Abstract
The tissues of the male reproductive tract are characterized by distinct morphologies, from highly coiled to un-coiled. Global gene expression profiles of efferent ducts, epididymis, and vas deferens were generated from embryonic day 14.5 to postnatal day 1 as tissue-specific morphologies emerge. Expression of homeobox genes, potential mediators of tissue-specific morphological development, was assessed. Twenty homeobox genes were identified as either tissue-enriched, developmentally regulated, or both. Additionally, ontology analysis demonstrated cell adhesion to be highly regulated along the length of the reproductive tract. Regulators of cell adhesion with variable expression between the three tissues were identified including Alcam, various cadherins, and multiple integrins. Immunofluorescence localization of the cell adhesion regulators POSTN and CDH2 demonstrated cell adhesion in the epithelium and mesenchyme of the epididymis may change throughout development. These results imply cell adhesion may be modulated in a tissue-specific manner, playing an important role in establishing each tissue’s final morphology.
Keywords: Microarray, epididymis, vas deferens, efferent duct, male reproductive tract
Introduction
The efferent ducts, epididymis, and vas deferens comprise the reproductive excurrent duct system in the male. Each tissue is characterized by distinct morphologies and functions. There are three to five efferent ducts in the mouse, which are moderately coiled and give rise to a single common duct before transitioning into the epididymis (Joseph et al., 2009). One major role of the efferent ducts is the resorption of water (Clulow et al., 1994; Clulow et al., 1998), under the control of both testosterone and estrogen (Hansen et al., 1997; Hess et al., 1997). The epididymis; a highly coiled, unbranched duct system is segmented into four gross anatomical regions, from cranial to caudal: the initial segment, caput, corpus, and cauda and has three major functions. Like the efferent ducts, the cranial portion of the epididymis concentrates spermatozoa. Additionally, the epididymis plays a key role in the maturation of spermatozoa to fully mature, motile sperm. Lastly, the caudal portion of the epididymis provides storage and protection for mature sperm prior to ejaculation. Distally, the epididymis terminates as the vas deferens. The vas deferens has the least complex ultrastructure, being uncoiled and unbranched, and serves primarily to store and protect mature sperm (Robaire and Hinton, 2002). During ejaculation, contractile waves in both the epididymis and vas deferens facilitate sperm expulsion from the excurrent ducts (Coolen et al., 2004; Vignozzi et al., 2008).
The majority of studies on the male excurrent duct system (reproductive tract) have focused on the expression and function of genes and proteins along the normal adult epididymal duct (Robaire and Hinton, 2002), however, proper development of all three tissues of the upper reproductive tract is essential for the maintenance of fertility. While there are very few genes that have been shown to be essential for male fertility in the adult epididymis, e.g. the forkhead box I1 (Foxi1) transcription factor (Blomqvist et al., 2006), there are several genes that likely play an important role during development of the epididymis, which in turn appear to be important for male fertility, e.g. Ros1 proto-oncogene (Ros1 or c-Ros) (Sonnenberg-Riethmacher et al., 1996) and leucine-rich repeat-containing G protein-coupled receptor 4 (Lgr4) (Mendive et al., 2006).
Although the final adult morphology and full cellular differentiation of the male reproductive tract is not complete until several weeks after birth (Robaire and Hinton, 2002), the period of late embryonic to early postnatal development is particularly important as it is during this time that the Wolffian duct undergoes regionalization and the individual tissues of the tract undergo the morphological changes that will eventually give rise to fully mature tissue (see Supp. Fig. 1 for a brief overview and (Joseph et al., 2009) for a detailed review). At E14.5, the upper reproductive tract consists of three distinct tube systems: the Wolffian duct, the mesonephric tubules, and the Mullerian duct. The upper portion of the Wolffian duct will give rise to the epididymis while the lower portion will form the vas deferens. The mesonephric tubules will eventually give rise to the efferent ducts. At E14.5, there is no morphological distinction between the upper and lower portion of the Wolffian duct, however by E16.5, the upper portion of the Wolffian duct has initiated the process of coiling while the Mullerian duct has fully regressed. During this period, the mesonephric tubules (putative efferent ducts) have also initiated coiling. From E16.5 to P1, coiling continues in the efferent ducts and moves caudally from the initial segment to the cauda of the developing epididymis. At no point in development does the vas deferens undergo coiling. Thus, regionalization of the Wolffian duct can be thought of as predominantly morphological with the upper portion diverging from the lower portion. However, increasing similarities in morphology are observed between the developing efferent ducts and epididymis, tissues derived from distinct embryonic structures.
Several factors have been shown to be important in the regionalization, elongation, or coiling of the developing reproductive tract. For example, animals lacking homeobox A10 (Hoxa10) or homeobox A11 (Hoxa11) have homeotic or partial homeotic transformations of the vas deferens to the epididymis (Hsieh-Li et al., 1995; Podlasek et al., 1999), linking the expression of Hox genes to regionalization of the Wolffian duct. Hox genes encode transcription factors that regulate anterior-posterior patterning and have been demonstrated to regulate the regionalization or patterning of multiple embryonic tube systems including the developing cardiac tube (Monier et al., 2007) and neural tube (Hidalgo-Sanchez et al., 2005; Ramos and Robert, 2005). Additionally, altered expression of Hox genes is known to have significant effects on the final morphology of moderately coiled tube systems, including the intestine (Wolgemuth et al., 1989; Zacchetti et al., 2007) and the kidney (Di-Poi et al., 2007). Unfortunately, a detailed analysis of Hox gene expression along the developing male reproductive tract has been unavailable.
Distinct morphologies are observed in the tissues of the male reproductive tract prior to and shortly after birth. To date, microarray analysis of this system has been limited to normal and treated adult epididymis (Wagenfeld et al., 2002; Ezer and Robaire, 2003; Xu et al., 2003; Chauvin and Griswold, 2004; Johnston et al., 2005; Yamazaki et al., 2006; Dube et al., 2007; Jelinsky et al., 2007; Turner et al., 2007) and treated adult efferent ducts (Snyder et al., 2009). There has been no global analysis examining the factors that govern the late embryonic and early postnatal development of the Wolffian and efferent ducts as well as those that regulate regionalization of the Wolffian duct. Tissue-specific global gene expression analysis throughout development provides a valuable resource for examining potential regulators of these processes. The global expression databases generated here will provide the opportunity for the research community to intensely study a dynamic period of development in three unique, but closely related reproductive tissues required for optimal fertility. Key factors involved in the development of these tissues are likely to serve homologous functions in other tissues and may provide insight into the mechanisms governing tubular and organ morphogenesis in the mammal.
Results
The Affymetrix microarray platform was used to determine global gene expression in three distinct tissues of the reproductive tract and their associated mesenchyme; the efferent ducts, the epididymis, and the vas deferens at four time points during development (E14.5, E16.5, E18.5, and P1). The resulting microarray output was used to determine transcripts that varied between tissues and throughout the developmental period. For the purpose of this work, the term tissue refers to both the ductal epithelium and the associated mesenchyme. Additionally, for clarity the terms efferent duct, epididymis, and vas deferens refer to the tissues as well as the structures from which they are derived. However, prior to E16.5 the epididymis and vas deferens combined would be more properly termed the Wolffian duct and prior to E18.5 the efferent ducts are more correctly called mesonephric tubules. Transcripts were considered enriched or reduced if the raw score was greater than 50 in all samples of the tissue/age of interest, the normalized value fold-change between the comparisons was greater than 2, and the comparison was found to be significant by two-way ANOVA (p < 0.05). Selected comparisons were further screened by ontology analysis to determine common or enriched biological process, molecular functions, or cellular components (Dennis et al., 2003; Huang da et al., 2009). Microarray data was submitted to GEO under accession numbers GSM 560923 through 560946.
Validation of Tissue Collection and Array Output
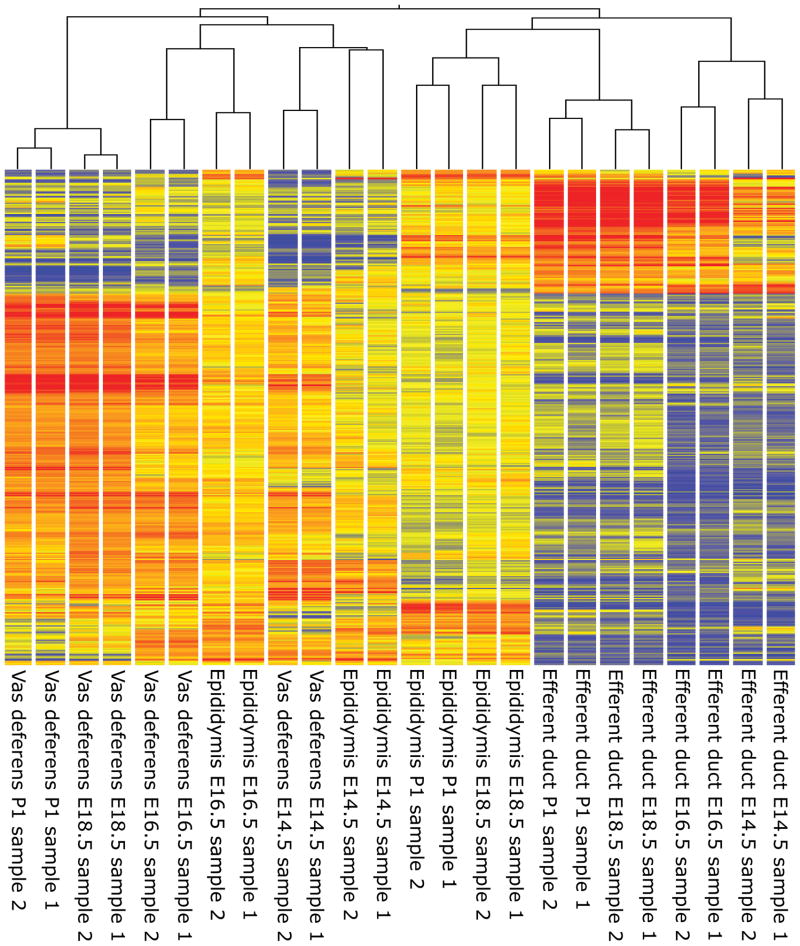
To determine the accuracy and reproducibility of tissue collection, a heat map and hierarchical clustering analysis was generated using 18,845 probe IDs determined to be significantly different by two-way ANOVA (p < 0.05) (Fig.1). Clustering analysis revealed that duplicate samples for each time point were more similar to one another than other samples in all cases. Efferent duct samples were found to be more similar to one another than to all other tissues regardless of age, while early (E14.5 and E16.5) epididymis and vas deferens were more similar to each other than to late (E18.5 and P1) epididymis and vas deferens. Notably, late epididymis clustered more closely with efferent duct samples than with late vas deferens.
FIG. 1.
Hierarchical clustering of duplicate samples from the developing reproductive tract, E14.5 to P1.
Isolation of the putative epididymis from the vas deferens at E14.5 is complicated by the lack of morphological markers to distinguish the two tissues while efferent ducts are easily isolated from the Wolffian duct based on tubule morphology (Supp. Fig. 2). As a result of this, isolation of epididymis from vas deferens was confirmed by expression analysis of known tissue-enriched genes (Fig. 2). Few tissue-enriched transcripts have been identified for the tissues derived from the Wolffian duct. To overcome this issue, a database of expression data for the urogenital tract, GUDMAP (www.gudmap.org) (Little et al., 2007) was queried to determine if transcripts identified as tissue-enriched at E14.5 for the epididymis and vas deferens had the expected message localization as demonstrated by in situ hybridization. Tissue-enrichment for decorin (Dcn) (GUDMAP: 7307), integrin alpha 4 (Itga4) (GUDMAP: 10897), and maternally expressed 3 (Meg3) (GUDMAP: 9022) was observed in the putative epididymis while angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 (Ace2) (GUDMAP: 9604), stanniocalcin 1 (Stc1) (GUDMAP: 10864), and transmembrane protein 100 (Tmem100) (GUDMAP: 7424) were observed in the putative vas deferens in this array analysis.
FIG. 2.
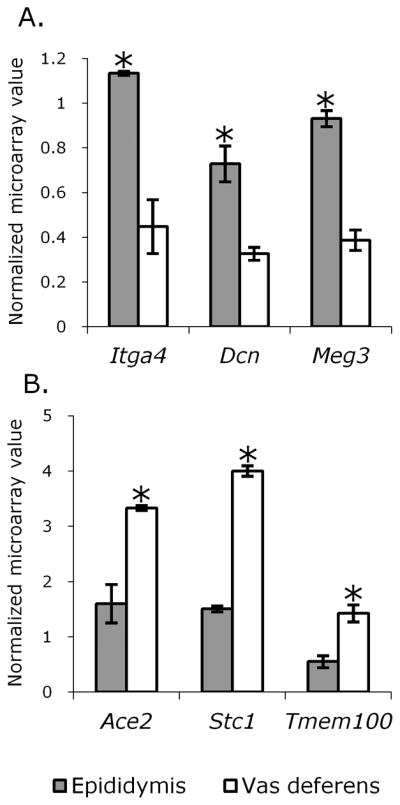
Expression profile of known tissue-enriched genes at E14.5 as determined by microarray. A.) Transcripts enriched in the epididymis at E14.5. B.) Transcripts enriched in the vas deferens at E14.5. Error bars represent standard deviation. Asterisks indicate significance (p > 0.05), determined by Tukey’s HSD.
Hox Gene Expression in the Developing Reproductive Tract
As key regulators of anterior-posterior patterning and tube regionalization, the expression profiles of Hox genes in the developing reproductive tract were examined. In order to define the appropriate data set for analysis, probe IDs of all genes with the word “homeobox” or “hox” in either the gene symbol, full name(s), or synonyms in the Gene Ontology (AmiGO) database were determined and used to query the reproductive tract developmental profile. The resulting probe ID list contained 254 probe IDs and targeted 172 Hox genes. Hox genes of interest included genes with tissue-enriched expression (Fig. 3) and those with unique expression profiles within a given tissue or set of tissues throughout development. Of the seven transcripts identified in this analysis as tissue-enriched throughout development (at least two-fold higher expression in one or more tissues as compared to the others at all time points and significantly greater at all time points as determined by two-way ANOVA with a p-value < 0.05), two (Hoxd10 and Meis2) displayed a unique expression profile with increasing expression from the anterior (epididymis) to the posterior (vas deferens) of the Wolffian duct throughout development.
FIG. 3.
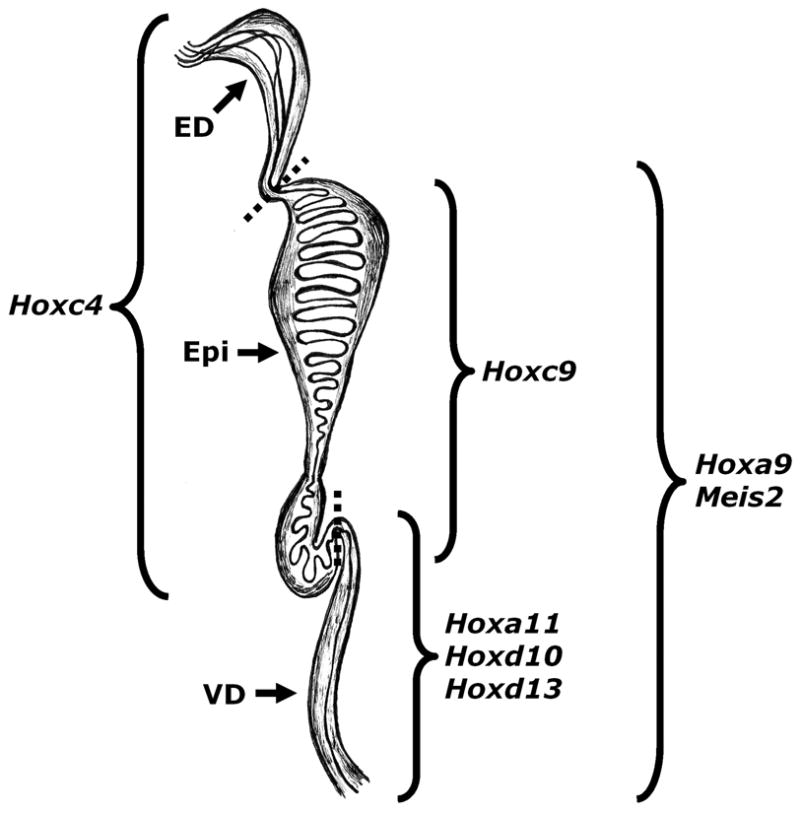
Tissue-enriched Hox transcripts in the developing reproductive tract. Dotted lines represent boundaries between tissues. ED (efferent ducts), Epi (epididymis), and VD (vas deferens).
Developmental regulation of Hox gene expression is observed in many tissues, thus analysis of Hox genes with distinct expression profiles throughout development in one or more tissues of the reproductive tract was undertaken. Expression profiles were only considered of interest if the expression of a given gene varied at least 2-fold throughout development in a given tissue and the change in expression throughout development was considered significant by two-way ANOVA (p < 0.05). Fifteen Hox genes were identified in this analysis (Supp. Fig. 3), one of which (Hoxd13) was also observed to have vas deferens-enriched expression. Eight Hox genes were observed to be developmentally regulated in only one of the three tissues: Dlx5, Hoxc10, and Hoxd13 in the vas deferens; Hoxa2, Hoxa4, Rhox2a, and Tlx2 in the efferent ducts; and Cux2 in the epididymis. The remaining six Hox genes were developmentally regulated in two or more of the tissues examined. Of these genes, developmental regulation between tissues was observed to be either similar or opposing in a gene-specific manner. Of particular interest were two genes with opposing developmental regulation (up in one tissue with age and down in another). These were Hoxd4 and Lhx1, both of which increased with age in the efferent ducts and decreased with age in the vas deferens.
Total Transcriptome Comparison of the Developing Efferent Ducts, Epididymis, and Vas Deferens
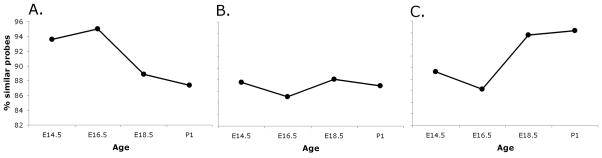
The total transcriptome for each tissue at each time point was determined by selecting for probe IDs having a raw score greater than 50 in both samples of a given tissue/age and being significantly different in at least one comparison across the entire experiment by two-way ANOVA (p <0.05). Comparisons between tissues at each time point were then used to determine the percent similarity between the two transcriptomes, reported as % similar probes (Fig. 4). This analysis determined the epididymis and vas deferens transcriptomes were initially very similar but diverged (decreased in similarity) with age, the vas deferens and efferent ducts began and remained relatively dissimilar throughout the developmental period examined, and the epididymis and efferent ducts were relatively dissimilar at E14.5 but increased in similarity with age. As the total transcriptome comparisons model the changes in tissue morphology occurring within the tract, i.e. increasing similarity between the epididymis and efferent ducts (coiling) and decreasing similarity between the epididymis and the vas deferens (presence or absence of coiling) two comparisons were chosen to shed light on this phenomenon; epididymis-vs.-vas deferens and epididymis-vs.-efferent ducts.
FIG. 4.
Total transcriptome comparisons of efferent ducts, epididymis, and vas deferens throughout development. A.) Epididymis versus vas deferens. B.) Vas deferens versus efferent ducts. C.) Epididymis versus efferent ducts.
Tissue to Tissue Comparisons throughout Development
Epididymis and Vas Deferens Comparisons
Transcripts enriched in either the epididymis or vas deferens, as compared to one another, were identified and their associated biological functions, molecular processes, and cellular components determined via functional annotation clustering. Sixteen transcripts were enriched in the epididymis at all time points while 18 were enriched in the vas deferens at all time points. Functional annotation clustering demonstrated these genes were associated predominantly with three annotation clusters. The first included GO terms for regionalization and pattern specification process. Genes associated with this cluster consisted almost entirely of Hox genes. The second cluster included the GO term for tube morphogenesis and the last cluster consisted of GO terms for the cellular component extracellular matrix. The five transcripts with the highest tissue-enrichment throughout development when comparing epididymis to vas deferens are reported in Table 1.
TABLE 1.
Genes enriched in the epididymis or vas deferens throughout development
| Genes enriched in the epididymis as compared to the vas deferens at all time points | |||
|---|---|---|---|
| Symbol | Name | RefSeq | Average* fold- change** |
| Hoxc6 | homeo box C6 a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin | NM_010465 | 6.6 |
| Adamts16 | type 1 motif, 16 | NM_172053 | 6.2 |
| Lrrc17 | leucine rich repeat containing 17 | NM_028977 | 5.7 |
| Ppp1r14a | protein phosphatase 1, regulatory (inhibitor) subunit 14A | NM_026731 | 5.1 |
| Arhgap20 | Rho GTPase activating protein 20 | NM_175535 | 4.9 |
| Genes enriched in the vas deferens as compared to the epididymis at all time points | |||
| Symbol | Name | RefSeq | Average* fold- change** |
| Hoxd13 | homeo box D13 ELAV (embryonic lethal, abnormal | NM_008275 | 24.5 |
| Elavl2 | vision, Drosophila)-like 2 (Hu antigen B) | NM_207685 | 8.0 |
| Kcnd2 | potassium voltage-gated channel, Shal-related family, member 2 | NM_019697 | 7.7 |
| 6330436F06Rik | RIKEN cDNA 6330436F06 gene | 7.6 | |
| Cxxc4 | CXXC finger 4 | NM_001004367 | 7.2 |
Average across all time points.
Fold-changes calculated by using the average of multiple probes, where applicable.
Transcripts enriched in the epididymis or vas deferens at E14.5, the time prior to significant morphological changes in the tissue and thus potentially involved in early Wolffian duct regionalization, were identified. One-hundred and eighty-nine transcripts in the epididymis and 65 transcripts in the vas deferens were determined to be tissue-enriched at E14.5. Functional annotation clustering determined many of these genes were associated with the cellular component extracellular matrix and the biological processes tube development and skeletal system development. The top five tissue-enriched transcripts at E14.5 when comparing epididymis to vas deferens are reported in Table 2.
TABLE 2.
Genes enriched in the epididymis or vas deferens at E14.5
| Genes enriched in the epididymis at E14.5 as compared to the vas deferens | |||
|---|---|---|---|
| Symbol | Name | RefSeq | Fold-change versus Vas deferens* |
| 2810474O19Rik | RIKEN cDNA 2810474O19 gene | 9.6 | |
| Shisa3 | shisa homolog 3 (Xenopus laevis) similar to nuclear pore complex- associated intranuclear coiled-coil protein TPR /// translocated promoter | NM_001033415 | 7.2 |
| LOC100043998 /// Tpr | region | 5.6 | |
| Eif2c2 | eukaryotic translation initiation factor 2C, 2 | NM_153178 | 5.4 |
| Itih5 | inter-alpha (globulin) inhibitor H5 | NM_172471 | 5 |
| Genes enriched in the vas deferens at E14.5 as compared to the epididymis | |||
| Symbol | Name | RefSeq | Fold-change versus Epididymis* |
| Hoxd13 | homeo box D13 | NM_008275 | 10.4 |
| Hoxa11 | homeo box A11 | NM_010450 | 8.2 |
| Nts | neurotensin | NM_024435 | 8.1 |
| Hoxd11 | homeo box D11 | NM_008273 | 6.7 |
| A2m | alpha-2-macroglobulin | NM_175628 | 4.8 |
Fold-changes calculated by using the average of multiple probes, where applicable.
As significant morphological differentiation of the epididymis and the vas deferens occurs from E14.5 to after birth, genes associated with this process were of interest. When comparing epididymis to the vas deferens, genes associated with tissue-specific morphological changes would be expected to be similar in expression early in development but diverge in expression later. To that end, genes whose expression was similar in the two tissues (fold-change of < 2) at E14.5 but different (fold-change > 2) at later time points were identified. Of the genes with similar expression levels in the epididymis and vas deferens at E14.5; 75, 1770, and 2079 had higher transcript levels in the epididymis at E16.5, E18.5 and P1, respectively and 79, 390, and 559 had higher transcript levels in the vas deferens at E16.5, E18.5, and P1, respectively. GO terms associated with cell adhesion were enriched in the epididymis at all time points and in the vas deferens at the later two time points. Terms associated with extracellular matrix were enriched in both tissues at E16.5 and in the vas deferens at E18.5. Notably, terms associated with apoptosis were enriched at E18.5 in the epididymis and at P1 in the vas deferens. GO terms associated with clusters having an enrichment score of greater than 2.0 in each tissue at each time point are reported in Supp. Table 1.
Epididymis and Efferent Ducts Comparisons
Transcripts enriched in either the epididymis or efferent ducts throughout development, as compared to one another, were identified and their associated biological functions, molecular processes, and cellular components determined via functional annotation clustering. Sixty-two transcripts were enriched in the epididymis at all time points while 26 were enriched in the efferent ducts. Functional annotation clustering demonstrated these genes were most commonly associated with the cellular component membrane and the biological processes of pattern specification and tube development. The five transcripts with the most enriched expression for each tissue throughout development are listed in Table 3.
TABLE 3.
Genes enriched in the epididymis or efferent ducts throughout development
| Genes enriched in the epididymis as compared to the efferent ducts at all time points | |||
|---|---|---|---|
| Symbol | Name | RefSeq | Average* fold- change** |
| Cldn8 | claudin 8 | NM_018778 | 29.4 |
| Spink8 | serine peptidase inhibitor, Kazal type 8 | NM_183136 | 16.7 |
| Hoxc10 | homeo box C10 | NM_010462 | 16.4 |
| Rprm | reprimo, TP53 dependent G2 arrest mediator candidate | NM_023396 | 15.9 |
| Hoxd10 | homeo box D10 | NM_013554 | 11.5 |
| Genes enriched in the efferent ducts as compared to the epididymis at all time points | |||
| Symbol | Name | RefSeq | Average* fold- change** |
| Pdzk1 | PDZ domain containing 1 | NM_021517 | 23.5 |
| Keg1 | kidney expressed gene 1 | NM_029550 | 16.6 |
| Cldn2 | claudin 2 | NM_016675 | 16.5 |
| Vil1 | villin 1 | NM_009509 | 10.9 |
| Aldh1l1 | aldehyde dehydrogenase 1 family, member L1 | NM_027406 | 8.7 |
Average across all time points.
Fold-changes calculated by using the average of multiple probes, where applicable.
In order to identify genes associated with the increasing morphological similarity between the epididymis and efferent ducts, genes with different (> 2-fold difference) expression in the epididymis and efferent ducts at E14.5 but similar (< 2-fold difference) expression at later time points were identified. Of the genes expressed differently at E14.5; 430, 737, and 758 transcripts were expressed similarly at E16.5, E18.5, and P1, respectively, in the two tissues. These genes, like the genes identified in the epididymis and vas deferens comparisons, were commonly associated with the GO terms cell adhesion and extracellular matrix. Additionally, the genes expressed in a similar manner in the epididymis and efferent ducts with increasing age were also associated with the GO terms cell projection organization and cell motility. A summary of the functional annotation results for the epididymis and efferent ducts comparisons can be found in Supp. Table 2.
Ontology Analysis Identification of Potential Morphological Regionalization Genes
In order to identify specific genes that may be playing a role in the morphological regionalization of the male reproductive tract, genes associated with the highly enriched GO term cell adhesion were identified and their tissue distribution and temporal expression patterns determined. This GO term was selected for further study for two reasons, it was identified as enriched throughout development in both tissue comparisons and regulation of cell adhesion is a potential mechanism for producing highly coiled tube structures within a mesenchymal tissue. In all, 92 individual genes were identified as associated with cell adhesion, 13 of which were identified in both the epididymis versus vas deferens and epididymis versus efferent ducts comparisons (Table 4). Of the 92 individual genes identified as associated with cell adhesion, potential players in establishing tissue-specific morphology were determined. A gene was considered a potential regulator of tissue-specific morphology if its expression change throughout development was found to be significant by two-way ANOVA (p < 0.05) and greater than 2-fold in at least one tissue. Additionally, it was required to display either vas deferens enriched or epididymis and efferent duct enriched (significant by two-way ANOVA and 2-fold or greater) expression at a minimum of one time point (Supp. Table 3). Of the genes passing criteria, 21 were vas deferens enriched and 9 epididymis and efferent duct enriched. Vas deferens enriched cell adhesion genes included three contactin genes, three genes encoding integrins, and five cadherin or protocadherin genes. Four well characterized cell adhesion genes were of particular interest: activated leukocyte cell adhesion molecule (Alcam), cadherin 2 (Cdh2), integrin alpha 1 (Itga1), and integrin alpha 8 (Itga8). The epididymis and efferent duct enriched genes included several additional genes of interest with clearly defined roles in cell adhesion: cysteine-rich protein 61 (Cyr61); integrin beta 1 binding protein 1 (Itgb1bp1); matrix metallopeptidase 14 (Mmp14); and periostin, osteoblast specific factor (Postn). Both Alcam and Postn expression can be observed histologically in the embryonic reproductive tract as early as E15.5 (GUDMAP: 11645 and 13280). The full expression profiles of all eight genes of interest can be found in Supp. Fig. 4.
TABLE 4.
Cell adhesion genes identified by ontology analysis of both tissue to tissue comparisons
| Symbol | Name | RefSeq |
|---|---|---|
| Astn1 | astrotactin 1 | NM_007495 |
| Bcl2 | B-cell leukemia/lymphoma 2 | NM_009741 |
| Cdh10 | cadherin 10 | NM_009865 |
| Cdh2 | cadherin 2 | NM_007664 |
| Col8a1 | collagen, type VIII, alpha 1 | NM_007739 |
| Cyr61 | cysteine rich protein 61 | NM_010516 |
| Itga1 | integrin alpha 1 | NM_001033228 |
| Itga4 | integrin alpha 4 | NM_010576 |
| Lef1 | lymphoid enhancer binding factor 1 | NM_010703 |
| Nrxn1 | neurexin I | NM_020252 |
| Pard3 | par-3 (partitioning defective 3) homolog (C. elegans) | NM_033620 |
| Pcdh9 | protocadherin 9 | NM_001081377 |
| Spp1 | secreted phosphoprotein 1 | NM_009263 |
Periostin and cadherin 2 protein localization in the developing epididymis
The high expression of Postn in coiled tissues (the epididymis and efferent duct) relative to the vas deferens and its increasing expression with age implied a potentially important role in reproductive tract tissue remodeling. In order to determine which cell populations were potentially undergoing POSTN mediated tissue remodeling, immunofluorescence was used to localize POSTN in the distal, developing epididymis (Fig. 5). Antibody specificity was confirmed by immunofluorescent detection of POSTN in POSTN positive control tissue (Supp. Fig. 5). At E14.5, POSTN is observed in both the epidermal and mesenchymal populations but is increasingly associated with the mesenchymal cell populations with age. By P1, POSTN is observed exclusively in the mesenchymal population of the epididymis. A notable shift from predominantly cytoplasmic to membrane or extracellular matrix associated is also observed with increasing age in the mesenchymal population as well (Supp. Fig. 5).
FIG. 5.
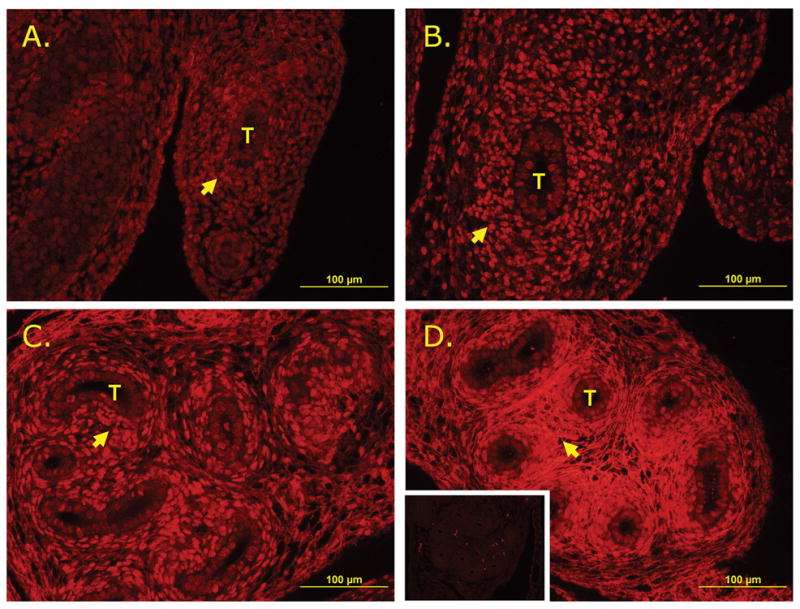
Immunofluorescent localization of POSTN in developing epididymis. A.) E14.5. B.) E16.5. C.) E18.5. D.) P1 (inset: negative control). Mesenchymal cells marked by arrows and epididymal tubules denoted by T.
The rapidly decreasing expression of Cdh2 in the epididymis implied it may also be mediating tissue remodeling events. In order to determine the CDH2-positive cell populations in the epididymis and confirm a general reduction of CDH2 in the developing epididymis, immunofluorescence against CDH2 was performed on the distal, developing epididymis (Fig. 6). Throughout development, CDH2 is associated predominantly with the epithelial cells of the epididymis with some observable signal in the mesenchymal cells early in development, however by P1, CDH2 is observed exclusively in a small subpopulation of epithelial cells in the distal epididymis.
FIG. 6.
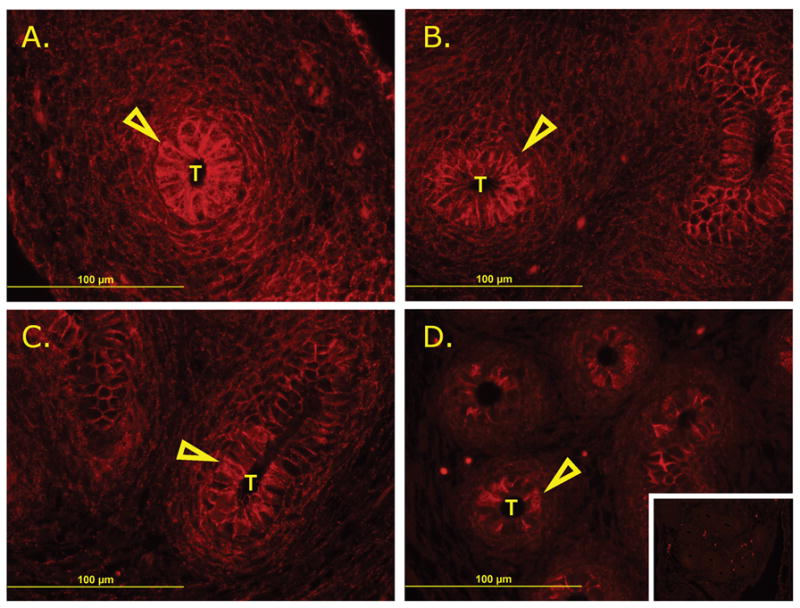
Immunofluorescent localization of CDH2 in developing epididymis. A.) E14.5. B.) E16.5. C.) E18.5. D.) P1 (inset: negative control). Epithelial cells marked by open arrows and epididymal tubules denoted by T.
Discussion
The reproductive tract is composed of a series of tissues that undergo dramatic morphological changes during embryonic development. The molecular mechanisms underpinning these changes are not clearly understood. Global gene expression studies via microarray analysis can identify potential mediators of morphological regionalization. This work represents the first global gene analysis of the developing reproductive tract in a tissue-specific manner and aimed to achieve two goals. First, to provide access to gene expression profiles of the developing reproductive tract for the research community and present strong evidence supporting the validity of the dataset. And second, to demonstrate how the provided expression profiles can be used as an initiation point for hypothesis driven research.
Validation of Tissue Collection and Array Output
This study utilized developing reproductive tract tissues that required careful dissection from many embryos. Initial evaluations of the array output using hierarchical clustering revealed the collections were performed with accuracy and the data were representative. Clustering analysis of genes identified as differentially expressed within or between tissues, demonstrated duplicate quality to be exceptional with all samples clustering with their respective duplicate. Clustering analysis also revealed an underlying relationship between the three tissues examined which would be observed throughout the analysis. Early in development, the epididymis and the vas deferens cluster together, however as development progresses the transcriptomes of the two Wolffian duct derived tissues diverge, resulting in late (E18.5 and P1) epididymis samples clustering separately from late vas deferens. And, although throughout development the efferent ducts transcriptome remains independent of the other two transcriptomes, late epididymis clusters more closely to late efferent duct tissue than to late vas deferens. This implies the epididymis and efferent ducts increase in similarity with age while the epididymis and vas deferens decrease in similarity with age.
In addition to clustering analysis, the expression of tissue-enriched markers early in development was examined to ensure proper isolation of the two Wolffian duct derived tissues. These markers included Dcn, Itga4, and Meg3 in the epididymis and Ace2, Stc1, and Tmem100 in the vas deferens. Each was observed to have the expected pattern of expression within this microarray analysis. Collectively, the expression profile of known tissue-enriched genes and the clustering analysis provide a high level of confidence in the array output and demonstrate the importance of tissue-specific transcriptome analysis in the developing reproductive tract.
Hox Gene Expression in the Developing Reproductive Tract
Regionalization is required for proper development and eventual function of the reproductive tract. Loss of function mutations in various Hox genes result in partial or complete transformation of one tissue of the reproductive tract to another and in most cases, a loss or decrease in fertility (Hsieh-Li et al., 1995; Podlasek et al., 1999). Thus, identification of previously uncharacterized Hox genes with tissue-enriched or temporally-regulated patterns of expression may reveal novel mediators of morphological regionalization within the reproductive tract.
Two classes of Hox genes were examined in this study: those expressed in a tissue-enriched manner throughout development and those with expression profiles indicative of developmental regulation. Of these, two expression profiles are of particular interest: those mediating the possible regionalization of the Wolffian duct and those involved in the diverging morphology of the epididymis and the vas deferens (and conversely the converging morphology of the efferent ducts and epididymis). Potential Wolffian duct regionalization candidates would include those Hox genes with opposing or diverging expression profiles in the epididymis and vas deferens. Of the 20 Hox genes examined in detail, 12 have expression profiles indicative of Wolffian duct regionalization mediators (Hoxc9, Hoxd10, Hoxa11, Hoxc4, Hoxc10, Hoxd13, Meis2, Cux2, Hoxd4, Hoxd8, Lhx1, and Mkx). Additionally, three Hox genes (Dlx5, Msx2, and Prrx2) are initially highly expressed in the vas deferens but less so in the epididymis, suggesting they may play very early roles in establishing Wolffian duct regions. The expression of all three decreases rapidly after E14.5, implying their role may be limited to a very specific time frame. Notably, this analysis identified Hoxa11 as a potential regionalization factor in the Wolffian duct. Previous work using knockout animals demonstrated a fundamental role for this gene in regulating the epididymal/vas deferens boundary (Hsieh-Li et al., 1995), further validating the approach taken for this analysis.
Genes associated with the morphological regionalization of the reproductive tract would be expected to have similar expression profiles in the epididymis and efferent ducts but dissimilar or diverging expression profiles between the epididymis and vas deferens. Four of the 20 fully analyzed Hox genes have this type of expression profile, including Hoxc4, Hoxd4, Hoxd8, and Lhx1. Of note, four Hox genes were identified as efferent duct-enriched for at least two developmental frames: Rhox2a and Tlx2 early in development and Hoxa2 and Hoxa4 later in development. To the authors’ best knowledge this represents the first description of efferent duct-enriched Hox gene expression.
Total Transcriptome Comparisons
As hierarchical clustering indicated a dynamic relationship between the transcriptomes of the developing reproductive tract tissues, total transcriptome comparisons between tissues and across development were performed to quantify this observation. The results from this comparison demonstrate that the transcriptomes of the epididymis and vas deferens are increasingly different with age, whereas the transcriptomes of the epididymis and efferent ducts become more similar with age. The transcriptomes of the vas deferens and efferent ducts begin and remain dissimilar throughout development. These results fit well with what is known about the biology of the three tissues. The epididymis and vas deferens are derived from the same embryonic tissue but diverge morphologically early in development while the epididymis and efferent ducts are derived from different embryonic tissue but become morphologically similar with age. The vas deferens and efferent ducts are derived from different embryonic tissue and have distinct morphologies throughout development. Results of the transcriptome analysis further support the premise that morphological convergence between the epididymis and efferent ducts is driven by similar transcriptional networks while morphological divergence between the epididymis and vas deferens is due, at least in part, to increasing differences in the transcriptomes of the tissues.
Tissue Specific Comparisons – Epididymis to Vas deferens
As distinct morphological changes occur between E14.5 and P1 in the epididymis and vas deferens and total transcriptome analysis indicated the two tissues are increasingly different, this work aimed to identify genes whose expression is similar in the two tissues early in development but differed later. These genes represent potential mediators of the observed morphological differences between the two tissues. Ontology analysis of the resulting probe lists demonstrated the GO term cell adhesion was commonly enriched in both tissue types. Additionally, the GO term extracellular matrix was enriched in both tissues early in development, the period in which major morphological differentiation is initiated. These results imply that the morphological differences between the epididymis and vas deferens may be, in part, a result of tissue-specific regulation of the extracellular matrix and cell adhesion. The major morphological difference between the two tissues is the presence of a high degree of tube coiling initiated after E14.5 in the epididymis but completely absent in the vas deferens. Thus, tissue-specific regulation of extracellular matrix-related genes and cell adhesion-related genes may play an important role in the development of the coiled or non-coiled phenotypes.
Tissue Specific Comparisons – Epididymis to Efferent ducts
Unlike the epididymis and vas deferens, the epididymis and efferent ducts morphologically converge over the course of development. This convergence is also evident in the transcriptomes of the two tissues as they become increasingly similar with age. Genes whose expression is initially different in the two tissues but becomes similar over time may mediate the increasingly similar morphology in the different tissues. This analysis demonstrated genes with the aforementioned expression pattern were enriched for several GO terms, including cell adhesion, extracellular matrix, cell projection organization, and cell motility. The demonstration that these GO terms are increasingly similar between the epididymis and efferent ducts and increasingly dissimilar between the epididymis and vas deferens imply these processes may represent the mechanisms by which these tissues arrive at their final morphology.
Ontology Analysis Identification of Potential Morphological Regionalization Genes
As ontology analysis of both epididymis to vas deferens and epididymis to efferent ducts comparisons implied cell adhesion may be tightly regulated along the length of the reproductive tract, expression profiles of genes associated with cell adhesion were examined. Eight genes of interest were identified from this analysis: four expressed most highly in the vas deferens (Alcam, Cdh2, Itga1, and Itga8) and four expressed most highly in the epididymis and efferent ducts (Cyr61, Itgb1bp1, Mmp14, and Postn). ALCAM-mediated cell adhesion plays an important role in diverse biological processes including development and cancer metastasis (Swart, 2002). Cdh2 encodes the very well characterized adhesion component N-cadherin, important in the formation of cell/cell adhesion complexes (Borghi and James Nelson, 2009). Itga1 is known to mediate cell adhesion via its interaction with collagen (Eble et al., 2006) and Itga8 has pro-adhesive properties in vitro (Bieritz et al., 2003). The increased expression with age and significantly higher expression of pro-adhesive genes in the vas deferens relative to the epididymis and efferent ducts imply a greater capacity for cell adhesion in the vas deferens relative to the other tissues of the reproductive tract.
Of the cell adhesion genes enriched in the epididymis and efferent ducts, each has been associated with decreases in or instability of cell adhesion. Loss of Itgb1bp1 in osteoblasts results in enhanced adhesion to extracellular matrices (Bouvard et al., 2007) while Mmp14 and Postn expression is associated with increased metastatic potential and invasiveness in many cancers (Ip et al., 2005; Moss et al., 2009; Ruan et al., 2009). Epididymal and efferent duct enriched expression of Cyr61 is of particular interest as its expression is induced with mechanical stress (Chaqour and Goppelt-Struebe, 2006). The downstream result of Cyr61 expression is modulation of cell adhesion and the extracellular matrix. The relatively high expression of genes associated with decreased cell adhesion and mechanical stress in the epididymis and efferent ducts relative to the vas deferens implies epididymal and efferent duct tissue may have an overall decrease in cell to cell adhesion, however it does not address the question of whether this decrease is occurring in the epithelium of the tubes or the surrounding mesenchyme.
To address this issue, immunofluorescence localization of POSTN and CDH2 was undertaken in the distal epididymis. These analyses demonstrated an increasingly mesenchymal localization for POSTN within the developing epididymis and an increasingly epithelial localization for CDH2. The high level of Postn expression in the epididymis and efferent ducts and its shift in localization within the mesenchyme of the epididymis implies POSTN may induce an overall decrease in cell cohesion in the mesenchyme, perhaps producing an environment more permissive for coiling phenotypes in the epididymis and efferent ducts. This correlates well with the observed general reduction of CDH2 with age in the epididymal mesenchyme implying CDH2 loss may also decrease cell adhesion in the mesenchyme. CDH2 maintenance in the epithelial population indicates CDH2 may be important in maintaining epididymal cell associations during the coiling process. The immunofluorescent results not only confirm the microarray-observed expression patterns but also demonstrate potential modulation of cell adhesion in both cell populations (the epithelial and mesenchymal), further supporting the notion that cell adhesion may be a key mechanism driving the coiling phenotype. Taken together, the expression profile of cell adhesion-associated genes indicates differential regulation of the same mechanism (cell adhesion) between the tissues of the reproductive tract may be responsible for their final coiled or uncoiled morphology.
Comparison of the three tissues examined in this work highlights the importance of both divergent and convergent expression patterns in the final morphology of the developing male reproductive tract. More importantly, the availability of tissue-specific datasets describing the entire transcriptome of the developing reproductive tract provides an invaluable opportunity to address questions regarding regionalization in a complex tissue system that is vital to mammalian reproduction.
Methods
Animal Procedures and Sample Processing
All animal experiments complied with the regulations set forth by the Animal Welfare Act (Public Law 91–579), the Guide for the Care and Use of Laboratory Animals (NRC, 1996) published by the Department of Health and Human Services, and the policies and procedures of the University of Virginia Institutional Animal Care and Use Committee. Animals were maintained in a humidity- and temperature-controlled environment with food and water provided ad libitum. Male embryos at E14.5, 16.5, 18.5 and P1 were collected from pregnant CD1 mice (Charles River, Wilmington, MA). The Wolffian ducts and the testes were removed and immediately placed into RNAlater® (Life Technologies Corp., Carlsbad, CA). The efferent ducts, the Wolffian duct and the vas deferens were separated from the testis and from each other and were placed into Trizol® reagent (Life Technologies Corp., Carlsbad, CA). Total RNA was extracted according to the manufacturer’s instructions. Three pools of samples were collected, with each individual animal contributing to only a single pool of a given tissue type. Thus, for all remaining experimental steps, pools were treated as biological replicate samples. In order to obtain sufficient RNA for analysis, the number of samples collected were as follows: E14.5; 50, 54 and 74 samples of each tissue type; E16.5; 62, 63 and 58 samples of each tissue type; E18.5; 56, 64 and 72 samples of each tissue type and P1; 37, 48 and 66 samples of each tissue type. Approximately 500 ng of RNA from each of the pooled samples were sent to the Biomolecular Research Facility Core at the University of Virginia School of Medicine where RNA was quantified on an ND-1000 Spectrometer (NanoDrop, Willmington, DE) and quality was assessed using a Bioanalyzer 2100 (Agilest, Palo Alto, CA). Only samples with an RNA integrity number, determined by Agilent’s 2100 Expert Software, of greater than 7.0 were used for further analysis. This measure of quality takes into account both the ratio of ribosomal bands as well as the presence of degraded products and as such is an accurate measure of quality.
Microarray Processing and Quality Control
Duplicate samples with the highest quality for each of the 12 sample types were selected and approximately 3 go of efferent duct, epididymis, or vas deferens total RNA was labeled using a Gene Chip One-cycle Target Labeling Kit (Affymetrix, Santa Clara, CA) to produce labeled crank which was then hybridized to the Affymetrix Gene Chip Mouse Genome M430 2.0 array. Hybridization quality was assessed using GCOS 1.4 (Affymetrix). Production of crank, hybridization to arrays and evaluation of hybridization quality were completed by the Laboratory for Biotechnology and Misanalysis I at Washington State University, Pullman, WA.
Microarray Analysis and Production of Gene Lists
Array output was normalized by the Robust Multichip Average (RMA) method with a per gene normalization to the median. Normalization and analysis of data was conducted using Gene Spring version 7.3.1 (Agilent Technologies, Palo Alto, CA). Probe IDs were considered enriched or reduced if 1.) the raw score was greater than 50 in both samples of a given sample type, 2.) the fold-change between comparisons was greater than 2-fold using the average normalized value, and 3.) the comparison was found to be significant by two-way ANOVA (p = 0.05) with a Benjamin and Hochberg False Discovery Rate multiple test correction.
Statistical analysis of selected genes
Normalized array values for each sample and each gene of interest were extracted from Gene Spring version 7.3.1 and averaged across multiple probe IDs, where appropriate. Averaged and normalized values were then imported into JMP 7.0.1 (SAS Institute Inc.; Cary, NC). Two-way ANOVA was performed on each sample for each gene individually. Connecting letters report was generated using least squared means differences (Turkey’s HSD) against the interaction term (tissue by age).
Ontology Analysis
Probe IDs identified in the microarray analysis as of interest were uploaded to DAVID Bioinformatics Resources (http://david.abcc.ncifcrf.gov/) and their biological process, cellular component, and molecular function annotation defined by using the functional clustering annotation tools. Clusters with associated GO terms and enrichment scores greater than 2.0 were selected for further study. Cluster names are derived from the most biologically relevant GO term assigned to that cluster.
Immunofluorescent detection of periostin and cadherin 2 protein
Epididymal tissue was immersion-fixed in 4% paraformaldehyde overnight at 4 C. Tissues were sent to the Center for Research in Reproduction Cell Science Core at the University of Virginia for paraffin embedding and sectioning. Slide-mounted sections were deparaffinized, rehydrated then microwaved in antigen unmasking solution (Vector Laboratories, Burlingame, CA) for 10 minutes on high in a 1300 W microwave and cooled for 1 hour at room temperature. Slides were blocked with 10% normal goat serum (Vector Laboratories) and 0.5% (v/v) gelatin from cold-water fish skin (FSG; Sigma) in TBS for 1.5 hours. Slides were then incubated overnight at 4 C in 1:50 dilution of POSTN primary antibody (Santa Cruz biotech sc-67233) or in a 1:400 dilution of CDH2 primary antibody (Cat#2019-1 Epitomics, Inc., California) in the blocking solution. After washing in TBS, the slides were incubated with a 1:200 dilution of Alexa Fluoro-goat anti-rabbit (Molecular Probe, Oregon) in the blocking solution for 1.5 hours at room temperature. All slides were washed in TBS, mounted with Prolong Anti-fade reagent (Molecular Probe, Oregon), and viewed in a Zeiss microscope equipped with epifluorescence.
Supplementary Material
SUPP. FIG. 1. Schematic of the developing reproductive tract from E14.5 to P1. Dotted lines represent boundaries between tissues. At E14.5, the efferent ducts, epididymis, and vas deferens are uncoiled and the Mullerian duct is still fully intact. By E16.5, the Mullerian duct has undergone significant degeneration and coiling has been initiated in the efferent ducts and proximal portion of the epididymis. The Mullerian duct is no longer observed by E18.5. Coiling continues in the efferent ducts, and moves distally in the epididymis from E18.5 to birth. After birth, coiling is essentially complete in both the efferent ducts and epididymis. Throughout development and into adulthood, coiling is not observed in the vas deferens. ED (efferent ducts), Epi (epididymis), VD (vas deferens), WD (Wolffian duct) and MD (Mullerian duct).
SUPP. FIG. 2. Light image of E14.5 dissected reproductive tract. Dotted lines represent boundaries between tissues. ED (efferent ducts), Epi (epididymis), VD (vas deferens), and MD (Mullerian duct).
SUPP. FIG. 3. Tissue and temporal expression profiles of Hox transcripts in the developing reproductive tract. A.) Tissue-enriched Hox transcripts. B.) Tissue-enriched and developmentally regulated Hox transcript. C.) Developmentally regulated Hox transcripts. Error bars represent standard deviation. Normalized microarray value represents the average of multiple probes where applicable.
SUPP. FIG. 4. Tissue and temporal expression profiles of ontology identified cell adhesion genes in the developing reproductive tract. A.) Vas deferens enriched. B.) Epididymis and efferent duct enriched. Error bars represent standard deviation. Microarray raw score represents the average of multiple probes where applicable.
SUPP. FIG. 5. Membrane-associated POSTN in adult heart and P1 epididymis. A.) Control tissue (adult heart (Kii et al., 2010)). B.) P1 epididymis. POSTN (red) overlaid with nuclear counter stain (DAPI, blue).
SUPP. TABLE 1. Functional annotation clustering of genes identified by comparing epididymis and vas deferens across development.
SUPP. TABLE 2. Functional annotation clustering of genes identified by comparing epididymis and efferent ducts across development.
SUPP. TABLE 3. Ontology identified cell adhesion genes passing criteria for developmental regulation and either vas deferens or epididymis and efferent duct enrichment.
Acknowledgments
The authors would like to thank Derek Pouchnik and the Laboratory for Bioanalysis and Biotechnology I (LBBI) for GeneChip processing and Jennifer Hilton for artistic contributions. This work supported by grants NIH-NICHD HD 10808 (ES and MG), NIH-NICHD KO8 award HD42058 (DB), NIH-NICHD HD52035 (BTH), and The Eunice Kennedy Shriver Institute for CHHD, SCCPIR program U54 28934 (DB and BTH).
Abbreviations
- E
embryonic day
- P
postnatal day
- Hox
homeobox gene
- DAVID
Database for Annotation, Visualization, and Integrated Discovery
- GEO
Gene Expression Omnibus
- GO
Gene Ontology
- GUDMAP
Genitourinary Database Molecular Anatomy Project
References
- Bieritz B, Spessotto P, Colombatti A, Jahn A, Prols F, Hartner A. Role of alpha8 integrin in mesangial cell adhesion, migration, and proliferation. Kidney Int. 2003;64:119–127. doi: 10.1046/j.1523-1755.2003.00057.x. [DOI] [PubMed] [Google Scholar]
- Blomqvist SR, Vidarsson H, Soder O, Enerback S. Epididymal expression of the forkhead transcription factor Foxi1 is required for male fertility. EMBO J. 2006;25:4131–4141. doi: 10.1038/sj.emboj.7601272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghi N, James Nelson W. Intercellular Adhesion in Morphogenesis: Molecular and Biophysical Considerations. In: Thomas L, editor. Current Topics in Developmental Biology. Chapter 1. Academic Press; 2009. pp. 1–32. [DOI] [PubMed] [Google Scholar]
- Bouvard D, Aszodi A, Kostka G, Block MR, Albiges-Rizo C, Fassler R. Defective osteoblast function in ICAP-1-deficient mice. Development. 2007;134:2615–2625. doi: 10.1242/dev.000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaqour B, Goppelt-Struebe M. Mechanical regulation of the Cyr61/CCN1 and CTGF/CCN2 proteins. FEBS J. 2006;273:3639–3649. doi: 10.1111/j.1742-4658.2006.05360.x. [DOI] [PubMed] [Google Scholar]
- Chauvin TR, Griswold MD. Androgen-regulated genes in the murine epididymis. Biol Reprod. 2004;71:560–569. doi: 10.1095/biolreprod.103.026302. [DOI] [PubMed] [Google Scholar]
- Clulow J, Jones RC, Hansen LA. Micropuncture and cannulation studies of fluid composition and transport in the ductuli efferentes testis of the rat: comparisons with the homologous metanephric proximal tubule. Exp Physiol. 1994;79:915–928. doi: 10.1113/expphysiol.1994.sp003817. [DOI] [PubMed] [Google Scholar]
- Clulow J, Jones RC, Hansen LA, Man SY. Fluid and electrolyte reabsorption in the ductuli efferentes testis. J Reprod Fertil Suppl. 1998;53:1–14. [PubMed] [Google Scholar]
- Coolen LM, Allard J, Truitt WA, McKenna KE. Central regulation of ejaculation. Physiology & Behavior. 2004;83:203–215. doi: 10.1016/j.physbeh.2004.08.023. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- Di-Poi N, Zakany J, Duboule D. Distinct roles and regulations for HoxD genes in metanephric kidney development. PLoS Genet. 2007;3:e232. doi: 10.1371/journal.pgen.0030232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube E, Chan PT, Hermo L, Cyr DG. Gene expression profiling and its relevance to the blood-epididymal barrier in the human epididymis. Biol Reprod. 2007;76:1034–1044. doi: 10.1095/biolreprod.106.059246. [DOI] [PubMed] [Google Scholar]
- Eble JA, Kassner A, Niland S, Morgelin M, Grifka J, Grassel S. Collagen XVI harbors an integrin alpha1 beta1 recognition site in its C-terminal domains. J Biol Chem. 2006;281:25745–25756. doi: 10.1074/jbc.M509942200. [DOI] [PubMed] [Google Scholar]
- Ezer N, Robaire B. Gene expression is differentially regulated in the epididymis after orchidectomy. Endocrinology. 2003;144:975–988. doi: 10.1210/en.2002-220705. [DOI] [PubMed] [Google Scholar]
- Hansen LA, Clulow J, Jones RC. Perturbation of fluid reabsorption in the efferent ducts of the rat by testosterone propionate, 17beta-oestradiol 3-benzoate, flutamide and tamoxifen. Int J Androl. 1997;20:265–273. doi: 10.1046/j.1365-2605.1997.00069.x. [DOI] [PubMed] [Google Scholar]
- Hess RA, Bunick D, Lee KH, Bahr J, Taylor JA, Korach KS, Lubahn DB. A role for oestrogens in the male reproductive system. Nature. 1997;390:509–512. doi: 10.1038/37352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo-Sanchez M, Millet S, Bloch-Gallego E, Alvarado-Mallart RM. Specification of the meso-isthmo-cerebellar region: the Otx2/Gbx2 boundary. Brain Res Brain Res Rev. 2005;49:134–149. doi: 10.1016/j.brainresrev.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Hsieh-Li HM, Witte DP, Weinstein M, Branford W, Li H, Small K, Potter SS. Hoxa 11 structure, extensive antisense transcription, and function in male and female fertility. Development. 1995;121:1373–1385. doi: 10.1242/dev.121.5.1373. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ip YC, Cheung ST, Leung KL, Fan ST. Mechanism of metastasis by membrane type 1-matrix metalloproteinase in hepatocellular carcinoma. World J Gastroenterol. 2005;11:6269–6276. doi: 10.3748/wjg.v11.i40.6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinsky SA, Turner TT, Bang HJ, Finger JN, Solarz MK, Wilson E, Brown EL, Kopf GS, Johnston DS. The rat epididymal transcriptome: comparison of segmental gene expression in the rat and mouse epididymides. Biol Reprod. 2007;76:561–570. doi: 10.1095/biolreprod.106.057323. [DOI] [PubMed] [Google Scholar]
- Johnston DS, Jelinsky SA, Bang HJ, DiCandeloro P, Wilson E, Kopf GS, Turner TT. The mouse epididymal transcriptome: transcriptional profiling of segmental gene expression in the epididymis. Biol Reprod. 2005;73:404–413. doi: 10.1095/biolreprod.105.039719. [DOI] [PubMed] [Google Scholar]
- Joseph A, Yao H, Hinton BT. Development and morphogenesis of the Wolffian/epididymal duct, more twists and turns. Dev Biol. 2009;325:6–14. doi: 10.1016/j.ydbio.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kii I, Nishiyama T, Li M, Matsumoto K, Saito M, Amizuka N, Kudo A. Incorporation of tenascin-C into the extracellular matrix by periostin underlies an extracellular meshwork architecture. J Biol Chem. 2010;285:2028–2039. doi: 10.1074/jbc.M109.051961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little MH, Brennan J, Georgas K, Davies JA, Davidson DR, Baldock RA, Beverdam A, Bertram JF, Capel B, Chiu HS, Clements D, Cullen-McEwen L, Fleming J, Gilbert T, Herzlinger D, Houghton D, Kaufman MH, Kleymenova E, Koopman PA, Lewis AG, McMahon AP, Mendelsohn CL, Mitchell EK, Rumballe BA, Sweeney DE, Valerius MT, Yamada G, Yang Y, Yu J. A high-resolution anatomical ontology of the developing murine genitourinary tract. Gene Expr Patterns. 2007;7:680–699. doi: 10.1016/j.modgep.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendive F, Laurent P, Van Schoore G, Skarnes W, Pochet R, Vassart G. Defective postnatal development of the male reproductive tract in LGR4 knockout mice. Dev Biol. 2006;290:421–434. doi: 10.1016/j.ydbio.2005.11.043. [DOI] [PubMed] [Google Scholar]
- Monier B, Tevy MF, Perrin L, Capovilla M, Semeriva M. Downstream of homeotic genes: in the heart of Hox function. Fly (Austin) 2007;1:59–67. doi: 10.4161/fly.3993. [DOI] [PubMed] [Google Scholar]
- Moss NM, Barbolina MV, Liu Y, Sun L, Munshi HG, Stack MS. Ovarian cancer cell detachment and multicellular aggregate formation are regulated by membrane type 1 matrix metalloproteinase: a potential role in I.p. metastatic dissemination. Cancer Res. 2009;69:7121–7129. doi: 10.1158/0008-5472.CAN-08-4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podlasek CA, Seo RM, Clemens JQ, Ma L, Maas RL, Bushman W. Hoxa-10 deficient male mice exhibit abnormal development of the accessory sex organs. Dev Dyn. 1999;214:1–12. doi: 10.1002/(SICI)1097-0177(199901)214:1<1::AID-DVDY1>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Ramos C, Robert B. msh/Msx gene family in neural development. Trends Genet. 2005;21:624–632. doi: 10.1016/j.tig.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Robaire HL, Hinton BT, editors. The Epididymis: From Molecules to Clinical Practice, A Comprehensive Survey of the Efferent ducts, the Epididymis, and the Vas Deferens. New York, NY: Kluwer Academic/Plenum Publisher; 2002. [Google Scholar]
- Ruan K, Bao S, Ouyang G. The multifaceted role of periostin in tumorigenesis. Cell Mol Life Sci. 2009;66:2219–2230. doi: 10.1007/s00018-009-0013-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder EM, Small CL, Li Y, Griswold MD. Regulation of gene expression by estrogen and testosterone in the proximal mouse reproductive tract. Biol Reprod. 2009;81:707–716. doi: 10.1095/biolreprod.109.079053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg-Riethmacher E, Walter B, Riethmacher D, Godecke S, Birchmeier C. The c-ros tyrosine kinase receptor controls regionalization and differentiation of epithelial cells in the epididymis. Genes Dev. 1996;10:1184–1193. doi: 10.1101/gad.10.10.1184. [DOI] [PubMed] [Google Scholar]
- Swart GW. Activated leukocyte cell adhesion molecule (CD166/ALCAM): developmental and mechanistic aspects of cell clustering and cell migration. Eur J Cell Biol. 2002;81:313–321. doi: 10.1078/0171-9335-00256. [DOI] [PubMed] [Google Scholar]
- Turner TT, Johnston DS, Finger JN, Jelinsky SA. Differential gene expression among the proximal segments of the rat epididymis is lost after efferent duct ligation. Biol Reprod. 2007;77:165–171. doi: 10.1095/biolreprod.106.059493. [DOI] [PubMed] [Google Scholar]
- Vignozzi L, Filippi S, Morelli A, Luconi M, Jannini E, Forti G, Maggi M. Regulation of epididymal contractility during semen emission, the first part of the ejaculatory process: a role for estrogen. J Sex Med. 2008;5:2010–2016. doi: 10.1111/j.1743-6109.2008.00914.x. quiz 2017. [DOI] [PubMed] [Google Scholar]
- Wagenfeld A, Yeung CH, Lehnert W, Nieschlag E, Cooper TG. Lack of glutamate transporter EAAC1 in the epididymis of infertile c-ros receptor tyrosine-kinase deficient mice. J Androl. 2002;23:772–782. [PubMed] [Google Scholar]
- Wolgemuth DJ, Behringer RR, Mostoller MP, Brinster RL, Palmiter RD. Transgenic mice overexpressing the mouse homoeobox-containing gene Hox-1.4 exhibit abnormal gut development. Nature. 1989;337:464–467. doi: 10.1038/337464a0. [DOI] [PubMed] [Google Scholar]
- Xu Y, Yeung CH, Setiawan I, Avram C, Biber J, Wagenfeld A, Lang F, Cooper TG. Sodium-inorganic phosphate cotransporter NaPi-IIb in the epididymis and its potential role in male fertility studied in a transgenic mouse model. Biol Reprod. 2003;69:1135–1141. doi: 10.1095/biolreprod.103.018028. [DOI] [PubMed] [Google Scholar]
- Yamazaki K, Adachi T, Sato K, Yanagisawa Y, Fukata H, Seki N, Mori C, Komiyama M. Identification and characterization of novel and unknown mouse epididymis-specific genes by complementary DNA microarray technology. Biol Reprod. 2006;75:462–468. doi: 10.1095/biolreprod.105.048058. [DOI] [PubMed] [Google Scholar]
- Zacchetti G, Duboule D, Zakany J. Hox gene function in vertebrate gut morphogenesis: the case of the caecum. Development. 2007;134:3967–3973. doi: 10.1242/dev.010991. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPP. FIG. 1. Schematic of the developing reproductive tract from E14.5 to P1. Dotted lines represent boundaries between tissues. At E14.5, the efferent ducts, epididymis, and vas deferens are uncoiled and the Mullerian duct is still fully intact. By E16.5, the Mullerian duct has undergone significant degeneration and coiling has been initiated in the efferent ducts and proximal portion of the epididymis. The Mullerian duct is no longer observed by E18.5. Coiling continues in the efferent ducts, and moves distally in the epididymis from E18.5 to birth. After birth, coiling is essentially complete in both the efferent ducts and epididymis. Throughout development and into adulthood, coiling is not observed in the vas deferens. ED (efferent ducts), Epi (epididymis), VD (vas deferens), WD (Wolffian duct) and MD (Mullerian duct).
SUPP. FIG. 2. Light image of E14.5 dissected reproductive tract. Dotted lines represent boundaries between tissues. ED (efferent ducts), Epi (epididymis), VD (vas deferens), and MD (Mullerian duct).
SUPP. FIG. 3. Tissue and temporal expression profiles of Hox transcripts in the developing reproductive tract. A.) Tissue-enriched Hox transcripts. B.) Tissue-enriched and developmentally regulated Hox transcript. C.) Developmentally regulated Hox transcripts. Error bars represent standard deviation. Normalized microarray value represents the average of multiple probes where applicable.
SUPP. FIG. 4. Tissue and temporal expression profiles of ontology identified cell adhesion genes in the developing reproductive tract. A.) Vas deferens enriched. B.) Epididymis and efferent duct enriched. Error bars represent standard deviation. Microarray raw score represents the average of multiple probes where applicable.
SUPP. FIG. 5. Membrane-associated POSTN in adult heart and P1 epididymis. A.) Control tissue (adult heart (Kii et al., 2010)). B.) P1 epididymis. POSTN (red) overlaid with nuclear counter stain (DAPI, blue).
SUPP. TABLE 1. Functional annotation clustering of genes identified by comparing epididymis and vas deferens across development.
SUPP. TABLE 2. Functional annotation clustering of genes identified by comparing epididymis and efferent ducts across development.
SUPP. TABLE 3. Ontology identified cell adhesion genes passing criteria for developmental regulation and either vas deferens or epididymis and efferent duct enrichment.