1. Introduction
Influenza virus is one of the most widespread respiratory viral pathogens currently known to man. It has three transmembrane proteins expressed on the surface of both virions and infected cells. Two of these, hemagglutinin (HA) and neuraminidase (NA), are glycoproteins and are highly variable whereas the third, the matrix 2 protein (M2), has remained relatively conserved. Current influenza vaccines depend on HA as the primary antigen but researchers are exploring using both the NA and M2 as potential alternate antigens for vaccine development.
Yearly epidemics and the more infrequent pandemics occur due to two separate mechanisms, both relating to the potential for changes in HA and NA. Yearly epidemics are caused by antigenic drift, which is the accumulation of mutations in the hemagglutinin or neuraminidase genes in the currently circulating virus allowing it to escape the immunity generated by the original virus. With current egg-based technologies the process of producing a matching vaccine takes 6–8 months. Because of this delay vaccine manufacturers must predict the antigenic features of the strain(s) that will predominate the next season. Therefore, the vaccine’s effectiveness depends heavily on the accuracy of this prediction. The more infrequent pandemics can be caused by antigenic shift, the replacement of either HA or NA subtypes with novel ones, resulting in influenza subtypes to which the general population may be totally naive [1]. Even with new technologies such as cell based influenza vaccines and chimeric virus-like particles (VLPs) [2] immunity will still be dependent upon the similarity of the HA between the circulating virus and the formulation of the vaccine. In light of this, conserved proteins such as the extracellular domain of the matrix 2 (M2) protein have been investigated as a potential “universal” vaccine for influenza [3].
The M2 protein of influenza A virus is a homotetramer which forms a proton channel and is required for viral entry into the cell [4] as well as for packaging viral genomes into newly formed virions [5, 6]. Although it is expressed only at low levels on viral particles, it is expressed at much higher levels on infected host cells [3, 7]. It is for this reason, in addition to its highly conserved nature, that the extracellular domain of the M2 protein (M2e) has been studied as a target for effective vaccine development for the past twenty years. Monoclonal antibodies against M2e have been demonstrated to reduce viral titers [3], providing the basis for the use of this protein as a vaccine candidate. The entire M2 protein produced in baculoviruses provided heterosubtypic protection from lethal challenge [8], bacterially-produced M2 lacking the transmembrane region was found to decrease lung titers across virus subtypes [9], and a DNA vaccine construct causing expression of the M proteins in host cells prevented mortality in mice [10]. The M2e portion of the molecule has been used in various forms including single or multiple peptide carrier conjugates or fusions [11–13], liposome encapsulated multiple antigens [14], fusions with TLR ligands [15], oligerimized M2e domains [16] and fusion with VLPs derived from the Qβ RNA phage [17], hepatitis B virus (HBV)[18, 19], papaya mosaic virus (PapMV) [20], and human papillomavirus (HPV) [21], all resulting in induction of both immunity and protection from lethal influenza virus challenge. Recently, the core antigen from the woodchuck hepatitis virus (WHc) has been developed as an epitope carrier platform [22] with specific advantages over the HBV platform such as ease of insertion of sequences and antigenic dissimilarity to the human hepatitis virus thereby avoiding any pre-existing immunity to the carrier [23, 24].
Without exception the above vaccine platforms are designed for either intranasal or needle assisted delivery (subcutaneous or intra-muscular) and require purification of the antigen or particle prior to delivery [15, 17–21, 23, 24]. With the exception of the flagellin linked M2e epitope [15] the above methods also require some form of adjuvant to be fully effective. Orally administered recombinant attenuated Salmonella vaccines (RASVs) are able to colonize the gut-associated lymphoid tissue (GALT) and the secondary lymphatic tissue, eliciting mucosal, humoral, and cellular immune responses against both itself and heterologous antigens [25, 26]. Over 50 different bacterial, viral, and protozoan antigens have been expressed in these RASVs in preclinical and clinical trials [25–27] making RASVs an attractive and a reliable platform for delivering antigens against various pathogens.
A number of factors influence the immunogenicity of antigens delivered by Salmonella, the most important being the method of attenuation [28] and the intracellular location of antigens [29]. Previous work has demonstrated that regulated delayed attenuation can increase antigen specific antibodies as well as protection from a lethal challenge [30] and that secretion or release of the antigen also improves the immune response [31]. As macromolecules such as VLPs cannot be secreted by Salmonella we took the advantage of the recently developed regulated lysis system to deliver VLPs to the host. [32].
The regulated lysis system consists of two components, the strain and the plasmid. The first component is Salmonella Typhimurium strain χ8937, with deletion of asdA and insertion of arabinose-regulated expression of murA. The asdA and murA genes are required for peptidoglycan synthesis. Additional mutations are also present to enhance complete lysis and antigen delivery. The second component is a derivative of plasmid pYA3681, which encodes arabinose regulated murA and asdA expression, and C2-regulated synthesis of antisense asdA and murA mRNA transcribed from the P22 PR promoter [32]. To create a strain exhibiting a delayed lysis phenotype ΔaraBAD and ΔaraE mutations were included in the original lysis strain χ8937. The ΔaraBAD denotes the deletion of structural genes for catabolism of arabinose thereby preventing the use of arabinose retained in the cell cytoplasm at the time of immunization. The ΔaraE mutation, which deletes the gene for arabinose transport and thus enhances retention of arabinose, precludes leakage of internalized arabinose. This inability to use arabinose prolongs time to lysis in vivo by one to two cell divisions increasing cell numbers and thus enhancing antigen delivery [33]. S. Typhimurium strain χ8888 (Table 1) used in the current study is the prototype strain exhibiting all the above mentioned features and we hypothesize that lysis of the vaccine strain would result in better accessibility of the target antigen.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or Relevant Characteristics | Source or reference |
|---|---|---|
| Strains | Invitrogen | |
| E. coli Top10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL(StrR) endA1 λ− |
|
| S. enterica serovar Typhimurium |
||
| χ8025 | Δcya -27 Δcrp -27 ΔasdA16 | Lab Collection |
| χ8888 | ΔaraE25 ΔaraBAD1923 ΔendA2311 ΔasdA19::TTaraC PBAD c2 TT ΔPmurA7:: TT araC PBAD murA Δ(gmd-fcl)-26 ΔrelA1123 |
Lab Collection |
| Plasmids | ||
| pUC HyW2 AM2A19 78 | Truncated woodchuck core with H7N7 derived M2e in immunodominant spike | Dr. Jean Noel Billaud |
| pYA4036 | Codon optimized HyW2 AM1A19 78 in pUC19 | This work |
| pYA3341 | Asd+ expression vector Ptrc promoter pUC ori | Lab collection |
| pYA3342 | Asd+ expression vector Ptrc promoter pBR ori | [31] |
| pYA3681 | Asd+ expression vector Ptrc promoter pBRori | [32] |
| pYA4037 | Codon optimized HyW2AM2A19 78 in pYA3341 | This work |
| pYA4038 | Codon optimized HyW2 AM2A19 78 in pYA3342 | This work |
| pYA4151 | HyW2 AM2A19 78 in pYA3681 | This work |
| pYA3664 | Codon optimized HyW2 AM2A19 78 in pYA3681 | This work |
In contrast to χ8888 the strain χ8025 (Table 1) is constitutively attenuated in vivo through the addition of the Δcya-27 and Δcrp-27 mutations which together eliminate the catabolite repressor protein. The ΔasdA16 mutation prevents synthesis of diaminopimelic acid (DAP), a unique essential constituent of the peptidoglycan layer of the cell wall. In the absence of DAP, cells undergo cell wall-less death and lyse. The mutation can be complemented either by an external supply of DAP in the growth medium or by complementing with the wild-type copy of asd on a plasmid, resulting in a balanced-lethal system [34, 35].
Attenuated S. Typhimurium strains have been used to deliver both viral and protozoan epitopes on a hybrid Hepatitis B virus (HBV) core antigen (HBc) [36]. In the present work we demonstrate that the WHcAg platform can be expressed in S. Typhimurium and when fused to an immunogenic viral epitope, can induce a strong immune response in mice. We further examine this model by comparing the effectiveness of delivery by the above two RASV strains as well as multiple routes of administration of the RASV.
2. Materials and Methods
2.1. Bacterial strains, plasmids, media, and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli Top10 strain was used for all cloning experiments. E. coli or S. Typhimurium cultures were grown at 37°C in LB broth or on LB agar plates [37]. When required, antibiotics were added to growth media at the following concentrations: ampicillin, 100 µg/ml; kanamycin, 50 µg/ml; and tetracycline, 12.5 µg/ml. Diaminopimelic acid (DAP) (50 µg/ml) was added for the growth of ΔasdA strains [34] and 0.2% arabinose was added in addition to DAP for the growth of strain χ8888 [32].
2.2. General DNA procedures
DNA manipulations were carried out as described by Sambrook et. al. [38]. Transformations into E. coli or S. Typhimurium were done by electroporation (Bio-Rad, Hercules, CA). Transformants containing Asd+ plasmids were selected on LB agar plates without DAP. S train χ8888 transformants containing derivatives of pYA3681 were selected on LB agar plates without DAP and with 0.1% arabinose. Only clones containing the recombinant plasmids were able to grow under these conditions [32, 34, 39].
2.3. Construction of plasmids pYA3664, pYA4037, and pYA4038
Plasmid pUCHyW2 AM2A19 78 (Table 1), which codes for an altered WHc lacking DNA/RNA binding site at its C-terminus, was used as the template for construction of WHc-M2e hybrid particles. The M2e sequence MSLLTEVETPTRNGWECSASDSSD was inserted at amino acid position 76 of the original sequence with the following changes: we deleted the leading methionine (M1) and the cysteine in position 19 was changed to alanine (C19A) to prevent formation of disulfide bonds [22]. To improve expression of the heterologous gene in Salmonella, we replaced certain rare codons in the gene with codons that are compatible with the tRNA pool in S. Typhimurium [40] specifically those encoding aa 3 (ATA to ATT), aa 5 (CCC to CCG), aa 27 (CTT-CTG), aa 42 (CTA to CTG), aa 45 (AGG to CGC), aa 56 (AGA to CGT), aa 76 (ATA to ATC), aa 81 (CTA to CTG), aa 83 (CTA to CTG), aa 84 (CTA to CTG), aa 92 (AGG to CGC), aa 109 (AGA to CGC), aa 125 (AGA to CGC), aa 153 (ATC to ATT), aa 154 (AGG to CGT), aa 160 (AGA to CGT), aa 165 (CCC to CCG), and aa 180 (AGG to CGT) resulting in pYA4036. The intermediate plasmid pYA4036 was digested with the restriction enzymes NcoI and BamHI and the insert was sub-cloned individually into expression vectors pYA3341 (pUC ori) to obtain pYA4037; pYA3342 (pBR ori) to obtain pYA4038 and into the lysis vector pYA3681 (pBR ori) to obtain pYA3664 (Fig. 1).
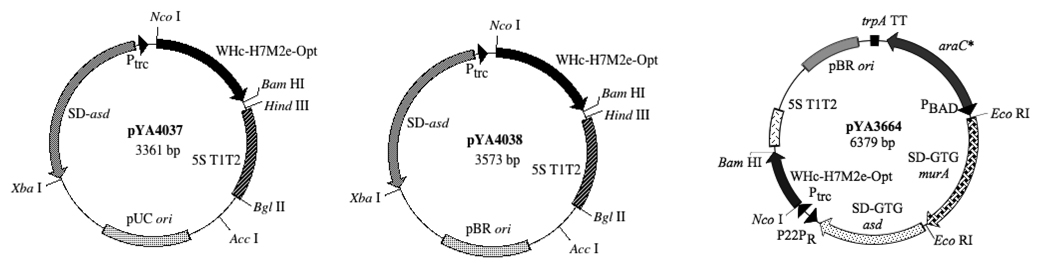
Fig. 1.
Plasmid maps of pYA4037, pYA4038, and pYA3664. The 569 bp WHc-H7M2e-opt fusion fragment flanked by NcoI and BamHI on either sides is in control of the Ptrc-5S T1T2 promoter-terminator unit in Asd+ vectors pYA4037 and pYA4038 and in control of P22PR-5S T1T2 promoter-terminator unit in pYA3664 to finally yield a ~20 kDa fusion protein.
Nucleotide sequencing reactions were performed by the sequencing facility at Arizona State University, using ABI Prism fluorescent BigDye terminators according to the instructions of the manufacturer (PE Biosystems, Norwalk, CT).
2.4 SDS and native PAGE gels and immunoblot analysis
To evaluate expression of hybrid core monomers and assembled particles, cells were grown in LB medium, with 0.1% arabinose added for the growth of the strain χ8888(pYA3664) overnight. The supernatant was removed and cultures were lysed in PBS with complete protease inhibitors (Roche, Indianapolis, IN) at 10 ml per liter by passing the culture twice through a French Pressure Press Cell (Thermo Scientific, Waltham, MA) at a pressure of 13,000 psi. Cell lysates were then cleared by centrifugation at 10,000 × g for 30 min and the soluble protein was normalized for protein concentration using absorbance at 280 nm. Samples for SDS-polyacrylamide gel electrophoresis (SDS-PAGE) were mixed with 1/5 volume of 5 × sodium dodecyl sulfate (SDS) loading buffer, boiled for 10 min and centrifuged at 10,000 × g for 10 min. Samples were loaded on a 12.5% SDS-PAGE gel and separated by electrophoresis as previously described [41]. Samples for native PAGE electrophoresis were mixed with 1/5 volume native loading buffer loaded onto a NativePAGE™ Novex® 3–12% Bis-Tris Gel (Invitrogen, Carlsbad, CA). Reduced samples were transferred to nitrocellulose membranes and native samples to polyvinylidene (PVDF) membranes. The membranes were blocked with 5% skim milk in PBS for 1 h at room temperature and incubated with a 1:5000 dilution of rabbit polyclonal antibody specific for the WHcAg overnight at 4°C. Membranes were then washed three times with PBS- Tween 20. Alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (IgG) (Southern Biotech, Birmingham, AL) diluted 1:10,000 was then added in PBS-milk for 1 h. After washing with PBS-T three times immunoreactive bands were visualized by the addition of nitroblue tetrazolium-5-bromo-4-chloro-3-indolyphosphate (BCIP) (Sigma, St. Louis, MO). The reaction was stopped after 5 min by washing with several large volumes of deionized water.
2.5. Purification and electron microscopy of core particles
Crude lysates were semi-purified by ultracentrifugation through a 30 % (v/v) sucrose cushion in an Airfuge (Beckman Coulter, Brea, CA) at 95,000 × g for15 min. The pellet was washed once with PBS (pH 7.2) and centrifuged at 95,000 × g for 15 min. The pellet was resuspended in 10 µl PBS and stained with an equal volume of 1% phosphotungstic acid (PTA) for 10 min and then adsorbed on carbon-coated formvar grids. Excess liquid was removed and the grids were air dried and observed by electron microscopy (Philips CM12, Hillsboro, OR).
2.6. Immunization of Mice and Sample Collection
All animal experiments were conducted as per protocols approved by the ASU IACUC. Mice were kept 1 week after arrival to acclimate them to our animal facility prior to immunization. Each group of 5 or 10 inbred 7 week old female BALB/c mice (Charles Rivers Laboratories, Wilmington, MA) was deprived of food and water for 6 hours before oral immunization. Rec ombinant S. Typhimurium strains χ8025(pYA3341) and χ8025(pYA4037) were grown in LB and strains χ8888(pYA3681) and χ8888(pYA3664) were grown in LB with 0.1% arabinose to an optical density at 600 nm of 0.9. The cultures were centrifuged at 4,000 × g at room temperature and suspended in buffered saline containing 0.01% gelatin (BSG) [42] to give a final concentration of 5 × 1010 or 1 × 108 CFU/ml. Then, groups of mice were inoculated orally with 1×109 CFU of bacteria in 20 µl or intranasally with 1×107 CFU of bacteria in 10 µl. Food and water was returned to the orally immunized mice 30 min after immunization. A separate group of mice was immunized subcutaneously with 50 µg of purified WHc antigen mixed 1:1 with Imject® Alum Adjuvant (Pierce, Rockford, IL). Booster immunization was given to all immunization groups three weeks later. Blood was drawn by cheek pouch bleeding prior to immunization and then on three separate occasions at 2 week intervals after the booster immunization and stored at room temperature overnight. Following centrifugation at 4,000 × g for 5 minutes the serum was removed and stored at −20°C. Vaginal washes were also collected at the same time points in 50 µl PBS and stored at −20°C [43].
2.7. Influenza A strain
The extracellular domain of the M2 protein of A/WSN/33 differs from that found in the avian influenza virus A/Weybridge (H7N7) at three positions: I11T, E14G, and N20S. PCR-mediated mutagenesis was utilized to change the DNA coding sequence of a plasmid encoding the M segment from A/WSN/33 [44] to that found in A/Weybridge at these three positions. The DNA sequence of the mutagenized plasmid was determined to confirm the presence of the introduced nucleotide changes and the absence of any other nucleotide changes within the M segment coding region. The recombinant influenza A virus (rWSN M2 avian) was rescued via plasmid based reverse genetics as described previously [5, 44]. Virus stocks were propagated on Madin Darby canine kidney (MDCK) cells, sequenced via RT-PCR to confirm the M2 extracellular domain sequence. Infectious virus titers were determined via the median tissue culture infectious dose (TCID50) method or plaque assay as described previously [5, 6].
2.8. ELISA
IgG, IgG1, IgG2a and IgA responses against each of M2e, LPS, and WHcAg in mouse sera or in vaginal secretions were determined by ELISA. Polystyrene 96-well flat-bottom microtiter plates (Dynatech Laboratories Inc., Chantilly, VA) were coated with synthetic M2e peptide (Sigma Genosys) suspended in PBS, purified WHcAg, or LPS suspended in carbonate coating buffer. Plates were coated with 100 ng/well of each antigen and incubated at 4°C overnight. Free binding sites were blocked with PBS-0.05% Tween 20 containing 0.5% bovine serum albumin. Sera were serially diluted from an initial dilution of 1:50 and the vaginal washes were serially diluted from an initial dilution of 1:10. A 50-µl volume of diluted sample was added to duplicate wells and incubated for 1 hour at room temperature. Plates were treated with biotinylated goat anti-mouse IgG, IgG1, IgG2a, or IgA (Southern Biotechnology Inc., Birmingham, AL.). Wells were developed with streptavidin-alkaline phosphatase conjugate (Southern Biotechnology Inc., Birmingham, AL.) follwed by p-nitrophenyl phosphate (pNPP) (Sigma). Color development (absorbance) was recorded at 405 nm using an automated ELISA plate reader (SpectraMax, Molecular Devices, Sunnydale, CA). Endpoint titers were expressed a s the reciprocal log2 values of the last positive sample dilution. Absorbance reading two times higher than preimmune serum baseline values was considered positive.
2.9. Challenge experiments
To assess vaccine efficacy groups of 5 or 10 mice were lightly anesthetized with 0.05 ml/ 20 gram body weight of a ketamine cocktail (21.0 mg ketamine, 2.4 mg xylazine, and 0.3 mg acepromazine) administered intraperitonealy seven weeks after the boost. They were challenged intranasally with either 1 × 103 or 1 × 104 TCID50 of rWSN M2 avian (30 µl). Animals were monitored daily for 14 days following the challenge for survival and weight loss. The percent weight loss was calculated for each individual animal per group by comparing the daily weight to the pre-challenge weight of the animal.
2.10. In vivo viral burden assay
To assess viral titer in vivo, five mice from each group were euthanized on day 5 post-challenge, and lungs were aseptically collected and homogenized to give a 10% (w/v) solution in PBS (pH=7.2). The homogenates were frozen at −70°C. Solid debris was pelleted by centrifugation, and clear homogenates were serially diluted ten-fold in RPMI-1640 (Gibco, Grand Island, NY) and titrated on confluent MDCK cells. The limits of virus detection were 101.3 TCID50/ml and titers were calculated by the method described by Reed and Muench [45].
2.11. Statistical Analysis
Differences in antibody titers were determined using analysis of variance (ANOVA) and statistically different means (P<0.05) were further separated using Tukeys methods. Viral lung titers were analyzed using the Kruskal Wallis test and different means (P<0.05) were determined using Dunns test. Survival analysis was analyzed using the Kaplan-Meier method (GraphPad Prism; GraphPad Software).
3. Results
3.1 Construction of RASVs
The codon optimized WHc-M2e fusion construct (569 bp) was individually ligated into three vectors in order to determine optimal expression conditions. The gene encoding the fusion protein was cloned into the Asd+ expression vectors pYA3341 (pUC ori), pYA3342 (pBR ori) and the lysis vector pYA3681 (pBR ori) to yield plasmids pYA4037, pYA4038 and pYA3664, respectively (Fig. 1). Plasmids were transformed into appropriate S. Typhimurium strains to yield χ8025(pYA4037), χ8025(pYA4038), and χ8888(pYA3664).
3.2 Synthesis of monomers and formation of virus-like particles
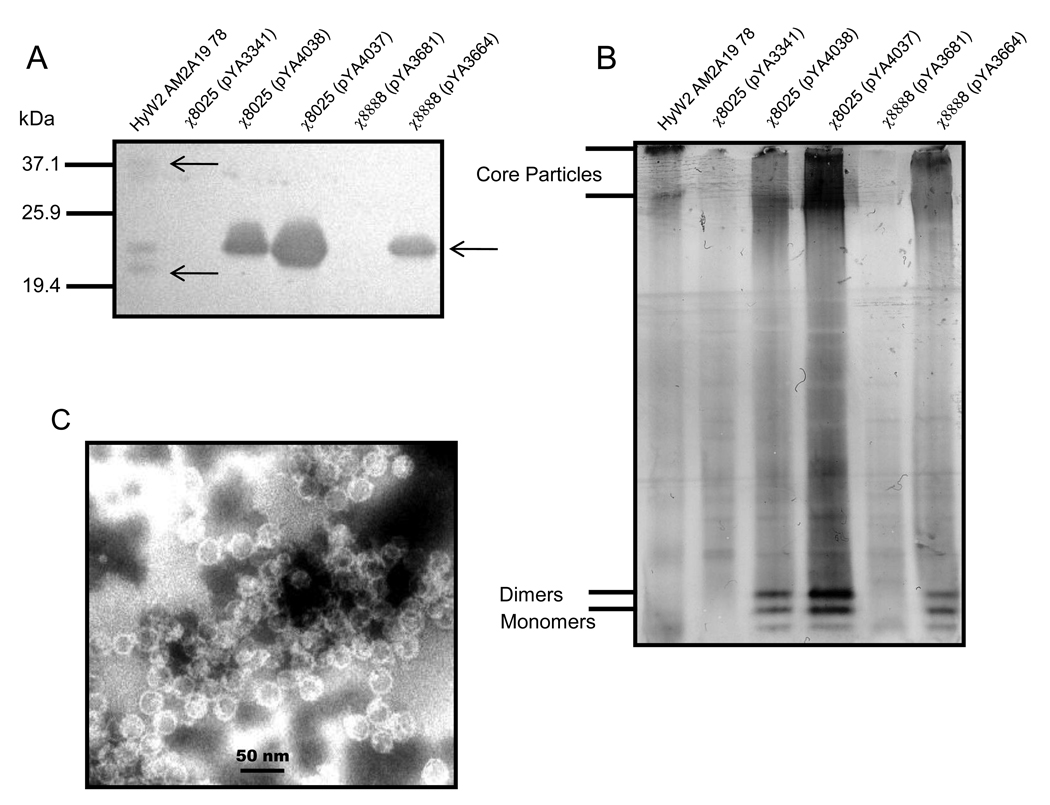
The relative amounts of fusion protein produced in S. Typhimurium χ8025(pYA4037), χ8025(pYA4038), and χ8888(pYA3664) were evaluated by immunoblot under both reducing (Fig. 2A) and non-reducing (Fig. 2B) conditions. Under reducing conditions cell lysates from the strains carrying the fusion protein reacted strongly and specifically with the anti-WHc antibody, as indicated by the presence of a single band. The same antibody reacted with two separate bands when the purified core particle HyW2 AM2A19 78 was run under the same conditions. One of the bands was of comparable size with the cell lysates and the smaller band probably indicates some proteolytic degradation during purification or storage. Strains harboring the empty vectors pYA3341 and pYA3681 had no positive bands. The lysate from strain χ8025(pYA4037) containing a pUCori had a stronger positive band than either of the other lysates that have pBRori plasmid replicon, although equal amounts of protein were loaded into the gel.
Fig. 2.
Synthesis and visualization of WHc-M2e cores by staining with polyclonal rabbit anti-WHc antibody under (A) reducing or (B) native conditions and (C) electron micrograph of semi-purified WHc-M2e core particles from lysates of χ8025/pYA4037. Molecular mass (kDa) are given. Arrow indicates protein in each lane.
Under native conditions immune-reactive protein bands were also detected only in lanes containing the purified core particle (served as control) or in lanes containing the lysates from strains χ8025(pYA4037), χ8025(pYA4038) or χ8888(pYA3664) (Fig. 2B), but not in lanes with lysates from the empty vector control χ8025(pYA3681) confirming that the staining was WHc-specific. Two distinct high molecular weight bands were observed probably representing the two different sized core particles, 180 and 240 subunits, commonly seen in hepatitis virus core particles [46] as well as two low molecular weight bands, probably representing monomers and dimers (Fig. 2B). Interestingly, these lower molecular weight bands were seen only in the Salmonella cell lysates, but not in the purified core particles suggesting that while core formation does occur in the Salmonella strain, only a portion of the monomers are incorporated into particles. Lysates were also examined for core particle formation via transmission electron microscopy (TEM). Particles of the expected size (~26 nm) were observed in all strains synthesizing the fusion protein (Fig. 2C).
As χ8025(pYA4037) synthesized more of the WHc-M2e fusion than χ8025(pYA4038) and as increased protein production generally correlates with increased immune response, the strain χ8025(4037) was chosen for in vivo testing. To test our hypothesis that a lysis phenotype could better deliver a core particle than one that did not lyse we chose to compare the constitutively attenuated strain χ8025(pYA4037) with the strain χ8888(pYA3664) displaying programmed delayed lysis.
3.3 Immune responses in orally immunized mice
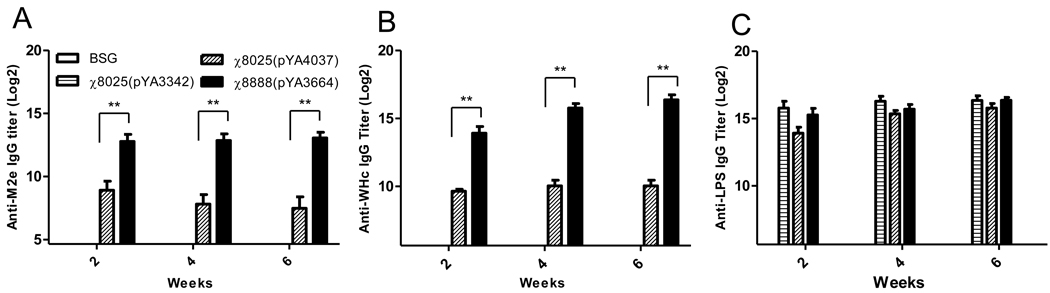
To investigate the immunogenicity of the WHc-M2e fusions delivered by RASV, we compared the immunogenicity of strains χ8025(pYA4037) and χ8888(pYA3664) in mice orally immunized on days 1 and 21. Serum IgG responses to M2e, the woodchuck hepatitis virus core particle and purified S. Typhimurium LPS were measured by ELISA. All of the vaccinated groups had significantly (P <0.01) higher anti-M2e titers than mice immunized with the vector control strain χ8025(pYA3342) and BSG control mice. Mice immunized with strain χ8888(pYA3664) achieved significantly higher anti-M2e titers than those immunized with χ8025(pYA4037) at all three time points (P<0.01) (Fig. 3A).
Fig. 3.
Induction of IgG to (A) M2e peptide, (B) woodchuck hepatitis core and (C) purified Salmonella Typhimurium LPS after second oral immunization of mice with recombinant attenuated Salmonella expressing WHc-M2e cores. Data presented here are representative data of two independent experiments and are the geometric mean ± SE of 10 mice. ** P<0.01
Similar results were seen with regards to the antibody response to WHc particles (Fig. 3B). All of the vaccinated groups had significantly (P <0.01) higher titers than mice immunized with the vector control strain χ8025(pYA3342) and BSG control mice. Mice immunized with strain χ8888(pYA3664) achieved higher anti-WHc titers than those immunized with χ8025(pYA4037) at all three time points (P<0.01). In contrast, LPS titers did not significantly differ between any Salmonella vaccinated group (Fig. 3C) (P>0.05).
These results indicate that the WHc-M2e fusion protein delivered by strain χ8888 induced higher antibody titers in mice than when delivered by strain χ8025. Additionally, since the anti-LPS IgG responses in all groups, including the vector control, were not significantly different this difference is most likely due to better access to the antigen rather than a difference in fitness between the strains. This is reinforced by the fact that the superiority of χ8888(pYA3664) is observed even though it synthesized less WHcM2e fusion than strain χ8025(pYA4037) (Fig.2).
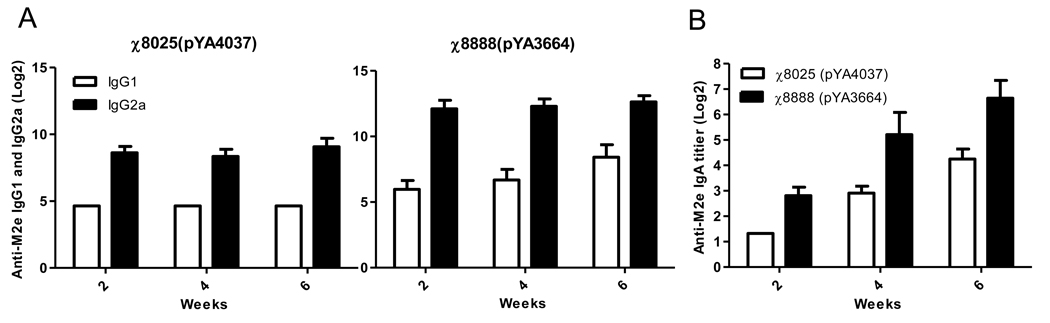
3.4. IgG isotype and mucosal IgA analysis
The immune responses to the M2e epitope were further examined by measuring the levels of IgG2a and IgG1 in the serum 2, 4 and 6 weeks post vaccination. The IgG2a titers to M2e in both vaccinated groups were higher than IgG1 and groups vaccinated with χ8888(pYA3664) induced higher titers of both isotypes than groups vaccinated with χ8025(pYA4037) (Fig. 4A). This indicates that all of the Salmonella vaccines induced a strong Th1 response against M2e regardless of whether the RASV strain was designed to lyse. Th1-type dominant responses are often observed after administration with attenuated Salmonella [47, 48]
Fig. 4.
Comparison of (A) serum IgG isotypes and (B) vaginal IgA levels for mice immunized twice with χ8025(pYA4037) and χ8888(pYA3664). Data presented here are representative data of two independent experiments and are the geometric mean ± SE of 10 mice.
Both strains also induced secretion of mucosal IgA in vaginal fluids (Fig. 4B). As with IgG levels, groups vaccinated with χ8888(pYA3664) had higher levels than those vaccinated with χ8025(pYA4037).
3.5. Evaluation of protection from influenza challenge: Trial 1
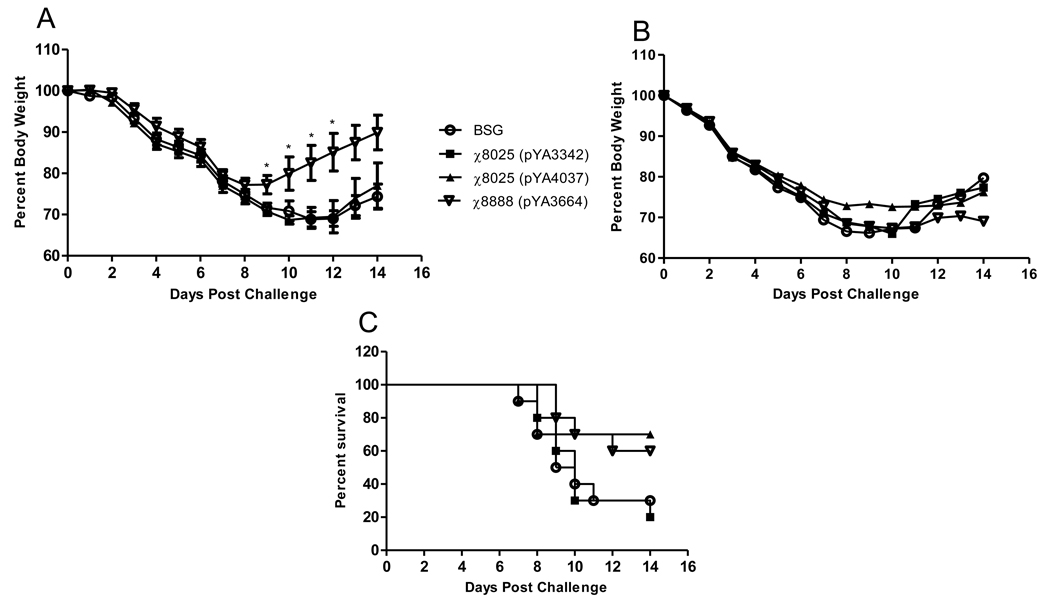
To determine whether the RASV delivered WHc-M2e fusions provided protection against influenza, we challenged immunized mice with either 1×103 or 1×104 TCID50 of rWSN M2 avian. At the low dose challenge we observed weight loss in all groups through day 8 post-infection. On days 8 through 12 the groups vaccinated with χ8888(pYA3664) had significantly higher (P>0.05) average weight than the other groups, signifying an earlier recovery from infection than the BSG control, vector control or χ8025(pYA4037) groups (Fig. 5A). At the high challenge dose we observed no difference in average weight throughout the 14 days period (Fig. 5B) and the survival rates were 70% and 60% for χ8025(pYA4037) and χ8888(pYA3664), respectively (Fig. 5C). The BSG and vector control groups had survival rates of 30% and 20% respectively (Fig. 5C). In summary, lower dose groups receiving χ8888(pYA3664) had a more rapid increase in body weight compared to both χ8025(pYA4037) and the controls, whereas at a higher dose groups receiving either vaccine had a relatively higher rate of survival as compared to the controls.
Fig. 5.
Weight loss after challenge of immunized mice with (A) 1 ×103 or (B) 1 × 104 TCID50 of rWSN M2 avian virus. (C) Mortality curve after challenge with 1 ×104 TCID50 rWSN M2 avian virus. * P<0.05. Data presented here are mean ± SE of 10 mice.
3.6. Comparison of administration routes
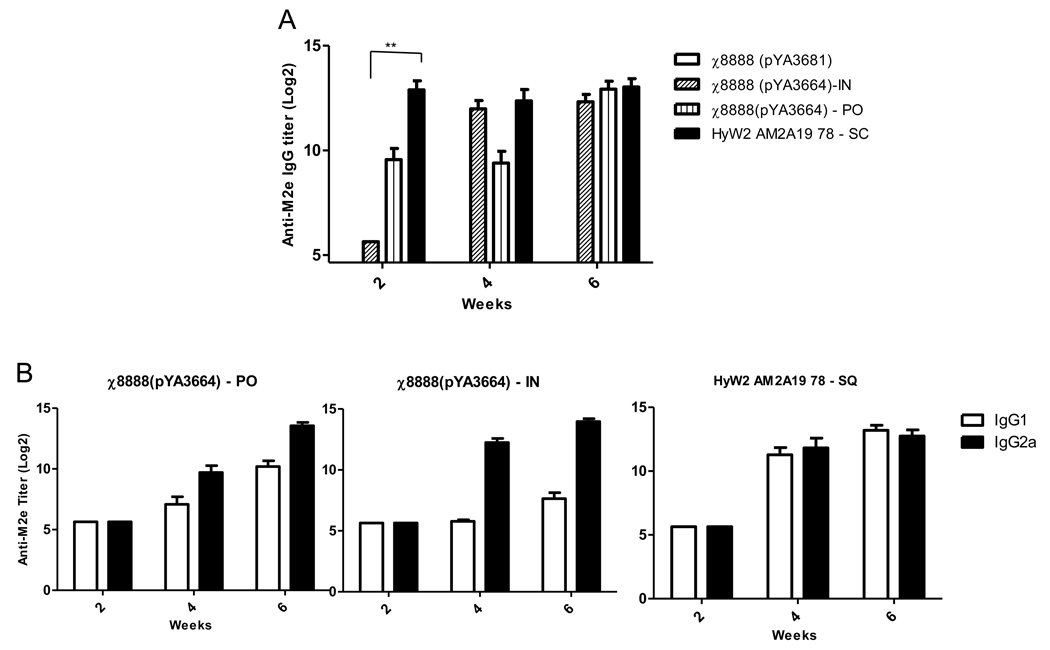
The data so far indicated that strain χ8888(pYA3664) was the most effective of all three in conferring protective immunity against virulent influenza challenge. Therefore we decided to compare the efficacy of this vaccine strain when administered to mice by alternate routes. χ8888(pYA3664) was given to mice orally (1×109 CFU) or intranasally (1×107 CFU) on days 1 and 21 and induction of protective immune response in each case were compared with that induced by purified HyW2AMA19 78 recombinant cores given subcutaneously. All vaccinated groups of mice had significantly higher anti-M2e titers than mice immunized with the vector control strain χ8025(pYA3342) (P<0.01) at all time points tested, with the only exception being the intranasal administration group at two-week time point (Fig. 6A). Both RASV administered groups exhibited relatively higher IgG2a as compa red to IgG1 whereas the subcutaneous injection had approximately equivalent titers of both IgG1 and IgG2a (Fig. 6B).
Fig. 6.
Induction of (A) IgG, and (B) IgG isotype antibodies to M2e peptide after oral or intranasal immunization of mice (n=10) with χ8888(pYA3664) as compared to subcutaneous injection of purified core particle. *P<0.01
3.7. Evaluation of protection from influenza challenge: Trial 2
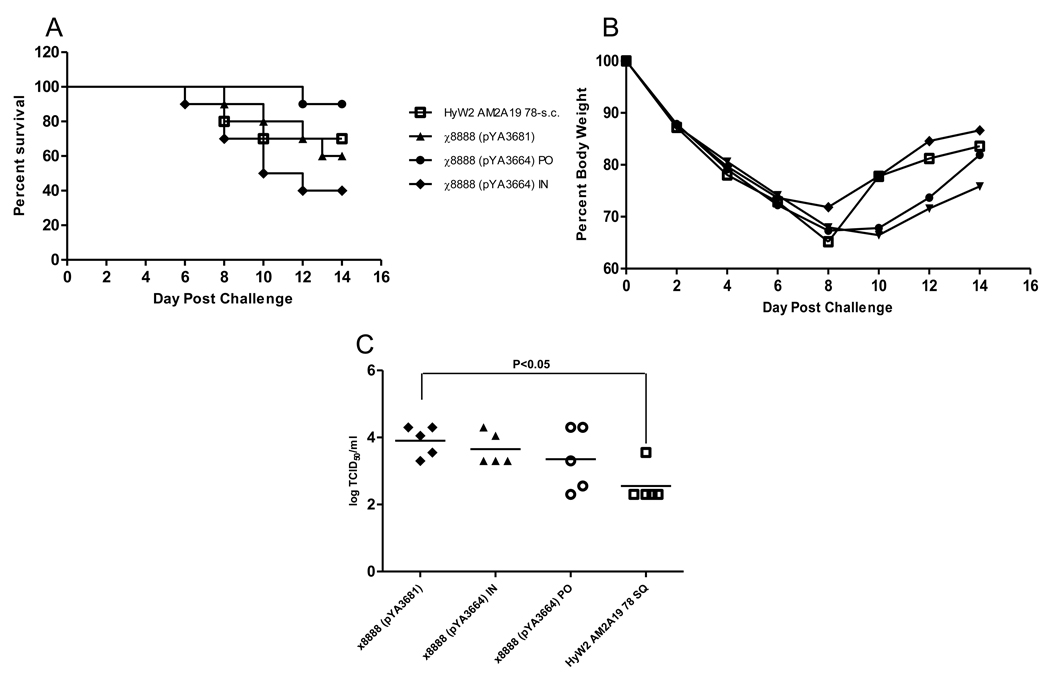
To evaluate protection in immunized mice we challenged 15 mice per group with 1×104 TCID50 of rWSN M2 avian and monitored body weight and mortality in 10 of them. Although not significant, survival was lowest at 40% in the group receiving the RASV intranasally, followed by the vector control group at 60%, the purified antigen administered subcutaneously provided 70% protection, and the oral administration gave the best protection at 90% (Fig. 7A). There was no difference between groups with regard to average body weight through 14 days (Fig 7B). The differences in lung titers between the RASV immunized mice were marginal (Fig. 7C). Despite the lower titers in two of five mice the geometric mean of the oral RASV group did not differ significantly from the vector control, however, the mean lung titers in s.c. group were significantly (P<0.05) lower than the vector control group (Fig.7C) suggesting that the difference between the two routes may have influenced the degree of protection.
Fig. 7.
Mortality (A) and weight loss (B) after challenge of immunized mice with 1 ×104 TCID50 of rWSN-avian virus. (C) individual and geometric mean lung titers 5 days post challenge after immunization.
4. Discussion
Vaccines based on the M2 protein of influenza may provide a novel and much needed alternative or addition to conventional influenza vaccines. Studies have defined the immunogenic epitope as being within the N-terminal 23 amino acids of the protein and determined that it is highly conserved among influenza serotypes. The differences that do exist, particularly between avian and humans strains, are located primarily between amino acids 6 and 13 [49]. Since this epitope is only 23 amino acids in length, two major factors in inducing a strong antibody response are associating it with an appropriate carrier and delivering it with an effective adjuvant. Three basic types of carriers have been used to deliver M2e to mammals; protein or peptide carrier conjugates [8, 9, 11–14, 16], genetic or chemical fusions to VLPs [17–21] and fusion with the Toll-like receptor (TLR) ligand flagellin [15]. With the exception of the flagellin fused construct, all other fusions required adjuvants to achieve maximum protection. This protection includes a decrease in viral lung titers, mortality, and body weight loss [12, 15, 16, 18, 21].
Previously our group has demonstrated the presentation and delivery of both viral and protozoan epitopes on the HBc platform using S. Typhimurium [36]. In this study we demonstrate that a related platform, WHc, with the avian M2e epitope genetically fused into the immunodominant region can be expressed in S. Typhimurium as assembled VLPs (Fig. 2). As responses to Salmonella presented antigens are influenced by a number of factors we compared two strains, the constitutively attenuated strain χ8025 and a strain exhibiting delayed lysis χ8888. Differences in the responses to each of the strains are directly linked to these characteristics. Lysis presumably allows the immune system better accessibility to the core particle as opposed to its release after self-destruction of the bacteria and delaying this event allows Salmonella time to invade and express certain virulence factors prior to lysis, thus inducing a more robust immune response [26, 30, 50].
Humoral immune responses (IgG) to the M2e epitope and the VLP itself were higher when expressed in the strain exhibiting the delayed-lysis phenotype (χ8888) than in the one constitutively attenuated (χ8025), while the response to Salmonella LPS did not differ between the two (Fig. 3). Although antibody levels were generally as high as in previously reported studies using M2e, complete protection from weight loss or mortality was not achieved with either construct given orally or by the strain exhibiting delayed lysis when given intranasally. We did observe a reduction, although not significant, in lung viral titers in mice given the delayed lysis construct orally as compared to both the intranasal and vector control groups. Interestingly, injecting the purified VLP with an alum adjuvant increased this reduction with a geometric mean significantly different (P<0.05) from that of the vector control. Given the observation that the injected particle with adjuvant induced a more balanced Th1/Th2 response as opposed to the Salmonella delivered particle (Fig. 6B), there is the possibility that the type of response induced may also be influencing the outcome (Fig. 7C).
Microbial pathogens and vaccines can drive CD4+ T cells to differentiate into Th1 or Th2 cells. Th1 helper cells direct cell-mediated immunity and promote IgG isotype switching to IgG2a while Th2 helper cells drive B-cell antibody production and promote isotype switching to IgG1[51, 52]. Salmonella vectored vaccines generally induce a Th1 biased response, as is the case with the present constructs (Fig. 4A&C). When the same antigen as presented by Salmonella was delivered s.c. with an alum adjuvant this response was more balanced with regards to the Th1–Th2 response (Fig. 6B). As most previous methods for delivering M2e have involved multiple injections using adjuvant, it may be assumed that they also induced either a balanced Th1–Th2 response or one more biased towards Th2. The single exception of adjuvant use also supports this as flagellin, when used as an adjuvant, has been reported to induce a Th2-biased response [53]. Therefore, despite the strong antibody response to M2e, one explanation for this delivery system’s difference in protection as compared to previous studies may be that a Th1 biased response is less effective against influenza virus than a more balanced response. As the inclusion of a sopB mutation into Salmonella can shift it to a mixed Th1–Th2 response [54], this could easily be tested in future studies.
This explanation is supported by a recent trial involving Salmonella vectored delivery of M2e to poultry. Despite a strong antibody response, vaccination against M2e using Salmonella could only decrease morbidity and early viral shed against low pathogenic avian influenza (LPAI) viruses. There was no protection from mortality or viral shed when vaccinates were challenged with highly pathogenic avian influenza (HPAI) virus [55]. Despite the apparent lack of different IgG isotypes in chickens, both Th1 and Th2 dominant cytokines have been identified and biases towards one or the other have been associated with specific pathogens [56]. Therefore it is possible that failure against the HPAI challenge may be due more to the type of response induced than the inability of the antigen to induce protection, as M2e has been demonstrated to be able to protect against lethal challenges in mice.
In summary we have shown that the WHc fused to an epitope can be expressed in attenuated Salmonella vectors, form cores, and be delivered in an immunogenic manner to the host. Two strains were compared and the strain exhibiting a mechanism for release of cores by lysis induced higher antibody responses than one exhibiting constitutive attenuation. The type of response induced by the constructs was also discussed including possibilities for improvement by incorporation of the sopB mutation. Although the goal to completely eliminate sy mptoms was not met, this represents a significant step towards an affordable, universal vaccine against an important viral pathogen.
Acknowledgments
We thank Kenneth Roland and Praveen Alamuri for their assistance in reviewing this manuscript, J. Kilbourne for her expert assistance with animal experiments, Hua Mo and Xiaoying Kuang for help with ELISA and Darrel Peterson for supplying the WHc purified core particles and polyclonal antibody. This work was supported by NIH grant AI065779-04 to R.C.III.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cox NJ, Subbarao K. Global epidemiology of influenza: past and present. Annu Rev Med. 2000;51:407–421. doi: 10.1146/annurev.med.51.1.407. [DOI] [PubMed] [Google Scholar]
- 2.Latham T, Galarza JM. Formation of wild-type and chimeric influenza virus-like particles following simultaneous expression of only four structural proteins. J Virol. 2001 Jul;75(13):6154–6165. doi: 10.1128/JVI.75.13.6154-6165.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zebedee SL, Lamb RA. Influenza A virus M2 protein: monoclonal antibody restriction of virus growth and detection of M2 in virions. J Virol. 1988 Aug;62(8):2762–2772. doi: 10.1128/jvi.62.8.2762-2772.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pinto LH, Lamb RA. Influenza virus proton channels. Photochem Photobiol Sci. 2006 Jun;5(6):629–632. doi: 10.1039/b517734k. [DOI] [PubMed] [Google Scholar]
- 5.McCown MF, Pekosz A. The influenza A virus M2 cytoplasmic tail is required for infectious virus production and efficient genome packaging. J Virol. 2005 Mar;79(6):3595–3605. doi: 10.1128/JVI.79.6.3595-3605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCown MF, Pekosz A. Distinct domains of the influenza a virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production. J Virol. 2006 Aug;80(16):8178–8189. doi: 10.1128/JVI.00627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamb RA, Zebedee SL, Richardson CD. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell. 1985 Mar;40(3):627–633. doi: 10.1016/0092-8674(85)90211-9. [DOI] [PubMed] [Google Scholar]
- 8.Slepushkin VA, Katz JM, Black RA, Gamble WC, Rota PA, Cox NJ. Protection of mice against influenza A virus challenge by vaccination with baculovirus-expressed M2 protein. Vaccine. 1995;13(15):1399–1402. doi: 10.1016/0264-410x(95)92777-y. [DOI] [PubMed] [Google Scholar]
- 9.Frace AM, Klimov AI, Rowe T, Black RA, Katz JM. Modified M2 proteins produce heterotypic immunity against influenza A virus. Vaccine. 1999 May 4;17(18):2237–2244. doi: 10.1016/s0264-410x(99)00005-5. [DOI] [PubMed] [Google Scholar]
- 10.Okuda K, Ihata A, Watabe S, Okada E, Yamakawa T, Hamajima K, et al. Protective immunity against influenza A virus induced by immunization with DNA plasmid containing influenza M gene. Vaccine. 2001 Jun 14;19(27):3681–3691. doi: 10.1016/s0264-410x(01)00078-0. [DOI] [PubMed] [Google Scholar]
- 11.Liu W, Peng Z, Liu Z, Lu Y, Ding J, Chen YH. High epitope density in a single recombinant protein molecule of the extracellular domain of influenza A virus M2 protein significantly enhances protective immunity. Vaccine. 2004 Dec 2;23(3):366–371. doi: 10.1016/j.vaccine.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 12.Tompkins SM, Zhao ZS, Lo CY, Misplon JA, Liu T, Ye Z, et al. Matrix protein 2 vaccination and protection against influenza viruses, including subtype H5N1. Emerg Infect Dis. 2007 Mar;13(3):426–435. doi: 10.3201/eid1303.061125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu F, Huang JH, Yuan XY, Huang WS, Chen YH. Characterization of immunity induced by M2e of influenza virus. Vaccine. 2007 Dec 17;25(52):8868–8873. doi: 10.1016/j.vaccine.2007.09.056. [DOI] [PubMed] [Google Scholar]
- 14.Ernst WA, Kim HJ, Tumpey TM, Jansen AD, Tai W, Cramer DV, et al. Protection against H1, H5, H6 and H9 influenza A infection with liposomal matrix 2 epitope vaccines. Vaccine. 2006 Jun 12;24(24):5158–5168. doi: 10.1016/j.vaccine.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Huleatt JW, Nakaar V, Desai P, Huang Y, Hewitt D, Jacobs A, et al. Potent immunogenicity and efficacy of a universal influenza vaccine candidate comprising a recombinant fusion protein linking influenza M2e to the TLR5 ligand flagellin. Vaccine. 2008 Jan 10;26(2):201–214. doi: 10.1016/j.vaccine.2007.10.062. [DOI] [PubMed] [Google Scholar]
- 16.De Filette M, Martens W, Roose K, Deroo T, Vervalle F, Bentahir M, et al. An influenza a vaccine based on tetrameric ectodomain of matrix protein 2. J Biol Chem. 2008 Feb 5; doi: 10.1074/jbc.M800650200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bessa J, Schmitz N, Hinton HJ, Schwarz K, Jegerlehner A, Bachmann MF. Efficient induction of mucosal and systemic immune responses by virus-like particles administered intranasally: implications for vaccine design. Eur J Immunol. 2008 Jan;38(1):114–126. doi: 10.1002/eji.200636959. [DOI] [PubMed] [Google Scholar]
- 18.De Filette M, Fiers W, Martens W, Birkett A, Ramne A, Lowenadler B, et al. Improved design and intranasal delivery of an M2e-based human influenza A vaccine. Vaccine. 2006 Nov 10;24(44–46):6597–6601. doi: 10.1016/j.vaccine.2006.05.082. [DOI] [PubMed] [Google Scholar]
- 19.Fiers W, De Filette M, Birkett A, Neirynck S, Min Jou W. A "universal" human influenza A vaccine. Virus Res. 2004 Jul;103(1–2):173–176. doi: 10.1016/j.virusres.2004.02.030. [DOI] [PubMed] [Google Scholar]
- 20.Denis J, Acosta-Ramirez E, Zhao Y, Hamelin ME, Koukavica I, Baz M, et al. Development of a universal influenza A vaccine based on the M2e peptide fused to the papaya mosaic virus (PapMV) vaccine platform. Vaccine. 2008 Jun 25;26(27–28):3395–3403. doi: 10.1016/j.vaccine.2008.04.052. [DOI] [PubMed] [Google Scholar]
- 21.Ionescu RM, Przysiecki CT, Liang X, Garsky VM, Fan J, Wang B, et al. Pharmaceutical and immunological evaluation of human papillomavirus viruslike particle as an antigen carrier. J Pharm Sci. 2006 Jan;95(1):70–79. doi: 10.1002/jps.20493. [DOI] [PubMed] [Google Scholar]
- 22.Billaud JN, Peterson D, Barr M, Chen A, Sallberg M, Garduno F, et al. Combinatorial approach to hepadnavirus-like particle vaccine design. J Virol. 2005 Nov;79(21):13656–13666. doi: 10.1128/JVI.79.21.13656-13666.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Billaud JN, Peterson D, Lee BO, Maruyama T, Chen A, Sallberg M, et al. Advantages to the use of rodent hepadnavirus core proteins as vaccine platforms. Vaccine. 2007 Feb 19;25(9):1593–15606. doi: 10.1016/j.vaccine.2006.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Billaud JN, Peterson D, Schodel F, Chen A, Sallberg M, Garduno F, et al. Comparative antigenicity and immunogenicity of hepadnavirus core proteins. J Virol. 2005 Nov;79(21):13641–13655. doi: 10.1128/JVI.79.21.13641-13655.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Curtiss R., III . Antigen delivery systems: development of live recombinant attenuated bacterial antigen and DNA vaccine delivery vector vaccines. In: Mestecky J, Strober W, Bienenstock J, McGhee JR, Mayer L, editors. Mucosal Immunology. 3rd ed. San Diego, CA: Academic Press; 2005. [Google Scholar]
- 26.Curtiss R, III, Zhang X, Wanda SY, Kang HY, Konjufca V, Li Y, et al. Induction of host immune responses using Salmonella-vectored vaccines. In: Brogden KA, Cornick N, Stanton TB, Zhang Q, Nolan LK, Wannemueler MJ, editors. Virulence Mechanisms of Bacterial Pathogens. 4th ed. Washington, D.C.: ASM Press; 2007. p. 297. [Google Scholar]
- 27.Curtiss R, III, Kelly SM, Hassan JO. Live oral avirulent Salmonella vaccines. Vet Microbiol. 1993;37:397–405. doi: 10.1016/0378-1135(93)90038-9. [DOI] [PubMed] [Google Scholar]
- 28.Dunstan SJ, Simmons CP, Strugnell RA. Comparison of the abilities of different attenuated Salmonella typhimurium strains to elicit humoral immune responses against a heterologous antigen. Infect Immun. 1998 Feb;66(2):732–740. doi: 10.1128/iai.66.2.732-740.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang HY, Curtiss R., III Immune responses dependent on antigen location in recombinant attenuated Salmonella typhimurium vaccines following oral immunization. FEMS Immunol Med Microbiol. 2003 Jul 15;37(2–3):99–104. doi: 10.1016/S0928-8244(03)00063-4. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Wang S, Scarpellini G, Gunn B, Xin W, Wanda SY, et al. Evaluation of new generation Salmonella enterica serovar Typhimurium vaccines with regulated delayed attenuation to induce immune responses against PspA. Proc Natl Acad Sci U S A. 2009 Jan 13;106(2):593–598. doi: 10.1073/pnas.0811697106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang HY, Srinivasan J, Curtiss R., III Immune responses to recombinant pneumococcal PspA antigen delivered by live attenuated Salmonella enterica serovar typhimurium vaccine. Infect Immun. 2002 Apr;70(4):1739–1749. doi: 10.1128/IAI.70.4.1739-1749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kong W, Wanda SY, Zhang X, Bollen W, Tinge SA, Roland KL, et al. Regulated programmed lysis of recombinant Salmonella in host tissues to release protective antigens and confer biological containment. Proc Natl Acad Sci U S A. 2008 Jul 7; doi: 10.1073/pnas.0803801105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kong W, Wanda SY, Curtiss R., III Construction and application of host vector systems for DNA vaccine vector delivery. American Society for Microbiology 103rd Annual Meeting; 2003; Washington DC. 2003. p. 677. [Google Scholar]
- 34.Galan JE, Nakayama K, Curtiss R., III Cloning and characterization of the asd gene of Salmonella typhimurium: use in stable maintenance of recombinant plasmids in Salmonella vaccine strains. Gene. 1990 Sep 28;94(1):29–35. doi: 10.1016/0378-1119(90)90464-3. [DOI] [PubMed] [Google Scholar]
- 35.Nakayama K, Kelly SM, Curtiss R., III Construction of an Asd+ expression-cloning vector: stable maintenance and high level expression of cloned genes in a Salmonella vaccine strain. Nat Biotech. 1988;6(6):693–697. [Google Scholar]
- 36.Schodel F, Kelly S, Tinge S, Hopkins S, Peterson D, Milich D, et al. Hybrid hepatitis B virus core antigen as a vaccine carrier moiety. II. Expression in avirulent Salmonella spp. for mucosal immunization. Adv Exp Med Biol. 1996;397:15–21. [PubMed] [Google Scholar]
- 37.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951 Sep;62(3):293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning; A Laboratory Manual. Second ed. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Curtiss R, III, Nakayama K, Kelly SM. Recombinant avirulent Salmonella vaccine strains with stable maintenance and high level expression of cloned genes in vivo. Immunol Invest. 1989 Jan–May;18(1–4):583–596. doi: 10.3109/08820138909112265. [DOI] [PubMed] [Google Scholar]
- 40.Henaut A, Danchin A. Analysis and Predictions from Escherichia coli sequences. In: Neidhardt FCea., editor. Escherichia coli and Salmonella. Washington, D.C.: ASM press; 1996. pp. 2047–2066. [Google Scholar]
- 41.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 42.Curtiss R., III Chromosomal Ab errations Associated With Mutations To Bacteriophage Resistance In Escherichia Coli. J Bacteriol. 1965 Jan;89:28–40. doi: 10.1128/jb.89.1.28-40.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X, Kelly SM, Bollen WS, Curtiss R., III Characterization and immunogenicity of Salmonella typhimurium SL1344 and UK-1 delta crp and delta cdt deletion mutants. Infect Immun. 1997 Dec;65(12):5381–5387. doi: 10.1128/iai.65.12.5381-5387.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, et al. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci U S A. 1999 Aug 3;96(16):9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reed JD, Muench A simple method for estimating fifty percent endpoints. Am J Hyg. 1938;27:493–496. [Google Scholar]
- 46.Crowther RA, Kiselev NA, Bottcher B, Berriman JA, Borisova GP, Ose V, et al. Three-dimensional structure of hepatitis B virus core particles determined by electron cryomicroscopy. Cell. 1994 Jun 17;77(6):943–950. doi: 10.1016/0092-8674(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 47.Lo-Man R, Langeveld JP, Deriaud E, Jehanno M, Rojas M, Clement JM, et al. Extending the CD4(+) T-cell epitope specificity of the Th1 immune response to an antigen using a Salmonella enterica serovar typhimurium delivery vehicle. Infect Immun. 2000 Jun;68(6):3079–3089. doi: 10.1128/iai.68.6.3079-3089.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okahashi N, Yamamoto M, Vancott JL, Chatfield SN, Roberts M, Bluethmann H, et al. Oral immunization of interleukin-4 (IL-4) knockout mice with a recombinant Salmonella strain or cholera toxin reveals that CD4+ Th2 cells producing IL-6 and IL-10 are associated with mucosal immunoglobulin A responses. Infect Immun. 1996 May;64(5):1516–1525. doi: 10.1128/iai.64.5.1516-1525.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu W, Zou P, Ding J, Lu Y, Chen YH. Sequence comparison between the extracellular domain of M2 protein human and avian influenza A virus provides new information for bivalent influenza vaccine design. Microbes Infect. 2005 Feb;7(2):171–177. doi: 10.1016/j.micinf.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 50.Curtiss R, III, Wanda SY, Gunn BM, Zhang X, Tinge SA, Ananthnarayan V, et al. Salmonella enterica serovar typhimurium strains with regulated delayed attenuation in vivo. Infect Immun. 2009 Mar;77(3):1071–1082. doi: 10.1128/IAI.00693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spellberg B, Edwards JE., Jr Type 1/Type 2 immunity in infectious diseases. Clin Infect Dis. 2001 Jan;32(1):76–102. doi: 10.1086/317537. [DOI] [PubMed] [Google Scholar]
- 52.O'Garra A, Arai N. The molecular basis of T helper 1 and T helper 2 cell differentiation. Trends Cell Biol. 2000 Dec;10(12):542–550. doi: 10.1016/s0962-8924(00)01856-0. [DOI] [PubMed] [Google Scholar]
- 53.Cunningham AF, Khan M, Ball J, Toellner KM, Serre K, Mohr E, et al. Responses to the soluble flagellar protein FliC are Th2, while those to FliC on Salmonella are Th1. Eur J Immunol. 2004 Nov;34(11):2986–2995. doi: 10.1002/eji.200425403. [DOI] [PubMed] [Google Scholar]
- 54.Li Y, Wang S, Xin W, Scarpellini G, Shi Z, Gunn B, et al. A sopB deletion mutation enhances the immunogenicity and protective efficacy of a heterologous antigen delivered by live attenuated Salmonella enterica vaccines. Infect Immun. 2008 Nov;76(11):5238–5246. doi: 10.1128/IAI.00720-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Layton SL, Kapczynski DR, Higgins S, Higgins J, Wolfenden AD, Liljebjelke KA, et al. Vaccination of chickens with recombinant Salmonella expressing M2e and CD154 epitopes increases protection and decreases viral shedding after low pathogenic avian influenza challenge. Poult Sci. 2009 Nov;88(11):2244–2252. doi: 10.3382/ps.2009-00251. [DOI] [PubMed] [Google Scholar]
- 56.Degen WG, Daal N, Rothwell L, Kaiser P, Schijns VE. Th1/Th2 polarization by viral and helminth infection in birds. Vet Microbiol. 2005 Feb 25;105(3–4):163–167. doi: 10.1016/j.vetmic.2004.12.001. [DOI] [PubMed] [Google Scholar]