Abstract
The zyxin family of proteins function as cytoskeletal regulators in adhesion, actin assembly, and cell motility. Though fibroblasts derived from zyxin-null mice show striking defects in motility and response to mechanical stimuli, the mice are viable and fertile. In Drosophila melanogaster, the family is represented by a single homologue, Zyx102. To study the role of zyxin during development, we generated a zyx102 RNA-interference transgenic line that allows for the conditional knockdown of Zyx102. When UAST-zyx102-dsRNAi expression is driven broadly by Actin5C-GAL4, loss of Zyx102 results in lethality during the pharate adult stage, a narrow developmental window during which the fly must molt, resorb molting fluid, fill adult trachea with air and execute a behavioral program to eclose. Zyx102 knockdown animals attempt to emerge, but their adult trachea do not fill with air. If dissected from the pupal case, knockdown individuals appear morphologically normal, but remain inviable.
Keywords: Zyx102, zyxin, cytoskeleton, pupation, RNA-interference
Introduction
In vertebrates, the zyxin family of proteins consists of zyxin, Thyroid Hormone Receptor Interacting Protein 6 (Trip6), and Lipoma Preferred Partner (LPP). These cytoskeletal proteins have roles in actin assembly, cell motility, and adhesion and share the molecular signature of three C-terminal LIM domains, which make up about half of the molecular mass of each species. LIM domains are capable of mediating protein-protein interactions (Kadrmas and Beckerle, 2004), such as those between zyxin and p130Cas (Yi et al., 2002) or CRP (Schmeichel and Beckerle, 1994; Schmeichel and Beckerle, 1997), and between Trip6 and hormone-activated thyroid hormone receptor (Lee et al., 1995) or glucocorticoid receptor (Kassel et al., 2004). The C-terminal tails of both LPP and Trip6 have been reported to interact with the PDZ domain of Scrib (Petit et al., 2005a; Petit et al., 2005b), the vertebrate homologue of Drosophila scribble, which is involved in cell polarity (reviewed in (Bilder, 2004)). The non-LIM portion of these proteins includes other functional domains, such as nuclear export motifs of the CRM1-mediated export type, polyproline motifs in zyxin and LPP that mediate interactions with the EVH1-domains of VASP and Mena (reviewed in (Renfranz and Beckerle, 2002)); and α-actinin binding sites present in both zyxin (Reinhard et al., 1999; Li and Trueb, 2001; Nix et al., 2001) and LPP (Li et al., 2003). Proteins of the zyxin family typically localize to sites of cell adhesion in vertebrate cells: in cultured fibroblasts, zyxin and LPP are primarily detected at focal adhesions, sites of integrin-mediated adhesion of the actin cytoskeleton to the extracellular matrix. They also localize to cell-cell junctions and appear to affect the rate at which cell-cell junctions form (Hansen and Beckerle, 2006). Zyxin, LPP, and Trip6 have all been detected in cell nuclei as well (Nix and Beckerle, 1997; Wang et al., 1999; Petit et al., 2000; Wang and Gilmore, 2001), indicating that they may play a role in nuclear responses to changes in cell adhesion and/or the cytoskeleton.
Fibroblast cells derived from zyxin-null mice show abnormalities in cell adhesion, migration, and response to mechanical stress (Yoshigi et al., 2005; Hoffman et al., 2006). Zyxin-null fibroblasts are more adherent to various extracellular matrices than cells derived from wild-type mice; these cells are also more migratory than wild-type cells in a wound-healing assay. When zyxin-null cells are subjected to mechanical, unidirectional stretch, they, like wild-type fibroblasts, orient their actin cytoskeleton perpendicular to the direction of stretch. However, while wild-type cells thicken the reoriented actin filaments, and show a redistribution of zyxin and VASP from focal contacts to actin filaments, zyxin-null cells display impaired actin thickening (Yoshigi et al., 2005). Additional evidence of zyxin mechanosensitivity was provided by laser severing of actin stress fibers in endothelial cells, which increased dissociation of zyxin as tension was released (Lele et al., 2006). Similar to mammalian cells in culture, in Drosophila egg chambers zyxin distribution along actin-rich bundles and at cell edges declined after laser surgery to release tension (Colombelli et al., 2009). These results have led to the hypothesis that zyxin may function in linking mechanical stimuli to cytoskeletal dynamics and cell motility, and raise the possibility that ablation of zyxin may have deleterious consequences under conditions of physiological stress.
Despite the observations discussed above that members of this family have a large repertoire of binding partners, are evolutionarily conserved, and, when absent in cultured cells, disrupt normal cellular functions, genetic deletion of the zyxin gene in the mouse has no overt effect on viability and fertility and elicits no obvious morphological or behavioral consequences (Hoffman et al., 2003). Although the steady-state expression level of the other family members is not increased in the zyxin-null mice (Hoffman et al., 2003), analysis of the murine Hox gene cluster shows that ablation of multiple members of a gene family may be required to result in a phenotype (Wellik and Capecchi, 2003). LPP-null mice were recently reported to be viable and fibroblasts from those mice exhibit changes in cell motility (Vervenne et al., 2009). Generation of LPP/zyxin double-null mice would address some functional redundancy questions, but could not be fully resolved until TRIP6-null mice become available.
In an effort to gain insight into the role of zyxin in vivo, we have undertaken the study of the zyxin protein in the fruit fly, Drosophila melanogaster, in which only one representative gene exists, zyx102 (Renfranz et al., 2003). The zyx102 gene is located at the tip of the 4th chromosome, and is predicted to code for a protein with high similarity to zyxin, LPP, and Trip6. We have pursued a heritable, inducible RNA-interference approach to eliminate Zyx102 protein in the fly. Here we show that reduction of Zyx102 protein levels using RNA-interference results in lethality at the pharate adult stage.
Materials and Methods
Sequences and strains
Genomic DNA sequences referred to in the text are from Release 5.1 of the Drosophila melanogaster genomic sequence (chromosome 4) and from Genbank accession number AF219947 (Renfranz et al., 2003). The sequence of cDNA clone zyx102-44 (AF219948; (Renfranz et al., 2003) provides the reference for cDNA sequences. Drosophila strains carrying the following GAL4 drivers or mutations were obtained from the Bloomington stock center: P{GawB}how24B, P{Act5C-GAL4}25FO1, witA12 st1, and Df(3L)C175. P{Act5C-GAL4}25FO1 was balanced with one of two different GFP-expressing balancer chromosomes for sorting purposes: CyO, P{ActGFP}JMR1 or CyO, P{GAL4-twi.G}2.2, P{UAS-2xEGFP}AH2.2.
The terminal cell marker pAS7 was constructed by Mark Metzstein (University of Utah) using the 490 bp terminal cell enhancer (Nussbaumer et al., 2000), which was multimerized and used to drive expression of GAP::GFP (Mark Metzstein, personal communication).
RT-PCR
The Micro Poly(A) Purist mRNA purification kit (Ambion) was used to isolate mRNA from 30 adult female flies and from ~39 mg (wet weight) of 0–16 hr embryos, according to the manufacturer’s instructions. Reverse transcription and PCR was performed on 50 ng mRNA using the Access RT-PCR kit (Promega). Controls included not incubating the mRNA template at 48°C, no template added, and alternative templates like specific cDNA clones or genomic DNA. When possible, gel-purified bands were sequenced directly using the PCR primers as sequencing primers. To increase yield for sequencing of products generated around Exons 1-2-3 and Exons 4–5, 1.0 μL of the RT-PCR reaction was subjected to further rounds of PCR using the same primer sets (Invitrogen Platinum Taq polymerase and dNTPs). Products were separated by agarose gel electrophoresis and bands purified using the Qiagen QIAquick Gel Extraction Kit. Individual products were cloned using the TOPO TA Cloning Kit with pCR 2.1 (Invitrogen) into the TOPO vector, plasmid DNA was prepared using the Qiagen QIAprep Spin Miniprep Kit, and inserts sequenced using vector-specific primers.
Nested primer sets were used to look for splice variants in Exon 6. Standard PCR using Invitrogen reagents was performed on 1.0 μL of the RT-PCR. Gel-purified reaction products (Qiagen QIAquick Gel Extraction Kit) were cloned, and plasmids prepared for sequencing as described above. All sequencing was performed at the DNA Sequencing Core Facility at the University of Utah Health Sciences Center.
Construction and transformation of UAST-zyx102-44::EGFP
The ORF of cDNA clone zyx102-44 (AF219948) was amplified by PCR with appropriate linkers and cloned into the mammalian expression vector pEGFP-N1 (BD Biosciences Clontech). The region encoding the zyx102-44::EGFP fusion was excised with Bgl II and Not I, and cloned into the transformation vector pUAST (Brand and Perrimon, 1993). One stable homozygous viable transgenic stock was established, P{UAST-zyx102-44::EGFP}6.1, which carries the insert on chromosome III.
Monoclonal antibody production and usage
The mouse monoclonal antibody P1D8 was produced at the University of Illinois at Urbana-Champaign Biotechnology Center, Protein Sciences Facility and Immunological Resource Center. A 30 amino acid peptide (K-INPRPCVADTLPRESYNLHNSYVNDNNPNI; AF219948: aa 299–328), common to all predicted splice variants from Zyx102, was used as immunogen. Only one MAb, P1D8, identified bacterially-expressed Zyx102 on western blots. Immunostaining Drosophila samples with P1D8 (1:2) was not successful following standard 4% formaldehyde fix with methanol devitellinization or with 4% paraformaldehyde protocols as described (Patel, 1994; Rothwell and Sullivan, 2000).
To test the specificity of antibody P1D8 on immunoblots, lysates were prepared from control embryos (w1118) and from embryos expressing EGFP-tagged Zyx102 using the GAL4 driver P{GawB}how24B. Immunoblots were probed with P1D8 and with Living Colors mouse anti-GFP (1:1000, Clontech). P1D8 was incubated with 50 μM peptide immunogen in PBS, or with PBS alone as a control, at 37°C for 60′. The antibody samples were then diluted 1:100 into TBS-T (0.1% Tween) with 5% dried milk before final use. For the developmental western, lysates from different developmental stages or from dissected tissues were prepared in RIPA buffer (1% NP40, 0.5% DOC, 0.1% SDS, 50 mM Tris pH 8, 150 mM NaCl); 20 μg protein was used per sample. Peptide incubation was as above, except that primary antibody MAb P1D8 was used at 1:50 final dilution.
RNA interference hairpin construct
The forward direction was amplified from Drosophila genomic DNA using TAQ polymerase (Invitrogen) and standard conditions with the following primer pair: Zyx GE 5′ (5′-CTA CTG TCT AGG TAC CGT TGT AGG AAA ACG GAA TCG; Kpn I site) and Zyx GE 4b 3′ (5′-CAG TAT AAG CGG CCG CCT CAT CTT TTT CGT TGT CTG; Not I site). This pair amplifies 1370 bp of genomic DNA, beginning in exon 2a and extending through the first 17 bp of exon 4b. The region to be used as the inverted repeat was selected after careful examination of the reverse complement of zyx102 cDNA sequences for the presence of potential splice donors or acceptors, which could result in a loss of portions of the inverted region upon transcription in vivo. The region selected (AF219948: 721-144; 578 bp) was amplified from cDNA clone pEXlox 44 using TAQ polymerase (Invitrogen) with primers Zyx C 5′ (5′-GTT ATA TTG CGG CCG CCT ATT TCT TTG CGA ACA G; Not I) and Zyx C 3′-2Xba (5′-CAA GTT CTA GAG TTG TAG GAA AAC GGA ATC G; Xba I). The region amplified corresponds to part of exon 3 and the same region of exon 2 as the forward genomic product. The reaction products were purified using Qiagen QIAquick PCR Purification Kit, and treated with the appropriate pair of restriction enzymes (New England Biolabs). The fragments were shuttled through the vector pUASP (Rorth, 1998) to generate the inverted repeat, which was then subcloned into the Drosophila transformation vector pUAST (Brand and Perrimon, 1993). Final product was subjected to diagnostic restriction enzyme digests and sequencing prior to generating transformant flies using standard techniques (Ashburner et al., 2005). One independent transformant, UAST-zyx102-dsRNA(16c), was identified. Inverse PCR showed that the insertion event occurred within either or both of the Juan transposable elements on chromosome 3R (Kaminker et al., 2002), and thus is unlikely to give rise to an overt phenotype.
The P{UAST-zyx102-dsRNAi}16c line was crossed to the enhancer trap line P{GawB}how24B, in which GAL4 is strongly expressed in muscle cells (Brand and Perrimon, 1993). To enrich for tissue in which GAL4 is expressed, larval muscle fillets were prepared from mid-third instar F1 larvae. Individual fillets were solubilized in SDS-PAGE sample buffer, and western immunoblots prepared. Filters were first probed with MAb P1D8 to detect Zyx102, then stripped and probed for Drosophila Lamin C (LC28.26, DSHB; (Riemer et al., 1995)). The level of Zyx102 signal relative to Lamin C was determined by measuring pixel intensity using NIH Image and Adobe Photoshop.
The National Institutes of Genetics (NIG) in Japan reported two RNA interference constructs that covered a 500 bp DNA similar to the P{UAST-zyx102-dsRNAi}16c construct but without introns (www.shigen.nig.ac.jp/fly/nigfly/index.jsp). Their DNA starts at the 3′ end of exon 2 (184 bp downstream of the start of P{UAST-zyx102-dsRNAi}16c construct) and ends at the 5′ end of exon 4b (21 bp downstream of the end of the P{UAST-zyx102-dsRNAi}16c construct). One of the NIG RNAi stocks (32018R-3) carries the insertion on the third chromosome and the other stock (32018R-1) carries the insertion on the second chromosome and both are pharate lethal.
Act5C-GAL4 expression pattern
Flies carrying Act5C-GAL4 25 were crossed to w1118; P{UAST-zyx102-44::EGFP}. Green-fluorescent, late third-instar larvae were identified, pinned in a Sylgard-coated dish in Ringer’s solution, and bissected longitudinally. The tissues not firmly connected to the epidermis, including imaginal discs, the optic lobes and nerve cord, gut, salivary gland, malpighian tubule, fat body, and associated trachea, were transferred to a microcentrifuge tube and fixed for 30 min in 4% formaldehyde in PBS. The epidermis with attached body wall muscle (fillet) was fixed for 5 min while still pinned in the dish, then transferred to a tube of fixative. After fixation, the samples were rinsed 2 × 2 min in an excess of PBS, equilibrated with 50% glycerol in PBS then with 70% glycerol in PBS. The samples were mounted in 70% glycerol in PBS with 2.5% DABCO. Images were collected using an Olympus Fluoview confocal system (FV300).
Characterization of trachea
Pupae were lined dorsal-side up on double-stick tape on a coverslip, which was then placed on a grape juice plate to prevent dessication. The animals were allowed to develop at 25°C. Approximately 12 hrs prior to eclosion, the operculum was dissected away from the case in order to observe the pupal-pharate adult transition (Kimura and Truman, 1990). At the desired stage, flies were dissected out of the pupal case in Ringer’s at RT. Samples were kept at RT for the entire procedure, in order to maintain any air present in the trachea. The ventral nerve cord (VNC) was dissected, transferred into fixative (4% formaldehyde in PBS), and incubated O/N at RT. Samples were then rinsed twice for 30 min each in PBS and mounted in 100% glycerol on poly-lysine-coated coverslips. Air-filled tracheae were easily observed by simple light microscopic methods, because air induces differential contrast.
To label the terminal cells, F2 progeny labeled using the pAS7 marker were used. Half of the resulting F2 progeny are expected to be pharate lethal (as they carry Act5C-GAL4 and P{UAST-zyx102-dsRNAi}), with half of these carrying the terminal cell marker. Viable F2 progeny should all carry at least one copy of the terminal cell marker. VNCs from ten F2 pharate lethals and seven F2 viable flies, as well as from five homozygous P{UAST-zyx102-dsRNAi}16c flies, were dissected in Ringer’s solution and fixed for 30 min in 4% formaldehyde in PBS. GFP was detected via indirect immunofluorescence using Rabbit anti-GFP (Invitrogen) and Goat anti-rabbit IgG conjugated to Alexa Fluor 568 (Molecular Probes). Samples were imaged on an Olympus confocal microscope FV300. Because we could not be sure of genotype, collected images were stacked and compared by a naive viewer; results are shown in Table 1.
Table 1.
Terminal cell labeling
| Sample | # VNC prepared | # expected to carry pAS7 | # showing labeled terminal cell processes | # showing no labeling | # showing background labeling | # with isolated cells labeled |
|---|---|---|---|---|---|---|
| Positive controla | 7 | 7 | 7 | 0 | 0 | 0 |
| Negative controlb | 5 | 0 | 0 | 2 | 3 | 1d |
| Test samplesc | 10 | 5 | 3e | 0 | 4f | 3g |
Positive control: P{UAST-zyx102-dsRNA}16c + pAS7 (1 or 2 copies)
Negative control: P{UAST-zyx102-dsRNA}16c
Test samples: Pharate adult lethal (Act5C-GAL4 + P{UAST-zyx102-dsRNA}16c) +/− 1 copy pAS7
One negative sample (from previous column) showed background labeling that resembles an isolated cell (Fig. 8B)
Samples likely to be zyx102-dsRNAi + Act5C-GAL4 + pAS7 (Fig. 8C–D)
Samples likely to be zyx102-dsRNAi + Act5C-GAL4 (Fig. 8F)
Label appears to be expressed in cells that do not have labeled processes (Fig. 8E)
Results
Molecular architecture of the Drosophila zyxin homolog, Zyx102
The predicted protein sequence and motif structure of Zyx102 implies that it is the functional orthologue of the vertebrate zyxin family (Fig. 1). The three C-terminal LIM domains, each of which consists of a double-zinc finger that can serve as a protein-protein binding interface (Schmeichel and Beckerle, 1994; Schmeichel and Beckerle, 1997; Kadrmas and Beckerle, 2004), are highly conserved between Zyx102 and murine zyxin, LPP, and Trip6 (average 60% identity) (Renfranz et al., 2003). Other conserved moieties include a poly-proline EVH1 domain-binding motif, which can serve as a binding site for members of the Ena/VASP family of actin cytoskeleton regulators (Renfranz and Beckerle, 2002). Previous work in our lab has shown that, at least in vitro, the Zyx102 protein interacts with Drosophila Ena, the representative of this family in the fly (Renfranz et al., 2003).
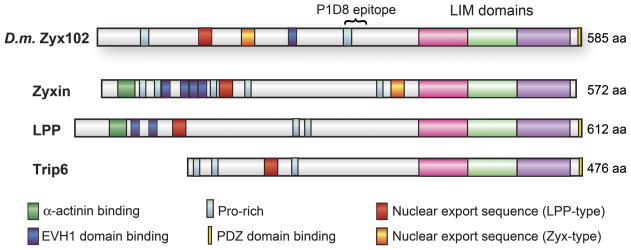
Figure 1. Schematic representation of Zyx102.
Zyx102 of Drosophila melanogaster is schematically depicted in comparison to human zyxin, Lipoma Preferred Partner (LPP), and Thyroid Hormone Receptor Interacting Protein 6 (Trip 6). Drosophila Zyx102 shows proline-rich domains, including one that matches the consensus for binding the EVH1 domains of Ena/VASP proteins; both a zyxin-type and a LPP-type nuclear export sequence; three C-terminal LIM domains; and a C-terminal PDZ domain-binding region. The zyx102 LIM domains exhibit an average of 60% identity with LIM domains from zyxin, LPP and Trip6. The α-actinin binding sites found in vertebrate zyxin and LPP are not conserved in the fly protein. The zyx102 gene spans about 3.6 kb on chromosome 4.
Alternative splicing of zyx102 transcripts predicts diverse protein products
Sequencing of zyx102 cDNA products indicated that alternative splicing of zyx102 transcripts occurs (Renfranz et al., 2003), potentially giving rise to alternative forms of the protein. To further examine splice complexity at the locus, we used RT-PCR to amplify splice variants from three different gene regions (Fig. 2). PolyA+-mRNAs isolated from adult females or 0–16 hr embryos were used as templates for RT-PCR. After gel purification and cloning into the TOPO vector, amplification products were sequenced. Alternative splicing was detected between Exon 1 and either Exon 2 or Exon 3; we observed five different 3-prime ends of Exon 1, three different 5-prime ends of Exon 2, and two different 3-prime ends of Exon 2 (Fig. 2B), necessitating the use of three alternative start codons. We were also able to identify amplification products that represented the use of either Exon 4a or 4b (Figure 2C).
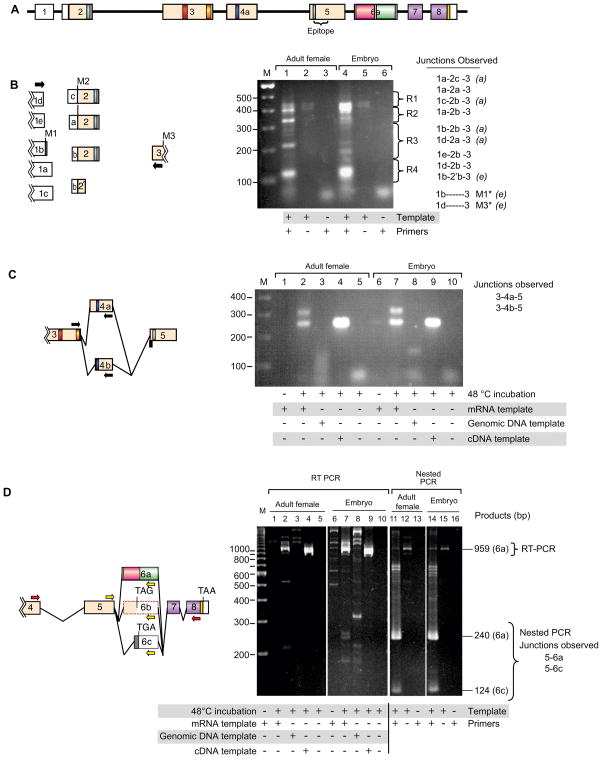
Figure 2. Transcript complexity generated by differential splicing.
A. The zyx102 gene structure as reported in (Renfranz et al., 2003). Sequence analysis of cDNA clones and genomic regions indicated that splicing between Exons 1 and 2 was highly variable, and that alternative splice variation likely occurred in Exons 4 and 6. Exons: numbered boxes; predicted introns: black lines. Untranslated regions: white boxes; coding regions: colored boxes. Regions that encode conserved motifs are colored as in Figure 1.
B–D. Analysis of splicing complexity. RT-PCR was performed on mRNA isolated from adult females and mixed-stage embryos, or on control DNAs. Templates, primers (arrows), and junctions observed are indicated. Products were separated by electrophoresis. Bands were isolated and either directly sequenced or subcloned prior to sequencing.
B. Complexity in splicing between exons 1 and 2. Agarose gel regions comprising lanes 1 and 4 were cut into four regions (R1–R4) prior to subcloning and sequencing. Observed alternative junctions are indicated. Splice variants detected only in adult female or in embryonic mRNA samples are indicated with (a) or (e), respectively. Lines joining schematized exons are not drawn because the pattern is too complex.
Most of the observed heterogeneity in splicing between Exons 1 and 2 does not alter the coding region. For all transcripts that include Exon 2, the first in-frame AUG codon is indicated M2. Two different products (*) skip Exon 2, and would utilize either of two different start methionines, indicated M1 and M3. One observed product utilizes Exon 2’b, which abbreviates the coding region of Exon 2 by 126 nt (42 amino acids), then splices in-frame to Exon 3. C. Analysis of Exon 4 splicing. RT-PCR experiments using mRNA, genomic, and cDNA templates (clone LD06023) were performed. Two different 5-prime ends of Exon 4 were observed: Exons 3-4a-5 (314 bp) and Exons 3-4b-5 (251 bp). In the genomic DNA reaction, no product was expected, as the 3′ primer spans Exons 4 and 5.
D. Variation in splicing of Exon 6, analyzed by RT-PCR (red primers) followed by nested PCR (yellow primers). RT-PCR results are shown in Lanes 1–10. Nested PCR reactions using RT-PCR reaction product from lane 2 (adult female) or 7 (embryo) as template and appropriate primers are shown in lanes 11–16. Markers at right indicate migration of products utilizing Exon 6a or 6c. Predicted Exon 6b (Renfranz et al., 2003) was not detected in these experiments.
We used RT-PCR followed by nested PCR to examine alternative splicing in the exons that encode the three LIM domains, Exons 6–8. In this analysis (Fig. 2D), two potential splice acceptor sites in Exon 6 were identified: 6a and 6c. A transcript utilizing the 6a splice acceptor site would encode the LIM domains; one using the 6c splice acceptor site would result in a change of reading frame and a truncated protein that lacks the LIM domains. Thus, although only a single gene encoding zyxin is present in Drosophila, expanded complexity at the protein level might be achieved by alternative splicing.
Temporal expression of Zyx102 protein
Previous northern blot analysis indicated that zyx102 RNA is expressed throughout embryonic development (Renfranz et al., 2003). In order to describe the temporal expression of the protein, we developed a monoclonal antibody, MAb P1D8, directed against a Zyx102 peptide (see Materials and Methods) common to all predicted splice variants. To test whether P1D8 identifies Zyx102, we used the GAL4/UAS binary expression system (Brand and Perrimon, 1993) to express a tagged form of the protein in an otherwise normal background. Transgenic flies were generated such that a Zyx102::EGFP fusion protein could be expressed under the control of the GAL4 enhancer trap line P{GawB}how24B (Brand and Perrimon, 1993; Sweeney et al., 1995; Schuster et al., 1996), which expresses in embryonic mesodermal derivatives. Progeny expressing Zyx102::EGFP via this GAL4 line are viable and show strong expression of the fusion protein in late embryonic and larval body wall muscle (data not shown). Lysates were prepared from embryos expressing both endogenous Zyx102 and the tagged fusion protein, and from control embryos (w1118), in which only endogenous Zyx102 would be expressed. The EGFP tag is predicted to shift the apparent molecular mass of Zyx102 by approximately 27 kD, from ~66 kD to ~93 kD.
As shown in Figure 3A, P1D8 identifies species present in both control and transgenic samples. Both samples exhibit a product migrating at approximately 66 kD, which disappears on preincubation of P1D8 with its peptide antigen. A product of about 90 kD is specific to the transgenic flies; this species co-migrates with a product identified specifically in transgenics by an anti-GFP antibody, and corresponds to GFP-tagged Zyx102. Thus, P1D8 appears to specifically identify Zyx102 protein.
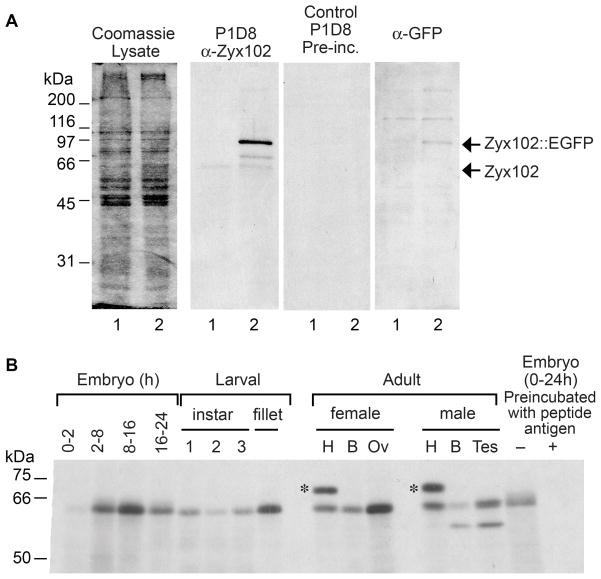
Figure 3. Zyx102 expression pattern.
A. Zyx102 is detected by the P1D8 monoclonal antibody on western blots. Lysates were prepared from control embryos (w1118; lane 1 in each panel) or from those expressing EGFP-tagged Zyx102 (w1118; P{GawB}how24B/UAS-zyx102::EGFP; lane 2 in each panel). Lysate -Coomassie blue-stained gel; P1D8 - western immunoblot probed with monoclonal antibody P1D8 directed against Zyx102 (diluted 1:100). Control P1D8-Preinc – a matched blot probed with P1D8 preincubated with peptide antigen to show specificity. Anti-GFP-matched blot probed with Living Colors mouse anti-GFP (1:1000, Clontech). Arrows indicate the predicted identity of the observed bands.
B. Zyx102 is expressed throughout fly development. Lysates from different developmental stages of Drosophila (w1118) were prepared in RIPA buffer; 20 μg total protein was loaded per lane. Shown is a protein immunoblot probed for Zyx102 using the P1D8 antibody (diluted 1:50). At right, P1D8 was incubated with peptide antigen prior to use. Samples were prepared from embryos (hours of development indicated), the three larval instars, a fillet sample from the third larval instar, and adult female and male tissues: heads (H), bodies (B), ovaries (Ov), and testes (Tes). The upper band in the two H lanes (*) is not consistently observed and does not match the migration pattern predicted for any Zyx102 gene product.
Protein lysates were prepared from the w1118 line at various developmental stages, and probed with P1D8 on immunoblots (Fig. 3B). A protein migrating at approximately 66 kD was detected at all developmental stages tested. The presence of protein in 0–2 hr embryos indicates that Zyx102 is maternally inherited. Consistent with this result, the protein was detected in ovaries dissected from adult females. We detected an additional band of about 63 kD in adult male bodies and dissected testes, indicating the presence of a male-specific form of the protein.
Zyx102 knockdown strategy
The zyx102 gene is located at the tip of chromosome 4. Because of its significant heterochromatin, lack of recombination, and small size, the fourth chromosome of Drosophila has not been extensively studied. As a result, few traditional genetic reagents exist for the study of genes localized to the fourth chromosome. Although our previous chromosomal in situ results suggested that the terminal deficiency Df(4)G removed the zyx102 gene (Renfranz et al., 2003), a recent molecular study (Podemski et al., 2004) indicated that the Df(4)G breakpoint occurs just distal to zyx102. At present, there are no other known deficiencies or transposable element insertions on the fourth chromosome that affect the zyx102 gene. The lack of classic genetic tools to probe Zyx102 function led us to employ an RNA-interference approach.
Several factors were taken into account in our design of an RNA-interference construct (Fig. 4A; Materials and Methods). One important consideration was the inclusion of intron sequences in the target construct, since the presence of introns has been shown to facilitate both cloning and efficacy of knockdown by RNA-interference constructs in Drosophila (Kalidas and Smith, 2002). We used a genomic DNA-cDNA inverted repeat hybrid to encode a double-stranded RNA hairpin targeted to zyx102.
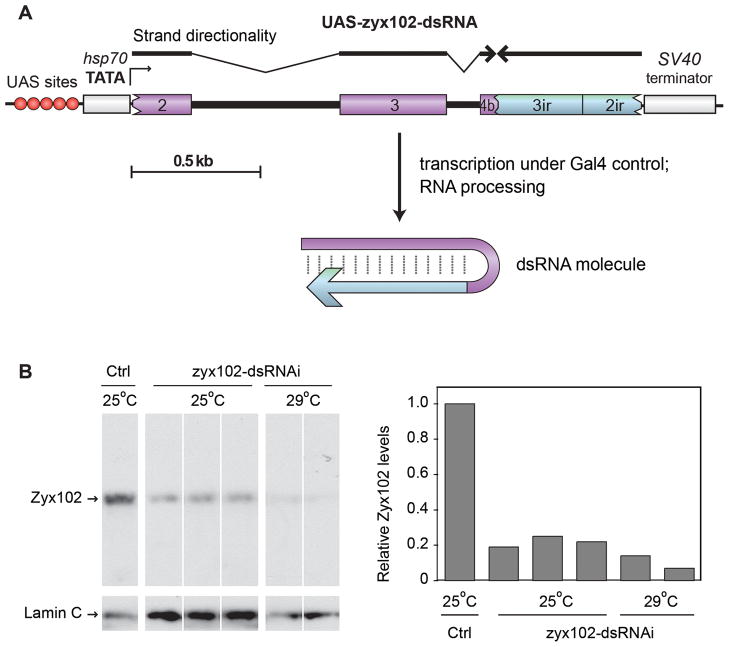
Figure 4. RNA-interference experiments using zyx102-dsRNA decrease Zyx102 protein levels.
A. The zyx102 RNA-interference construct used. The UAS-zyx102-dsRNA construct consists of an inverted repeat of zyx102-coding sequence, with endogenous introns present in the forward strand. Numbers correspond to exons indicated in Figure 2.
B. Lanes show protein from individual fillet preparations of 3rd instar larvae extracted in SDS-PAGE sample buffer. Samples were control (w1118) or w1118; P{GawB}how24B/P{UAS-zyx102-dsRNA}16c raised at either 25° or 29°C. Shown is an immunoblot probed with antibody P1D8 for Zyx102. The same blot was reprobed for Lamin C, and relative levels of Zyx102 to Lamin C were determined using pixel density. No detectable larval phenotype is associated with the observed decrease in Zyx102 protein level resulting from RNA-interference.
The use of a long dsRNA, in this case, 578 nt of base-pairing in the hairpin structure, runs the risk of interfering with the expression of non-target genes that share a region of identity of at least 19 nt to zyx102 (Echeverri and Perrimon, 2006; Kulkarni et al., 2006; Ma et al., 2006). We used the Off-Target Search Tool available from the Drosophila RNAi Screening Center (<http://flyrnai.org/>) to search for regions of identity between the exon portions of the forward direction of the zyx102-dsRNA molecule and all other predicted transcripts in Drosophila melanogaster. No regions of identity ≥ 19 nt were found other than in zyx102 itself, indicating that an off-target effect is not likely to occur. Thus, any phenotype we observe due to the expression of zyx102-dsRNAi is likely to be specific to the knockdown of Zyx102 protein levels alone.
The hairpin-encoding fragment was subcloned into the pUAST Drosophila transformation vector (Brand and Perrimon, 1993); using this construct, we recovered the transgenic fly line, P{UAST-zyx102-dsRNAi}16c. The insertion segregates with chromosome 3R and is homozygous viable.
RNA interference results in decreased levels of Zyx102 protein
To assess inducible knockdown of Zyx102 protein levels, the P{UAST-zyx102-dsRNAi}16c line was crossed to the enhancer trap line P{GawB}how24B, expressed in muscle. Larval muscle isolates were prepared from F1 progeny of this cross. As shown in Figure 4B, RNA interference results in reduced levels of Zyx102 protein relative to Lamin C, with levels ranging from 11–33% at 25°C and 5–19% at 29°C. These results are consistent with the fact that expression from UAST-based constructs often increases with increasing temperature due to the presence of heat shock promoter elements in the vector (Brand and Perrimon, 1993). Residual Zyx102 protein could result from incomplete RNA interference in muscle, where the GAL4 driver is expressed, and/or from Zyx102 expression in cells where the driver is not expressed. In these experiments, the tissue specificity of the P{GawB}how24B driver, coupled with the preparation of protein from larval muscle isolates, likely contributed to our success in observing knockdown of Zyx102 protein on a Western blot. Western blots using whole pupal extracts as a source of Zyx102 after crossing more broadly expressed GAL4 drivers to P{UAST-zyx102-dsRNAi}16c were less conclusive in showing knockdown of Zyxin102 protein levels. Extracts were also prepared from progeny of crosses between more broadly expressed GAL4 drivers and P{UAST-zyx102-dsRNAi}16c; however, western blots using pupal-stage extracts were less conclusive in showing any decrease of Zyx102 protein levels (data not shown).
Knockdown of Zyx102 is lethal
Despite the strong decrease of Zyx102 protein levels when P{GawB}how24B was used to drive expression of zyx102-dsRNAi in mesodermal derivatives, viability and fertility were unaffected and no other consequences were observed. We reasoned that, akin to vertebrate zyxin (Hoffman et al., 2003), zyx102 may be broadly expressed, and that using a more broadly-expressed GAL4 driver to knock down Zyx102 might have a phenotypic consequence. We therefore tested Act5C-GAL4 (Ito et al., 1997), in which GAL4 expression is driven by the promoter of the Drosophila Actin 5C gene.
The Act5C-GAL4 driver is reportedly expressed ubiquitously (Ito et al., 1997). Here, we independently characterized the expression pattern of this driver using EGFP-tagged Zyx102 [P{UAST-zyx102(44)::EGFP}] as a reporter. While broadly expressed in mid- to late-third instar larvae, Act5C-GAL4 does not drive the expression of the marker ubiquitously in all tissues (Fig. 5). EGFP-tagged Zyx102 protein was detected in salivary gland, foregut, proventriculous, gastric cecae, and mid-gut. The tagged protein was also found in the most anterior region of the hindgut and in the Malpighian tubules. We detected strong expression in the tracheal system, ventral nerve cord, and imaginal discs. Examination of larval body wall muscle and the underlying epidermis showed that the driver was not expressed in muscle tissue and is expressed nonuniformly in the larval epidermis, where it is most apparent in tendon cells. Expression of EGFP-tagged Zyx102 using the Act5C-GAL4 driver did not lead to deleterious effects on the fly.
Figure 5. Expression pattern of the Act5C-GAL4 driver.
Tissues prepared from mid-3rd instar larvae carrying Act5C-GAL4 and UAS-zyx102(44)::EGFP are shown. The marker for Act5C-GAL4 expression is thus the fluorescence of EGFP-tagged Zyx102. Z-projections of confocal stacks (left) and corresponding transmitted light micrographs (right). In A–B and D–F, bar = 100 μm; in C, bar = 20 μm.
A. Salivary gland and trachea. B. Proventriculus (junction of foregut and midgut). C. Malpighian tubule. D. Ventral nerve cord and trachea. E. En face view of fillet preparation, showing larval body wall muscle and underlying epidermis. F. En face view of fillet preparation, showing larval body wall muscle and trachea.
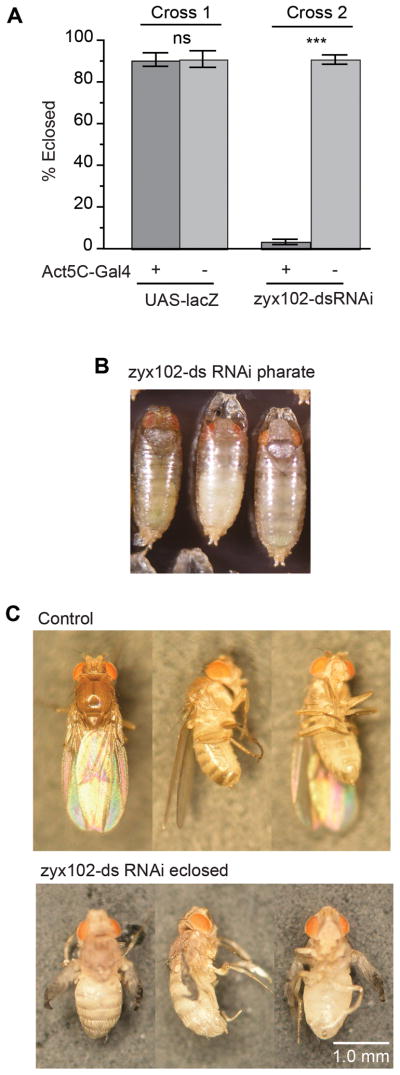
In contrast, when Act5C-GAL4 was used to drive expression of zyx102-dsRNA, reproducible lethality occurred (Fig. 6A). Presence of the driver alone did not result in appreciable lethality, even in combination with another transformation vector construct, such as UAS-lacZ (Brand and Perrimon, 1993). To determine the lethal period, we collected staged embryos, and determined that the frequency of hatching into larvae of Zyx102-knockdown embryos was within the normal range (data not shown). We then separated larvae based on markers that indicated genotype and observed siblings during the remainder of development. Control and Zyx102-knockdown larvae were able to molt through successive larval instars, pupariate normally, and progress through pupation at the same frequency. The duration of each developmental period did not vary between the two populations. However, Zyx102-knockdown flies did not emerge fully from the pupal case, and largely died as pharate adults (Fig. 6B–C). Upon inspection, there were no obvious malformations of the eyes or bristles; thoracic and abdominal morphogenesis appeared normal. The few Zyx102-knockdown flies that were able to emerge did not undergo wing inflation, contraction of the abdomen, or cuticle hardening, and were, for the most part, unable to move properly. They died within a day or two of eclosion.
Figure 6. Loss of Zyx102 expression by RNA interference is pharate adult lethal when using Act5C-GAL4.
A. F1 progeny larvae of Drosophila crosses carrying the Act5C-GAL4 driver and either UAS-zyx102-dsRNA or UAS-lacZ were sorted via genetic markers. Nearly 100% of larvae pupated (data not shown). Fewer than 5% of those carrying Act5C-GAL4 and UAS-zyx102-dsRNA eclosed as adult flies, whereas greater than 90% of those carrying either one of those elements, whether or not UAS-lacZ was present, eclosed as adults. ns=not significant.***p<0.001.
B. Shown are pharate adult flies carrying Act5C-GAL4 and UAS-zyx102-dsRNA. The vast majority of these flies were incapable of escaping from the pupal case.
C. Rare knockdown flies (Act5C-GAL4 and UAS-zyx102-dsRNA) were able to escape the pupal case. Though the legs of these flies could twitch and their external morphology was normal, they did not undergo post-eclosion maturation (the wings did not inflate, the body did not tan or harden, and flies were unable to walk), and soon died. Control flies (w1118; P{UAS-zyx102-dsRNA}16c) at 24 hr post-eclosion are shown for comparison.
The tracheolar network in Zyx102-knockdown flies is not properly air-filled
A number of important events occur during the lethal phase associated with Zyx102 knockdown. During the last nine hours of pupation, the fruit fly undergoes a stereotyped series of events that culminate in its emergence from the pupal case (Kimura and Truman, 1990). Some events occur as a result of steroid hormone signaling, including deposition of adult cuticle, resorption of molting fluid, and tracheal air-filling. Other events occur in response to neuropeptide signals, such as activation of a motor program to shed the pupal cuticle and extension of the ptilinum, which “pops open the hatch” of the pupal case. In comparing control and knockdown animals, we observed that Zyx102-knockdown flies did not progress through these final stages properly. Molting fluid within the pupal case was not absorbed and the trachea did not whiten, an indicator of adult tracheal air filling (Fig. 7). By direct observation using light microscopy, we determined that Zyx102-knockdown flies extended the ptilinum to rupture the pupal cuticle at the operculum. Some flies attempted to execute eclosion, though most were trapped halfway out of the case, where they died. Even when dissected free from the pupal case, however, Zyx102-knockdown flies died; in contrast, liberated control flies easily exited the case and survived.
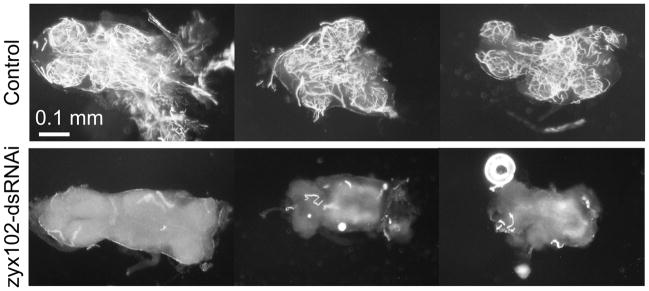
Figure 7.
The tracheolar network of Zyx102-knockdown adults is not properly air-filled Ventral nerve cords (VNCs) were dissected from responsive pharate adults and prepared for differential-interference contrast (DIC) microscopy using techniques that preserve air present in the trachea (Kimura and Truman, 1990). The air-filled tracheal network is bright relative to the surrounding tissue. VNCs from three control flies are compared to three dissected from Act5C-GAL4 + UAS-zyx102-dsRNA flies. Just prior to eclosion, the tracheal networks present in each lobe of the nerve cord are air-filled (white) in the control, but not in the Zyx102-knockdown, animals. Bar = 100 μm.
In wild-type flies, adult trachea begin to fill with air after molting fluid is resorbed. The fine tracheolar processes present in the ventral nerve cord (VNC) fill during the last hour prior to eclosion (Kimura and Truman, 1990). VNCs from control and Zyx102-knockdown pharate adult stage animals were dissected prior to fluid resorption, as well as at the time the animals attempted to eclose. Prior to molting fluid resorption, both control and Zyx102-knockdown flies exhibit a similar lack of air-filled, adult tracheoles (data not shown). When dissected from eclosing adult flies, however, one can see that each lobe of control VNCs comprises a fine meshwork of tracheolar processes (Fig. 7, Control), visible in these micrographs because the air in the tubes is in high contrast with the surrounding aqueous tissue. Zyx102-knockdown animals, however, do not exhibit this air-filled network (Fig. 7, zyx102-dsRNAi). The lack of air-filled tracheolar processes in the knockdown is not unique to the VNC, as it can also be observed in ovaries dissected from pharate female flies (data not shown). We observed that the degree of tracheolar air-filling in VNCs dissected from knockdown flies was in proportion to the degree to which the fly attempted to eclose.
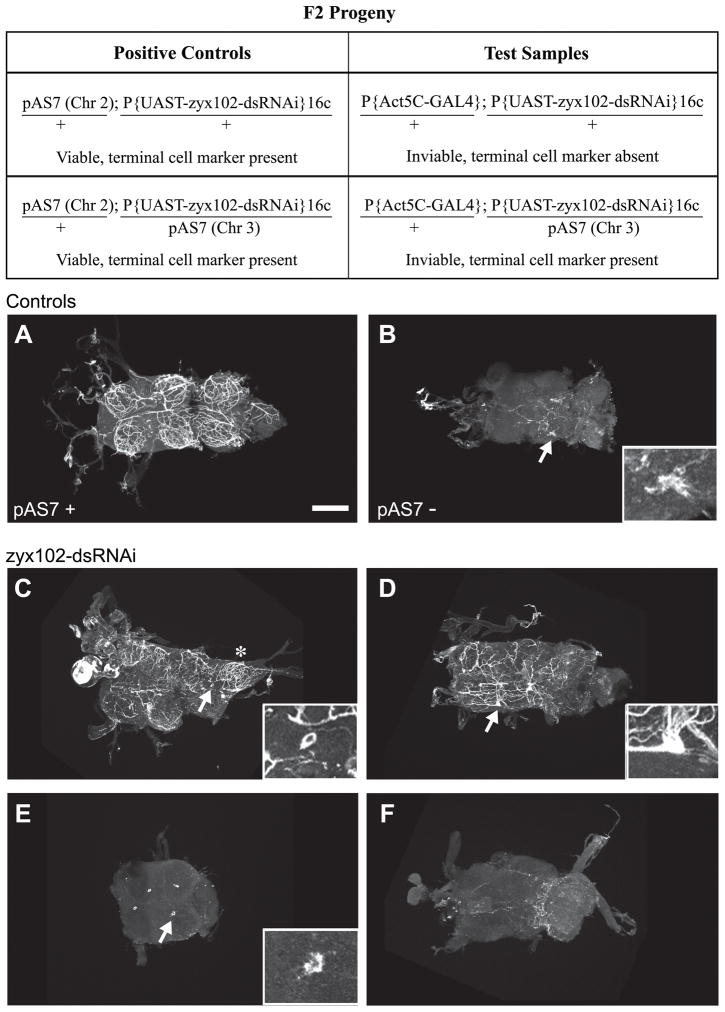
A tracheal terminal cell marker was used to investigate whether the tracheal cells that give rise to the fine meshwork in the VNC were present in the knockdown, and to what extent these cells developed processes that could undergo air-filling (Fig. 8; Table 1). In flies carrying pAS7, a membrane-associated form of GFP is expressed in tracheal terminal cells via a specific enhancer element (Mark Metzstein, personal communication; (Nussbaumer et al., 2000)). VNCs were dissected from pharate or freshly emerged adults, fixed, and the GFP signal stabilized by immunostaining. As shown in Figure 8, VNCs were prepared from four different genotypes: Positive controls carried pAS7 and P{UAST-zyx102-dsRNAi}16c. As a negative control for GFP immunostaining, we used the P{UAST-zyx102-dsRNAi}16c stock. Test samples were dissected from pharate-adult lethals (thus, Act5C-GAL4 and P{UAST-zyx102-dsRNAi}16c) that either did or did not carry the terminal cell marker; these adults were still responsive, however, when poked.
Figure 8. Tracheal terminal cells elaborate processes in the Zyx102-knockdown flies.
To investigate whether tracheal cells in the VNC are present if RNA-interference is used to reduce the levels of Zyx102, and to see to what extent these tracheal cells develop processes that undergo normal air-filling, a GFP-tagged marker for differentiated terminal cells of the tracheal system (pAS7) was used. VNCs were prepared from pharate or freshly eclosed adults and processed for immunostaining using anti-GFP. Top panel: F2 progeny from a single cross were used as test samples and positive controls. Genotypes for chromosomes 2 and 3 are shown; viability and presence or absence of the pAS7 marker are indicated. Half of the inviable pharate adult flies should be negative for GFP, half positive for GFP. These flies cannot a priori be distinguished from each other. The P{UAS-zyx102-dsRNA}16c stock was used as a negative control. Refer to Table 1 for further description of experimental observations. Arrows indicate areas shown in insets. Anterior is to the left. Bar = 100 μm.
A. Positive control: a VNC dissected from a viable pharate adult carrying the tracheal system terminal cell marker, pAS7 and P{UAS-zyx102-dsRNA}16c. The fine network of processes tagged with the terminal cell marker is nearly identical to the tracheal network observable because of air-filling (Fig. 7).
B. Negative control: a VNC dissected from a viable pharate adult carrying P{UAS-zyx102-dsRNA}16c only. Note the background evident as either fine processes or a larger aggregate (arrow and inset). Although two out of five samples showed no labeling, three out of five showed patterns like this one.
C–F. VNCs dissected from inviable pharate adults (thus carrying both Act5C-GAL4 and UAS-zyx102-dsRNA; half are expected to carry the terminal cell marker, pAS7, half should not). Some lobes exhibit a normal tracheolar network (asterisk in C), while others do not. In C and D, terminal cell bodies can be detected (arrow and inset), some with (D) and some without (C) labeled processes. In some VNCs dissected from inviable adults, single cells were strongly labeled with the terminal cell marker (arrows and insets, E), a pattern not seen in negative controls (data not shown). The VNC in E lost two of the six lobes during the staining procedure. The labeling pattern in F is strongly akin to that seen in the negative controls, and is likely from a specimen not marked with pAS7.
All of the positive controls showed terminal cell labeling, as expected (Fig. 8A). Three out of five VNCs prepared as negative controls showed some fluorescence signal, but it was clearly distinguishable from the positive samples; one example is shown (Fig. 8B). Ten test samples were prepared from inviable but still responsive pharate adults. Half of these are expected to carry the tracheal terminal cell marker. Consistent with this expectation, 40% of the samples had labeling like the negative control; the remainder expressed the terminal cell marker. Thus, the tracheal terminal cells differentiate in the Zyx102 knockdown flies.
Of the Zyx102-knockdown samples that showed terminal cell labeling, elaborations of the terminal cells were often abnormal. A range of terminal cell ramifications was noted. In certain cases, strong labeling was observed and morphologically normal terminal processes were detected (Fig. 8C, D). In many cases, however, labeling was observed but either the terminal cell branching was defective or a single, labeled cell devoid of labeled processes was associated with each lobe of the nerve cord (Fig 8E). We did not observe labeled cells like this in the negative control. Thus, Zyx102-knockdown adults appear to have a range in the degree of process ramification from tracheal terminal cells. However, terminal cells in Zyx102-knockdown animals that have ramified processes are likely unable to undergo air-filling, since air-filled tracheal processes, like those in Figure 7, were not observed in VNCs dissected from dying knockdown flies. Thus, these results indicate that, while Zyx102 is not essential for specification of terminal cells of the adult tracheal system, it may be required for these cells to complete morphological and functional differentiation.
Discussion
In this report, we demonstrate that RNA interference-mediated knockdown of the zyxin family representative in the fly, Zyx102, results in lethality. Knockdown animals die at the pharate adult stage, exhibiting defects in tracheal air filling and tracheal terminal cell organization when zyx102-specific RNA-interference is driven by Act5C-GAL4. Knockdown animals are able to complete embryonic and larval stages of development successfully, pupariate normally, and undergo metamorphosis without engendering external morphological abnormalities.
Several lines of evidence support the conclusion that the observed lethality is attributable to reduced Zyx102 expression. As shown in Figure 4, when the RNA-interference construct P{UAS-zyx102-dsRNAi}16c is expressed in muscle tissue using the GAL4 driver P{GawB}how24B, a marked reduction in the level of Zyx102 protein is detected in that tissue. A robust and reproducible lethal effect was observed when zyx102-dsRNA expression was driven by the broadly-expressed GAL4 driver, Act5C-GAL4. In third-instar larvae, the Act5C-GAL4 construct is predominantly expressed in trachea, imaginal discs, nervous system, and gut epithelium (Fig. 5). Since we observed no external morphological defects in Zyx102-knockdown pharate adult or escapers, Zyx102 is unlikely to be involved in imaginal disc differentiation.
Two independently isolated fly strains carrying a similar inducible RNA-interference construct directed towards zyx102 were recently made available from the National Institute of Genetics (NIG) in Japan. When tested both by the NIG and by ourselves using the Act5C-GAL4 driver, these lines exhibited the same lethal period as the line derived in our laboratory, i.e., late pupal/pharate adult stage (data not shown). The similar phenotype obtained using three independently isolated knockdown lines supports the conclusion that the phenotype is due to knockdown of zyx102 and not the insertion site.
Our RT-PCR analysis extends our understanding of the gene structure and splicing patterns predicted by Zyx102 cDNA sequences (Renfranz et al., 2003; Wilson et al., 2008). The developmental western blot presented here (Fig. 3) showed a faster-migrating, male-specific species of Zyx102 with estimated molecular mass of approximately 63,000 Da, which could be accounted for by differential usage of Exon 2 and/or Exon 4. RT-PCR also identified a variant of Exon 6, termed Exon 6c, which would give rise to a Zyx102 isoform lacking the C-terminal LIM domains, shifting the molecular mass down by approximately 21,000 Da while still being recognized by MAb P1D8 used in this study. Transcripts encoding Drosophila Paxillin, which displays four C-terminal LIM domains, also undergo alternative splicing that affects the LIM region (Yagi et al., 2001).
The tubule morphogenesis of Drosophila tracheal development involves transcriptional regulation, cell fate specification, cytoskeletal dynamics with cell shape changes and cell migration, and culminates in tracheal air filling (Zelzer and Shilo, 2000; Myat, 2005; Kerman et al., 2006). Our results suggest that the lethal phenotype observed in Zyx102-knockdown flies is attributable to the lack of tracheal air filling, a process that normally occurs just prior to eclosion (Kimura and Truman, 1990). Experiments using the terminal tracheal cell marker pAS7, which in wild-type flies reveals tracheal processes that have ramified into adult tissues like the ventral nerve cord, showed that tracheal terminal cells in Zyx102-knockdown animals are correctly specified and, in many cases, appear to extend processes as in the wild type flies. However, we have observed abnormal terminal cell morphology in the Zyx102-knockdown animals. This phenotype provides a potential link between Zyx102 function and a number of genes involved in the differentiation of terminal cells and/or air-filling of the tracheolar branches. For example, Levi et al. (2006) showed that the cytoskeletal protein talin, as well as heterodimeric integrins, are required for the maintenance of tracheal terminal cell branches. Furthermore, Drosophila serum response factor, encoded by the blistered/pruned gene, is directly involved in the extension of processes from tracheal system terminal cells (Guillemin et al., 1996), as is Drosophila Myocardin-related transcription factor, the myocardin family homolog (Han et al., 2004). Tracheal air-filling also requires such apparently disparate elements as epithelial Na+ channel-related genes (Liu et al., 2003), clathrin-mediated endocytosis proteins (Behr et al., 2007), and mitochondrial carrier proteins (Hartenstein et al., 1997). Further analysis will be required to elucidate the mechanism by which Zyx102 contributes to tracheal structure and function.
A tracheolar defect similar to the one we observed in Zyx102-knockdown animals is seen in mutants of the Drosophila BMP Type II Receptor, wishful thinking (wit). Marques et al. (2002) showed that, while about half of wit mutants die during larval and early pupal stages, the other half die as pharate adults, with no obvious external abnormalities and with a similar behavioral repertoire as the Zyx102-knockdown animals. wit was initially identified in a screen for mutants with abnormal morphology of the larval neuromuscular junction (NMJ) (Aberle et al., 2002; Marques et al., 2002). Although wit mutants display abnormal motor neuron branching patterns at the NMJ, they, like Zyx102-knockdown animals, are largely able to complete adult morphogenesis. We examined ventral nerve cords from wit-mutant pharate adults and noted a lack of air-filled tracheoles, similar to our observations in Zyx102-knockdown flies (data not shown). The pharate adult lethality of wit mutants has been tied directly to decreased levels of a systemic neuropeptide, FMRFa (Marques et al., 2003), proper expression of which requires a BMP-mediated retrograde signaling pathway.
To conclude, analysis of zyxin function in Drosophila demonstrates a requirement of this highly conserved gene family for viability that had not been previously recognized. Our work hints at the involvement of zyxin family members in the outgrowth of processes from cells, which has been shown to be dependent on the regulation of actin filament dynamics in tracheal cells (Oshima et al., 2006) and in neurons (reviewed in (Gallo and Letourneau, 2004; Kalil and Dent, 2005)), as well as on integrin-mediated cell adhesion (Beumer et al., 1999; Boube et al., 2001; Nakamoto et al., 2004; Levi et al., 2006; Huang et al., 2007; Inoue and Hayashi, 2007). Analysis of the zyxin family in an organism where ambiguity, introduced by redundancy among gene family members, is absent reemphasizes the importance of this LIM domain-containing protein and indicates possible avenues for future experiments in Drosophila and in vertebrates.
Acknowledgments
We thank Mark Metzstein for allowing us to use the pAS7 stock prior to publication and for helpful discussions; the Bloomington Drosophila Stock Center at Indiana University and the National Institute of Genetics Fly Stock Center in Japan for other Drosophila stocks; Maura McGrail for generation of the UAS-zyx102(44)::EGFP stock; Kathleen Clark for the larval fillet protocol; the Immunological Resource Center at the University of Illinois for monoclonal antibodies; the Developmental Studies Hybridoma Bank at the University of Iowa for the anti-Lamin C antibody developed by D. Riemer; and the University of Utah Core Facilities. We are especially grateful to members of the Beckerle Lab for helpful suggestions, and to Laura Hoffman, Diana Lim and Ellen T. Wilson for assistance with preparing the manuscript. Supported by NIH Grant GM50877 and the Huntsman Cancer Foundation.
Grant Sponsor: NIH: GM50877 and Huntsman Cancer Foundation.
References
- Aberle H, Haghighi AP, Fetter RD, McCabe BD, Magalhaes TR, Goodman CS. wishful thinking encodes a BMP type II receptor that regulates synaptic growth in Drosophila. Neuron. 2002;33:545–58. doi: 10.1016/s0896-6273(02)00589-5. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Golic KG, Hawley RS. Drosophila: A Laboratory Handbook. 2. Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- Behr M, Wingen C, Wolf C, Schuh R, Hoch M. Wurst is essential for airway clearance and respiratory-tube size control. Nat Cell Biol. 2007;9:847–53. doi: 10.1038/ncb1611. [DOI] [PubMed] [Google Scholar]
- Beumer KJ, Rohrbough J, Prokop A, Broadie K. A role for PS integrins in morphological growth and synaptic function at the postembryonic neuromuscular junction of Drosophila. Development. 1999;126:5833–46. doi: 10.1242/dev.126.24.5833. [DOI] [PubMed] [Google Scholar]
- Bilder D. Epithelial polarity and proliferation control: links from the Drosophila neoplastic tumor suppressors. Genes Dev. 2004;18:1909–25. doi: 10.1101/gad.1211604. [DOI] [PubMed] [Google Scholar]
- Boube M, Martin-Bermudo MD, Brown NH, Casanova J. Specific tracheal migration is mediated by complementary expression of cell surface proteins. Genes Dev. 2001;15:1554–62. doi: 10.1101/gad.195501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–15. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Colombelli J, Besser A, Kress H, Reynaud EG, Girard P, Caussinus E, Haselmann U, Small JV, Schwarz US, Stelzer EH. Mechanosensing in actin stress fibers revealed by a close correlation between force and protein localization. J Cell Sci. 2009;122:1665–79. doi: 10.1242/jcs.042986. [DOI] [PubMed] [Google Scholar]
- Echeverri CJ, Perrimon N. High-throughput RNAi screening in cultured cells: a user’s guide. Nat Rev Genet. 2006;7:373–84. doi: 10.1038/nrg1836. [DOI] [PubMed] [Google Scholar]
- Gallo G, Letourneau PC. Regulation of growth cone actin filaments by guidance cues. J Neurobiol. 2004;58:92–102. doi: 10.1002/neu.10282. [DOI] [PubMed] [Google Scholar]
- Guillemin K, Groppe J, Ducker K, Treisman R, Hafen E, Affolter M, Krasnow MA. The pruned gene encodes the Drosophila serum response factor and regulates cytoplasmic outgrowth during terminal branching of the tracheal system. Development. 1996;122:1353–62. doi: 10.1242/dev.122.5.1353. [DOI] [PubMed] [Google Scholar]
- Han Z, Li X, Wu J, Olson EN. A myocardin-related transcription factor regulates activity of serum response factor in Drosophila. Proc Natl Acad Sci U S A. 2004;101:12567–72. doi: 10.1073/pnas.0405085101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen MD, Beckerle MC. Opposing Roles of Zyxin/LPP ACTA Repeats and the LIM Domain Region in Cell-Cell Adhesion. J Biol Chem. 2006;281:16178–88. doi: 10.1074/jbc.M512771200. [DOI] [PubMed] [Google Scholar]
- Hartenstein K, Sinha P, Mishra A, Schenkel H, Torok I, Mechler BM. The congested-like tracheae gene of Drosophila melanogaster encodes a member of the mitochondrial carrier family required for gas-filling of the tracheal system and expansion of the wings after eclosion. Genetics. 1997;147:1755–68. doi: 10.1093/genetics/147.4.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman LM, Jensen CC, Kloeker S, Wang CL, Yoshigi M, Beckerle MC. Genetic ablation of zyxin causes Mena/VASP mislocalization, increased motility, and deficits in actin remodeling. J Cell Biol. 2006;172:771–82. doi: 10.1083/jcb.200512115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman LM, Nix DA, Benson B, Boot-Hanford R, Gustafsson E, Jamora C, Menzies AS, Goh KL, Jensen CC, Gertler FB, Fuchs E, Fassler R, Beckerle MC. Targeted disruption of the murine zyxin gene. Mol Cell Biol. 2003;23:70–9. doi: 10.1128/MCB.23.1.70-79.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Yazdani U, Thompson-Peer KL, Kolodkin AL, Terman JR. Crk-associated substrate (Cas) signaling protein functions with integrins to specify axon guidance during development. Development. 2007;134:2337–47. doi: 10.1242/dev.004242. [DOI] [PubMed] [Google Scholar]
- Inoue Y, Hayashi S. Tissue-specific laminin expression facilitates integrin-dependent association of the embryonic wing disc with the trachea in Drosophila. Dev Biol. 2007;304:90–101. doi: 10.1016/j.ydbio.2006.12.022. [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–71. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5:920–31. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- Kalidas S, Smith DP. Novel genomic cDNA hybrids produce effective RNA interference in adult Drosophila. Neuron. 2002;33:177–84. doi: 10.1016/s0896-6273(02)00560-3. [DOI] [PubMed] [Google Scholar]
- Kalil K, Dent EW. Touch and go: guidance cues signal to the growth cone cytoskeleton. Curr Opin Neurobiol. 2005;15:521–6. doi: 10.1016/j.conb.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Kaminker JS, Bergman CM, Kronmiller B, Carlson J, Svirskas R, Patel S, Frise E, Wheeler DA, Lewis SE, Rubin GM, Ashburner M, Celniker SE. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 2002;3:RESEARCH0084. doi: 10.1186/gb-2002-3-12-research0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassel O, Schneider S, Heilbock C, Litfin M, Gottlicher M, Herrlich P. A nuclear isoform of the focal adhesion LIM-domain protein Trip6 integrates activating and repressing signals at AP-1- and NF-kappaB-regulated promoters. Genes Dev. 2004;18:2518–28. doi: 10.1101/gad.322404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerman BE, Cheshire AM, Andrew DJ. From fate to function: the Drosophila trachea and salivary gland as models for tubulogenesis. Differentiation. 2006;74:326–48. doi: 10.1111/j.1432-0436.2006.00095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura KI, Truman JW. Postmetamorphic cell death in the nervous and muscular systems of Drosophila melanogaster. J Neurosci. 1990;10:403–1. doi: 10.1523/JNEUROSCI.10-02-00403.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni MM, Booker M, Silver SJ, Friedman A, Hong P, Perrimon N, Mathey-Prevot B. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat Methods. 2006;3:833–8. doi: 10.1038/nmeth935. [DOI] [PubMed] [Google Scholar]
- Lee JW, Choi HS, Gyuris J, Brent R, Moore DD. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol Endocrinol. 1995;9:243–254. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- Lele TP, Pendse J, Kumar S, Salanga M, Karavitis J, Ingber DE. Mechanical forces alter zyxin unbinding kinetics within focal adhesions of living cells. J Cell Physiol. 2006;207:187–94. doi: 10.1002/jcp.20550. [DOI] [PubMed] [Google Scholar]
- Levi BP, Ghabrial AS, Krasnow MA. Drosophila talin and integrin genes are required for maintenance of tracheal terminal branches and luminal organization. Development. 2006;133:2383–93. doi: 10.1242/dev.02404. [DOI] [PubMed] [Google Scholar]
- Li B, Trueb B. Analysis of the alpha-actinin/zyxin interaction. J Biol Chem. 2001;276:33328–35. doi: 10.1074/jbc.M100789200. [DOI] [PubMed] [Google Scholar]
- Li B, Zhuang L, Reinhard M, Trueb B. The lipoma preferred partner LPP interacts with alpha-actinin. J Cell Sci. 2003;116:1359–66. doi: 10.1242/jcs.00309. [DOI] [PubMed] [Google Scholar]
- Liu L, Johnson WA, Welsh MJ. Drosophila DEG/ENaC pickpocket genes are expressed in the tracheal system, where they may be involved in liquid clearance. Proc Natl Acad Sci U S A. 2003;100:2128–33. doi: 10.1073/pnas.252785099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Creanga A, Lum L, Beachy PA. Prevalence of off-target effects in Drosophila RNA interference screens. Nature. 2006;443:359–63. doi: 10.1038/nature05179. [DOI] [PubMed] [Google Scholar]
- Marques G, Bao H, Haerry TE, Shimell MJ, Duchek P, Zhang B, O’Connor MB. The Drosophila BMP type II receptor Wishful Thinking regulates neuromuscular synapse morphology and function. Neuron. 2002;33:529–43. doi: 10.1016/s0896-6273(02)00595-0. [DOI] [PubMed] [Google Scholar]
- Marques G, Haerry TE, Crotty ML, Xue M, Zhang B, O’Connor MB. Retrograde Gbb signaling through the Bmp type 2 receptor wishful thinking regulates systemic FMRFa expression in Drosophila. Development. 2003;130:5457–70. doi: 10.1242/dev.00772. [DOI] [PubMed] [Google Scholar]
- Myat MM. Making tubes in the Drosophila embryo. Dev Dyn. 2005;232:617–32. doi: 10.1002/dvdy.20293. [DOI] [PubMed] [Google Scholar]
- Nakamoto T, Kain KH, Ginsberg MH. Neurobiology: New connections between integrins and axon guidance. Curr Biol. 2004;14:R121–3. [PubMed] [Google Scholar]
- Nix DA, Beckerle MC. Nuclear-cytoplasmic shuttling of the focal contact protein, zyxin: a potential mechanism for communication between sites of cell adhesion and the nucleus. J Cell Biol. 1997;138:1139–47. doi: 10.1083/jcb.138.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix DA, Fradelizi J, Bockholt S, Menichi B, Louvard D, Friederich E, Beckerle MC. Targeting of zyxin to sites of actin membrane interaction and to the nucleus. J Biol Chem. 2001;276:34759–67. doi: 10.1074/jbc.M102820200. [DOI] [PubMed] [Google Scholar]
- Nussbaumer U, Halder G, Groppe J, Affolter M, Montagne J. Expression of the blistered/DSRF gene is controlled by different morphogens during Drosophila trachea and wing development. Mech Dev. 2000;96:27–36. doi: 10.1016/s0925-4773(00)00373-7. [DOI] [PubMed] [Google Scholar]
- Oshima K, Takeda M, Kuranaga E, Ueda R, Aigaki T, Miura M, Hayashi S. IKK epsilon regulates F actin assembly and interacts with Drosophila IAP1 in cellular morphogenesis. Curr Biol. 2006;16:1531–7. doi: 10.1016/j.cub.2006.06.032. [DOI] [PubMed] [Google Scholar]
- Patel NH. Imaging Neuronal Subsets and Other Cell Types in Whole-Mount Drosophila Embryos and Larvae Using Antibody Probes. Methods in Cell Biology. 1994;44:445–488. doi: 10.1016/s0091-679x(08)60927-9. [DOI] [PubMed] [Google Scholar]
- Petit MM, Crombez KR, Vervenne HB, Weyns N, Van de Ven WJ. The tumor suppressor Scrib selectively interacts with specific members of the zyxin family of proteins. FEBS Lett. 2005a;579:5061–8. doi: 10.1016/j.febslet.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Petit MM, Fradelizi J, Golsteyn RM, Ayoubi TA, Menichi B, Louvard D, Van de Ven WJ, Friederich E. LPP, an actin cytoskeleton protein related to zyxin, harbors a nuclear export signal and transcriptional activation capacity. Mol Biol Cell. 2000;11:117–29. doi: 10.1091/mbc.11.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit MM, Meulemans SM, Alen P, Ayoubi TA, Jansen E, Van de Ven WJ. The tumor suppressor Scrib interacts with the zyxin-related protein LPP, which shuttles between cell adhesion sites and the nucleus. BMC Cell Biol. 2005b;6:1. doi: 10.1186/1471-2121-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podemski L, Sousa-Neves R, Marsh JL, Locke J. Molecular mapping of deletion breakpoints on chromosome 4 of Drosophila melanogaster. Chromosoma. 2004;112:381–8. doi: 10.1007/s00412-004-0286-4. [DOI] [PubMed] [Google Scholar]
- Reinhard M, Zumbrunn J, Jaquemar D, Kuhn M, Walter U, Trueb B. An alpha-actinin binding site of zyxin is essential for subcellular zyxin localization and alpha-actinin recruitment. J Biol Chem. 1999;274:13410–8. doi: 10.1074/jbc.274.19.13410. [DOI] [PubMed] [Google Scholar]
- Renfranz PJ, Beckerle MC. Doing (F/L)PPPPs: EVH1 domains and their proline-rich partners in cell polarity and migration. Curr Opin Cell Biol. 2002;14:88–103. doi: 10.1016/s0955-0674(01)00299-x. [DOI] [PubMed] [Google Scholar]
- Renfranz PJ, Siegrist SE, Stronach BE, Macalma T, Beckerle MC. Molecular and phylogenetic characterization of Zyx102, a Drosophila orthologue of the zyxin family that interacts with Drosophila Enabled. Gene. 2003;305:13–26. doi: 10.1016/s0378-1119(02)01173-3. [DOI] [PubMed] [Google Scholar]
- Riemer D, Stuurman N, Berrios M, Hunter C, Fisher PA, Weber K. Expression of Drosophila lamin C is developmentally regulated: analogies with vertebrate A-type lamins. J Cell Sci. 1995;108(Pt 10):3189–98. doi: 10.1242/jcs.108.10.3189. [DOI] [PubMed] [Google Scholar]
- Rorth P. Gal4 in the Drosophila female germline. Mech Dev. 1998;78:113–8. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- Rothwell WF, Sullivan W. Fluorescent Analysis of Drosophila Embryos. In: Sullivan W, Ashburner M, Hawley RS, editors. Drosohila Protocols. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 141–157. [Google Scholar]
- Schmeichel KL, Beckerle MC. The LIM domain is a modular protein-binding interface. Cell. 1994;79:211–219. doi: 10.1016/0092-8674(94)90191-0. [DOI] [PubMed] [Google Scholar]
- Schmeichel KL, Beckerle MC. Molecular dissection of a LIM domain. Mol Biol Cell. 1997;8:219–230. doi: 10.1091/mbc.8.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster CM, Davis GW, Fetter RD, Goodman CS. Genetic dissection of structural and functional components of synaptic plasticity. I. Fasciclin II controls synaptic stabilization and growth. Neuron. 1996;17:641–54. doi: 10.1016/s0896-6273(00)80197-x. [DOI] [PubMed] [Google Scholar]
- Sweeney ST, Broadie K, Keane J, Niemann H, O’Kane CJ. Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron. 1995;14:341–51. doi: 10.1016/0896-6273(95)90290-2. [DOI] [PubMed] [Google Scholar]
- Vervenne HB, Crombez KR, Delvaux EL, Janssens V, Van de Ven WJ, Petit MM. Targeted disruption of the mouse Lipoma Preferred Partner gene. Biochem Biophys Res Commun. 2009;379:368–73. doi: 10.1016/j.bbrc.2008.12.074. [DOI] [PubMed] [Google Scholar]
- Wang Y, Dooher JE, Koedood Zhao M, Gilmore TD. Characterization of mouse TRIP6: a putative intracellular signaling protein. Gene. 1999;234:403–409. doi: 10.1016/s0378-1119(99)00168-7. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gilmore TD. LIM domain protein Trip6 has a conserved nuclear export signal, nuclear targeting sequences, and multiple transactivation domains. Biochim Biophys Acta. 2001;1538:260–72. doi: 10.1016/s0167-4889(01)00077-5. [DOI] [PubMed] [Google Scholar]
- Wellik DM, Capecchi MR. Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton. Science. 2003;301:363–7. doi: 10.1126/science.1085672. [DOI] [PubMed] [Google Scholar]
- Wilson RJ, Goodman JL, Strelets VB. FlyBase: integration and improvements to query tools. Nucleic Acids Res. 2008;36:D588–93. doi: 10.1093/nar/gkm930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Ishimaru S, Yano H, Gaul U, Hanafusa H, Sabe H. A novel muscle LIM-only protein is generated from the paxillin gene locus in Drosophila. EMBO Rep. 2001;2:814–20. doi: 10.1093/embo-reports/kve178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J, Kloeker S, Jensen CC, Bockholt S, Honda H, Hirai H, Beckerle MC. Members of the Zyxin family of LIM proteins interact with members of the p130Cas family of signal transducers. J Biol Chem. 2002;277:9580–9. doi: 10.1074/jbc.M106922200. [DOI] [PubMed] [Google Scholar]
- Yoshigi M, Hoffman LM, Jensen CC, Yost HJ, Beckerle MC. Mechanical force mobilizes zyxin from focal adhesions to actin filaments and regulates cytoskeletal reinforcement. J Cell Biol. 2005;171:209–15. doi: 10.1083/jcb.200505018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelzer E, Shilo BZ. Cell fate choices in Drosophila tracheal morphogenesis. Bioessays. 2000;22:219–26. doi: 10.1002/(SICI)1521-1878(200003)22:3<219::AID-BIES3>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]