Abstract
The power of proteomics allows unparalleled opportunity to query the molecular mechanisms of a malignant cell and the tumor microenvironment in patients with ovarian cancer and other solid tumors. This information has given us insight into the perturbations of signaling pathways within tumor cells and has aided the discovery of new drug targets for the tumor and possible prognostic indicators of outcome and disease response to therapy. Proteomics analysis of serum and ascites has also given us sources with which to discover possible early markers for the presence of new disease and for the progression of established cancer throughout the course of treatment. Unfortunately, this wealth of information has yielded little to date in changing the clinical care of these patients from a diagnostic, prognostic, or treatment perspective. The rational examination and translation of proteomics data in the context of past clinical trials and the design of future clinical trials must occur before we can march forward into the future of personalized medicine.
Keywords: ovarian cancer, proteomics, clinical trials
Introduction
Epithelial ovarian cancer is the second leading gynecologic malignancy in the United States and remains the leading cause of death from gynecologic malignancies and fifth leading cause of death among all cancers in women. Approximately 20,000 new cases of ovarian cancer are expected to be diagnosed in 2010 with approximately 15,000 deaths from the disease [1]. The age-adjusted incidence has been decreasing slightly over the last 35 years but remains at 10–15 cases per 100,000 women with a mean age at diagnosis in the sixth decade (SEER database, accessed 8/2009). The majority of women present initially with advanced stage disease (Stage III/IV) for which cure is rare, with the 5-year median survival approximately 40% for stage III and less than 20% for stage IV [2]. Early cooperative group trials showed that treatment of stage I disease can be limited to surgical management for well differentiated carcinoma while poorly differentiated and locally more advanced tumors do benefit from systemic or localized chemotherapy [3]. Women diagnosed with advanced stage disease are managed with a combination of cytoreductive surgery followed by intravenous or, in selected patients, intraperitoneal platinum-based chemotherapy [4, 5]. Unfortunately, less than 40% of all patients are cured with the current available regimens [1]. The –omics revolution, proteomics, genomics, lipomics, and the various subsidiaries of each, has ushered in a new era that promises to aid in the early diagnosis, guidance of treatment modalities, and monitoring of response in treated patients, all tools that have been lacking thus far in ovarian cancer.
The application of proteomics technologies has pushed the boundaries of diagnosis and personalized medicine [6]. Fundamentally, cancer is a genetic defect that drives abnormal cellular proliferation. The genetic defect is transcribed and translated into proteins, the pawns that drive this abnormal phenotype [7]. Capturing and mining the information change in the proteome of the tumor cells and those cells in the local environment using advancing proteomics technologies can yield information that can be applied to develop early diagnosis, treatment, and patient monitoring decision trees.
Proteomics techniques can be applied in two general areas: the screening of tissue or fluid for indicators of the presence of disease and the characterization of known malignant samples. Traditionally, mass spectroscopy (MS) based techniques have been used to screen for diagnostic circulating biomarkers. This technique has used various front-end interfaces including liquid chromatography (LC), surface enhanced laser desorption and ionization (SELDI), and matrix-assisted laser desorption and ionization (MALDI) to screen serum, urine, and ascites for possible markers of early ovarian cancer or recurrence of disease [8–10]. More recent work has analyzed CSF for the presence of unique markers for glioblastoma and stool samples for the presence of colon cancer markers [11, 12]. Tumor samples have been analyzed by traditional MS techniques as well, but more recent analysis has focused on reverse phase protein arrays (RPPA). RPPAs examine serially diluted protein lysate samples in an array format and analyze with standardized specific antibodies. Serial samples can then be compared for response to treatment or, in the case of clinical trials, target validation and inhibition [13].
Due to the high relative mortality of ovarian cancer patients, effective screening for early stage disease could increase the detection of treatable early disease relative to late stage cancers. Unfortunately, diagnosis is all too often delayed due the lack of specific clinical symptoms in early stage disease [14]. Patients with a strong family history of ovarian or breast cancer (e.g. BRCA mutation carriers) or cancer syndromes with an ovarian component (e.g. Lynch syndrome) have an increased risk, but not all will develop ovarian cancer, which suggests a role for genetic and environmental interactions. Epidemiological studies have demonstrated the possible influence of age, racial, gynecological, geographical and hormonal factors [15, 16]. Environmental factors including diet, cigarette smoking and obesity may also be important but there is still debate [17–19]. Proteomics analysis of serum or tissue samples may fill the niche of a highly sensitive technique that is able to aid in the diagnosis of early stage disease in high risk populations or in screening the general population.
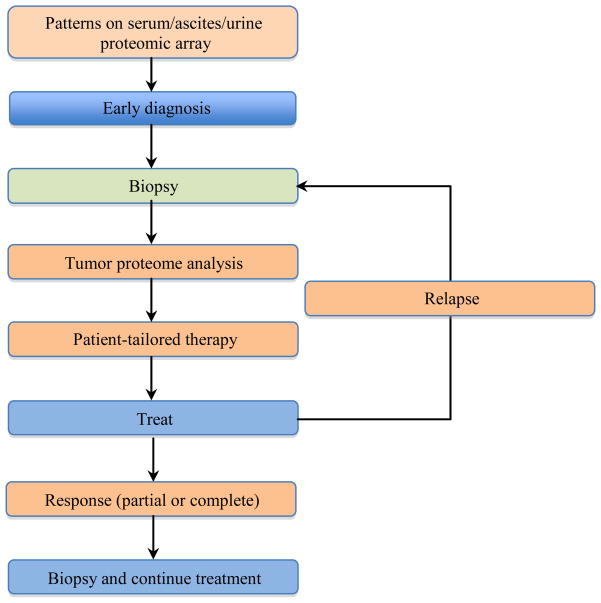
Many have suggested that each patient’s cancer may have derived from a unique subset of pathogenic molecular mutations within the spectrum of generalized mutations common to solid tumors [6]. These modulations result in changes in protein amount or function and may affect the flow of information that occurs within a cell and its microenvironment. This has been shown to deregulate the normal cellular phenotype. Since each patient may present a unique fingerprint within a known cancer subtype, one would expect differing responses to standard therapy. The workflow associated with the use of clinical proteomics in the guidance of targeted therapies and monitoring requires the input of multiple clinical practitioners and laboratories across many disciplines. The promise of this new emerging paradigm is to tailor therapy to a particular patients disease and perhaps reap beneficial results by guiding the use of selected targeted therapies throughout the course of treatment for a particular patient (Figure 1).
Figure 1. Schematic representation of the integration of proteomics into clinical trials.
Each tissue sample obtained allows for the measurement of information contained within the proteome that may be used to guide therapeutic decisions. Proteomic arrays are currently limited by the need to develop readily accessible assays for diagnostic and treatable targets. Patient-tailored therapy is limited by the need for targeted therapies that are safe, effective, show correlation to diagnostic biomarkers and have been validated in prospective clinical trials. Additionally, we need to develop rationally designed clinical trials that connect biomarkers to targeted agents.
There are three main limitations to this new paradigm. First, we need to know what targets to look for in a given patient using an assay that is readily accessible. Second, we need to have targeted therapies that are safe, effective, and show correlation to the markers studied for which they were developed. Third, we have to have rationally designed clinical trials that connect biomarkers with the targeted agent. In breast cancer, we currently test for hormone receptor and HER2 status to guide treatment with anti-hormonal therapy or HER2 targeted agents as well as to provide prognostic information to the patient. In colon cancer, K-ras mutation status helps define possible therapeutic decisions due to a lack of response to EGFR targeted therapies in patients with the mutation. It is not unreasonable to think that perhaps similar markers for the presence or prognosis of disease, or proteins that can be targeted can be found in ovarian cancer through the applied use of proteomics technologies.
Ovarian Cancer Screening and Diagnosis
Screening techniques must meet certain characteristics to be considered useful. Ideally, the biomarker must be readily measurable in an easily accessible specimen (body fluid, serum, urine). It has been estimated that a diagnostic test for ovarian cancer must also be highly specific (>99%) and reasonably sensitive with a suggested minimum positive predictive value (PPV) of 10% to first ensure no harm by limiting the number of surgeries necessary per case of ovarian cancer diagnosed [20, 21]. As has been demonstrated for other types of more prevalent cancer, including lung and prostate, a high PPV does not necessarily translate into mortality-associated benefits and prospective trials must be performed for validation of any new predictive marker.
CA125, approved in the Unites States only for monitoring response to therapy and suggestion of recurrence, is the most extensively studied predictive marker for ovarian cancer. Unfortunately, CA125 is only elevated in about 50–60% of early stage ovarian cancer at diagnosis and can be elevated in many other malignant or benign conditions [22], which limits its usefulness as an early screening tool when used alone. HE4 was discovered by comparative screening of cDNA arrays from normal ovarian epithelium and ovarian cancer tissue [23]. HE4 has a higher sensitivity for early stage disease than CA125, and the combination may be more sensitive and specific than either marker alone, but it still does not reach the level necessary to advance as a screening diagnostic biomarker [24, 25]. HE4 has been approved recently, like CA125, as a recurrence monitoring biomarker. Studies of these markers were limited to selected patient populations of women with pelvic masses and in both high- and low-risk patient populations still showed less than ideal specificity and sensitivity (~75% and 74–90% respectively).
Visintin et al. proposed a six marker panel (leptin, prolactin, OPN, IGF-II, MIF, CA125) that originally was reported to have a sensitivity of 95%, specificity of >99% and a PPV of >99% [26]. Reanalysis yielded a PPV of 6.5%, which would still lead to about 15 unnecessary surgeries for each patient with ovarian cancer [27]. A recently reported major study that is still maturing (UKCTOCS) is comparing no screening to transvaginal ultrasound with or without CA125 [28]. About 200,000 post-menopausal women were assigned in a 2:1:1 fashion with 58 invasive ovarian cancers detected in the screening arms to date, 24 of which were Stage I (41%). This is a higher percentage of early stage tumors compared to reported historical controls. The number of surgeries per cancer detected was 2.9, which would be acceptable. Maturation of this study is awaited to determine if there is a mortality benefit in these populations or if either test in combination with pelvic ultrasonography can be applied to the general population. This is especially important in light of the ongoing controversy over whether many serous ovarian cancers originate in the ovarian epithelium or in the distal fallopian tube, which may be a potential stumbling block for traditional imaging-based technologies [29–31].
Proteomics technology has increased the diagnostic sensitivity of screening samples for putative tumor markers. The use of MS can allow visualization of a snapshot of the serum proteome and may allow the early detection of circulating tumor markers long before the presentation of clinical symptoms. Problems from a technical standpoint include the concentration range of proteins within the plasma proteome, where likely significant markers for early detection may exist at concentrations 106–109 below those of abundant plasma proteins [32, 33]. There have been many attempts to deplete the plasma proteome of abundant proteins, but this may also deplete the concentration of desirable markers given the likelihood that a fraction of potential biomarkers may also be bound to abundant serum proteins like albumin [33, 34]. Early work by Petricoin et al. demonstrated that the serum MS protein signature derived from ovarian cancer and healthy patients predicted the presence of ovarian cancer in a blinded test group with 100% sensitivity and 95% specificity [35]. While this particular report has been controversial, it has stimulated significant discussion and brought many other investigators into the fray to help advance the concept towards early detection. More recently, others have obtained similar results using various MS based methodologies for multiplexed screening of healthy vs. cancer serum [36–39] as well classification information on specific ovarian cancer sub-types [40]. Many of the studies present sensitivity and specificity values that exceed published results for traditional ELISA or imaging-based assays. One of the pitfalls of all of these studies has been the lack of similarity between the general population and the validation data set, making specific claims about statistical importance of the data difficult. Unfortunately, no prospective clinical trials have yet been published that report the predictive value of any of these modalities in ovarian cancer patients.
A large Gynecologic Oncology Group trial, GOG-220, was designed to generate and then validate a proteomic signature pattern that can diagnose ovarian carcinoma on the background of patients presenting with a pelvic mass. Serum samples have been collected from over 2000 women undergoing surgical intervention for an undiagnosed pelvic mass. The objectives are to create protein pattern signatures that differentiate malignant ovarian disease from benign or non-ovarian malignancy in presurgical specimens using mass spectrometry. This trial was designed to yield independent discovery and validation cohorts and was prospectively powered to give a 96% specificity and sensitivity in the validation stage. Secondary objectives include differential analysis of early- versus late-stage disease and histotypes, post-operative residual disease, and prognostic outcome. We await these experimental results.
Another recent pathway has been the evaluation of ascites for hallmarks of ovarian cancer [41]. Ascites may be considered a surrogate representative of the tumor microenvironment in ovarian cancer and may contain tumor cells, secreted growth factors and tumor or vascular derived exosomes. A recent report published the first comprehensive ascites proteome in advanced ovarian cancer [42]. This study focused on ascites from a small number of Stage III/IV ovarian cancer patients and was able to use an integrated approach that combined the identification power of multidimensional protein identification techniques (MudPIT) technology with the high throughput capabilities of LC-MS, known urine and plasma proteomes and gene expression array data from multiple ovarian cancer cell lines. They report a list of 80 candidate markers, 18 of which had previously been reported in the serum or urine proteome and were then mapped against microarray data for multiple ovarian cancer cell lines [42]. Further studies will need to validate these biomarkers prospectively with high throughput screening assays on large populations.
The large number and variability of the candidates identified in MS based proteome-wide screening coming from a limited number of samples has many inherent biases. These include high false positive rates, high frequency of identification of nonspecific acute phase reactants, and a dataset complexity making it unlikely that data mining would repetitively yield the same outcome. Further, patient and tumor heterogeneity can yield insufficient true signal to be identified against the complex background. Some investigators have traversed these issues by taking only those peaks that “survive” across multiple patient samples, use enrichment techniques creating different bias, or subtraction techniques to remove abundant protein/peptides that are more likely to be physiologic or acute phase, leaving the low abundance signals.
With the relative scarcity of patient samples for analysis (in relation to the total number of patients with the disease), this leads to a backlog of potential markers that need to be validated. A concerted effort needs to be made to bank ovarian cancer tissue from a large number of patients beyond the current large but limited banks available to help both with increasing the pool size for the discovery phase but also the available samples for validation data sets.
Proteomics and Treatment Decisions
Another application of proteomics technologies is the guidance of choices for clinical care in newly diagnosed patients and the continued monitoring of these patients during their treatment and in subsequent follow-up visits. In other types of cancer, analysis of the tumor proteome and gene expression profile has led to the use of targeted therapies with definite clinical benefit. A study in patients with non-small cell lung cancer (NSCLC) demonstrated an important stepping-stone in proteomics research: the ability to risk-stratify patients based on a proteomic pattern. Yanagisawa et al. showed not only that their training set-derived multiplex pattern could accurately diagnose and classify their validation set, it also demonstrated a proteomic pattern that could predict survival with current best care practices [43]. Current clinical trials are underway in analyzing whether using proteomics data to make clinical decisions is beneficial in NSCLC patients at multiple institutions [44]. In breast cancer, routine screening of tumor samples for hormone receptor status and HER2/neu status as well as newer genomic technologies such as the Oncotype-DX and Mammaprint have aided in decisions regarding both targeted and traditional cytotoxic chemotherapy [45]. It is not unreasonable to think that similar technologies may aid in the treatment decisions relating to ovarian cancer.
The three main limitations to this new paradigm of patient-selected treatment, the targets, the therapies, and rationally designed clinical trials, apply here as well. However, a fourth limitation must also be considered, that modulation of the target is causative to the clinical response to the agent. Important to the first two limitations is a validated and reliable assay that can be applied to a readily attainable specimen. When targeted therapies first became available with the introduction of EGFR tyrosine kinase inhibitors, there was much hope for ovarian cancer patients. EGFR has been shown to be over-expressed in multiple ovarian cancer cell lines and primary tumor samples, and the disregulation of the EGFR signaling pathway has been shown to contribute to the progression of ovarian cancer [46]. Another recent large study used MudPIT analysis to examine the proteomic profile of six ovarian cancer cell lines and revealed many proteins that had not been previously described by gene microarray analysis [47]. These proteins were correlated with an in vitro invasive phenotype and many of the up-regulated proteins were associated with extra-cellular matrix (ECM)-modifying enzymes and their families and signaling partners. Di Michele et al. demonstrated a differential expression profile in cell lines that were sensitive or resistant to paclitaxel therapy [48]. This observation may be important now that there are agents available targeted to proteins involved in modulation of the ECM. Others have shown a time-dependent change in proteomic profiles of IGROV1 cells in response to cisplatin treatment, as well as a differential proteomic profile between sensitive and resistant IGROV1 cell lines [49, 50].
The validity of two-dimensional cell cultures being representative of three-dimensional tumors has been questioned. Grun et al. found measurable differences in proteomic profiles of traditional two-dimensional cultures and the same cell lines grown in a three-dimensional cell culture system [51]. An integrated analysis of primary tumor tissue, including proteomics and genomic array data by Pan et al., revealed a set of proteins that may help define chemotherapy response [52]. A major limitation of this study is the use of only two patient samples: one who immediately progressed through frontline chemotherapy and one who remained stable for 23 months after primary therapy. All of these studies demonstrate possible markers that can be used to guide treatment decisions if active therapies are available and if prospective trials continue to confirm the preclinical results. Other proteomic studies have demonstrated the activation of specific pathways in ovarian cancer cell lines [47, 53] or tumor specimens [54, 55] through detection of phosphorylated intermediates or downstream markers.
Of the second issue regarding integrating proteomics into clinical practice, we have an unfortunate dearth of clinically active targeted therapies. Some studies have sought to assign molecular signatures of sensitive and resistant cell lines and tumor samples to traditional cytotoxic chemotherapy [48–50, 52]. While this process may identify a subset of patients likely to respond to traditional cisplatin or paclitaxel-based therapy, it does not address what treatments are suitable for the remainder of the patient population. Many single agent targeted therapies have been tried for ovarian cancer including agents targeted at angiogenesis, platelet derived growth factor receptor (PDGFR), EGFR, phosphoinositide-3-kinase pathway (PI3K), and Ras/MEK/ERK pathway with varying levels of success (for a recent review on targeted therapies see ref [56]). Many of the studies involving targeted therapies did not involve screening or stratification of patients, to look for particular mutations or pathway activation. This might have missed the benefit of the agent by limiting enrichment for expression of the target but may allow determination of on- vs. off-target activity and address the question of the role of the target in clinical behavior. Even in studies where pathways were dissected, target inhibition did not necessarily correlate with treatment success [54, 55]. Reasons for treatment failure may be non-selected patient populations or that inhibition of a single node is not sufficient to block the signaling network. Some promising studies combined targeted therapies, resulting in enhancement of both toxicity and clinical response. Proteomic analysis of these tumor samples is ongoing [57].
How can proteomics guide us further and predict patient responses to specific therapies? Some tumors are known to demonstrate oncogene addiction to a particular mutation or disregulation within the cell. Most notably, this is true with the BCR-ABL fusion protein and chronic myelogenous leukemia (CML). Considerable treatment success has been observed with the tyrosine kinase inhibitor imatinib inhibiting the ABL tyrosine kinase, PDGFR, and c-kit [58]. Signaling networks are organized into distinct modules and the existence of interchangeable and redundant modules helps lead to complexity in signaling pathways that may overcome the inhibition of single selected targets. This points to the need for multiple targeted therapies in a given patient. Fortunately, there are many inhibitors to kinases and accessory proteins within these modules that are well on their way through clinical trials. New assays that query which of these modules are activated may help guide us to a more effective personalized therapy.
The three main limitations to this new paradigm of patient-selected treatment, the targets, the therapies, and targeted therapy clinical trials have presented clinical investigators and bench researchers with treatment decision challenges. Targets have been associated and validated in a subset of carcinomas, with few validated in ovarian cancer. More trials with well designed and executed translational endpoints to evaluate putative protein biomarkers/endpoints are needed. The value of these endpoints in a trial without clinical benefit may still be present as a mechanism with which to dissect the failure of response. Most prospective clinical trials of targeted agents in ovarian cancer to date have had limited benefit, including when there is evident modulation of the therapeutic target [54, 55, 57]. The first step in expanding our ability to benefit from targeted therapies is the design of trials with proteomic entry and endpoints that will help to understand response, target modulation and effect of combination therapies in ovarian cancer patients.
Proteomics and Clinical Trials
Identifying which patients will benefit from targeted therapy remains a crucial problem for ovarian cancer and other solid tumors. In ovarian cancer, patients who are considered platinum resistant or refractory are often offered single agent cytotoxic chemotherapy with a small but measurable benefit. This patient population, for whom there is little hope of prolonged clinical response, should be the focus of clinical trials investigating targeted therapies. A logical first step in this process should be the proteomic analysis of archived tissue samples from previous clinical trials and samples obtained from ongoing trials. We and others have looked at tumor samples and ascites from treated patients enrolled in clinical trials, but at this time little correlation has been demonstrated between target inhibition and clinical response [39, 41, 54, 55]. Others have also demonstrated unique signatures from ovarian cancer cell lines that may be beneficial in monitoring response to front line therapies [48–50]. A complete list of active clinical trials with proteomic endpoints as the primary objective for the trial is given in Table 1.
Table 1.
Open trials incorporating primary proteomic endpoints.a
| Trial Name | Primary Investigator | Trial Number | Primary Endpoint | Inclusion Criteria | Status |
|---|---|---|---|---|---|
| A Multi-Institutional Study of Proteomic Evaluation of Epithelial Ovarian Cancer, Primary Peritoneal Cancer, and Fallopian Tube Cancer Patients in First Clinical Remission: Development of a Protein Fingerprint Profile of Relapse | Kohn, E.C. | NCT00119353 | Serum proteomic profile with direct comparison to CA125 and creation of a serum repository | Histologically confirmed ovarian epithelial, primary peritoneal cavity, or fallopian tube cancer in first complete remission | Closed to Accrual |
| ACT: Analyzing the Composition of Tears to Identify Cancer (Breast, Ovarian, Colon) | Klimberg, V.S. | NCT00574678 | This study will further evaluate the use of tumor markers (substances in body fluids that may be elevated as a consequence of certain diseases or conditions) in the diagnosis of breast and/or other cancers. | - Female or male, 18–100 years old with newly diagnosed tumor with some part of the amss still present | Currently recruiting |
| Whole Blood Collection Protocol For Ovarian Assay Clinical Trial In Women With Ovarian Tumors | Ueland, F., and Fung, E. | NCT00436189 | Evaluation of OvaRl test in patients with an ovarian mass. | - Subject is female and age 18 years or older with a documented adnexal tumor with planned surgical intervention | Completed |
| Cancer-Related Protein Biomarkers in Blood and Tumor Tissue of Patients With Cancer | Hanash, S.M. | NCT00900094 | Identification of tumor antigens, secreted proteins in tissue and fluid samples and early detection markers | - Scheduled to undergo primary surgical resection or first debulking surgery (prior to any anticancer treatment) for suspected or newly diagnosed cancer, and metastatic or unresectable cancer of multiple cancer types including epithelial ovarian cancer | Open but not recruiting |
| Proteomic Analysis of Serology and Peptides Presented on the HLA Complex of Breast, Ovarian, Colon, Rectal, Gastric and Pancreatic Adenocarcinoma | Ashkenazi, I. | NCT00419627 | Evaluation of proteins presented by the HLA complex of solid tumors. | Patients with adenocarcinoma of breast, colon, rectum, ovary, stomach, pancreas | Open but not recruiting |
| Urine PGE-M in Ovarian Cancer | Rao, G.G. | NCT00900523 | Measurement of urine PGEM Correlation of urine PGEM with tissue COX-1 and COX-2 levels | Diagnosis of known or suspected ovarian cancer | Open but not recruiting. |
| Pelvic Mass Study to Develop Serum Proteomic Profiles (SIGNATURES) for Epithelial Ovarian Cancer Diagnosis and Prognosis | Kohn, E.C., Khleif, S.N., Copeland, L.J. | NCT00238342 | Generate and validate a serum proteomic profile that can predict the presence of invasive ovarian epithelial cancer. | Patients with an abnormal pelvic mass by physical examination or imaging test planning to undergo surgical evaluation within the next 3 weeks | Open but not recruiting |
Trial information from clinicaltrials.gov database filtered for search terms Ovarian Cancer and/or Proteomics with a proteomics endpoint as a primary endpoint.
The next step in the process of developing targeted therapies benefiting selected populations is the evaluation of protein function in a tissue specific context. Genetic aberrations in malignant cells result in alterations of protein expression and function that drive cancer development and progression. Recent identification of these driving forces has allowed cancer therapy to target pathways critical to cell survival. Germline mutations of BRCA 1 and BRCA 2 have been strongly associated with the development of breast, ovarian, and other cancers [59]. Deleterious mutations result in loss of function of BRCA 1 or BRCA 2 which are critically involved in the homologous recombination pathway of double-stranded DNA damage repair [60]. A somewhat paradoxical approach to targeting this pathway is to further impair the ability of a cell to repair single-stranded DNA damage forcing cells to undergo apoptosis. Poly-(ADP-ribose) polymerase (PARP) is a nuclear enzyme that signals single-stranded DNA damage and plays a critical role in base excision repair (BER) pathway [61]. Preclinical data demonstrate that selective sensitivity of BRCA mutation associated cancer cells and cells with defects in other repair pathways such as mismatch repair pathways and microsatellite instability, may also be susceptible to PARP inhibition [62, 63]. Genomic profiling of tumor samples from familial and sporadic BRCA 1 and BRCA 2 mutation carriers as well as non-BRCA associated tumors revealed significant similarities in genomic alterations between the BRCA groups and a subset of non-BRCA mutation carriers [64]. This implies, at a genomic level, there may be other groups of patients that benefit from PARP inhibition.
These results have led for a call for the development of assays for functionality of these repair pathways which can reliably predict those patients who would most likely benefit from the administration of drugs that can inhibit PARP [65]. Recent work by Pirnia et al. looked at the combination of ex-vivo culturing of normal colon and colon cancer tissue from selected patients coupled with RPPA analysis [66] and demonstrated differential expression of PARP and cleaved PARP. Future clinical trials assessing proteomics data from patient samples should move beyond assessment of simple expression and would take into consideration the genetic, epigenetic, and proteomic derangements that drive cancer progression. Successful targeting of these derangements may result in paradigm shifting treatment options for those afflicted by cancer.
Angiogenesis is crucial for tumors to grow beyond a certain size. Single agent anti-angiogenic therapy with bevacizumab or combined therapy with cytotoxic agents has shown early promise [67–69] with response rates in the 20% range even for platinum-resistant patients and with progression-free survival of about 4 months for single agents and 7 months for combined therapy. While there have been many studies implicating VEGFA concentrations being associated with extent of disease, time to progression, and overall survival after therapy with bevacizumab [70], none have addressed the role of patient selection in the trials process. Could a phosphoproteomics pattern emerge that might better select patients who would respond to anti-angiogenic therapy while excluding patients unlikely to benefit and only receive the much higher than expected risk of GI perforation? This is a critical area for further investigation. Genomic analysis of patients treated with bevacizumab and continuous low dose cyclophosphamide demonstrated that a polymorphism within the IL8 gene may predict response to bevacizumab while polymorphisms within the CXCR2 gene and VEGF gene may be predictive of progression-free survival [71]. Targeting angiogenesis with multiple inhibitors also demonstrated early benefit in a phase I trial of bevacizumab with sorafenib [57], albeit at the cost of enhanced toxicity. Forty-six percent of patients with ovarian cancer attained a partial response (compared to 16–21% with single agent bevacizumab [67, 68]). A phase II trial of this combination therapy in relapsed or refractory ovarian cancer is now underway with proteomics studies integrated as a secondary outcome into the trial. A similar trial combined bevacizumab with the EGFR targeted agent erlotinib [72]. This study was terminated secondary to increased incidence of fatal bowel perforations (2/13 patients) and a response rate of ~15%. While proteomic analysis from this study has not been performed, previous studies have demonstrated that inhibition of the EGFR axis was not sufficient to produce a clinically meaningful response [54, 55].
Conclusions and Future Directions
Potential roles for proteomics in the clinical management of ovarian cancer include biomarker discovery and validation, molecular target discovery, therapy decisions, and patient monitoring. Each of these areas presents unique challenges. The discovery of new biomarkers that assist in the screening of at risk populations must take into account the rarity of the disease, as well as the potential complications associated with invasive diagnostic procedures. To date, no reliable screening tests have been confirmed, although CA125 has been used to monitor response to treatment and possible relapse. New data may indicate that relying solely on CA125 concentration in recurrence monitoring may not change overall survival and may expose women to potentially harmful therapies long before their disease would become clinically significant [73]. The application of proteomics technologies to serum, ascites or urine samples has yielded some intriguing preclinical data, but prospective trials have not been undertaken or have not confirmed pre-clinical data. The field of ovarian cancer screening, even in preselected high-risk populations, remains an open field for development.
Proteomics technologies will not only be important in determining which targets and biomarkers should be applied for screening and followed, but will also help in making treatment decisions. Proteomics analysis will allow the testing of global or targeted signaling networks to determine the on-target efficacy of combination therapies that inhibit signaling pathways in series or in parallel. These results can then be correlated with clinical outcome to help drive the design of future studies. Figure 1 shows a general outline for integrating proteomics into clinical management. Each time a clinical specimen is obtained from a patient, the investigator has a unique opportunity to ask important questions about the tumor or body fluid proteome. These results may not only guide initial therapy, they can offer insight into the molecular perturbations caused by specific therapies. If a patient’s cancer were to recur, further proteomic analysis could determine whether similar treatments might still be effective, or it may warrant a change to a new therapeutic option. From the initial screening to treatment decisions through recurrence and monitoring, proteomics will play an important role.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. I. G. was supported by the Research Scholars Program of the Howard Hughes Medical Institute at the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Heintz AP, Odicino F, Maisonneuve P, Quinn MA, et al. Carcinoma of the ovary. FIGO 6th Annual Report on the Results of Treatment in Gynecological Cancer. Int J Gynaecol Obstet. 2006;95(Suppl 1):S161–192. doi: 10.1016/S0020-7292(06)60033-7. [DOI] [PubMed] [Google Scholar]
- 3.Young RC, Walton LA, Ellenberg SS, Homesley HD, et al. Adjuvant therapy in stage I and stage II epithelial ovarian cancer. Results of two prospective randomized trials. N Engl J Med. 1990;322:1021–1027. doi: 10.1056/NEJM199004123221501. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong DK, Bundy B, Wenzel L, Huang HQ, et al. Intraperitoneal cisplatin and paclitaxel in ovarian cancer. N Engl J Med. 2006;354:34–43. doi: 10.1056/NEJMoa052985. [DOI] [PubMed] [Google Scholar]
- 5.Markman M, Walker JL. Intraperitoneal chemotherapy of ovarian cancer: a review, with a focus on practical aspects of treatment. J Clin Oncol. 2006;24:988–994. doi: 10.1200/JCO.2005.05.2456. [DOI] [PubMed] [Google Scholar]
- 6.Petricoin EF, Zoon KC, Kohn EC, Barrett JC, Liotta LA. Clinical proteomics: translating benchside promise into bedside reality. Nat Rev Drug Discov. 2002;1:683–695. doi: 10.1038/nrd891. [DOI] [PubMed] [Google Scholar]
- 7.Tchabo NE, Liel MS, Kohn EC. Applying proteomics in clinical trials: assessing the potential and practical limitations in ovarian cancer. Am J Pharmacogenomics. 2005;5:141–148. doi: 10.2165/00129785-200505030-00001. [DOI] [PubMed] [Google Scholar]
- 8.Kuk C, Kulasingam V, Gunawardana CG, Smith CR, et al. Mining the ovarian cancer ascites proteome for potential ovarian cancer biomarkers. Mol Cell Proteomics. 2009;8:661–669. doi: 10.1074/mcp.M800313-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petri AL, Simonsen AH, Yip TT, Hogdall E, et al. Three new potential ovarian cancer biomarkers detected in human urine with equalizer bead technology. Acta Obstet Gynecol Scand. 2009;88:18–26. doi: 10.1080/00016340802443830. [DOI] [PubMed] [Google Scholar]
- 10.Simpkins F, Czechowicz JA, Liotta L, Kohn EC. SELDI-TOF mass spectrometry for cancer biomarker discovery and serum proteomic diagnostics. Pharmacogenomics. 2005;6:647–653. doi: 10.2217/14622416.6.6.647. [DOI] [PubMed] [Google Scholar]
- 11.Tadano T, Kanoh M, Kondoh H, Matsumoto M, et al. Kinetic analysis of bile acids in the feces of colorectal cancer patients by gas chromatography-mass spectrometry (GC-MS) Rinsho Byori. 2007;55:417–427. [PubMed] [Google Scholar]
- 12.Schuhmann MU, Zucht HD, Nassimi R, Heine G, et al. Peptide screening of cerebrospinal fluid in patients with glioblastoma multiforme. Eur J Surg Oncol. 2009 doi: 10.1016/j.ejso.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 13.Annunziata CM, Azad N, Dhamoon AS, Whiteley G, Kohn EC. Ovarian cancer in the proteomics era. Int J Gynecol Cancer. 2008;18(Suppl 1):1–6. doi: 10.1111/j.1525-1438.2007.01096.x. [DOI] [PubMed] [Google Scholar]
- 14.Goff BA, Mandel L, Muntz HG, Melancon CH. Ovarian carcinoma diagnosis. Cancer. 2000;89:2068–2075. doi: 10.1002/1097-0142(20001115)89:10<2068::aid-cncr6>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 15.Daly M, Obrams GI. Epidemiology and risk assessment for ovarian cancer. Semin Oncol. 1998;25:255–264. [PubMed] [Google Scholar]
- 16.Tworoger SS, Fairfield KM, Colditz GA, Rosner BA, Hankinson SE. Association of oral contraceptive use, other contraceptive methods, and infertility with ovarian cancer risk. Am J Epidemiol. 2007;166:894–901. doi: 10.1093/aje/kwm157. [DOI] [PubMed] [Google Scholar]
- 17.Jordan SJ, Whiteman DC, Purdie DM, Green AC, Webb PM. Does smoking increase risk of ovarian cancer? A systematic review. Gynecol Oncol. 2006;103:1122–1129. doi: 10.1016/j.ygyno.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 18.Olsen CM, Bain CJ, Jordan SJ, Nagle CM, et al. Recreational physical activity and epithelial ovarian cancer: a case-control study, systematic review, and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2007;16:2321–2330. doi: 10.1158/1055-9965.EPI-07-0566. [DOI] [PubMed] [Google Scholar]
- 19.Olsen CM, Green AC, Whiteman DC, Sadeghi S, et al. Obesity and the risk of epithelial ovarian cancer: a systematic review and meta-analysis. Eur J Cancer. 2007;43:690–709. doi: 10.1016/j.ejca.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 20.Nossov V, Amneus M, Su F, Lang J, et al. The early detection of ovarian cancer: from traditional methods to proteomics. Can we really do better than serum CA-125? Am J Obstet Gynecol. 2008;199:215–223. doi: 10.1016/j.ajog.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 21.Jacobs IJ, Menon U. Progress and challenges in screening for early detection of ovarian cancer. Mol Cell Proteomics. 2004;3:355–366. doi: 10.1074/mcp.R400006-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Sasaroli D, Coukos G, Scholler N. Beyond CA125: the coming of age of ovarian cancer biomarkers. Are we there yet? Biomark Med. 2009;3:275–288. doi: 10.2217/bmm.09.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schummer M, Ng WV, Bumgarner RE, Nelson PS, et al. Comparative hybridization of an array of 21,500 ovarian cDNAs for the discovery of genes overexpressed in ovarian carcinomas. Gene. 1999;238:375–385. doi: 10.1016/s0378-1119(99)00342-x. [DOI] [PubMed] [Google Scholar]
- 24.Moore RG, Brown AK, Miller MC, Skates S, et al. The use of multiple novel tumor biomarkers for the detection of ovarian carcinoma in patients with a pelvic mass. Gynecol Oncol. 2008;108:402–408. doi: 10.1016/j.ygyno.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 25.Moore RG, McMeekin DS, Brown AK, DiSilvestro P, et al. A novel multiple marker bioassay utilizing HE4 and CA125 for the prediction of ovarian cancer in patients with a pelvic mass. Gynecol Oncol. 2009;112:40–46. doi: 10.1016/j.ygyno.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Visintin I, Feng Z, Longton G, Ward DC, et al. Diagnostic markers for early detection of ovarian cancer. Clin Cancer Res. 2008;14:1065–1072. doi: 10.1158/1078-0432.CCR-07-1569. [DOI] [PubMed] [Google Scholar]
- 27.Greene MH, Feng Z, Gail MH. The importance of test positive predictive value in ovarian cancer screening. Clin Cancer Res. 2008;14:7574. doi: 10.1158/1078-0432.CCR-08-2232. author reply 7577–7579. [DOI] [PubMed] [Google Scholar]
- 28.Menon U, Gentry-Maharaj A, Hallett R, Ryan A, et al. Sensitivity and specificity of multimodal and ultrasound screening for ovarian cancer, and stage distribution of detected cancers: results of the prevalence screen of the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS) Lancet Oncol. 2009;10:327–340. doi: 10.1016/S1470-2045(09)70026-9. [DOI] [PubMed] [Google Scholar]
- 29.Crum CP, Drapkin R, Kindelberger D, Medeiros F, et al. Lessons from BRCA: the tubal fimbria emerges as an origin for pelvic serous cancer. Clin Med Res. 2007;5:35–44. doi: 10.3121/cmr.2007.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crum CP, Drapkin R, Miron A, Ince TA, et al. The distal fallopian tube: a new model for pelvic serous carcinogenesis. Curr Opin Obstet Gynecol. 2007;19:3–9. doi: 10.1097/GCO.0b013e328011a21f. [DOI] [PubMed] [Google Scholar]
- 31.Lee Y, Miron A, Drapkin R, Nucci MR, et al. A candidate precursor to serous carcinoma that originates in the distal fallopian tube. J Pathol. 2007;211:26–35. doi: 10.1002/path.2091. [DOI] [PubMed] [Google Scholar]
- 32.Tirumalai RS, Chan KC, Prieto DA, Issaq HJ, et al. Characterization of the low molecular weight human serum proteome. Mol Cell Proteomics. 2003;2:1096–1103. doi: 10.1074/mcp.M300031-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Faca V, Pitteri SJ, Newcomb L, Glukhova V, et al. Contribution of protein fractionation to depth of analysis of the serum and plasma proteomes. J Proteome Res. 2007;6:3558–3565. doi: 10.1021/pr070233q. [DOI] [PubMed] [Google Scholar]
- 34.Lopez MF, Mikulskis A, Kuzdzal S, Golenko E, et al. A novel, high-throughput workflow for discovery and identification of serum carrier protein-bound peptide biomarker candidates in ovarian cancer samples. Clin Chem. 2007;53:1067–1074. doi: 10.1373/clinchem.2006.080721. [DOI] [PubMed] [Google Scholar]
- 35.Petricoin EF, Ardekani AM, Hitt BA, Levine PJ, et al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359:572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 36.Hong YJ, Wang XD, Shen D, Zeng S. Discrimination analysis of mass spectrometry proteomics for ovarian cancer detection. Acta Pharmacol Sin. 2008;29:1240–1246. doi: 10.1111/j.1745-7254.2008.00861.x. [DOI] [PubMed] [Google Scholar]
- 37.Montazery-Kordy H, Miran-Baygi MH, Moradi MH. A data-mining approach to biomarker identification from protein profiles using discrete stationary wavelet transform. J Zhejiang Univ Sci B. 2008;9:863–870. doi: 10.1631/jzus.B0820163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu C, Shea N, Rucker S, Harvey L, et al. Proteomic patterns for classification of ovarian cancer and CTCL serum samples utilizing peak pairs indicative of post-translational modifications. Proteomics. 2007;7:4045–4052. doi: 10.1002/pmic.200601044. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Zhang X, Ge X, Guo H, et al. Proteomic studies of early-stage and advanced ovarian cancer patients. Gynecol Oncol. 2008;111:111–119. doi: 10.1016/j.ygyno.2008.06.031. [DOI] [PubMed] [Google Scholar]
- 40.Wu B, Abbott T, Fishman D, McMurray W, et al. Ovarian Cancer Classification based on Mass Spectrometry Analysis of Sera. Cancer Inform. 2007;2:123–132. [PMC free article] [PubMed] [Google Scholar]
- 41.Davidson B, Espina V, Steinberg SM, Florenes VA, et al. Proteomic analysis of malignant ovarian cancer effusions as a tool for biologic and prognostic profiling. Clin Cancer Res. 2006;12:791–799. doi: 10.1158/1078-0432.CCR-05-2516. [DOI] [PubMed] [Google Scholar]
- 42.Gortzak-Uzan L, Ignatchenko A, Evangelou AI, Agochiya M, et al. A proteome resource of ovarian cancer ascites: integrated proteomic and bioinformatic analyses to identify putative biomarkers. J Proteome Res. 2008;7:339–351. doi: 10.1021/pr0703223. [DOI] [PubMed] [Google Scholar]
- 43.Yanagisawa K, Shyr Y, Xu BJ, Massion PP, et al. Proteomic patterns of tumour subsets in non-small-cell lung cancer. Lancet. 2003;362:433–439. doi: 10.1016/S0140-6736(03)14068-8. [DOI] [PubMed] [Google Scholar]
- 44.Pao W, Kris MG, Iafrate AJ, Ladanyi M, et al. Integration of molecular profiling into the lung cancer clinic. Clin Cancer Res. 2009;15:5317–5322. doi: 10.1158/1078-0432.CCR-09-0913. [DOI] [PubMed] [Google Scholar]
- 45.Dunn L, Demichele A. Genomic predictors of outcome and treatment response in breast cancer. Mol Diagn Ther. 2009;13:73–90. doi: 10.1007/BF03256317. [DOI] [PubMed] [Google Scholar]
- 46.Maihle NJ, Baron AT, Barrette BA, Boardman CH, et al. EGF/ErbB receptor family in ovarian cancer. Cancer Treat Res. 2002;107:247–258. doi: 10.1007/978-1-4757-3587-1_11. [DOI] [PubMed] [Google Scholar]
- 47.Sodek KL, Evangelou AI, Ignatchenko A, Agochiya M, et al. Identification of pathways associated with invasive behavior by ovarian cancer cells using multidimensional protein identification technology (MudPIT) Mol Biosyst. 2008;4:762–773. doi: 10.1039/b717542f. [DOI] [PubMed] [Google Scholar]
- 48.Di Michele M, Della Corte A, Cicchillitti L, Del Boccio P, et al. A proteomic approach to paclitaxel chemoresistance in ovarian cancer cell lines. Biochim Biophys Acta. 2009;1794:225–236. doi: 10.1016/j.bbapap.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 49.Le Moguen K, Lincet H, Deslandes E, Hubert-Roux M, et al. Comparative proteomic analysis of cisplatin sensitive IGROV1 ovarian carcinoma cell line and its resistant counterpart IGROV1-R10. Proteomics. 2006;6:5183–5192. doi: 10.1002/pmic.200500925. [DOI] [PubMed] [Google Scholar]
- 50.Le Moguen K, Lincet H, Marcelo P, Lemoisson E, et al. A proteomic kinetic analysis of IGROV1 ovarian carcinoma cell line response to cisplatin treatment. Proteomics. 2007;7:4090–4101. doi: 10.1002/pmic.200700231. [DOI] [PubMed] [Google Scholar]
- 51.Grun B, Benjamin E, Sinclair J, Timms JF, et al. Three-dimensional in vitro cell biology models of ovarian and endometrial cancer. Cell Prolif. 2009;42:219–228. doi: 10.1111/j.1365-2184.2008.00579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pan S, Cheng L, White JT, Lu W, et al. Quantitative proteomics analysis integrated with microarray data reveals that extracellular matrix proteins, catenins, and p53 binding protein 1 are important for chemotherapy response in ovarian cancers. OMICS. 2009;13:345–354. doi: 10.1089/omi.2009.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dai L, Li C, Shedden KA, Misek DE, Lubman DM. Comparative proteomic study of two closely related ovarian endometrioid adenocarcinoma cell lines using cIEF fractionation and pathway analysis. Electrophoresis. 2009;30:1119–1131. doi: 10.1002/elps.200800505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Posadas EM, Kwitkowski V, Kotz HL, Espina V, et al. A prospective analysis of imatinib-induced c-KIT modulation in ovarian cancer: a phase II clinical study with proteomic profiling. Cancer. 2007;110:309–317. doi: 10.1002/cncr.22757. [DOI] [PubMed] [Google Scholar]
- 55.Posadas EM, Liel MS, Kwitkowski V, Minasian L, et al. A phase II and pharmacodynamic study of gefitinib in patients with refractory or recurrent epithelial ovarian cancer. Cancer. 2007;109:1323–1330. doi: 10.1002/cncr.22545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yap TA, Carden CP, Kaye SB. Beyond chemotherapy: targeted therapies in ovarian cancer. Nat Rev Cancer. 2009;9:167–181. doi: 10.1038/nrc2583. [DOI] [PubMed] [Google Scholar]
- 57.Azad NS, Posadas EM, Kwitkowski VE, Steinberg SM, et al. Combination targeted therapy with sorafenib and bevacizumab results in enhanced toxicity and antitumor activity. J Clin Oncol. 2008;26:3709–3714. doi: 10.1200/JCO.2007.10.8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krause DS, Van Etten RA. Tyrosine kinases as targets for cancer therapy. N Engl J Med. 2005;353:172–187. doi: 10.1056/NEJMra044389. [DOI] [PubMed] [Google Scholar]
- 59.Easton DF, Bishop DT, Ford D, Crockford GP. Genetic linkage analysis in familial breast and ovarian cancer: results from 214 families. The Breast Cancer Linkage Consortium. Am J Hum Genet. 1993;52:678–701. [PMC free article] [PubMed] [Google Scholar]
- 60.Ford D, Easton DF, Bishop DT, Narod SA, Goldgar DE. Risks of cancer in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Lancet. 1994;343:692–695. doi: 10.1016/s0140-6736(94)91578-4. [DOI] [PubMed] [Google Scholar]
- 61.Wooster R, Neuhausen SL, Mangion J, Quirk Y, et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science. 1994;265:2088–2090. doi: 10.1126/science.8091231. [DOI] [PubMed] [Google Scholar]
- 62.Sharan SK, Morimatsu M, Albrecht U, Lim DS, et al. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 63.Fong PC, Boss DS, Yap TA, Tutt A, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 64.Stefansson OA, Jonasson JG, Johannsson OT, Olafsdottir K, et al. Genomic profiling of breast tumours in relation to BRCA abnormalities and phenotypes. Breast Cancer Res. 2009;11:R47. doi: 10.1186/bcr2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tutt A, Garber MRJE, Domchek S, Audeh MW, Weitzel JN, Friedlander M, Carmichael J. Phase II trial of the oral PARP inhibitor olaparib in BRCA-deficient advanced breast cancer. J Clin Oncol. 2009;27 [Google Scholar]
- 66.Pirnia F, Pawlak M, Thallinger GG, Gierke B, et al. Novel functional profiling approach combining reverse phase protein microarrays and human 3-D ex vivo tissue cultures: expression of apoptosis-related proteins in human colon cancer. Proteomics. 2009;9:3535–3548. doi: 10.1002/pmic.200800159. [DOI] [PubMed] [Google Scholar]
- 67.Burger RA, Sill MW, Monk BJ, Greer BE, Sorosky JI. Phase II trial of bevacizumab in persistent or recurrent epithelial ovarian cancer or primary peritoneal cancer: a Gynecologic Oncology Group Study. J Clin Oncol. 2007;25:5165–5171. doi: 10.1200/JCO.2007.11.5345. [DOI] [PubMed] [Google Scholar]
- 68.Cannistra SA, Matulonis UA, Penson RT, Hambleton J, et al. Phase II study of bevacizumab in patients with platinum-resistant ovarian cancer or peritoneal serous cancer. J Clin Oncol. 2007;25:5180–5186. doi: 10.1200/JCO.2007.12.0782. [DOI] [PubMed] [Google Scholar]
- 69.Garcia AA, Hirte H, Fleming G, Yang D, et al. Phase II clinical trial of bevacizumab and low-dose metronomic oral cyclophosphamide in recurrent ovarian cancer: a trial of the California, Chicago, and Princess Margaret Hospital phase II consortia. J Clin Oncol. 2008;26:76–82. doi: 10.1200/JCO.2007.12.1939. [DOI] [PubMed] [Google Scholar]
- 70.Burger RA. Experience with bevacizumab in the management of epithelial ovarian cancer. J Clin Oncol. 2007;25:2902–2908. doi: 10.1200/JCO.2007.12.1509. [DOI] [PubMed] [Google Scholar]
- 71.Schultheis AM, Lurje G, Rhodes KE, Zhang W, et al. Polymorphisms and clinical outcome in recurrent ovarian cancer treated with cyclophosphamide and bevacizumab. Clin Cancer Res. 2008;14:7554–7563. doi: 10.1158/1078-0432.CCR-08-0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nimeiri HS, Oza AM, Morgan RJ, Friberg G, et al. Efficacy and safety of bevacizumab plus erlotinib for patients with recurrent ovarian, primary peritoneal, and fallopian tube cancer: a trial of the Chicago, PMH, and California Phase II Consortia. Gynecol Oncol. 2008;110:49–55. doi: 10.1016/j.ygyno.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rustin GJ, van ber Berg ME. A randomized trial in ovarian cancer (OC) of early treatment of relapse based on CA125 level alone versus delayed treatment based on conventional clinical indicators (MRC OV05/EORTC 55955 trials) Journal of Clinical Oncology. 2009;27 Abstract 1. [Google Scholar]