonvulsant adenosine. Overexpression of ADK, and the resulting adenosine deficiency, are pathological hallmarks of the sclerotic epileptic brain, and have been implicated in seizure generation. Therefore, RNAi-strategies aimed at reducing ADK (and increasing adenosine) are based on a direct neurochemical rationale that has recently been explored experimentally using ex vivo and in vivo gene therapy approaches. Technical issues and challenges remain before those promising tools can be developed into future therapeutics for epilepsy.
Keywords: adenosine, RNA interference, RNAi, gene therapy, glia
Despite the recent advent of additional antiepileptic drugs, the treatment of epilepsy remains a major challenge. There is currently no cure for epilepsy. Furthermore, the available pharmacotherapy is largely symptomatic; these approaches do not affect the underlying disease processes, are associated frequently with severe side effects, and are limited by a high incidence of pharmacoresistance (Vajda, 2007). Therefore, there is a need to develop innovative therapeutic strategies based on knowledge of underlying pathogenetic mechanisms. If a molecular pathway is known that ultimately leads to the expression of epileptic seizures, then silencing the expression of this pathway’s components should prevent the expression of seizures.
In 1998, Andrew Fire and Craig Mello discovered a novel mechanism – RNA interference (RNAi) – by which gene expression can be silenced in a homology-dependent manner (Fire et al., 1998). In this seminal study, the authors demonstrated that the injection of double-stranded RNA (dsRNA), but not antisense or sense RNA alone, led to an efficient loss of a target mRNA in C. elegans. In a follow-up study published in the same year by Fire’s group, the investigators demonstrated that mRNA was the target for dsRNA and that targeted mRNAs were degraded prior to translation (Montgomery et al., 1998). Within a year, RNAi was discovered in a wide range of organisms, including vertebrates (Tuschl et al., 1999). Due to the significance of their discovery, Fire and Mello were awarded the Nobel Prize in Physiology or Medicine in 2006. Today, RNAi is used as a powerful tool to test experimentally the function of almost any gene in a cell; this approach has major implications for biomedical research that are likely to be translated into scores of medical applications in the future (Davidson & Boudreau, 2007; Gonzalez-Alegre & Paulson, 2007; Lingor & Bahr, 2007).
This review article will provide background information on RNAi -mechanisms and -technology, highlight research data from use of RNAi in neurodegenerative diseases, and will then critically discuss whether RNAi might be a feasible strategy for the treatment of epilepsy.
PRINCIPLES OF RNA INTERFERENCE (RNAI)
RNA is a cellular process that has likely evolved to provide an innate defense mechanism against viral infection and to prevent the uncontrolled spread of transposable elements (McManus & Sharp, 2002). Mechanistically, dsRNA is processed into small inhibitory RNAs (siRNA) by an enzymatic complex called Dicer. The functionally active siRNAs usually have a length of around 21 nucleotides (Provost et al., 2002) and can bind in a sequence-specific manner to target mRNAs, causing gene silencing either by messenger degradation or translational inhibition (Lee et al., 2004b).
Regulation of gene expression by endogenous RNAi
Recently, the discovery of endogenous microRNA (miRNA or miR) has identified RNAi as a key physiological regulatory mechanism for gene function and introduced a new level of complexity in gene regulation (Bartel, 2004; Chen & Rajewsky, 2007; Pillai et al., 2007). Today hundreds of human genes have been identified that code for endogenous miRNAs (Bentwich et al., 2005), each capable of potentially regulating hundreds of target genes (Guarnieri & DiLeone, 2008). It is estimated that more than 30 percent of all human genes are regulated by miRNAs (Esquela-Kerscher & Slack, 2006).
Micro RNAs originate as larger primary gene transcripts called pre-miRNAs that form internal hairpin structures (Lee et al., 2004a). Those pre-miRNAs are then subjected to a sequence of tightly controlled processing steps that include cleavage and transport to the cytoplasm, where a mature miRNA duplex with a stem-loop of 19 to 25 base pairs is generated via Dicer cleavage (Provost et al., 2002). A single antisense strand of the duplex (complementary to a target mRNA) is then incorporated into an RNA Induced Silencing Complex (RISC), which is the final functional RNA/protein complex that identifies and silences target mRNAs (Khvorova et al., 2003; Schwarz et al., 2003).
The expression profile of miRNAs can be highly tissue-specific and can change in response to injury. Therefore, profiling changes in miRNA expression is of scientific and diagnostic value. For example, it was recently demonstrated that ischemic preconditioning regulates both the expression of miRNAs as well as a predicted target (Lusardi et al., 2009). In a recent study, miRNA expression profiles were compared from the hippocampus and blood of rats that were either naïve, sham-treated, or subjected to either ischemic stroke, intracerebral hemorrhage, or kainic acid-induced seizures (Liu et al., 2010). Remarkably, different patterns of miRNA expression were found in the brain and blood after each of those treatments. After a given injury, several miRNAs were up- or down-regulated in both brain and blood samples; in contrast, only a few specific miRNAs changes were common to the different injuries, a finding, which might have diagnostic value (Liu et al., 2010).
One particular endogenous miRNA (miR-146a) has recently been studied within the context of epileptogenesis (Aronica et al., 2010). miR-146a is a likely endogenous regulator of innate inflammatory responses that are mediated by toll-like receptors that depend on cytokine receptor signalling, suggesting a link between specific miRNAs and human inflammatory diseases (Taganov et al., 2006; Nahid et al., 2009). Importantly, Aronica et al demonstrated an upregulation of miR-146a, including prominent expression in reactive astrocytes during epileptogenesis in a rat model of temporal lobe epilepsy (TLE) as well as in human TLE (Aronica et al., 2010). This important new finding not only establishes a link between specific miRNAs and human inflammatory diseases, but also suggests that dysregulation of a particular miRNA implicated in the regulation of innate inflammatory responses might contribute to (or be a consequence of) epileptogenesis.
The basics of therapeutic RNAi
Since endogenous miRNAs are important regulators of gene function, it is not surprising that synthetic exogenous miRNAs or related RNAi approaches are widely considered to be very potent and promising therapeutics. Interestingly, experimental gene silencing approaches based on the use of long antisense RNA have already been used in the early 1990s prior to the discovery of RNAi (Nellen & Lichtenstein, 1993; Boison & Stoffel, 1994). Subsequently, several tools have been developed that make therapeutic use of RNA interference (Fig. 1). RNAi can be designed to mimic primary miRNA stem-loops (artificial miRNA), processed pre-miRNAs (short hairpin RNA, or shRNA), or mature miRNAs that perfectly match with their target mRNAs (small interfering RNA, or siRNA). Algorithms have been developed that predict effective binding sites for effective artificial miRNAs, shRNAs, or siRNAs; those binding sites do not necessarily need to be identical to the binding sites of endogenous miRNAs. Importantly, binding sites for artificial RNAi constructs are designed in a way to allow high specificity to only one gene, whereas endogenous miRNAs can target several genes simultaneously (see above).
Figure 1. Simplified representation of endogenous and exogenous pathways of inhibitory RNA processing.
Endogenous miRNAs are transcribed as primary miRNA stem cell loops from endogenous miRNA genes. Those are processed by Drosha into pre-miRNAs, which are a substrate for Dicer performing the final cleavage step into mature miRNA (please note that intracellular localizations and trafficking steps have been omitted for clarity). Artificial RNAi constructs (exogenous pathway) can feed into the endogenous pathways at different levels. Artificial miRNA and shRNA can be expressed from viral vectors and mimic primary miRNA stem loops and processed pre-miRNA, respectively. Synthetic siRNA mimics mature endogenous miRNA. In addition, long antisense transcripts derived from either endogenous antisense genes or from exogenous viral vector constructs can form long dsRNA, which can be processed by Dicer. All miRNAs, irrespective of origin and processing rout, can induce gene silencing by forming the RNA induced silencing complex RISC. For further explanations please refer to main text. Abbreviations: miRNA, micro-RNA; shRNA, short hairpin RNA; siRNA, small interfering RNA; dsRNA double-stranded RNA.
Small interfering RNA is a double-stranded molecule that is typically generated by direct chemical synthesis. After introduction into a cell, it might be further processed by Dicer or may be incorporated into a RISC (Elbashir et al., 2001). In contrast to siRNAs, shRNAs are usually expressed from vector systems using strong constitutive promoters (Paul et al., 2002; Sui et al., 2002) to transcribe sense and antisense sequences connected by an unpaired loop. Following transcription, shRNAs are exported from the nucleus and processed into mature siRNAs by Dicer using the endogenous RNAi processing machinery (Provost et al., 2002; Lund et al., 2004). To generate improved RNAi tools that more closely mimic endogenous RNAi substrates, and to improve efficacy and accuracy of downstream processing events, artificial miRNAs have been developed by embedding siRNA sequences into a more complex nucleic acid backbone (Silva et al., 2005). Those artificial miRNAs are transcribed in larger transcripts and amenable to RNA polymerase II-based expression systems that permit better control of tissue specificity. Furthermore, artificial miRNAs have been shown to mitigate shRNA-mediated toxicity in the brain (McBride et al., 2008). Based on these considerations, artificial miRNAs appear to be the system of choice to achieve long-term gene silencing in brain.
In addition to the use of synthetic siRNAs, or expressed artificial shRNA and miRNA constructs, the transcription of cloned antisense cDNAs is an effective approach for gene silencing. It was widely assumed that dsRNA is not normally expressed in cells but rather exclusively a consequence of viral infection - which typically elicits an antiviral response. However, this response is not the only response to dsRNA, and is probably not even the major one. The existence of endogenous RNAi mechanisms suggests that the physiological expression of antisense message, and the subsequent formation of dsRNA, play a major role in the regulation of gene expression. In fact, up to 5–10% of all transcripts are regulated by antisense transcription (Lipman, 1997). Interestingly, the majority of these antisense transcripts are located within the 5’ and 3’ untranslated regions of their target genes (Lipman, 1997). Expression of long antisense transcripts results in the formation of long dsRNAs within the nucleus. These long dsRNAs are initially cleaved by the RNAse III enzyme, Dicer, to form small 21–23 bp dsRNAs or siRNAs (Wang & Carmichael, 2004). The resulting siRNAs dissociate into single stranded RNAs and assemble into RISC and induce gene silencing as described above.
For scientific or therapeutic approaches a suitable method is needed to deliver the RNAi construct into the cell. Focal delivery approaches, involving direct tissue-injection of “naked” siRNAs (Wolff & Budker, 2005), can be used to achieve transient gene knockdown; however, the duration of the desired effect is limited by the half-life of the siRNA. To achieve long-term gene silencing, shRNAs or miRNAs are usually included in a viral gene expression vector (Raoul et al., 2005). Lentiviral vectors are particularly suited to transduce dividing cells in vivo or stem cells in vitro, whereas adeno-associated viruses (AAV) are highly suited to target quiescent cells of the CNS.
RNAI AS RESEARCH TOOL
In basic research applications, biological functions are frequently studied by inducing specific loss of function situations. If one component within a complex network is removed, any resulting deficiencies can be ascribed to the loss of function of the targeted component. Loss-of-function has been widely achieved by generating knockout animals that lack a specific gene. Although scientifically valid, this approach is experimentally laborious, costly, and low throughput. The advent of RNAi technologies has revolutionized loss-of-function screens. Using RNAi screens, investigators can easily and effectively test multiple different siRNAs in parallel, using model systems that allow high throughput analyses - such as cultured neurons, or C. elegans or D. melanogaster. RNAi screens have recently been used to identify molecules required for glutamatergic or GABAergic synapse development (Paradis et al., 2007) as well as genes that directly regulate GABA synapses (Vashlishan et al., 2008). In the latter study, which employed C. elegans to test the function of body muscles that receive direct input from GABAergic motor neurons, 90 genes were identified - 21 of which were previously implicated in epilepsy (Vashlishan et al., 2008). This example illustrates the power of RNAi screens as experimental tools that identify genes or functions implicated in seizure generation. RNAi approaches can also be used to examine endogenous anticonvulsant mechanisms in vivo. As an example, Xiao et al., observed a 137% increase in seizure duration following electrical stimulation in animals injected with AAV carrying an antisense construct for the GABAA α1 gene (Xiao et al., 1997).
RNAI AS THERAPEUTIC IN NEURODEGENERATIVE DISEASE
The use of RNAi as a therapy for neurodegenerative disease is based on a compelling rationale: Many neurodegenerative diseases are at least in part associated with toxic gain of function mutations. Therefore, an RNAi-mediated knockdown of gene expression should present a rational therapeutic strategy. Due to these considerations, the development of RNAi-based CNS-related therapeutics has largely focused on neurodegenerative diseases, as illustrated in the following examples:
Spinocerebellar ataxia type 1 (SCA1) and Huntington disease (HD) are both progressive, untreatable, neurodegenerative disorders that are caused by a dominant expansion of polyglutamine repeats (Paulson et al., 2000). SCA1 is mimicked in a transgenic mouse model that expresses high levels of a pathogenic form of a human ataxin-1 with 82 CAG repeats in cerebellar Purkinje cells (Burright et al., 1995). Mutant mice that received intracerebellar injections of an AAV1 expressing shRNA directed against human ataxin-1 shortly before disease onset have improved motor coordination, restored cerebellar morphology and resolved characteristic ataxin-1 inclusions in Purkinje cells (Xia et al., 2004).
CAG repeat expansion within the huntingtin gene is the causative agent for HD. In transgenic mice containing mutant human huntingtin with 82 or 144 CAG repeats, intrastriatal treatment with AAV expressing anti-human huntingtin shRNAs led to significant amelioration of disease pathology and extension of life-span (Harper et al., 2005).
Similar to SCA1 and HD, Alzheimer’s Disease (AD) is thought to be caused by toxic gain of function mutations that may lead to the accumulation of amyloid beta-containing plaques or tau-containing neurofibrillary tangles (Hardy & Selkoe, 2002). Therefore, amyloid precursor protein (APP) and its processing enzymes constitute rational therapeutic targets for RNAi (Singer et al., 2005).
In AD, the therapeutic potential of RNAi has been brought to a new level by accomplishing allele specific gene silencing (Rodriguez-Lebron & Paulson, 2006). In an elegant recent approach, shRNA was designed to selectively target mutant, but not normal, APP. Integration of this shRNA into an AAV5 vector and injection of this virus into the hippocampus of a transgenic APP-mutant mouse model of AD led to a significant reduction of mutant disease-causing APP, while the expression of wild type murine APP remained unaltered (Rodriguez-Lebron et al., 2009). Importantly, treated mice displayed improved performance in spatial learning and object recognition tasks (Rodriguez-Lebron et al., 2009). This example illustrates the high molecular specificity that can be achieved with RNAi strategies.
Amyotrophic lateral sclerosis (ALS) is a fatal disease of motor neuron degeneration in brain and spinal cord. Again, a toxic gain of function mutation (mutations in Cu/Zn superoxide dismutase 1, SOD1) is thought to be the underlying cause of the disease (Gurney et al., 1994). Consequently, viral RNAi approaches targeting SOD1, including an allele-specific strategy, were shown to retard symptomatology in a transgenic mouse model of ALS (caused by a mutant form of SOD1) and to increase the lifespan of the animals (Ralph et al., 2005; Raoul et al., 2005).
RATIONALE FOR USING RNAI AS THERAPEUTICS FOR EPILEPSY
The examples discussed above illustrate that targeting toxic gain of function mutations by RNAi is an effective strategy to ameliorate disease expression and progression in a variety of neurodegenerative diseases. Whereas those strategies are well advanced within the field of neurodegenerative diseases, this powerful technique has received less attention as a potential therapeutic for epilepsy. The putative reason for this discrepancy is the lack of well-defined targets in epilepsy. In contrast to neurodegenerative diseases with specific targets, potential targets for epilepsy still need to be identified and translated into therapeutic applications. Whereas gene silencing has largely been neglected as a therapeutic option for epilepsy, several promising gene therapy approaches have been developed with the aim to overexpress select anticonvulsant molecules, such as NPY, galanin, or a specific subunit of the GABAAR. Conventional gene therapies for epilepsy will not be further discussed here, since those therapeutic strategies have received wide coverage in the literature (Raol et al., 2006; Loscher et al., 2008; McCown, 2009; Noe et al., 2009; Riban et al., 2009).
Therapeutic targets for RNAi in epilepsy
What are suitable targets for gene silencing in epilepsy? Obviously, any gain of function mutation or pathological overexpression of any disease-specific contributor to seizure generation would be a suitable therapeutic target for RNAi. In contrast to conventional gene therapy approaches that aim to overexpress a desired gene product, gene silencing strategies aim to reduce the availability of a harmful gene product.
A promising group of targets are ion channels. Several gain of function mutations have been identified that cause so-called channelopathies (Catterall et al., 2008). One such candidate is the gain of function missense mutation in the neuronal voltage-gated sodium channel SCN1A (NAV1.1). SCN1A mutations are associated with generalized epilepsy with febrile seizures plus (GEFS+), severe myoclonic epilepsy of infancy, and familial hemiplegic migraine (Stafstrom, 2009). The introduction of a human GEFS+ -causing SCNA1 mutation into the orthologous mouse gene led to the expression of spontaneous generalized seizures and premature death (Martin et al., 2010). Mice heterozygous for the mutation exhibit infrequent spontaneous generalized seizures, reduced threshold and accelerated propagation of febrile seizures, and decreased threshold to flurothyl-induced seizures (Martin et al., 2010). Mutations in SCNA1 predominantly impaired sodium channel activity in interneurons, leading to decreased inhibition and increased seizure susceptibility (Martin et al., 2010). Together, these findings suggest that SCN1 mutations might be excellent candidates for allele-specific gene silencing.
Receptors or transporters for neurotransmitters constitute an alternative class of targets. However, in many cases, loss of function mutations or reduced expression has been associated with epilepsy. A notable exception is the N-methyl-D-aspartate receptor (NMDAR). Haberman et al., observed a decrease in seizure sensitivity following AAV-mediated delivery and in vivo expression of an NMDAR1 antisense construct (Haberman et al., 2002). Likewise, it was recently shown that an AAV-mediated knockdown of the NR1 subunit of the NMDAR protected against seizures (Kalev-Zylinska et al., 2009). However, reduction of the NMDAR was associated with impaired cognition and might therefore not be a reasonable therapeutic option (Kalev-Zylinska et al., 2009).
A different group of potential targets includes proteins implicated in the development of drug resistance in epilepsy. Several of those proteins, such as P-glycoprotein or multidrug resistance protein, were found to be overexpressed in epilepsy (Lazarowski et al., 2007; Loscher, 2007). RNAi-based therapeutic approaches have been tested in preliminary in vitro models (Tian et al., 2009) and might eventually be useful to reduce the development of pharmacoresistance in epilepsy. However, prior to validating the applicability of these approaches in vivo, further characterization of these approaches is needed.
In addition to the examples outlined above, RNAi approaches might be useful to intervene with pathogenetic mechanisms involved in ictogenesis and epileptogenesis. This rationale will be illustrated for adenosine, which is an endogenous anticonvulsant of the brain. It has been shown that adenosine deficiency can trigger seizures, and that dysregulation of adenosine metabolism and signalling is a pathological hallmark of the epileptic brain (Boison, 2008a). Therefore reconstitution of normal adenosine signalling is a neurochemical rationale for therapeutic intervention.
Adenosine kinase
The purine ribonucleoside adenosine exerts potent antiepileptic and neuroprotective functions, primarily by activating Gi-protein-coupled adenosine A1 receptors (A1Rs) that, among other functions, provide presynaptic inhibition and stabilization of the postsynaptic membrane potential (Fredholm et al., 2005; Boison et al., 2009; Sebastiao & Ribeiro, 2009). In adult brain, synaptic levels of adenosine are largely regulated by an astrocyte-based adenosine cycle (Boison et al., 2009). In contrast to classical neurotransmitters that have their own energy-driven transporter-mediated re-uptake systems to terminate their activity, no comparable system exists for adenosine. Instead, two types of equilibrative nucleoside transporters are located in the cell membrane of astrocytes and allow for the rapid equilibration of extra- and intracellular adenosine levels. In adult brain, synaptic levels of adenosine are largely controlled by the astrocyte-based enzyme, adenosine kinase (ADK), that effectively removes adenosine by phosphorylation into adenosine-5’-monophosphate (AMP) – and thereby drives the influx of adenosine into the cell (Boison, 2008b).
Astrogliosis is a pathological hallmark of the epileptic brain (Wieser, 2004). However, the epileptic brain is characterized by a complex histopathology that not only involves astrogliosis (e.g. hippocampal sclerosis) but also mossy fiber sprouting, granule cell dispersion, the loss of specific neuronal populations and the emergence of ectopic neurons with aberrant neuronal connections. In this complex situation, it is almost impossible to identify an individual pathological component with a causative role in seizure generation. A recent study by Li et al. separated astrogliosis from other epileptogenetic events, and identified the enzyme ADK in astrocytes as a molecular link between astrogliosis and neuronal dysfunction in epilepsy (Li et al., 2008b). In this study, a CA3-selective model of epileptogenesis was created by injecting the excitotoxin kainic acid (KA) into the amygdala of mice. This manipulation resulted in acute seizures and neuronal injury that was restricted to the ipsilateral CA3. Three weeks following the injury, the authors found astrogliosis, overexpression of ADK, and frequent spontaneous electrographic seizures all restricted to the ipsilateral CA3 (Li et al., 2008b). Astrogliosis, overexpression of ADK, and seizures not only coincided spatially (Li et al., 2008b) but also coincided temporally during epileptogenesis (Li et al., 2007). It is important to note that astrogliosis and overexpression of ADK were linked to spontaneous seizures in the absence of any other potentially epileptogenic event such as mossy fiber sprouting or granule cell dispersion. Importantly, the same type of seizures was recreated in transgenic mice overexpressing ADK (Adk-tg mice) or lacking the A1R (Li et al., 2007). In a subsequent study in which overexpression of ADK was molecularly uncoupled from astrogliosis, it was demonstrated that astrogliosis without concurrent overexpression of ADK was not sufficient to trigger seizures; in contrast, overexpression of ADK as such, and in the absence of astrogliosis, was sufficient to trigger seizures (Li et al., 2008a). Importantly, genetic reduction of ADK in the forebrain of mice (fb-Adk-def mice) rendered the animals resistant to acute seizures and to acute neuronal injury (Li et al., 2008b). When injury in those animals was paired with a transient block of A1Rs to recreate a wild-type like injury as trigger for subsequent epileptogenesis, the animals failed to develop astrogliosis and spontaneous seizures under conditions of reduced forebrain ADK (increased adenosine). These experiments suggest a potential antiepileptogenic role of reduced brain ADK (Li et al., 2008b).
Based on the above evidence for an ictogenic and possibly epileptogenic role of increased brain ADK, RNAi strategies to reduce ADK expression - and thereby to increase ambient levels of the endogenous anticonvulsant adenosine - appear to be a rational and promising therapeutic strategy. This therapeutic concept has been tested in recent ex vivo and in vivo gene therapy approaches.
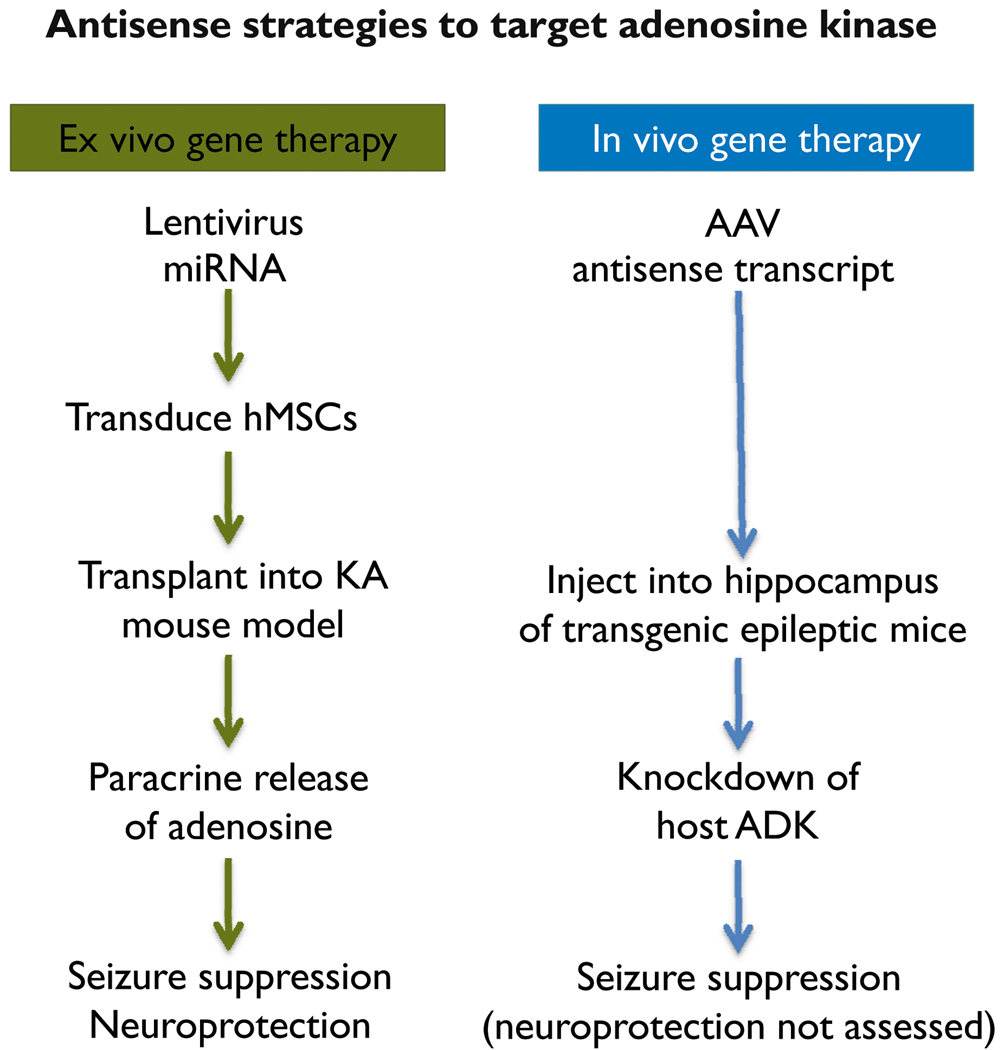
RNAI APPROACHES TARGETING ADENOSINE KINASE (FIGURE 2)
Figure 2. Comparison of ex vivo and in vivo gene therapy approaches targeting adenosine kinase.
Please refer to main text for details.
Ex vivo gene therapy: human stem cells for therapeutic adenosine delivery
As outlined above, reconstitution of adenosine signalling establishes a neurochemical rationale for seizure suppression. Indeed, focal adenosine augmentation therapies (AATs) that restrict adenosine treatment to the area of seizure genesis (i.e., an epileptogenic region characterized by astrogliosis, overexpression of ADK, and resulting adenosine deficiency) effectively suppress seizures in rodent models of epilepsy. Importantly, this suppression is accomplished without systemic side effects that are typical for pharmacological approaches to augment adenosine signalling (Boison, 2009a). One potential focal AAT is the transplantation of cells that have been engineered to release adenosine. It was previously demonstrated that the genetic disruption of ADK, but not of adenosine deaminase, in cultured cells induces the release of therapeutically relevant amounts of adenosine (Huber et al., 2001). Furthermore, adenosine release from human mesenchymal stem cells (hMSCs) was effective in attenuating epileptogenesis when implanted either prior to or after KA injury (Ren et al., 2007; Li et al., 2009).
Human MSCs were chosen for therapeutic adenosine delivery for the following reasons: (i) Readily accessible hMSCs would permit autologous cell transplantation. (ii) Therapeutic effects can be achieved by paracrine adenosine delivery, with the cellular differentiation fate of transplanted cells not being a critical factor (Boison, 2009b). (iii) Gene expression with lentiviral vectors can effectively be achieved in hMSCs (Totsugawa et al., 2002), with continuous stable gene expression demonstrated for at least 6 weeks and throughout differentiation (Sugiyama et al., 2005). (iv) Functional RNAi in hMSCs throughout differentiation is well documented (Hoelters et al., 2005).
To induce therapeutic adenosine release in human stem cells, artificial miRNAs directed against human ADK were cloned into a lentiviral expression vector to co-express the miRNA together with a green fluorescent protein (GFP) tracer under the control of a ubiquitous CAG promoter (Ren et al., 2007). Lentiviral transduction of hMSCs with anti-ADK miRNA expression cassettes yielded up to 80% downregulation of ADK and a concentration of 8.5 ng adenosine per ml of culture medium after incubating 105 cells for 8 hours (Ren et al., 2007). Adenosine-releasing hMSCs transplanted into the infrahippocampal fissure of mice one week prior to the injection of KA into the ipsilateral amygdala, formed dense infrahippocampal grafts that protected the hippocampus from KA-induced acute seizures and neuronal injury. Recipients of the ADK-knockdown cells were characterized by a 35% reduction in the duration of acute seizures and a 65% reduction of acute neuronal cell loss in CA3, compared to animals receiving a sham procedure or hMSCs that were transduced with a scrambled control sequence instead of the anti-ADK miRNA (Ren et al., 2007). When transplanted into the infrahippocampal fissure 24 hours after intraamygdaloid KA-injection, there was a significant reduction of spontaneous recurrent seizure frequency and duration 3 weeks post transplantation (a time point when control animals experienced frequent spontaneous seizures) in the animals receiving ADK-knockdown hMSCs (Li et al., 2009). Together these data demonstrate that lentiviral miRNAs directed against ADK is an effective tool to engineer human stem cells for therapeutic adenosine release. Additionally, these engineered cells, when transplanted close to the hippocampal formation, not only effectively reduce acute excitotoxin-induced seizures and neuronal injury, but also reduce seizures in a post status epilepticus model of spontaneous recurrent seizures.
In vivo gene therapy: knockdown of ADK with adenoassociated virus
In contrast to the ex vivo gene therapy approaches described above that rely on the paracrine delivery of adenosine by transplanted cells, an in vivo gene therapy approach aimed at directly reducing the expression of ADK in epileptic animals would constitute an alternate therapeutic strategy for seizure control. To achieve this goal, AAV8-based vector systems were generated to express a cDNA of ADK in either sense or antisense orientation, to overexpress or knockdown ADK, respectively (Theofilas et al.). To achieve astrocyte-selective expression, a truncated glial fibrillary acidic protein (GFAP) promoter was used. Mice that received Adk-sense virus injections into CA3 developed spontaneous electrographic seizures, indicating that overexpression of ADK in astrocytes per se is sufficient to trigger seizures. To determine whether an antisense-mediated knockdown of ADK can be used therapeutically to prevent seizures, Adk-tg mice that express bilateral spontaneous hippocampal seizures received a unilateral CA3-injection of an AAV8-Adk-antisense virus. Four weeks after virus injection, seizure activity was almost completely suppressed within the virus-injected hippocampus, whereas the unaffected contralateral hippocampus continued to generate seizures at the pre-treatment rate (Theofilas et al.). These results demonstrate that antisense strategies directed at reducing the expression of ADK might be a promising therapeutic alternative to prevent seizures by restoring anticonvulsant adenosine signalling. However, to demonstrate general applicability of this approach, future studies are needed to demonstrate efficacy of antisense strategies directed against ADK in a variety of epilepsy models.
TECHNICAL ISSUES AND CHALLENGES
As outlined above, synthetic siRNAs are very useful tools to address basic research questions. However, biological effects of siRNAs are always transient. Nevertheless, siRNAs are of therapeutic interest since they are chemically synthesized and can theoretically be applied repeatedly like a conventional drug. A major advantage of siRNA therapeutics is the possibility of local administration, e.g. via injection directly into a target organ or tissue. Whereas siRNA injections might be very useful for clinical pilot and proof-of-principle studies or in select cases where repeated administration is possible, any long-term therapeutic applications of RNAi technology will require a gene therapy approach to enable permanent expression of a specific shRNA, miRNA, or antisense cDNA. For those applications, AAV-based vector systems might be most suitable as they combine safety, tropism for brain cells, and long-term efficacy (McCown, 2009; Noe et al., 2009; Riban et al., 2009; Gray et al., 2010).
A recent database search performed on www.clinicaltrials.gov (March 2010) revealed 2056 studies with “gene therapy”. In contrast, a search with “RNA interference OR RNAi OR siRNA OR shRNA OR miRNA” yielded only few results. Most of the latter studies (35) assess endogenous miRNAs (miRs) as biomarkers for disease and for pharmacogenomics studies. Only a few safety and feasibility studies were found, all of them based on siRNA delivery for the following conditions: cancer (5), macular degeneration (5), kidney injury (2), autosomal dominant inherited diseases (1), hypercholesterolemia (1), and hepatitis (1).
This analysis reveals the following: (i) Whereas gene therapies appear to be fairly advanced and considered for a wide range of conditions, RNAi-based therapeutics are in its infancy. (ii) Clinical trials to date focus largely on siRNAs. Effects of siRNAs are transient and therefore safer than permanent (i.e. gene therapy–based) approaches. Further, siRNAs can be chemically synthesized, produced, and marketed like conventional drugs - thus providing an economic incentive to favor siRNA-based approaches. (iii) RNAi has not yet been considered for clinical application for any neurological condition.
What are the challenges that need to be met for translating RNAi-based approaches into clinical applications for the prevention of seizures in epilepsy? The clinical examples mentioned above rely on multiple injections of therapeutic siRNAs. In the case of macular degeneration, multiple intraocular injections of conventional therapeutics are the current standard of care. Therefore, intraocular injection of a clinical preparation of synthetically generated siRNA is a logical extension of current practice. For the treatment of neurological disease (including epilepsy), however, systemic application of siRNA is likely not a therapeutic option due to limited metabolic half life and poor penetration of the blood brain barrier. Repeated direct injections of siRNAs into the brain are not feasible and long-term infusion via pumps is associated with risks. Therefore, the combination of standard gene therapy methods with the stable and long-term expression of a suitable shRNA, miRNA or antisense construct appears to be a necessity.
Neurodegenerative diseases have well-defined targets (e.g. dominant negative mutations) that are thought to be the underlying pathogenetic cause for the disease. Therefore, experimental RNAi approaches are well advanced in the field of neurodegenerative disorders. In contrast, epilepsy is likely a multifactorial condition with multiple possible targets. Many of those targets (e.g. ion channels) can be modulated with conventional antiepileptic drugs; thus, the need to develop gene therapy-based RNAi strategies to knockdown those targets may not be necessary. However, RNAi might become a superior therapeutic tool for novel targets that cannot be modified with conventional AEDs, or for focal applications that are restricted to an epileptogenic focus.
OUTLOOK
Treatment of refractory TLE might be an ideal medical condition for the implementation of RNAi-based therapeutics for several reasons: (i) Seizures arise from a well-defined focal area; therefore treatment approaches restricted to the focal area of seizure generation might be superior to systemic therapy. Those approaches might avoid systemic side effects that frequently hamper the most optimal use of currently available AEDs. (ii) Molecular targets have been identified (e.g. overexpressed ADK) that are specific for an epileptogenic focus that can best be treated with a focal RNAi-based gene silencing approach based on a concise neurochemical rationale. (iii) Clinical feasibility studies could easily be accomplished with transient infusion of siRNA into an epileptogenic focus of a patient that is targeted for surgical resection of the epileptogenic focus. Those studies could safely be performed during the routine pre-surgical evaluation of a patient.
Eventually, to provide long-term seizure relief, shRNA, miRNA or antisense constructs would need to be delivered via gene therapy vectors. The parallel development of gene therapies for CNS disorders that are currently in development should facilitate this transition. At the end, there is hope that a rationally designed focal therapy for epilepsy might not only suppress symptomatology but also provide a cure, by selectively targeting (i.e. silencing) aberrantly-expressed regulators of neuronal excitability.
ACKNOWLEDGMENTS
The author thanks Ursula Sandau for critically reading and commenting on the manuscript. The work of the author is supported by grants R01NS058780, R01NS061844, and R01MH083973, from the National Institutes of Health (NIH).
Footnotes
Conflict of interest: The author has no conflict of interest to disclose.
Ethical publication: I confirm that I have read the Journal’s position on issues involved in ethical publication and affirm that this review is consistent with those guidelines.
REFERENCES
- Aronica E, Fluiter K, Iyer A, Zurolo E, Vreijling J, van Vliet EA, Baayen JC, Gorter JA. Expression pattern of Mir-146a, an inflammation-associated microRNA, in experimental and human temporal lobe epilepsy. Eur J Neurosci. 2010;31:1100–1107. doi: 10.1111/j.1460-9568.2010.07122.x. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- Boison D. Adenosine as a neuromodulator in neurological diseases. Curr Opin Pharmacol. 2008a;8:2–7. doi: 10.1016/j.coph.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boison D. The adenosine kinase hypothesis of epileptogenesis. Progress in Neurobiology. 2008b;84:249–262. doi: 10.1016/j.pneurobio.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boison D. Adenosine augmentation therapies (AATs) for epilepsy: Prospect of cell and gene therapies. Epilepsy Res. 2009a;85:131–141. doi: 10.1016/j.eplepsyres.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boison D. Engineered adenosine-releasing cells for epilepsy therapy: Human mesenchymal stem cells and human embryonic stem cells. Neurotherapeutics. 2009b;6:278–283. doi: 10.1016/j.nurt.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boison D, Chen JF, Fredholm BB. Adenosine signalling and function in glial cells. Cell Death Differ. 2009 September 18; doi: 10.1038/cdd.2009.131. 2009; doi:10.1038/cdd.2009.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boison D, Stoffel W. Disruption of the compacted myelin sheath of axons of the central nervous system in proteolipid protein-deficient mice. Proc. Natl. Acad. Sci. USA. 1994;91:11709–11713. doi: 10.1073/pnas.91.24.11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burright EN, Clark HB, Servadio A, Matilla T, Feddersen RM, Yunis WS, Duvick LA, Zoghbi HY, Orr HT. Sca1 transgenic mice: A model for neurodegeneration caused by an expanded CAG trinucleotide repeat. Cell. 1995;82:937–948. doi: 10.1016/0092-8674(95)90273-2. [DOI] [PubMed] [Google Scholar]
- Catterall WA, Dib-Hajj S, Meisler MH, Pietrobon D. Inherited neuronal ion channelopathies: New windows on complex neurological diseases. J Neurosci. 2008;28:11768–11777. doi: 10.1523/JNEUROSCI.3901-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- Davidson BL, Boudreau RL. RNA interference: A tool for querying nervous system function and an emerging therapy. Neuron. 2007;53:781–788. doi: 10.1016/j.neuron.2007.02.020. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. OncoMirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fredholm BB, Chen JF, Cunha RA, Svenningsson P, Vaugeois JM. Adenosine and brain function. Int Rev Neurobiol. 2005;63:191–270. doi: 10.1016/S0074-7742(05)63007-3. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Alegre P, Paulson HL. Technology insight: Therapeutic RNA interference--how far from the neurology clinic? Nat Clin Pract Neurol. 2007;3:394–404. doi: 10.1038/ncpneuro0551. [DOI] [PubMed] [Google Scholar]
- Gray SJ, Blake BL, Criswell HE, Nicolson SC, Samulski RJ, McCown TJ. Directed evolution of a novel adeno-associated virus (AAV) vector that crosses the seizure-compromised blood-brain barrier (BBB) Mol Ther. 2010;18:570–578. doi: 10.1038/mt.2009.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnieri DJ, DiLeone RJ. MicroRNAs: A new class of gene regulators. Ann Med. 2008;40:197–208. doi: 10.1080/07853890701771823. [DOI] [PubMed] [Google Scholar]
- Gurney ME, Pu H, Chiu AY, Dal Canto MC, Polchow CY, Alexander DD, Caliendo J, Hentati A, Kwon YW, Deng HX, et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- Haberman R, Criswell H, Snowdy S, Ming Z, Breese G, Samulski R, McCown T. Therapeutic liabilities of in vivo viral vector tropism: Adeno-associated virus vectors, nmdar1 antisense, and focal seizure sensitivity. Mol Ther. 2002;6:495–500. doi: 10.1006/mthe.2002.0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Harper SQ, Staber PD, He X, Eliason SL, Martins IH, Mao Q, Yang L, Kotin RM, Paulson HL, Davidson BL. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc Natl Acad Sci U S A. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelters J, Ciccarella M, Drechsel M, Geissler C, Gulkan H, Bocker W, Schieker M, Jochum M, Neth P. Nonviral genetic modification mediates effective transgene expression and functional RNA interference in human mesenchymal stem cells. Journal of Gene Medicine. 2005;7:718–728. doi: 10.1002/jgm.731. [DOI] [PubMed] [Google Scholar]
- Huber A, Padrun V, Deglon N, Aebischer P, Mohler H, Boison D. Grafts of adenosine-releasing cells suppress seizures in kindling epilepsy. Proc. Natl. Acad. Sci. USA. 2001;98:7611–7616. doi: 10.1073/pnas.131102898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalev-Zylinska ML, Symes W, Young D, During MJ. Knockdown and overexpression of nr1 modulates NMDA receptor function. Mol Cell Neurosci. 2009;41:383–396. doi: 10.1016/j.mcn.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Lazarowski A, Czornyj L, Lubienieki F, Girardi E, Vazquez S, D'Giano C. ABC transporters during epilepsy and mechanisms underlying multidrug resistance in refractory epilepsy. Epilepsia. 2007;48 Suppl 5:140–149. doi: 10.1111/j.1528-1167.2007.01302.x. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004a;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for drosophila dicer-1 and dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004b;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Li T, Lan JQ, Boison D. Uncoupling of astrogliosis from epileptogenesis in adenosine kinase (ADK) transgenic mice. Neuron Glia Biology. 2008a;4:91–99. doi: 10.1017/S1740925X09990135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Lan JQ, Fredholm BB, Simon RP, Boison D. Adenosine dysfunction in astrogliosis: Cause for seizure generation? Neuron Glia Biology. 2007;3:353–366. doi: 10.1017/S1740925X0800015X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Ren G, Kaplan DL, Boison D. Human mesenchymal stem cell grafts engineered to release adenosine reduce chronic seizures in a mouse model of ca3-selective epileptogenesis. Epilepsy Res. 2009;84:238–241. doi: 10.1016/j.eplepsyres.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Ren G, Lusardi T, Wilz A, Lan JQ, Iwasato T, Itohara S, Simon RP, Boison D. Adenosine kinase is a target for the prediction and prevention of epileptogenesis in mice. J Clin Inv. 2008b;118:571–582. doi: 10.1172/JCI33737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingor P, Bahr M. Targeting neurological disease with RNAi. Mol Biosyst. 2007;3:773–780. doi: 10.1039/b701169e. [DOI] [PubMed] [Google Scholar]
- Lipman DJ. Making (anti)sense of non-coding sequence conservation. Nucleic Acids Res. 1997;25:3580–3583. doi: 10.1093/nar/25.18.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DZ, Tian Y, Ander BP, Xu H, Stamova BS, Zhan X, Turner RJ, Jickling G, Sharp FR. Brain and blood microRNA expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J Cereb Blood Flow Metab. 2010;30:92–101. doi: 10.1038/jcbfm.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loscher W. Drug transporters in the epileptic brain. Epilepsia. 2007;48 Suppl 1:8–13. doi: 10.1111/j.1528-1167.2007.00993.x. [DOI] [PubMed] [Google Scholar]
- Loscher W, Gernert M, Heinemann U. Cell and gene therapies in epilepsy - promising avenues or blind alleys? Trends in Neurosciences. 2008;31:62–73. doi: 10.1016/j.tins.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Lusardi TA, Farr CD, Faulkner CL, Pignataro G, Yang T, Lan J, Simon RP, Saugstad JA. Ischemic preconditioning regulates expression of microRNAs and a predicted target, mecp2, in mouse cortex. J Cereb Blood Flow Metab. 2009 doi: 10.1038/jcbfm.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MS, Dutt K, Papale LA, Dube CM, Dutton SB, de Haan G, Shankar A, Tufik S, Meisler MH, Baram TZ, Goldin AL, Escayg A. Altered function of the scn1a voltage-gated sodium channel leads to gamma-aminobutyric acid-ergic (GABAergic) interneuron abnormalities. J Biol Chem. 2010;285:9823–9834. doi: 10.1074/jbc.M109.078568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, Gilmore BL, Burstein H, Peluso RW, Polisky B, Carter BJ, Davidson BL. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: Implications for the therapeutic development of RNAi. Proc Natl Acad Sci U S A. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCown TJ. Adeno-associated virus vector-mediated expression and constitutive secretion of galanin suppresses limbic seizure activity. Neurotherapeutics. 2009;6:307–311. doi: 10.1016/j.nurt.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus MT, Sharp PA. Gene silencing in mammals by small interfering RNAs. Nat Rev Genet. 2002;3:737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- Montgomery MK, Xu S, Fire A. RNA as a target of double-stranded RNA-mediated genetic interference in caenorhabditis elegans. Proc Natl Acad Sci U S A. 1998;95:15502–15507. doi: 10.1073/pnas.95.26.15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahid MA, Pauley KM, Satoh M, Chan EK. Mir-146a is critical for endotoxin-induced tolerance: Implication in innate immunity. J Biol Chem. 2009;284:34590–34599. doi: 10.1074/jbc.M109.056317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellen W, Lichtenstein C. What makes an mRNA anti-sense-itive? TIBS. 1993;18:419–423. doi: 10.1016/0968-0004(93)90137-c. [DOI] [PubMed] [Google Scholar]
- Noe F, Frasca A, Balducci C, Carli M, Sperk G, Ferraguti F, Pitkanen A, Bland R, Fitzsimons H, During M, Vezzani A. Neuropeptide y overexpression using recombinant adeno-associated viral vectors. Neurotherapeutics. 2009;6:300–306. doi: 10.1016/j.nurt.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis S, Harrar DB, Lin Y, Koon AC, Hauser JL, Griffith EC, Zhu L, Brass LF, Chen C, Greenberg ME. An RNAi-based approach identifies molecules required for glutamatergic and GABAergic synapse development. Neuron. 2007;53:217–232. doi: 10.1016/j.neuron.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul CP, Good PD, Winer I, Engelke DR. Effective expression of small interfering RNA in human cells. Nat Biotechnol. 2002;20:505–508. doi: 10.1038/nbt0502-505. [DOI] [PubMed] [Google Scholar]
- Paulson HL, Bonini NM, Roth KA. Polyglutamine disease and neuronal cell death. Proc Natl Acad Sci U S A. 2000;97:12957–12958. doi: 10.1073/pnas.210395797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Radmark O. Ribonuclease activity and RNA binding of recombinant human dicer. EMBO J. 2002;21:5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph GS, Radcliffe PA, Day DM, Carthy JM, Leroux MA, Lee DC, Wong LF, Bilsland LG, Greensmith L, Kingsman SM, Mitrophanous KA, Mazarakis ND, Azzouz M. Silencing mutant sod1 using RNAi protects against neurodegeneration and extends survival in an ALS model. Nat Med. 2005;11:429–433. doi: 10.1038/nm1205. [DOI] [PubMed] [Google Scholar]
- Raol YH, Lund IV, Bandyopadhyay S, Zhang G, Roberts DS, Wolfe JH, Russek SJ, Brooks-Kayal AR. Enhancing GABA(a) receptor alpha 1 subunit levels in hippocampal dentate gyrus inhibits epilepsy development in an animal model of temporal lobe epilepsy. J Neurosci. 2006;26:11342–11346. doi: 10.1523/JNEUROSCI.3329-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoul C, Abbas-Terki T, Bensadoun JC, Guillot S, Haase G, Szulc J, Henderson CE, Aebischer P. Lentiviral-mediated silencing of sod1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nature Medicine. 2005;11:423–428. doi: 10.1038/nm1207. [DOI] [PubMed] [Google Scholar]
- Ren G, Li T, Lan JQ, Wilz A, Simon RP, Boison D. Lentiviral RNAi-induced downregulation of adenosine kinase in human mesenchymal stem cell grafts: A novel perspective for seizure control. Exp Neurol. 2007;208:26–37. doi: 10.1016/j.expneurol.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riban V, Fitzsimons HL, During MJ. Gene therapy in epilepsy. Epilepsia. 2009;50:24–32. doi: 10.1111/j.1528-1167.2008.01743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Lebron E, Gouvion CM, Moore SA, Davidson BL, Paulson HL. Allele-specific RNAi mitigates phenotypic progression in a transgenic model of Alzheimer’s disease. Mol Ther. 2009;17:1563–1573. doi: 10.1038/mt.2009.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Lebron E, Paulson HL. Allele-specific RNA interference for neurological disease. Gene Therapy. 2006;13:576–581. doi: 10.1038/sj.gt.3302702. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Sebastiao AM, Ribeiro JA. Tuning and fine-tuning of synapses with adenosine. Curr Neuropharmacol. 2009;7:180–194. doi: 10.2174/157015909789152128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, Sheth N, Bradshaw J, Burchard J, Kulkarni A, Cavet G, Sachidanandam R, McCombie WR, Cleary MA, Elledge SJ, Hannon GJ. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- Singer O, Marr RA, Rockenstein E, Crews L, Coufal NG, Gage FH, Verma IM, Masliah E. Targeting bace1 with siRNAs ameliorates Alzheimer disease neuropathology in a transgenic model. Nat Neurosci. 2005;8:1343–1349. doi: 10.1038/nn1531. [DOI] [PubMed] [Google Scholar]
- Stafstrom CE. Severe epilepsy syndromes of early childhood: The link between genetics and pathophysiology with a focus on scn1a mutations. J Child Neurol. 2009;24:15S–23S. doi: 10.1177/0883073809338152. [DOI] [PubMed] [Google Scholar]
- Sugiyama O, An DS, Kung SPK, Feeley BT, Gamradt S, Liu NQ, Chen ISY, Lieberman JR. Lentivirus-mediated gene transfer induces long-term transgene expression of bmp-2 in vitro and new bone formation in vivo. Molecular Therapy. 2005;11:390–398. doi: 10.1016/j.ymthe.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Sui G, Soohoo C, Affar el B, Gay F, Shi Y, Forrester WC. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc Natl Acad Sci U S A. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappab-dependent induction of microRNA mir-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theofilas P, Brar S, Li T, Stewart K-A, Sandau U, Poulsen DJ, Boison D. Adenosine kinase is a therapeutic target for RNA interference in epilepsy. Epilepsia. 2010 doi: 10.1111/j.1528-1167.2010.02947.x. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Chen L, Yang T, Zhang Q, Zhou D. RNAi inhibits coriaria lactone-induced mdr1b overexpression in rat brain microvascular endothelial cells. J Mol Neurosci. 2009;39:284–293. doi: 10.1007/s12031-009-9198-3. [DOI] [PubMed] [Google Scholar]
- Totsugawa T, Kobayashi N, Okitsu T, Noguchi H, Watanabe T, Matsumura T, Maruyama M, Fujiwara T, Sakaguchi M, Tanaka N. Lentiviral transfer of the LacZ gene into human endothelial cells and human bone marrow mesenchymal stem cells. Cell Transplantation. 2002;11:481–488. [PubMed] [Google Scholar]
- Tuschl T, Zamore PD, Lehmann R, Bartel DP, Sharp PA. Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajda FJE. Pharmacotherapy of epilepsy: New armamentarium, new issues. Journal of Clinical Neuroscience. 2007;14:813–823. doi: 10.1016/j.jocn.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Vashlishan AB, Madison JM, Dybbs M, Bai J, Sieburth D, Ch'ng Q, Tavazoie M, Kaplan JM. An RNAi screen identifies genes that regulate GABA synapses. Neuron. 2008;58:346–361. doi: 10.1016/j.neuron.2008.02.019. [DOI] [PubMed] [Google Scholar]
- Wang Q, Carmichael GG. Effects of length and location on the cellular response to double-stranded RNA. Microbiol Mol Biol Rev. 2004;68:432–452. doi: 10.1128/MMBR.68.3.432-452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieser HG. ILAE commission report. Mesial temporal lobe epilepsy with hippocampal sclerosis. Epilepsia. 2004;45:695–714. doi: 10.1111/j.0013-9580.2004.09004.x. [DOI] [PubMed] [Google Scholar]
- Wolff JA, Budker V. The mechanism of naked DNA uptake and expression. Adv Genet. 2005;54:3–20. doi: 10.1016/S0065-2660(05)54001-X. [DOI] [PubMed] [Google Scholar]
- Xia H, Mao Q, Eliason SL, Harper SQ, Martins IH, Orr HT, Paulson HL, Yang L, Kotin RM, Davidson BL. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat Med. 2004;10:816–820. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- Xiao X, Li J, McCown TJ, Samulski RJ. Gene transfer by adeno-associated virus vectors into the central nervous system. Exp Neurol. 1997;144:113–124. doi: 10.1006/exnr.1996.6396. [DOI] [PubMed] [Google Scholar]