Abstract
PKC, β-arrestin 2, CARMA3, BCL10, MALT1, TRAF6 and MEKK3 are signaling proteins that have a key role in G protein-coupled receptor (GPCR)-mediated activation of nuclear factor-κB (NF-κB) pathway in nonhematopoietic cells in response to lysophosphatidic acid (LPA) stimulation. The PKC, β-arrestin 2, CARMA3-BCL10-MALT1-TRAF6 signalosome, and MEKK3 functions as a link between GPCR signaling and proinflammatory IKK-NF-κB activation. Here we briefly summarize recent progress in the understanding of the molecular and biological functions of these proteins in GPCR-mediated NF-κB activation in nonhematopoietic cells.
Keywords: NF-κB, LPA, GPCR
Introduction
Lysophosphatidic acid (LPA) is a potent bioactive phospholipid derivative that is capable of inducing diverse cellular responses, such as cell proliferation, migration, and cytokine release [1]. In vertebrate, significant amounts of LPA can be detected in various biological fluids, including serum, saliva, and bronchoalveolar lavage fluid. LPA exerts its biological effect through activation of three high-affinity G protein-coupled receptors (GPCRs), called LPA1, LPA2, and LPA3 (also known as EDG2, EDG4, and EDG7). Additional, three GPCRs have been identified newly as LPA receptors include LPA4 (p2y9/GPR23), LPA5 (GPR92) and LPA6 (GPR87). LPA-mediated signal transduction pathways regulate gene expression through activation of several transcriptional factors, such as nuclear factor-κB (NF-κB) and AP-1.
GPCRs transduce environmental signals across the plasma membrane by stimulating guanine nucleotide exchange by heterotrimeric G proteins [2]. Exchange of GDP for GTP results in activation of the Gα subunits and dissociation of the Gβγ subunits. The Gα subunits contain several subgroups, including Gi, Gs, Gq, G16, and G12/13. These G proteins can independently activate their downstream signaling cascades that lead to activation of various transcription factors, including NF-κB [3]. Previous studies have shown that the stimulation of GPCR ligands, such as LPA, endothelin-1 (ET-1), and angiotensin-II (Ang-II), induces NF-κB activation [4, 5].
Recent studies have revealed PKC, β-arrestin 2, CARMA3 (caspase recruitment domain, CARD, membrane-associated guanylate kinase, MAGUK, protein 3), BCL10 (B-cell lymphoma 10), MALT1 (mucosa-associated lymphoid tissue lymphoma translocation protein 1), TRAF6 (the tumor necrosis factor (TNF) receptor-associated factor 6), and MEKK3 (MAP kinase kinase kinase 3) as signaling components that have crucial and specific roles in LPA-GPCR-induced NF-κB activation. This review summarizes the presently available knowledge on the molecular and biological function of these proteins in LPA-induced NF-κB activation in nonhematopoietic cells.
NF-κB activation
A major challenge in the NF-κB field is to understand how distinct upstream stimuli activate IKK in a signal-specific manner [6, 7]. GPCRs-mediated NF-κB activation has been shown to be involved in the regulation of expression of genes that are essential for nonhematopoietic cells activation and the cellular responses. The NF-κB family comprises five mammalian members NF-κB1 (p50 and its precursor p105), NF-κB2 (p52 and its precursor p100), c-Rel, RelA (p65), and RelB [8–12]. In its active DNA-binding form, NF-κB exists as a heterogeneous collection of dimers composed of various combinations of members of the NF-κB/Rel family. In unstimulated cells, NF-κB is sequestered in the cytoplasm bound to inhibitory proteins, which are members of the IκB family. NF-κB activation is triggered by the activated IκB kinase (IKK) complex, which contains two catalytic subunits, IKKα and IKKβ, and a regulatory subunit, IKKγ (also known as NEMO). IKK complex is the convergence point for many NF-κB signaling pathways and its activity is regulated by phosphorylation [13, 14]. Activation of the IKK complex leads to phosphorylation and subsequent ubiquitination and proteolytic degradation of IκBα, and allows NF-κB to translocate to the nucleus and activate gene transcription and expression [11, 15, 16]. Various cell surface receptors, including receptors for proinflammatory cytokines such as TNFα and Interleukin-1 (IL-1β), Toll-like receptors (TLRs), antigen receptors, and GPCRs, activate NF-κB pathway using specific sets of signaling molecules.
A recent breakthrough in our understanding of GPCR-mediated NF-κB activation is the identification of several physically and functionally interacting proteins known as PKC, β-arrestin-2, CARMA3, BCL10, MALT1, TRAF6 and MEKK3 that act as crucial signaling compounds downstream of the GPCR and upstream of the IKK complex [17–23].
PKC family
PKC is a superfamily of kinases that phosphorylates protein substrates on serine and threonine residues and transduces the cellular signals. PKC was originally identified as a phospholipids and calcium-dependent protein kinase [24, 25]. The subsequent classification of the isoforms is based on structural and activational characteristics. There are ten PKC genes that encode for isozymes in mammals divided into 3 subgroups: classical or conventional PKCs (cPKCs; PKCα, PKCβI, PKXβII and PKCγ), which are calcium-dependent and activated by diacylglycerol and phospholipids; novel PKCs (nPKCs; PKCδ, PKCε, PKCη and PKCθ), which are calcium-independent and regulated by diacylglycerol and phospholipids; and atypical PKCs (aPKCs; PKCζ and PKCι (known as PKCλ in mice)), which are calcium independent and do not require diacylglycerol for activation [24, 26–29]. PKC isoforms differ in their structure, tissue distribution, subcellular localization, and substrate specificity, as well as biological functions, although some of these PKCs show overlapping substrate specificities [24, 29].
GPCR-mediated signaling leads to phosphorylation and activation of different PKC isoforms, followed by activating IKK-NF-κB. It has been shown that PKC is involved in LPA-induced NF-κB activation. PKCδ mediates NF-κB activation/IL-8 secretion in response to LPA stimulation in bronchial epithelial cells [30]. PKCβ and PKCθ mediate B- and T-cell antigen receptor-induced NF-κB activation, respectively [31, 32]. PKCζ is required for IL-1-induced NF-κB activation in articular chondrocytes [33]. LPA also activates PKCα and induced RAS-PKCα interaction, causing NF-κB activation via CARMA3-BCL10-MALT1 signaling complex in ovarian cancer cells [34].
CARMA3
CARMA3 belongs to the CARMA family that contains three proteins, CARMA1, CARMA2, and CARMA3 [35–37]. CARMA family proteins share similar structural regions, with a CARD domain at N terminus, followed by a coiled-coil (CC) domain, a PDZ domain, a SH3 domain, and a guanylate kinase-like (GUK) domain at C terminus. However, these CARMA proteins show different expression pattern with CARMA1 (CARD11) expressed in hematopoietic cells, CARMA2 (CARD14/Bimp2) in the placenta and CARMA3 (CARD10/Bimp1) in all nonhematopoietic cells [35–38]. Recent studies have shown CARMA1 plays an essential role in antigen receptor-induced NF-κB activation [39–45]. The overexpression of CARMA2 and CARMA3 could induce NF-κB activation in HEK293 cells [37, 38]. Furthermore, ~50% of Carma3-deficient mice have the neural tube defect (NTD) phenotype known as anencephaly before embryonic day 10.5 (E10.5) resulting in perinatal mortality of the mice due to either bleeding out from the skull or infanticide by the mother [22]. As CARMA3 associates with BCL10, when overexpressed in HEK293 cells, indicating CARMA3 and BCL10 may function in the same signal transduction pathway [36, 37].
CARMA3 is specifically required for GPCR-induced NF-κB activation in Carma3-deficient cells with LPA and ET-1 stimulation, while its defect does not effect NF-κB activation by other stimuli such as TNFα, lipopolysaccharide (LPS), and extracellular matrix proteins [22]. Although CARMA3 is required for LPA-induced IKK-NF-κB activation, it is not required for the LPA-induced IKK phosphorylation. The deficiency of either Carma1 or Carma3 impairs IKK activation without affecting the signal-induced IKKα/β phosphorylation after TCR, LPA, or PKC agonist stimulation, suggesting that the signal-induced phosphorylation of IKKα/β is not sufficient to activate the IKK complex, and other modifications such as ubiquitination may be required to do so [22, 46].
Defective NF-κB activation in Carma1/3, Bcl10 or Malt1-deficient T/B cells or murine embryonic fibroblasts (MEFs) show that CARMA1/3, BCL10 and MALT1 function as part of a signaling complex to bridge PKC to IKK-mediated NF-κB activation in both lymphocytes and nonhematopoietic cells [7, 21, 22, 31, 32]. CARMA3/BCL10/MALT1 complex has also been shown to act downstream of G(i) mediated RAS-PKCα interaction and upstream of NF-κB activation in LPA-induced urokinase plasminogen activator (uPA) upregulation in ovarian cancer cells [34, 47, 48].
β-arrestin 2
There are four members in the arrestin family in human genome. Visual arrestin (arrestin 1) is localized to retinal rods and cones, whereas X-arrestin (arrestin 4) is found exclusively in retinal cones, both arrestin 1 and arrestin 4 regulate opsin [49]. Arrestins 2 and 3, also named β-arrestin 1 and β-arrestin 2, respectively, are ubiquitously expressed in most tissues, and play important roles in regulating signal transduction by numerous GPCRs [49–52]. The amino acid sequences of the two β-arrestin isoforms are ≈70% identical, most of the coding differences appear in the C termini [53]. Genetics deficiency studies show that mice with either β-arrestin 1 or β-arrestin 2 deficiency are viable [53–55], whereas the double-knockout mice is embryonic lethal [56], indicating that each β-arrestin partially function as a substitute for the other isoform. Recent studies suggest that they have different functions in GPCR-induced signaling pathways [20, 57–72].
Several studies suggest that β-arrestins may function as negative regulators to suppress NF-κB activation [64–66, 73]. β-arrestins has been shown to regulate NF-κB activation by interacting with TRAF6 and preventing its autoubiquitination and activation of NF-κB [64]. Furthermore, GPCRs associate with β-arrestins upon stimulation by their ligands such as LPA [20, 57–63, 74]. Genetic evidence demonstrates that β-arrestin 2, but not β-arrestin 1, is required for LPA-induced NF-κB activation through recruiting CARMA3 to LPA receptor, functions as a positive regulator for LPA-induced IKK-NF-κB activation and cytokine production [20]. Proteomic study suggests that β-arrestin 2 is associated with the TAK1 and IKKα complexes in response to Ang-II stimulation [75].
BCL10 and MALT1
Molecular cloning of the breakpoint identified a novel gene, Bcl10, in MALT B lymphoma [76, 77]. The human Bcl10 encodes a protein of 233 amino acids with residues 13–101 forming an N-terminal CARD, whereas the C-terminal 132 amino acids contain no known motifs, is rich in serine and threonine residues, and can be phosphorylated [76–82]. The BCL10 CARD domain alone is sufficient and necessary for NF-κB activation [76, 78–80].
The human paracaspase MALT1 has been identified as a caspase-like BCL10-binding protein involved in NF-κB activation [83–85]. MALT1 contains an N-terminal death domain (DD), two immunoglobulin (Ig)-like domains, a caspase-like domain and a C-terminal region that contains another Ig-like domain [83, 84, 86]. Similar to BCL10, MALT1 has been found to be recurrently rearranged in chromosomal translocation in some MALT lymphomas, resulting in a chimeric fusion protein between the MALT1 C-terminal region and the N-terminal portion of cIAP2 (an anti-apoptotic protein), which activates NF-κB [84]. MALT1 contains three potential binding sites for TRAF6, by which MALT1 may regulate in coordination the recruitment and activation of TRAF6. TRAF6 then ubiquitinates itself and MALT1 on its multiple C-terminal lysine residues that can in turn recruit the IKK complex [83, 84, 86–92].
Using genetic deficiency mice, BCL10 and MALT1 were revealed have critical roles in both TCR and BCR-mediated NF-κB signaling pathway [16, 93–97]. BCL10 and MALT1 physically and functionally cooperate to relay antigen receptor-induced PKC signaling (PKC-θ in T cells or PKC-β in B cells) to IKK-NF-κB activaction [7, 31, 32, 93, 95, 96]. Similar to Carma3-deficient mice, 40% of Bcl10-deficeint mice show the NTD phenotype [93].
By using MEF cells from Bcl10- or Malt1-deficient mice as a genetic model, BCL10 and MALT1 are identified critically required for NF-κB activation and cytokine production in response to LPA stimulation in nonhematopoietic cells. BCL10 and MALT1 collaborate with PKCs specifically for LPA-induced NF-κB activation but are not required for the activation of the JNK, p38, ERK MAP kinase, and AKT signaling pathways [7, 21].
Therefore, the PKC-CARMA-BCL10/MALT1 module may constitute a common axis to transduce signals from PKC to IKK downstream of antigen receptors and GPCRs in multiple cell types and functions in a similar mechanism.
TRAF6
TRAFs are a family of adaptor proteins that couple the TNF receptor family to signaling pathways, and they are also shown to be signal transducers of Toll/IL-1 family members. Seven members of the TRAF family have been identified. All TRAF proteins share a conserved C-terminal TRAF-C domain that can interact with the cytoplasmic domain of receptors and other TRAF proteins, a CC TRAF-N domain, in additional, TRAFs 2–7 have RING and zinc finger motifs that are important for signaling downstream events [98–104].
TRAF6 was initially identified as a signal transducer for IL-1 [100]. Overexpression of TRAF6 activates NF-κB, and a dominant negative mutant of TRAF6 inhibits NF-κB activation by IL-1 but not TNF. The RING finger domain of TRAF6 can function as an E3 ubiquitin ligase, which, together with the Ubc13/Uev1A complex mediates another unidentified protein polyubiquitination involved in IKK activation [103, 105].
Using genetic deficiency mice model, Traf6-deficient mice show predominant abnormal phenotype relating to defective bone formation [106, 107]. CD40-mediated NF-κB activation and proliferation in splenic B cells, and IL-1-induced activation of both NF-κB and JNK/SAPK, and IL-1 induced thymocyte proliferation were all abolished in Traf6-deficient mice model [106].
TRAF6 and MALT1 may function as E3 ligases to induce lysine 63 (K63)-linked polyubiquitination of IKKγ, leading to activation of the IKK complex and subsequently NF-κB [88, 108, 109]. In addition, Traf6 deficiency also displays a similar defect of the neural tube closure as Carma3 deficiency does [22, 110]. TRAF6 is required for GPCR-induced NF-κB activation [22]. The activation of NF-κB induced by LPA or PKC agonist was completely defective in Traf6-deficient MEF cells, while similar to the role of CARMA3/BCL10/MALT1 in IKK activation, deficient of TRAF6 expression does not effect LPA- or PKC agonist-induce IKKβ phosphorylation. These studies indicate that CARMA3, BCL10, MALT1 and TRAF6 mediate LPA-induced NF-κB activation through an IKKβ phosphorylation-independent mechanism.
MEKK3
MEKK3 cDNA was first isolated from NIH3T3 cells [111]. MEKK3 is a member of the mitogen-activated protein kinase kinase kinase (MAP3K) family, it activates IKK and MAPK when overexpressed [111, 112].
The genetic inactivation of MEKK3 in mice gives rise to an embryonic lethal phenotype characterized by defects in angiogenesis and early cardiovascular development [113]. Endothelial cells from Mekk3-deficient embryos defects in cell proliferation, apoptosis, and interactions with myocardium in the heart [114]. In addition, MEKK3 is required for TCR-mediated IKK-NF-κB activation [115].
In Mekk3-deficient MEF cells, LPA and PKC-induced IKK phosphorylation and NF-κB activation is significantly impaired [23]. Phosphorylation of MEKK3 at Thr-516 and Ser-520 within the kinase activation loop is essential for LPA-induced MEKK3- mediated IKK-NF-κB activation [116]. Together, these data suggest that MEKK3 plays an essential role in LPA-induced NF-κB activation.
Perspectives
The data discussed here suggest that GPCR signaling are relayed to the NF-κB activating IKK complex by a pathway that depends on PKC, β-arrestin 2, CARMA3, BCL10, MALT1, TRAF6 and MEKK3. However, still much remains to be learned about how these signaling proteins are connected to GPCR, on the one hand, and to the downstream components controlling IKK-NF-κB activation, on the other hand.
PKC family has been identified including many members, actually which isoform(s) involved in LPA-induced NF-κB activation are still needs to be clearly defined. An intriguing question concerns the physical and functional relationship between PKC and CARMA3/BCL10/MALT1/TRAF6 complex and MEKK3. It remains to be determined whether these proteins are substrates or physical interaction partners of PKC to transduce the GPCR signals to the downstream IKK complex, and activate NF-κB signaling pathway.
Although MEKK3 has been identified as a relay to mediate upstream GPCR-PKC signals and downstream IKK complex activation, it remains obscure how MEKK3 plays this role in the whole GPCR-NF-κB signaling pathway. MEKK3 is a kinase with a PB1 domain. PB1 domain is a scaffold module that has been shown to adopt the topology of ubiquitin-like β-grasp fold that interacts with each other in a front-to-back mode to arrange heterodimers or homo-oligomers of PB1-containing proteins [117]. Human genome encodes several PB1-domain-containing proteins, including p62, aPKC, MEKK2/MEKK3, MEK5, and Par-6. The PB1 domain has been proposed to provide specificity for PB1-containing kinases to ensure the effective transmission of cellular signals. p62 specifically binds to PKCλ/ι through its PB1 domain to modulate aPKC activation and p62/aPKC cassette regulates TRAF6-mediated NF-κB activation [118]. PB1-containing MEKK2 and MEKK3 are involved in the regulation in different phases of IKK activation and MEK5 activation through their PB1 domain [119, 120, 121]. The aPKCs induce MEK5 activation through their PB1 domain interaction [122]. Recent studies have shown that p62-MEKK3 interaction through their PB1 domain is involved in the regulation of TRAF6-mediated ubiquitination of IKK complex and downstream NF-κB activation [123]. Base on these observations, it is likely that these PB1-containing molecules are involved in LPA-mediated MEKK3 activation, as well as MEKK3-mediated IKK-NF-κB activation. Therefore, more studies are needed to clarify the association among these PB1 domain molecules in GPCR-induced NF-κB signaling pathway, to determine whether PB1 domain is the essential region for the functional and physical interaction of these PB1 domain-containing proteins, in the LPA signal-induced NF-κB activation.
Although significant progress has been made on the mechanism of the LPA-induced NF-κB activation, it is unclear how LPA-induced NF-κB activation is negatively regulated. As the data discovered so far, IKKβ has been shown to be required for LPA-induced NF-κB activation [23]. Two protein serine/threonine phosphatases, PPM1A and PPM1B, have been identified as IKKβ phosphatases to dephosphorylate IKKβ and downregulate IKK-mediated NF-κB activation [124, 125]. Therefore, it is highly likely that PPM1A and PPM1B are involved in the downregulation of LPA-induced NF-κB activation by targeting on IKKβ. However, the identity of the protein serine/threonine phosphatases that dephosphorylate MEKK3 and inhibit its activity in LPA-induced NF-κB activation remains to be clearly defined.
Finally, an important issue that needs to be addressed is how CARMA3/BCL10/MALT1/TRAF6 complex-mediated IKK complex ubiquitination is coordinated with MEKK3-mediated IKK phosphorylation to activate IKK-NF-κB in LPA-induced NF-κB activation.
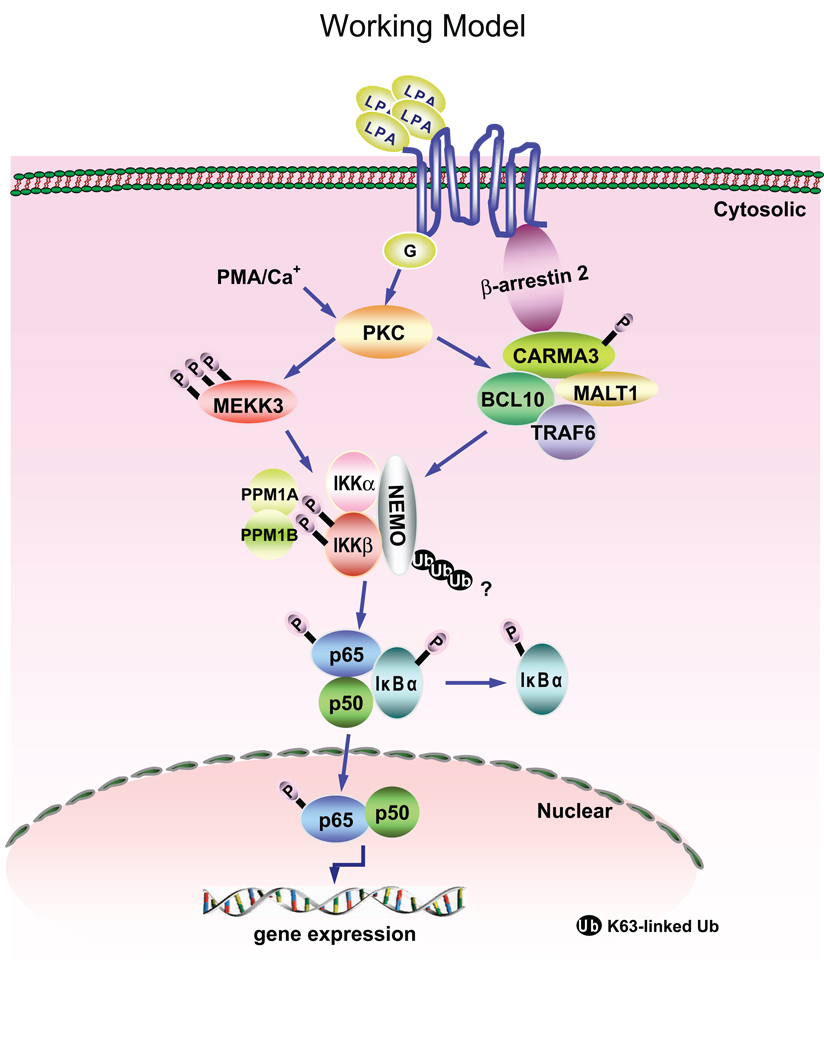
Based on the data reported thus far, we draw a working model (Fig. 1), in which binding of its cognate GPCR by LPA induces PKC activation that leads to MEKK3-mediated phosphorylation of IKKβ and β-arrestin 2-CARMA3-BCL10-MALT1-TRAF6-mediated ubiquitination of IKK complex that result in optimal IKKβ-mediated NF-κB activation. PPM1A and PPM1B phosphatases bind to the phosphorylated IKKβ and terminate NF-κB activation through dephosphorylation of IKKβ at Ser-177 and Ser-181 residues.
Figure 1.
A working model for LPA-induced NF-κB activation. LPA binding to its cognate GPCR induces PKC activation that leads to MEKK3-mediated phosphorylation of IKKβ and β-arrestin 2-CARMA3-BCL10-MALT1-TRAF6-mediated ubiquitination of IKK complex that result in optimal IKKβ-mediated NF-κB activation. PPM1A and PPM1B phosphatases bind to the phosphorylated IKKβ and terminate NF-κB activation through dephosphorylation of IKKβ at Ser-177 and Ser-181 residues
Acknowledgement
The research in Yang's laboratory is supported by the NIH/NCI grant 1R21CA106513-01A2 (to J.Y.) from the National Cancer Institute, a research scholar grant RSG-06-070-01-TBE (to J.Y.) from the American Cancer Society, the Virginia & L E Simmons Family Foundation Collaborative Research Fund (to J.Y.) and DLDCC Pilot study from NIH/NCI grant 5P30CA125123-03 (to J.Y.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Marinissen MJ, Gutkind JS. Trends Pharmacol Sci. 2001;22(7):368–376. doi: 10.1016/s0165-6147(00)01678-3. [DOI] [PubMed] [Google Scholar]
- 2.Gilman AG. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 3.Ye RD. J Leukoc Biol. 2001;70(6):839–848. [PubMed] [Google Scholar]
- 4.Shahrestanifar M, Fan X, Manning DR. J Biol Chem. 1999;274(6):3828–3833. doi: 10.1074/jbc.274.6.3828. [DOI] [PubMed] [Google Scholar]
- 5.Purcell NH, Tang G, Yu C, Mercurio F, DiDonato JA, Lin A. Proc Natl Acad Sci U S A. 2001;98(12):6668–6673. doi: 10.1073/pnas.111155798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dixit V, Mak TW. Cell. 2002;111(5):615–619. doi: 10.1016/s0092-8674(02)01166-2. [DOI] [PubMed] [Google Scholar]
- 7.Klemm S, Zimmermann S, Peschel C, Mak TW, Ruland J. Proc Natl Acad Sci U S A. 2007;104(1):134–138. doi: 10.1073/pnas.0608388103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karin M. Nature. 2006;441(7092):431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 9.Baldwin AS., Jr Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 10.May MJ, Ghosh S. Immunol Today. 1998;19(2):80–88. doi: 10.1016/s0167-5699(97)01197-3. [DOI] [PubMed] [Google Scholar]
- 11.Li Q, Verma IM. Nat Rev Immunol. 2002;2(10):725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 12.Karin M, Ben-Neriah Y. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 13.Zandi E, Karin M. Mol Cell Biol. 1999;19(7):4547–4551. doi: 10.1128/mcb.19.7.4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delhase M, Hayakawa M, Chen Y, Karin M. Science. 1999;284(5412):309–313. doi: 10.1126/science.284.5412.309. [DOI] [PubMed] [Google Scholar]
- 15.Karin M, Delhase M. Semin Immunol. 2000;12(1):85–98. doi: 10.1006/smim.2000.0210. [DOI] [PubMed] [Google Scholar]
- 16.Thome M. Nat Rev Immunol. 2004;4(5):348–359. doi: 10.1038/nri1352. [DOI] [PubMed] [Google Scholar]
- 17.Thome M, Tschopp J. Trends Immunol. 2003;24(8):419–424. doi: 10.1016/s1471-4906(03)00177-7. [DOI] [PubMed] [Google Scholar]
- 18.Jun JE, Goodnow CC. Nat Immunol. 2003;4(11):1057–1064. doi: 10.1038/ni1001. [DOI] [PubMed] [Google Scholar]
- 19.Ruland J, Mak TW. Immunol Rev. 2003;193:93–100. doi: 10.1034/j.1600-065x.2003.00049.x. [DOI] [PubMed] [Google Scholar]
- 20.Sun J, Lin X. Proc Natl Acad Sci U S A. 2008;105(44):17085–17090. doi: 10.1073/pnas.0802701105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang D, You Y, Lin PC, Xue L, Morris SW, Zeng H, Wen R, Lin X. Proc Natl Acad Sci U S A. 2007;104(1):145–150. doi: 10.1073/pnas.0601894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grabiner BC, Blonska M, Lin PC, You Y, Wang D, Sun J, Darnay BG, Dong C, Lin X. Genes Dev. 2007;21(8):984–996. doi: 10.1101/gad.1502507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun W, Li H, Yu Y, Fan Y, Grabiner BC, Mao R, Ge N, Zhang H, Fu S, Lin X, Yang J. Cell Signal. 2009;21(10):1488–1494. doi: 10.1016/j.cellsig.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mackay HJ, Twelves CJ. Nat Rev Cancer. 2007;7(7):554–562. doi: 10.1038/nrc2168. [DOI] [PubMed] [Google Scholar]
- 25.Takai Y, Kishimoto A, Iwasa Y, Kawahara Y, Mori T, Nishizuka Y. J Biol Chem. 1979;254(10):3692–3695. [PubMed] [Google Scholar]
- 26.Newton AC. J Biol Chem. 1995;270(48):28495–28498. doi: 10.1074/jbc.270.48.28495. [DOI] [PubMed] [Google Scholar]
- 27.Nishizuka Y. Science. 1992;258(5082):607–614. doi: 10.1126/science.1411571. [DOI] [PubMed] [Google Scholar]
- 28.Schenk PW, Snaar-Jagalska BE. Biochim Biophys Acta. 1999;1449(1):1–24. doi: 10.1016/s0167-4889(98)00178-5. [DOI] [PubMed] [Google Scholar]
- 29.Rosse C, Linch M, Kermorgant S, Cameron AJ, Boeckeler K, Parker PJ. Nat Rev Mol Cell Biol. 2010;11(2):103–112. doi: 10.1038/nrm2847. [DOI] [PubMed] [Google Scholar]
- 30.Cummings R, Zhao Y, Jacoby D, Spannhake EW, Ohba M, Garcia JG, Watkins T, He D, Saatian B, Natarajan V. J Biol Chem. 2004;279(39):41085–41094. doi: 10.1074/jbc.M404045200. [DOI] [PubMed] [Google Scholar]
- 31.Matsumoto R, Wang D, Blonska M, Li H, Kobayashi M, Pappu B, Chen Y, Lin X. Immunity. 2005;23(6):575–585. doi: 10.1016/j.immuni.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 32.Sommer K, Guo B, Pomerantz JL, Bandaranayake AD, Moreno-Garcia ME, Ovechkina YL, Rawlings DJ. Immunity. 2005;23(6):561–574. doi: 10.1016/j.immuni.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 33.LaVallie ER, Chockalingam PS, Collins-Racie LA, Freeman BA, Keohan CC, Leitges M, Dorner AJ, Morris EA, Majumdar MK, Arai M. J Biol Chem. 2006;281(34):24124–24137. doi: 10.1074/jbc.M601905200. [DOI] [PubMed] [Google Scholar]
- 34.Mahanivong C, Chen HM, Yee SW, Pan ZK, Dong Z, Huang S. Oncogene. 2008;27(9):1273–1280. doi: 10.1038/sj.onc.1210746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaide O, Martinon F, Micheau O, Bonnet D, Thome M. Tschopp J. FEBS Lett. 2001;496(2–3):121–127. doi: 10.1016/s0014-5793(01)02414-0. [DOI] [PubMed] [Google Scholar]
- 36.McAllister-Lucas LM, Inohara N, Lucas PC, Ruland J, Benito A, Li Q, Chen S, Chen FF, Yamaoka S, Verma IM, Mak TW, Nunez G. J Biol Chem. 2001;276(33):30589–30597. doi: 10.1074/jbc.M103824200. [DOI] [PubMed] [Google Scholar]
- 37.Wang L, Guo Y, Huang WJ, Ke X, Poyet JL, Manji GA, Merriam S, Glucksmann MA, DiStefano PS, Alnemri ES, Bertin J. J Biol Chem. 2001;276(24):21405–21409. doi: 10.1074/jbc.M102488200. [DOI] [PubMed] [Google Scholar]
- 38.Bertin J, Wang L, Guo Y, Jacobson MD, Poyet JL, Srinivasula SM, Merriam S, DiStefano PS, Alnemri ES. J Biol Chem. 2001;276(15):11877–11882. doi: 10.1074/jbc.M010512200. [DOI] [PubMed] [Google Scholar]
- 39.Gaide O, Favier B, Legler DF, Bonnet D, Brissoni B, Valitutti S, Bron C, Tschopp J, Thome M. Nat Immunol. 2002;3(9):836–843. doi: 10.1038/ni830. [DOI] [PubMed] [Google Scholar]
- 40.Pomerantz JL, Denny EM, Baltimore D. EMBO J. 2002;21(19):5184–5194. doi: 10.1093/emboj/cdf505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang D, You Y, Case SM, McAllister-Lucas LM, Wang L, DiStefano PS, Nunez G, Bertin J, Lin X. Nat Immunol. 2002;3(9):830–835. doi: 10.1038/ni824. [DOI] [PubMed] [Google Scholar]
- 42.Egawa T, Albrecht B, Favier B, Sunshine MJ, Mirchandani K, O'Brien W, Thome M, Littman DR. Curr Biol. 2003;13(14):1252–1258. doi: 10.1016/s0960-9822(03)00491-3. [DOI] [PubMed] [Google Scholar]
- 43.Hara H, Wada T, Bakal C, Kozieradzki I, Suzuki S, Suzuki N, Nghiem M, Griffiths EK, Krawczyk C, Bauer B, D'Acquisto F, Ghosh S, Yeh WC, Baier G, Rottapel R, Penninger JM. Immunity. 2003;18(6):763–775. doi: 10.1016/s1074-7613(03)00148-1. [DOI] [PubMed] [Google Scholar]
- 44.Jun JE, Wilson LE, Vinuesa CG, Lesage S, Blery M, Miosge LA, Cook MC, Kucharska EM, Hara H, Penninger JM, Domashenz H, Hong NA, Glynne RJ, Nelms KA, Goodnow CC. Immunity. 2003;18(6):751–762. doi: 10.1016/s1074-7613(03)00141-9. [DOI] [PubMed] [Google Scholar]
- 45.Newton K, Dixit VM. Curr Biol. 2003;13(14):1247–1251. doi: 10.1016/s0960-9822(03)00458-5. [DOI] [PubMed] [Google Scholar]
- 46.Shambharkar PB, Blonska M, Pappu BP, Li H, You Y, Sakurai H, Darnay BG, Hara H, Penninger J, Lin X. EMBO J. 2007;26(7):1794–1805. doi: 10.1038/sj.emboj.7601622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pustilnik TB, Estrella V, Wiener JR, Mao M, Eder A, Watt MA, Bast RC, Jr, Mills GB. Clin Cancer Res. 1999;5(11):3704–3710. [PubMed] [Google Scholar]
- 48.Li H, Ye X, Mahanivong C, Bian D, Chun J, Huang S. J Biol Chem. 2005;280(11):10564–10571. doi: 10.1074/jbc.M412152200. [DOI] [PubMed] [Google Scholar]
- 49.Gurevich EV, Gurevich VV. Genome Biol. 2006;7(9):236. doi: 10.1186/gb-2006-7-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Attramadal H, Arriza JL, Aoki C, Dawson TM, Codina J, Kwatra MM, Snyder SH, Caron MG, Lefkowitz RJ. J Biol Chem. 1992;267(25):17882–17890. [PubMed] [Google Scholar]
- 51.Reiter E, Lefkowitz RJ. Trends Endocrinol Metab. 2006;17(4):159–165. doi: 10.1016/j.tem.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 52.Ma L, Pei G. J Cell Sci. 2007;120(Pt 2):213–218. doi: 10.1242/jcs.03338. [DOI] [PubMed] [Google Scholar]
- 53.DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. Annu Rev Physiol. 2007;69:483–510. doi: 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]
- 54.Conner DA, Mathier MA, Mortensen RM, Christe M, Vatner SF, Seidman CE, Seidman JG. Circ Res. 1997;81(6):1021–1026. doi: 10.1161/01.res.81.6.1021. [DOI] [PubMed] [Google Scholar]
- 55.Bohn LM, Lefkowitz RJ, Gainetdinov RR, Peppel K, Caron MG, Lin FT. Science. 1999;286(5449):2495–2498. doi: 10.1126/science.286.5449.2495. [DOI] [PubMed] [Google Scholar]
- 56.Kohout TA, Lin FS, Perry SJ, Conner DA, Lefkowitz RJ. Proc Natl Acad Sci U S A. 2001;98(4):1601–1606. doi: 10.1073/pnas.041608198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shenoy SK, Lefkowitz RJ. Sci STKE. 2005;2005(308) doi: 10.1126/stke.2005/308/cm10. cm10. [DOI] [PubMed] [Google Scholar]
- 58.Lefkowitz RJ, Rajagopal K, Whalen EJ. Mol Cell. 2006;24(5):643–652. doi: 10.1016/j.molcel.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 59.McDonald PH, Lefkowitz RJ. Cell Signal. 2001;13(10):683–689. doi: 10.1016/s0898-6568(01)00203-0. [DOI] [PubMed] [Google Scholar]
- 60.Shenoy SK, Lefkowitz RJ. Biochem J. 2003;375(Pt 3):503–515. doi: 10.1042/BJ20031076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buchanan FG, DuBois RN. Cell Cycle. 2006;5(18):2060–2063. doi: 10.4161/cc.5.18.3212. [DOI] [PubMed] [Google Scholar]
- 62.Lefkowitz RJ, Whalen EJ. Curr Opin Cell Biol. 2004;16(2):162–168. doi: 10.1016/j.ceb.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 63.Lefkowitz RJ, Shenoy SK. Science. 2005;308(5721):512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- 64.Wang Y, Tang Y, Teng L, Wu Y, Zhao X, Pei G. Nat Immunol. 2006;7(2):139–147. doi: 10.1038/ni1294. [DOI] [PubMed] [Google Scholar]
- 65.Luan B, Zhang Z, Wu Y, Kang J, Pei G. EMBO J. 2005;24(24):4237–4246. doi: 10.1038/sj.emboj.7600882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gao H, Sun Y, Wu Y, Luan B, Wang Y, Qu B, Pei G. Mol Cell. 2004;14(3):303–317. doi: 10.1016/s1097-2765(04)00216-3. [DOI] [PubMed] [Google Scholar]
- 67.Shi Y, Feng Y, Kang J, Liu C, Li Z, Li D, Cao W, Qiu J, Guo Z, Bi E, Zang L, Lu C, Zhang JZ, Pei G. Nat Immunol. 2007;8(8):817–824. doi: 10.1038/ni1489. [DOI] [PubMed] [Google Scholar]
- 68.Wang P, Wu Y, Ge X, Ma L, Pei G. J Biol Chem. 2003;278(13):11648–11653. doi: 10.1074/jbc.M208109200. [DOI] [PubMed] [Google Scholar]
- 69.Wang P, Gao H, Ni Y, Wang B, Wu Y, Ji L, Qin L, Ma L, Pei G. J Biol Chem. 2003;278(8):6363–6370. doi: 10.1074/jbc.M210350200. [DOI] [PubMed] [Google Scholar]
- 70.Sun Y, Cheng Z, Ma L, Pei G. J Biol Chem. 2002;277(51):49212–49219. doi: 10.1074/jbc.M207294200. [DOI] [PubMed] [Google Scholar]
- 71.Kang J, Shi Y, Xiang B, Qu B, Su W, Zhu M, Zhang M, Bao G, Wang F, Zhang X, Yang R, Fan F, Chen X, Pei G, Ma L. Cell. 2005;123(5):833–847. doi: 10.1016/j.cell.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 72.Beaulieu JM, Marion S, Rodriguiz RM, Medvedev IO, Sotnikova TD, Ghisi V, Wetsel WC, Lefkowitz RJ, Gainetdinov RR, Caron MG. Cell. 2008;132(1):125–136. doi: 10.1016/j.cell.2007.11.041. [DOI] [PubMed] [Google Scholar]
- 73.Witherow DS, Garrison TR, Miller WE, Lefkowitz RJ. Proc Natl Acad Sci U S A. 2004;101(23):8603–8607. doi: 10.1073/pnas.0402851101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gesty-Palmer D, El Shewy H, Kohout TA, Luttrell LM. J Biol Chem. 2005;280(37):32157–32167. doi: 10.1074/jbc.M507460200. [DOI] [PubMed] [Google Scholar]
- 75.Xiao K, McClatchy DB, Shukla AK, Zhao Y, Chen M, Shenoy SK, Yates JR, 3rd, Lefkowitz RJ. Proc Natl Acad Sci U S A. 2007;104(29):12011–12016. doi: 10.1073/pnas.0704849104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Willis TG, Jadayel DM, Du MQ, Peng H, Perry AR, Abdul-Rauf M, Price H, Karran L, Majekodunmi O, Wlodarska I, Pan L, Crook T, Hamoudi R, Isaacson PG, Dyer MJ. Cell. 1999;96(1):35–45. doi: 10.1016/s0092-8674(00)80957-5. [DOI] [PubMed] [Google Scholar]
- 77.Zhang Q, Siebert R, Yan M, Hinzmann B, Cui X, Xue L, Rakestraw KM, Naeve CW, Beckmann G, Weisenburger DD, Sanger WG, Nowotny H, Vesely M, Callet-Bauchu E, Salles G, Dixit VM, Rosenthal A, Schlegelberger B, Morris SW. Nat Genet. 1999;22(1):63–68. doi: 10.1038/8767. [DOI] [PubMed] [Google Scholar]
- 78.Koseki T, Inohara N, Chen S, Carrio R, Merino J, Hottiger MO, Nabel GJ, Nunez G. J Biol Chem. 1999;274(15):9955–9961. doi: 10.1074/jbc.274.15.9955. [DOI] [PubMed] [Google Scholar]
- 79.Thome M, Martinon F, Hofmann K, Rubio V, Steiner V, Schneider P, Mattmann C, Tschopp J. J Biol Chem. 1999;274(15):9962–9968. doi: 10.1074/jbc.274.15.9962. [DOI] [PubMed] [Google Scholar]
- 80.Yan M, Lee J, Schilbach S, Goddard A, Dixit V. J Biol Chem. 1999;274(15):10287–10292. doi: 10.1074/jbc.274.15.10287. [DOI] [PubMed] [Google Scholar]
- 81.Srinivasula SM, Ahmad M, Lin JH, Poyet JL, Fernandes-Alnemri T, Tsichlis PN, Alnemri ES. J Biol Chem. 1999;274(25):17946–17954. doi: 10.1074/jbc.274.25.17946. [DOI] [PubMed] [Google Scholar]
- 82.Costanzo A, Guiet C, Vito P. J Biol Chem. 1999;274(29):20127–20132. doi: 10.1074/jbc.274.29.20127. [DOI] [PubMed] [Google Scholar]
- 83.Lucas PC, Yonezumi M, Inohara N, McAllister-Lucas LM, Abazeed ME, Chen FF, Yamaoka S, Seto M, Nunez G. J Biol Chem. 2001;276(22):19012–19019. doi: 10.1074/jbc.M009984200. [DOI] [PubMed] [Google Scholar]
- 84.Uren AG, O'Rourke K, Aravind LA, Pisabarro MT, Seshagiri S, Koonin EV, Dixit VM. Mol Cell. 2000;6(4):961–967. doi: 10.1016/s1097-2765(00)00094-0. [DOI] [PubMed] [Google Scholar]
- 85.Schulze-Luehrmann J, Ghosh S. Immunity. 2006;25(5):701–715. doi: 10.1016/j.immuni.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 86.Zhou H, Du MQ, Dixit VM. Cancer Cell. 2005;7(5):425–431. doi: 10.1016/j.ccr.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 87.Thome M. Nat Rev Immunol. 2008;8(7):495–500. doi: 10.1038/nri2338. [DOI] [PubMed] [Google Scholar]
- 88.Sun L, Deng L, Ea CK, Xia ZP, Chen ZJ. Mol Cell. 2004;14(3):289–301. doi: 10.1016/s1097-2765(04)00236-9. [DOI] [PubMed] [Google Scholar]
- 89.Noels H, van Loo G, Hagens S, Broeckx V, Beyaert R, Marynen P, Baens M. J Biol Chem. 2007;282(14):10180–10189. doi: 10.1074/jbc.M611038200. [DOI] [PubMed] [Google Scholar]
- 90.Coornaert B, Baens M, Heyninck K, Bekaert T, Haegman M, Staal J, Sun L, Chen ZJ, Marynen P, Beyaert R. Nat Immunol. 2008;9(3):263–271. doi: 10.1038/ni1561. [DOI] [PubMed] [Google Scholar]
- 91.Rebeaud F, Hailfinger S, Posevitz-Fejfar A, Tapernoux M, Moser R, Rueda D, Gaide O, Guzzardi M, Iancu EM, Rufer N, Fasel N, Thome M. Nat Immunol. 2008;9(3):272–281. doi: 10.1038/ni1568. [DOI] [PubMed] [Google Scholar]
- 92.Zhou H, Wertz I, O'Rourke K, Ultsch M, Seshagiri S, Eby M, Xiao W, Dixit VM. Nature. 2004;427(6970):167–171. doi: 10.1038/nature02273. [DOI] [PubMed] [Google Scholar]
- 93.Ruland J, Duncan GS, Elia A, del Barco Barrantes I, Nguyen L, Plyte S, Millar DG, Bouchard D, Wakeham A, Ohashi PS, Mak TW. Cell. 2001;104(1):33–42. doi: 10.1016/s0092-8674(01)00189-1. [DOI] [PubMed] [Google Scholar]
- 94.Xue L, Morris SW, Orihuela C, Tuomanen E, Cui X, Wen R, Wang D. Nat Immunol. 2003;4(9):857–865. doi: 10.1038/ni963. [DOI] [PubMed] [Google Scholar]
- 95.Ruland J, Duncan GS, Wakeham A, Mak TW. Immunity. 2003;19(5):749–758. doi: 10.1016/s1074-7613(03)00293-0. [DOI] [PubMed] [Google Scholar]
- 96.Ruefli-Brasse AA, French DM, Dixit VM. Science. 2003;302(5650):1581–1584. doi: 10.1126/science.1090769. [DOI] [PubMed] [Google Scholar]
- 97.Isaacson PG, Du MQ. Nat Rev Cancer. 2004;4(8):644–653. doi: 10.1038/nrc1409. [DOI] [PubMed] [Google Scholar]
- 98.Rothe M, Wong SC, Henzel WJ, Goeddel DV. Cell. 1994;78(4):681–692. doi: 10.1016/0092-8674(94)90532-0. [DOI] [PubMed] [Google Scholar]
- 99.Rothe M, Sarma V, Dixit VM, Goeddel DV. Science. 1995;269(5229):1424–1427. doi: 10.1126/science.7544915. [DOI] [PubMed] [Google Scholar]
- 100.Cao Z, Xiong J, Takeuchi M, Kurama T, Goeddel DV. Nature. 1996;383(6599):443–446. doi: 10.1038/383443a0. [DOI] [PubMed] [Google Scholar]
- 101.Takeuchi M, Rothe M, Goeddel DV. J Biol Chem. 1996;271(33):19935–19942. doi: 10.1074/jbc.271.33.19935. [DOI] [PubMed] [Google Scholar]
- 102.Yeh WC, Hakem R, Woo M, Mak TW. Immunol Rev. 1999;169:283–302. doi: 10.1111/j.1600-065x.1999.tb01323.x. [DOI] [PubMed] [Google Scholar]
- 103.Bradley JR, Pober JS. Oncogene. 2001;20(44):6482–6491. doi: 10.1038/sj.onc.1204788. [DOI] [PubMed] [Google Scholar]
- 104.Xu LG, Li LY, Shu HB. J Biol Chem. 2004;279(17):17278–17282. doi: 10.1074/jbc.C400063200. [DOI] [PubMed] [Google Scholar]
- 105.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Cell. 2000;103(2):351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 106.Lomaga MA, Yeh WC, Sarosi I, Duncan GS, Furlonger C, Ho A, Morony S, Capparelli C, Van G, Kaufman S, van der Heiden A, Itie A, Wakeham A, Khoo W, Sasaki T, Cao Z, Penninger JM, Paige CJ, Lacey DL, Dunstan CR, Boyle WJ, Goeddel DV, Mak TW. Genes Dev. 1999;13(8):1015–1024. doi: 10.1101/gad.13.8.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Naito A, Azuma S, Tanaka S, Miyazaki T, Takaki S, Takatsu K, Nakao K, Nakamura K, Katsuki M, Yamamoto T, Inoue J. Genes Cells. 1999;4(6):353–362. doi: 10.1046/j.1365-2443.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- 108.Chen ZJ. Nat Cell Biol. 2005;7(8):758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gao M, Karin M. Mol Cell. 2005;19(5):581–593. doi: 10.1016/j.molcel.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 110.Lomaga MA, Henderson JT, Elia AJ, Robertson J, Noyce RS, Yeh WC, Mak TW. J Neurosci. 2000;20(19):7384–7393. doi: 10.1523/JNEUROSCI.20-19-07384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Blank JL, Gerwins P, Elliott EM, Sather S, Johnson GL. J Biol Chem. 1996;271(10):5361–5368. doi: 10.1074/jbc.271.10.5361. [DOI] [PubMed] [Google Scholar]
- 112.Zhao Q, Lee FS. J Biol Chem. 1999;274(13):8355–8358. doi: 10.1074/jbc.274.13.8355. [DOI] [PubMed] [Google Scholar]
- 113.Yang J, Boerm M, McCarty M, Bucana C, Fidler IJ, Zhuang Y, Su B. Nat Genet. 2000;24(3):309–313. doi: 10.1038/73550. [DOI] [PubMed] [Google Scholar]
- 114.Deng Y, Yang J, McCarty M, Su B. Am J Physiol Cell Physiol. 2007;293(4):C1404–C1411. doi: 10.1152/ajpcell.00058.2007. [DOI] [PubMed] [Google Scholar]
- 115.Shinohara H, Yamasaki S, Maeda S, Saito T, Kurosaki T. Int Immunol. 2009;21(4):393–401. doi: 10.1093/intimm/dxp007. [DOI] [PubMed] [Google Scholar]
- 116.Sun W, Ge N, Yu Y, Burlingame S, Li X, Zhang M, Ye S, Fu S, Yang J. J Biol Chem. 285(11):7911–7918. doi: 10.1074/jbc.M109.051219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Moscat J, Diaz-Meco MT, Albert A, Campuzano S. Mol Cell. 2006;23(5):631–640. doi: 10.1016/j.molcel.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 118.Moscat J, Diaz-Meco MT. EMBO Rep. 2000;1(5):399–403. doi: 10.1093/embo-reports/kvd098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Huang Q, Yang J, Lin Y, Walker C, Cheng J, Liu ZG, Su B. Nat Immunol. 2004;5(1):98–103. doi: 10.1038/ni1014. [DOI] [PubMed] [Google Scholar]
- 120.Schmidt C, Peng B, Li Z, Sclabas GM, Fujioka S, Niu J, Schmidt-Supprian M, Evans DB, Abbruzzese JL, Chiao PJ. Mol Cell. 2003;12(5):1287–1300. doi: 10.1016/s1097-2765(03)00390-3. [DOI] [PubMed] [Google Scholar]
- 121.Nakamura K, Uhlik MT, Johnson NL, Hahn KM, Johnson GL. Mol Cell Biol. 2006;26(6):2065–2079. doi: 10.1128/MCB.26.6.2065-2079.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Moscat J, Rennert P, Diaz-Meco MT. Cell Death Differ. 2006;13(5):702–711. doi: 10.1038/sj.cdd.4401823. [DOI] [PubMed] [Google Scholar]
- 123.Nakamura K, Kimple AJ, Siderovski DP, Johnson GL. J Biol Chem. 285(3):2077–2089. doi: 10.1074/jbc.M109.065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Prajapati S, Verma U, Yamamoto Y, Kwak YT, Gaynor RB. J Biol Chem. 2004;279(3):1739–1746. doi: 10.1074/jbc.M306273200. [DOI] [PubMed] [Google Scholar]
- 125.Sun W, Yu Y, Dotti G, Shen T, Tan X, Savoldo B, Pass AK, Chu M, Zhang D, Lu X, Fu S, Lin X, Yang J. Cell Signal. 2009;21(1):95–102. doi: 10.1016/j.cellsig.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]