Abstract
Cationic polymers created through recombinant DNA technology have the potential to fill a void in the area of gene delivery. The recombinant cationic polymers to be discussed here are amino acid based polymers synthesized in E.coli with the purpose to not only address the major barriers to efficient gene delivery but offer safety, biodegradability, targetability and cost-effectiveness. This review helps the readers to get a better understanding about the evolution of recombinant cationic polymers; and the potential advantages that they could offer over viral and synthetic non-viral vectors for gene delivery. It also discusses some of the major challenges that must be addressed in future studies to turn recombinant polymers into clinically effective gene delivery systems. Recent advances with the biopolymer design suggest that this emerging new class of gene delivery systems has the potential to address some of the major barriers to efficient, safe and cost-effective gene therapy.
Keywords: Gene delivery, targeted therapy, biopolymer, non-viral, bioinspired, fusion peptides, nanoparticles
1. Introduction
A major hurdle in successful gene therapy is the lack of a suitable gene delivery vehicle (vector). The main features of a suitable vector for clinical applications are low cytotoxicity/immunogenicity, high efficiency, tissue specificity and cost effectiveness. While there has been significant progress in vector development for various gene therapy needs [1, 2], there is no single vector that is equipped with all of these essential features. As a result, the need for research into innovative and novel delivery vehicles remains.
In the past decade, there has been significant progress in the development of viral vectors which gives them a significant edge over the non-viral alternative [2]. Besides high transduction efficiency, one critically important advantage of viral vectors is the single DNA packaging capability. This level of control over DNA packaging process does not exist for non-viral vectors because they rely on an uncontrolled vector/DNA self-assembly process to package DNA into condensed nanosized particles. While viruses reproducibly package a single DNA chain at their core, non-viral vectors tend to form particles with multiple chains of DNA in each particle. Consequently, the reproducibility of the uniform nanosize particle formation and gene transfer process could be undermined due to batch to batch variations. This shortcoming may be resolved with the emergence of new technologies such as “porous nanocontainers” [3]. Despite these advantages, the use of viral vectors has been limited due to the difficulties associated with production in high titers, safety concerns, and constraints related to tissue targeting. To fulfill the deficiencies associated with viral vectors, synthetic non-viral vectors such as cationic lipids and polymers have emerged as potential safer alternatives.
While cationic lipids afford relatively high gene transfer efficiency, reproducible large scale production methods and vector related cytotoxicity remain as major points of concern [4]. One method to reduce the non-specific cytotoxicity of the lipid-based vectors is the attachment of targeting peptides or polyethylene glycol (PEG) to their surface. This will reduce the surface positive charge, minimize non-specific toxicity and enhance cellular uptake [5, 6]. Unfortunately, reproducible ligand attachment process is still a significant challenge, due primarily to the thermodynamically driven limitations of the chemical synthetic methods. Reproducibility of ligand attachment process is critically important as it impacts ligand density on the nanoparticle surface which in turn affects their binding affinity toward receptors. The importance of ligand density and its impact on receptor binding and gene transfer efficiency have been discussed in details elsewhere [7].
Another alternative to viral vectors are cationic polymers which are moderately biocompatible but suffer from poor gene-transfer efficiency [8]. One polymer-based non-viral system that has shown some degree of success is polyethyleneimine (PEI) [9]. PEI is considered a versatile polymeric vector that condenses plasmid DNA efficiently with the ability to deliver exogenes to various mammalian cells [10, 11]. Unfortunately, due to its non-biodegradable property, uncomplexed free PEI or free PEI generated after DNA delivery can cause immediate and delayed cytotoxicity [4].
In summary, non-viral gene delivery vectors are still in the developmental stage and remain marred by poor gene transfer rates, lack of reproducibility, heterogeneity and cytotoxicity. Despite immunogenicity, high costs and other safety concerns, viruses still remain the most widely used vectors for gene delivery owing primarily to their high rate of gene transfer efficiency and ability to bioengineer the architecture at the molecule level for various gene therapy needs.
2. Recombinant Polymers as Potential Invention Solution
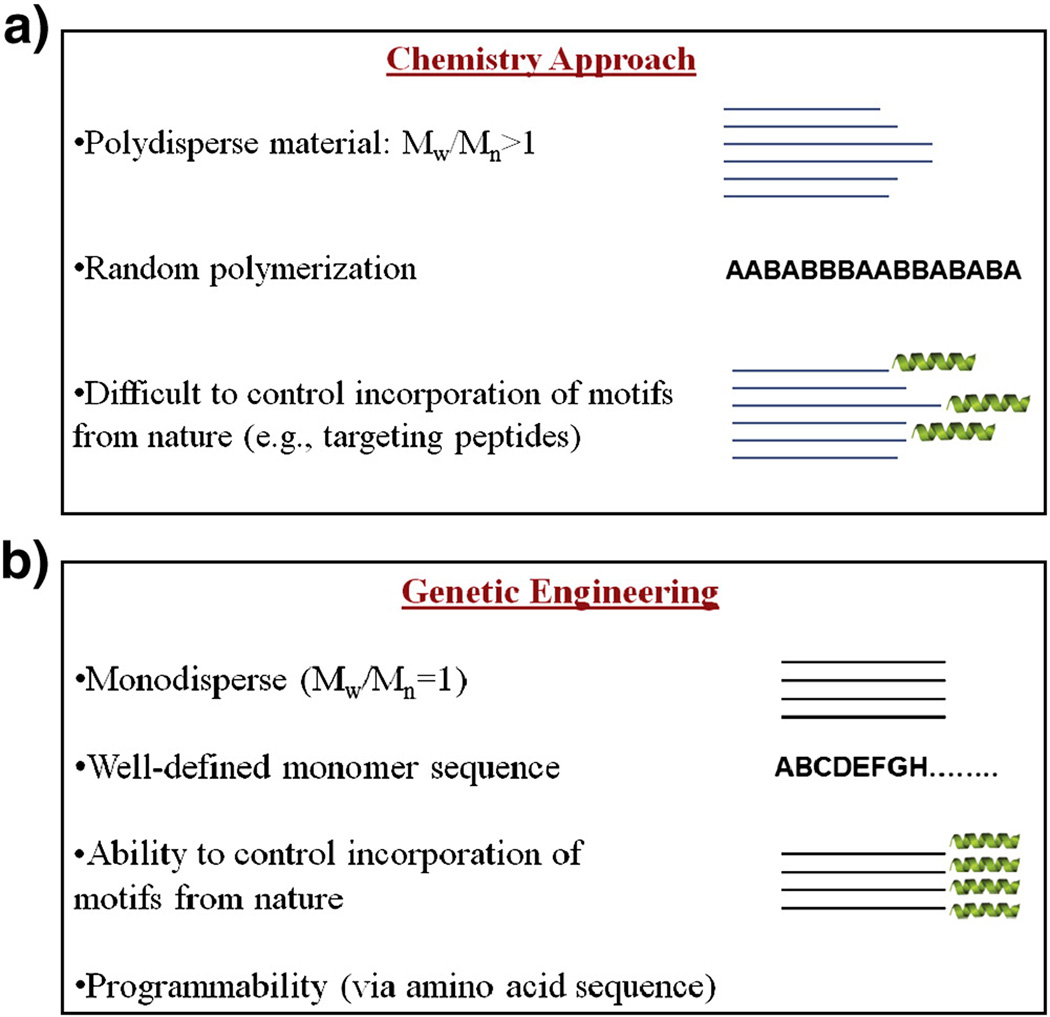
As the inability to explain or predict transfection efficiency of non-viral vectors results partly from insufficient understanding of the intracellular processes, development of a class of biomaterials is required that provides the possibility of performing reliable structure/activity relationship studies. Such studies would help the scientists to better understand the rate limiting steps to each specific vector, devise new approaches to overcome the deficiencies and develop non-viral vectors that could potentially be more efficient than the viruses. One class of biomaterials that allows precise correlation of structure with function is recombinant polymers (biopolymers). The advent of recombinant DNA technology allowed the design and development of recombinant polymers for use in drug/gene delivery offering several advantages over more conventional methods (Fig. 1). Synthetic methods of polymer production utilize conventional thermodynamically-driven chemical synthesis techniques which result in heterogeneous products manifested by molecular weight distribution. If biological motifs (e.g., targeting peptides, nuclear localization signals, etc.) are to be incorporated in the polymer structure, conjugation followed by purification steps are required which could significantly add to the costs. In contrast, amino acid based polymers are synthesized using genetic engineering techniques in biological systems (e.g., E.coli) resulting in homogeneous biopolymers with specific compositions where functions can be dictated via amino acid sequence (programmability) [12]. This allows multiple functionalities to be incorporated onto a single biopolymer backbone by merely changing the gene encoding the amino acid instructions. This concept is demonstrated in section 2.3 (below) where a single chain biopolymer can perform several distinct tasks sequentially. In terms of safety, the endotoxins (structural component in the bacteria cell wall) can be simply removed during the washing steps of the biopolymer purification process by affinity chromatography. Given the fact that there is no need for the removal of toxic solvents or un-reacted monomers, such biopolymers could be just as, if not more cost-effective than synthetic polymers.
Fig. 1.
Comparison between chemical and recombinant methods of targeted polymer production for gene delivery.
When compared to viral vectors, biopolymers can be made with significantly lower costs and safety concerns. For example, virus production processes must be certified and operated within Biosafety Level 2 and 3 (BSL2/3) guidelines, whereas recombinant biopolymers can be produced in large amounts in BSL1 facilities. Furthermore, biopolymers are produced in E.coli which is among the most efficient and cost-effective methods of protein production [13, 14]. This is in contrast to the methods of virus production which require painstaking and time-consuming processes to reach high titers (>108 pfu/ml) suitable for clinical applications.
An alternative to biological synthesis of biopolymers is synthetic peptide production. This approach generally relies on organic chemistry solid phase synthesis techniques. Solid phase synthesis is often limited by reaction yields and thus restrains the length of the peptide that can be made. Longer peptides can be made by chemical linkage of these shorter fragments but this is usually a prohibitively expensive process when large quantities are needed [15].
In the past, technological hurdles associated with gene cloning, sequencing and protein expression restricted the supply of readily available recombinant polymers. Due to the advancements in the field of recombinant DNA technologies in recent years, these technological obstacles are overcome and recombinant polymers can be made in a cost-effective manner to fill the void in the gene delivery arena. This review highlights the history and evolution of recombinant cationic polymers ranging from simple bi-functional to the more complex multifunctional constructs. For the purposes of this article, word “biopolymer” refers only to recombinant amino acid based polymers in order to differentiate from synthetic polymers (e.g., PEI) and natural polymers (e.g., chitosan).
2.1. Bi-functional Biopolymers
Aris et al. (2000) were among the first to report the genetic engineering of a gene delivery system, namely 249AL, composed of a cationic lysine oligomer (K10) fused to a β-galactosidase-derived protein displaying RGD cell attachment peptide [16]. The role of K10 was to condense plasmid DNA (pDNA) and the RGD was for binding to the αVβ3 integrins on the cell surfaces [17]. It was also speculated, but not shown in this report that the β-galactosidase could act as a DNA protector as well as a nuclear targeting motif.
The ability of the gene carrier to mediate gene expression was examined by complexing 249AL with pDNA encoding luciferase reporter gene and transfecting CaCo2 cells. While the total luciferase gene expression was reported, the percentage of transfected cells was not measured. Because the 249AL was designed to be targeted, the percent transfected cell in addition to the total gene expression could provide a better understanding of the efficiency of the gene delivery system. The transfection efficiency of 249AL was compared with lipofectamine and shown to be significantly less efficient. This was expected as 249AL is not well-equipped to escape from the endosomal compartments. One important point worth emphasizing is that comparing gene transfer efficiency of targeted vectors such as 249AL with non-targeted ones (e.g., lipofectamine) may not be appropriate as they internalize via entirely different pathways. As a result, the efficiency of non-targeted vectors should be discussed in the context of each cell type rather than generalization to all mammalian cells. This importance is discussed in more detail elsewhere [18]. Nonetheless, this study was among the early reports on the use of genetically engineering techniques to make a fusion vector with gene delivery application.
In a similar approach, Furgeson’s group (2008) reported the development of a recombinant elastin-based cationic diblock biopolymer for gene delivery [19]. The biopolymer consisted of a cationic oligomer block (VGK8G) fused to a thermoresponsive elastinlike polymer (ELP) with 60 repeats of (VPGXG) where X is V, A, or G in a 5:2:3 ratio (Fig. 2). They utilized a recursive directional ligation method to synthesize the gene which is a pseudo biosynthetic route achieved in bacterial cell culture [19, 20]. ELPs are biocompatible and undergo a rapid reversible phase transition at a transition temperature which is a function of the type of guest residue, the ionic state, the molecular weight among other factors [21, 22]. This system was specifically designed for use in hyperthermic gene therapy. The biopolymer [K8-ELP(1–60)] was genetically engineered in E.coli and characterized. It was shown that it can not only condense pDNA encoding green fluorescent protein (pEGFP) into nanosize particles but responds to heat and goes through thermal transition. The results of the MCF-7 cell transfection studies demonstrated the ability of the system to mediate gene expression. Unfortunately, the transfection of the cells was only visualized and no total gene expression or percent transfected cells were reported. Because the transfection studies were performed in the presence of chloroquine, it can be deduced that the gene delivery system was not able to escape from the endosomal compartments efficiently. This was expected as [K8-ELP(1–60)] was not equipped with any endosomolytic motif to facilitate its escape from endosomes. Before proceeding further in clinical administration as a gene therapy vector, the efficiency, ability to target, and the thermal transition point for this type of recombinant cationic polymer need to be optimized.
Fig. 2.
The chemical structure of biosynthesized K8-ELP(1–60) diblock copolymers and preparation of intelligent thermosensitive nonviral gene vectors. Reproduced with permission from [19].
2.2. Tri-functional Biopolymers
In an attempt to overcome the endosomal barrier and also provide targetability, Ghandehari’s group (2006) reported the structure of the first recombinant cationic biopolymer with tandem repeating units composed of lysine (K) and histidine (H) residues fused to fibroblast growth factor 2 (FGF2) [23]. The biopolymer with the general structure of (KHKHKHKHKK)6-FGF2 or in short dKH-FGF2, contains 36 lysine residues (K) in the dKH segment to condense pDNA, and 24 histidine residues (H) to promote endosomal escape via the proton sponge effect [24]. Addition of FGF2 to the biopolymer was expected to give affinity towards FGFR expressing cells such as T47D (breast cancer) and NIH3T3 (fibroblasts). As a starting point, the lysine residues in the dKH tail (i.e., KHKHKHKHKK) were arranged as dispersed, while keeping the lysine to histidine ratio constant at 60:40. The results demonstrated that the biopolymer was able to condense DNA into nanosize particles [23]. It was also shown that the FGF2 motif in dKH-FGF2 was functional and could induce significant cell proliferation, whereas the dKH segment alone did not show any cell proliferative activity. While the result of the transfection efficiency studies showed targeted gene transfer via FGFR, the biopolymer efficiency was suboptimal and required further development.
This suboptimal transfection efficiency prompted further characterization dKH-FGF2 and modification of its structure to improve efficiency. It was hypothesized that by changing the arrangement of KH residues in the KHKHKHKHKK repeating units and organizing them in clusters, the pDNA condensation efficiency will be improved resulting in more stable nanocarriers with higher transfection efficiency [25]. This hypothesis was motivated by the fact that natural motifs such as histones and adenovirus μ peptide which are known to condense DNA efficiently have their lysine and arginine residues arranged in clusters [26–28]. To test the hypothesis, the following vector was designed in which the lysine residues are organized in clusters: (KKKHHHHKKK)6-FGF, namely cKH-FGF2. To be consistent with dKH-FGF2, the K:H ratio was kept constant at 60:40. The results of this study demonstrated that cKH-FGF2 was able to condense pDNA into more compact nanoparticles in comparison to dKH-FGF2 resulting in higher gene transfer efficiency (Fig. 3) [25]. Although efficiency was improved, the cell transfection studies in the presence of chloroquine revealed that the biopolymer still could not efficiently escape from the endosomes highlighting the fact that even such large numbers of histidines in the biopolymer sequence are not sufficient to efficiently disrupt endosome membranes.
Fig. 3.
Qualitative and quantitative representation of the transfection efficiency of cKH-FGF2 and dKH-FGF2 in different cell lines. (a) Representative confocal images of NIH3T3 (left) and T47D (right) cells transfected with cKH-FGF2/pEGFP complexes. The green dots are the cells expressing green fluorescent protein (GFP). (b) Percentage of cells transfected with cKH-FGF2/ pEGFP (closed bar) and dKH-FGF2/pEGFP (open bar). Cells were transfected with vectors in DMEM supplemented with serum. The percent transfected cells with cKH-FGF2 in NIH3T3 and T47D was 41±4 and 28±5, respectively (mean±SD, n=9). The percentages of transfected cells with dKH-FGF2 in NIH3T3 and T47D were 9±3 and 7±2, respectively. Reproduced with permission from [25].
The two studies on dKH-FGF2 and cKH-FGF2 vectors demonstrate that the vector architecture and not just amino acid composition plays a significant role in gene transfer. This highlights an important point that random copolymerization methods used to make synthetic polymers results in a myriad of structures making structure activity relationships hard to elucidate. The homogeneous nature of recombinant polymers makes them a more appropriate tool for examining the contribution of architecture to gene delivery and its effect on efficiency.
One important point that needs to be mentioned is that the production of dKH-FGF2 and cKH-FGF2 peptides in soluble form in E. coli was problematic. For example, expression of ca. 800 µg of soluble cKH-FGF2 in 1 liter of cell culture yielded ca. 100 µg of purified biopolymer after purification process. While this level of biopolymer expression in E. coli can support the in vitro structure/activity relationship studies, it is not sufficient for in vivo studies. To overcome the large scale biopolymer production problem as well as improving escape from the endosomal compartments, the next generation of biopolymers was designed which is discussed in the following section.
2.3. Multi-functional Biopolymers
For a gene carrier to successfully overcome intracellular barriers and reach the nucleus of target cells, it must accurately mimic viral vectors. This includes protecting the DNA from endonucleases by condensation, binding to the surface receptors on the target cells followed by internalization, escape from endosomes into cytosol, rapid shuttling of DNA toward the nucleus via microtubules, entering the cell nucleus and mediating gene expression. From the studies discussed above, each biopolymer overcame some of the abovementioned barriers. However, no single system had yet been designed to systematically overcome all hurdles.
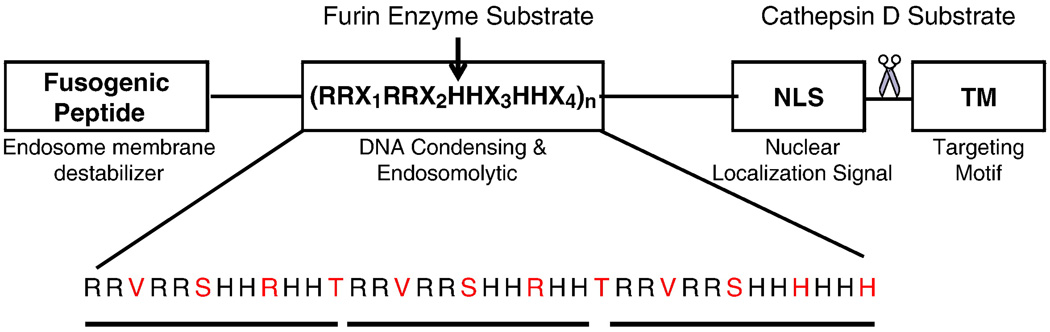
To date, there has been only one report on the biosynthesis and characterization of a cationic biopolymer with ability to perform multiple functions. The next generation of biopolymers was designed to push the boundaries of biopolymer development even further and solve the problems associated with production, formulation development, biodegradability, targetability and efficient intracellular trafficking toward the cell nucleus. Hatefi’s group recently described the genetic engineering of a prototype multi-functional vector that incorporates multiple domains, each with a distinct function, onto a single biopolymer backbone [29] (Fig. 4). This biopolymer, in contrast to viruses which allot different functions to multiple peptide subunits, embeds several functional domains onto a single module (biopolymer chain). The reason behind fusing all the functional domains into one single biopolymer chain is to drastically reduce the number of variables that needs to be optimized in structure/activity and formulation development studies. It features a unique cationic domain with repeating units of arginine-histidine (RH) with the general structure of (RRXRRXHHXHHX)n, where X is any amino acid except D and E, and n is the number of repeating units. In the repeating units, serine and threonine residues were engineered in between histidine clusters, to enhance solubility and facilitate large scale production in E.coli. As a result, up to ca. 10mg of pure biopolymer from 1 liter of E.coli culture was produced which is approximately 100 times higher than the one reported for dKH/cKH-FGF2 vectors. This high level of biopolymer production is sufficient to support all the preclinical studies in a cost-effective manner.
Fig. 4.
Schematic representation of the a multifunctional biopolymer composed of fusogenic peptide, DNA condensing and endosomyltic motif (DCE), nuclear localization signal, cathepsin substrate and targeting motif. In the RRX1RRX2HHX3HHX4 domain, residue X1 was designed to be valine in order to generate RVRR sequences along the DCE unit to enhance biodegradability by ubiquitous intracellular furin enzyme. This was intended to promote biodegradation of the cationic domain intracellularly resulting in less biopolymer related toxicity. Residues X2 and X4 are designed to be serine (S) and threonine (T). T and S were selected to increase the solubility of the biopolymer and yield of production. Residue X3 in the first and second repeating unit is R and in the third repeating unit is H. This is to incorporate an intrinsic histag into the biopolymer sequence which facilitates its purification via Ni-NTA chromatography. Reproduced with permission from [29].
As shown in Fig. 4, the biopolymer is composed of four major domains: 1) a DNA condensing and endosomolytic (DCE) motif comprised of repeating units of RRVRR (where R is arginine and V is valine) to condense pDNA, and histidine (H) to disrupt endosomes via the proton sponge effect; 2) a targeting motif (TM) for biorecognition by cancer cells over-expressing HER2 [30]; 3) an endosome destabilizing motif, namely fusogenic peptide (FP) to destabilize endosome membranes and synergistically enhance the endosome disrupting activity of the histidine residues; and 4) a nuclear localization signal (NLS), known as M9-NLS [31], to assist in active translocation of the genetic material towards the cell nucleus. The semi-clustered arrangement of the DCE was based upon the previous work done with the cKH-FGF2 biopolymer which is discussed above. In addition, a cathepsin D enzyme substrate (CS) was engineered in between NLS and TM to facilitate dissociation of the targeting motif from the biopolymer inside late endosomes and increase the possibility of exposure of the NLS to the cell’s transportin machinery. The expected intracellular trafficking of the biopolymer is schematically shown in Fig. 5. While this study demonstrates the role of NLS in the biopolymer structure and its interaction with microtubules to enhance gene transfer, no evidence of direct interaction between the NLS and transportin protein was provided. This could be an interesting subject to examine in future studies.
Fig. 5.
The positively charged DCE is used to complex with pDNA and form condensed nanosize particles (step 1). The targeting motif binds to HER2 over-expressed on HER2 positive cells allowing the internalization of the complexes via receptor mediated endocytosis (step 2). TM separates from the complexes inside endosomes with the help of endogenous cathepsin D (step 3). The FP fuses with the endosome membrane and forms a pore. In cooperation with histidine residues, FP facilitates escape of the vector/pDNA complex into the cytosol (step 4). The NLS motif in the vector structure interacts with microtubules with the help of transportin and this complex shuttles the pDNA towards the nucleus (step 5). If small enough (~30nm), the complexes may pass through the nuclear pore complex (NPC) (step 6). If the size is more than 30nm, the complex may end up in the nucleus at the mitosis (M) phase of the cell cycle where the nuclear membrane dissolves (step 7). Once inside the nucleus, the pDNA will be released for transcription (step 8). The unpacking of the complexes may occur in the nucleus with the help of transcription factors. Note: The 3D structure of the targeting motif is predicted by the SWISS-MODEL program.
The results of this study demonstrated that each motif in the vector structure was active and had an impact on its intended function. For instance, a control biopolymer which did not contain the FP motif was constructed and when compared to the full length biopolymer, it was apparent that the biopolymer that lacked FP was unable to efficiently escape from endosomes resulting in seven fold lower transfection efficiency (Fig. 6). This considerable decrease in ability to escape from endosomes was despite the presence of significant number of histidine residues in both biopolymers. This is an interesting observation and could be an important reason behind the fact that no known viruses have utilized the proton sponge effect as a means to escape from endosomes. The same significant reduction in gene expression was observed when cells were transfected with biopolymer without NLS indicating the important role of the NLS in trafficking of the nanoparticles toward the cell nucleus. Again, this is another advantage of working with biopolymers which allow accurate structure/activity relationship studies in order to evaluate the impact of each domain on the overall performance of the multi-functional vectors. It was also observed that the biopolymer could be recognized by the ubiquitous intracellular furin enzyme and no biopolymer related toxicity was detectable in the range tested. Overall, the experimental data showed that the NLS, FP and TM in the biopolymer structure are functioning at their optimum level of efficiency.
Fig. 6.
Evaluation of the functionality of fusogenic peptide and NLS in the biopolymer structure. SKOV-3 cell transfection with biopolymer, biopolymer plus chloroquine, biopolymer plus bafilomycin, biopolymer without fusogenic peptide [biopolymer (−) FP] and biopolymer without nuclear localization signal [biopolymer (−) NLS].
One detriment of any non-self material that is introduced into a biological system is the possibility for immunogenicity. As none of the above studies specifically addressed this question, this will be an important parameter to consider in future work. The ultimate judgment with regard to the usefulness of recombinant polymers for gene therapy should be made after the in vivo studies where in vivo stability, tissue targetability as well as low immunogenicity are examined.
3. Conclusions
While great strides in non-viral gene delivery have been made, as illustrated by the work reviewed here, there is a vast room for improvement. In the pursuit of a new and improved gene delivery vector a better understanding of the molecular interactions between gene delivery system and surrounding biological environment is required. Utilization of biopolymers to elucidate these interactions will result in not only understanding of the cellular pathways but also an improved gene delivery vector. The described bi-functional vectors illustrate the use of poly-lysine sequences for DNA condensation in combination with a thermoresponsive peptide. The tri-functional vectors show the utility of using biopolymers to elucidate structure activity relationships by comparing architectural changes of two content equivalent KH-FGF2 vectors and the impact on efficiency and stability. The multi-functional vectors take the next logical step to overcome the cellular barriers by incorporating multiple domains with diverse functions. The molecular interactions of particular interest to gene delivery are at both intracellular and extracellular level. While the intracellular interactions impact the efficiency of gene delivery system in reaching the cell nucleus, the extracellular interactions determine the level of immune system response to these foreign objects. Such interactions can be studied more reliably than ever before with the help of biopolymers. With a more complete understanding of these interactions development of more advanced, efficient and safe gene delivery vehicles is at horizon.
Acknowledgements
The studies in Hatefi’s lab are supported by the American Cancer Society, and Department of Defense Breast and Prostate Cancer Programs. We also acknowledge the support provided by the NIH biotechnology training fellowship (T-32 GM008336) to Canine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Akita H, Harashima H. Advances in non-viral gene delivery: using multifunctional envelope-type nano-device. Expert Opin Drug Deliv. 2008;5:847–859. doi: 10.1517/17425247.5.8.847. [DOI] [PubMed] [Google Scholar]
- 2.Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 3.Cisse I, Okumus B, Joo C, Ha T. Fueling protein DNA interactions inside porous nanocontainers. Proc Natl Acad Sci U S A. 2007;104:12646–12650. doi: 10.1073/pnas.0610673104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lv H, Zhang S, Wang B, Cui S, Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J Control Release. 2006;114:100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 5.Hayes ME, et al. Increased target specificity of anti-HER2 genospheres by modification of surface charge and degree of PEGylation. Mol Pharm. 2006;3:726–736. doi: 10.1021/mp060040v. [DOI] [PubMed] [Google Scholar]
- 6.Yang T, et al. Antitumor effect of paclitaxel-loaded PEGylated immunoliposomes against human breast cancer cells. Pharm Res. 2007;24:2402–2411. doi: 10.1007/s11095-007-9425-y. [DOI] [PubMed] [Google Scholar]
- 7.Varga CM, Wickham TJ, Lauffenburger DA. Receptor-mediated targeting of gene delivery vectors: insights from molecular mechanisms for improved vehicle design. Biotechnol Bioeng. 2000;70:593–605. doi: 10.1002/1097-0290(20001220)70:6<593::aid-bit1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 8.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 9.Huang H, Yu H, Tang G, Wang Q, Li J. Low molecular weight polyethylenimine cross-linked by 2-hydroxypropyl-gamma-cyclodextrin coupled to peptide targeting HER2 as a gene delivery vector. Biomaterials. 2009 doi: 10.1016/j.biomaterials.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 10.Boussif O, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Florea BI, Meaney C, Junginger HE, Borchard G. Transfection efficiency and toxicity of polyethylenimine in differentiated Calu-3 and nondifferentiated COS-1 cell cultures. AAPS pharmSci. 2002;4:E12. doi: 10.1208/ps040312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urry DW. Physical chemistry of biological free energy transduction as demonstrated by elastic protein-based polymers. J Phys Chem B. 1997;101:11007–11028. [Google Scholar]
- 13.Li Y. Carrier proteins for fusion expression of antimicrobial peptides in Escherichia coli. Biotechnol Appl Biochem. 2009;54:1–9. doi: 10.1042/BA20090087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demain AL, Vaishnav P. Production of recombinant proteins by microbes and higher organisms. Biotechnol Adv. 2009;27:297–306. doi: 10.1016/j.biotechadv.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 15.Tickler AK, Wade JD. Overview of solid phase synthesis of "difficult peptide" sequences Chapter 18: Unit 18. Curr Protoc Protein Sci. 2007:18. doi: 10.1002/0471140864.ps1808s50. [DOI] [PubMed] [Google Scholar]
- 16.Aris A, Feliu JX, Knight A, Coutelle C, Villaverde A. Exploiting viral cell-targeting abilities in a single polypeptide, non-infectious, recombinant vehicle for integrin-mediated DNA delivery and gene expression. Biotechnol Bioeng. 2000;68:689–696. doi: 10.1002/(sici)1097-0290(20000620)68:6<689::aid-bit13>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 17.Villaverde A, Feliu JX, Harbottle RP, Benito A, Coutelle C. A recombinant, arginine-glycine-aspartic acid (RGD) motif from foot-and-mouth disease virus binds mammalian cells through vitronectin and, to a lower extent, fibronectin receptors. Gene. 1996;180:101–106. doi: 10.1016/s0378-1119(96)00413-1. [DOI] [PubMed] [Google Scholar]
- 18.Hatefi A, Canine BF. Perspectives in vector development for systemic cancer gene therapy. Gene Ther Mol Biol. 2009;13:15–19. [PMC free article] [PubMed] [Google Scholar]
- 19.Chen TH, Bae Y, Furgeson DY. Intelligent biosynthetic nanobiomaterials (IBNs) for hyperthermic gene delivery. Pharm Res. 2008;25:683–691. doi: 10.1007/s11095-007-9382-5. [DOI] [PubMed] [Google Scholar]
- 20.Meyer DE, Chilkoti A. Genetically encoded synthesis of protein-based polymers with precisely specified molecular weight and sequence by recursive directional ligation: examples from the elastin-like polypeptide system. Biomacromolecules. 2002;3:357–367. doi: 10.1021/bm015630n. [DOI] [PubMed] [Google Scholar]
- 21.Urry DW. Physical chemistry of biological free energy transduction as demonstrated by elastic protein-based polymers. J Phys Chem B. 1997:11007–11028. [Google Scholar]
- 22.Girotti AJRR, Arias FJ, Alonso M, Testera AM, Rodiguez-Cabello JC. Influence of the molecular weight on the inverse temperature transition of a model genetically engineered elastin-like pH-responseive polymer. Macromolecules. 2004;37:3396–3400. [Google Scholar]
- 23.Hatefi A, Megeed Z, Ghandehari H. Recombinant polymer-protein fusion: a promising approach towards efficient and targeted gene delivery. J Gene Med. 2006;8:468–476. doi: 10.1002/jgm.872. [DOI] [PubMed] [Google Scholar]
- 24.Behr JP. The proton sponge: A trick to enter cells the viruses did not exploit. Chimia. 1997;51:34–36. [Google Scholar]
- 25.Canine BF, Wang Y, Hatefi A. Evaluation of the effect of vector architecture on DNA condensation and gene transfer efficiency. J Control Release. 2008;129:117–123. doi: 10.1016/j.jconrel.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brewer L, Corzett M, Balhorn R. Condensation of DNA by spermatid basic nuclear proteins. J Biol Chem. 2002;277:38895–38900. doi: 10.1074/jbc.M204755200. [DOI] [PubMed] [Google Scholar]
- 27.Khadake JR, Rao MR. Condensation of DNA and chromatin by an SPKK-containing octapeptide repeat motif present in the C-terminus of histone H1. Biochemistry. 1997;36:1041–1051. doi: 10.1021/bi961617p. [DOI] [PubMed] [Google Scholar]
- 28.Tecle M, Preuss M, Miller AD. Kinetic study of DNA condensation by cationic peptides used in nonviral gene therapy: analogy of DNA condensation to protein folding. Biochemistry. 2003;42:10343–10347. doi: 10.1021/bi034325e. [DOI] [PubMed] [Google Scholar]
- 29.Canine BF, Wang Y, Hatefi A. Biosynthesis and characterization of a novel genetically engineered polymer for targeted gene transfer to cancer cells. J Control Release. 2009;138:188–196. doi: 10.1016/j.jconrel.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orlova A, et al. Tumor imaging using a picomolar affinity HER2 binding affibody molecule. Cancer Res. 2006;66:4339–4348. doi: 10.1158/0008-5472.CAN-05-3521. [DOI] [PubMed] [Google Scholar]
- 31.Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]