Abstract
Rationale and Objective
Assessment of the breast tissue pattern asymmetry depicted on bilateral mammograms is routinely used by radiologists when reading and interpreting mammograms. The purpose of this study is to develop an automated scheme to detect breast tissue asymmetry depicted on bilateral mammograms and use the computed asymmetric features to predict the likelihood (or the risk) of women having or developing breast abnormalities or cancer.
Materials and Methods
A testing dataset was selected from a large and diverse full-field digital mammography image database, which includes 100 randomly selected negative cases (not recalled during the screening) and 100 positive cases for having or developing breast abnormalities or cancer. Among these positive cases 40 were recalled (biopsy) due to suspicious findings in which 8 were determined as high-risk with the lesions surgically removed and the remaining were proven to be benign, and 60 cases were acquired from examinations that were interpreted as negative (without dominant masses or micro-calcifications) but the cancers were detected 6 to 18 months later. A computerized scheme was developed to detect asymmetry of mammographic tissue density represented by the related feature differences computed from bilateral images. Initially, each of 20 features was tested to classify between the positive and the negative cases. To further improve the classification performance, a genetic algorithm (GA) was applied to select a set of optimal features and build an artificial neural network (ANN). The leave-one-case-out validation method was used to evaluate the ANN classification performance.
Results
Using a single feature, the maximum classification performance level measured by the area under the receiver operating characteristic curve (AUC), was 0.681±0.038. Using the GA-optimized ANN, the classification performance level increased to an AUC = 0.754±0.024. At 90% specificity, the ANN classifier yielded 42% sensitivity in which 42 positive cases were correctly identified. Among them, 30 were the “prior” examinations of the cancer cases and 12 were recalled benign cases, which represent 50% and 30% sensitivity levels in these two sub-groups, respectively.
Conclusions
This study demonstrated that using the computerized detected feature differences related to the bilateral mammographic breast tissue asymmetry, an automated scheme is able to classify a set of testing cases into the two groups of positive or negative of having or developing breast abnormalities and/or cancer. Hence, further development and optimization of this automated method may eventually help radiologists identify a fraction of women at high-risk of developing breast cancer and ultimately detect cancer at an early stage.
Keywords: Breast cancer, Mammography, Mammographic breast tissue asymmetry, Computer-aided detection (CAD), Risk assessment
I. INTRODUCTION
Mammographic breast tissue density has been widely considered as a useful risk indicator of women having or developing breast abnormalities and cancer [1, 2]. Several models have been developed to describe and classify mammographic breast tissue density including the Wolfe model that classifies mammographic parenchymal patterns into four categories (N1, P1, P2, and DY) based on the radiographic appearance of prominent ducts and dysplasia [1]. The American College of Radiology has established a Breast Imaging Reporting and Data System (BI-RADS) as the current standard in breast tissue density reporting [3]. Thus, in the clinical practice, radiologists rate and classify breast tissue composition or density into one of four BI-RADS categories based on the ratio of fibro-glandular tissue to the total breast areas. The four BI-RADS categories are: (1) almost entirely fatty (<25% fibro-glandular), (2) scattered fibro-glandular densities (25% – 50% fibro-glandular), (3) heterogeneously dense breast tissue (51% – 75% fibro-glandular), and (4) extremely dense breast tissue (>75% fibro-glandular). Based on the visual assessment of breast tissue density, a number of studies have been performed to investigate the relationship between reported breast tissue density BI-RADS rating and the likelihood of developing breast cancer. The studies reported that, in general, women with dense breast tissue had a higher risk of developing breast cancer [4–7]. Despite the great efforts made in these laboratory based studies, using visual assessment results to predict risk of developing cancer is difficult and often unreliable in the clinical practice due to the considerable inter- and intra-observer variability in the visual assessment of breast tissue density (i.e., BI-RADS) [8].
To overcome the limitation of visual assessment of breast tissue, several research groups developed and tested different computerized schemes to segment and compute breast tissue density or patterns depicted on mammograms [9–15]. Many of these studies reported high correlation with the average visual assessment or consensus rating of a group of radiologists in segmenting or rating dense breast tissue using computerized schemes. For examples, one study used an adaptively determined threshold method that was based on the image characteristic of the gray level histogram to segment dense tissue from the breast areas and compared the automatically segmented results with the average results of manual segmentation of five radiologists. The study reported that in a dataset with four-view mammograms acquired from 65 women, the correlation between the automated and the manual segmentation of dense tissue was 0.94 and 0.91 for craniocaudal (CC) and mediolateral oblique (MLO) views, respectively [10]. Another study applied a computerized scheme to compute eight image texture-based features and derived a computerized index that attempted to mimic the radiologists’ BI-RADS rating of breast tissue composition. The study reported that in a dataset of 200 mammograms, the correlation between the computerized index and the average rating of three radiologists was 0.87, which was also very comparable to the correlations of individually provided ratings between these radiologists [11]. In addition, another previous study [13] demonstrated high correlation (0.9) between breast tissue density levels estimated from mammograms and magnetic resonance (MR) images.
Due to the proven feasibility of automatically computing and assessing breast tissue density or patterns, developing computer-aided detection schemes to identify women with a higher risk of having and/or developing breast cancer based on the breast tissue density or pattern analysis has been attracting research interest. For example, one group reported a series of studies that focused on classification between women with and without the BRCA1/BRCA2 gene mutation based on the computed image features related to the mammographic tissue patterns [16–18]. Using a dataset that includes 30 high-risk women with BRCA1/BRCA2 gene mutation and 142 low-risk women, a computerized scheme which was based on the power spectral analysis of breast tissue patterns depicted on manually selected regions of interest of mammograms achieved a classification performance level of 0.9, measured by the area under the receiver operating characteristic (ROC) curve [18].
Although women with dense breast tissue may have a relatively high-risk (i.e., four to five times higher) of developing breast cancer [4–7], measuring or computing the global tissue density depicted on each mammogram alone has actually a rather low positive prediction value for the individual woman because cancer can be detected in women with breast tissue density in any of the four BI-RADS categories. However, in the clinical practice, radiologists routinely assess the global and local asymmetry of breast tissue patterns depicted on bilateral mammograms acquired from the left and right breast and use such information to identify women with a higher risk of having or developing breast abnormalities and/or cancer (i.e., recalled for additional imaging examination and recommended for biopsy). Thus, instead of computing breast tissue density and comparing the difference among different women, we investigated a new approach that focuses on applying a computerized scheme to analyze and compare the mammographic breast tissue difference between the bilateral images acquired from the same woman. Our approach is based on three well-known facts: (1) the asymmetry of mammographic tissue patterns between bilateral images is an important and practical risk indicator of developing breast abnormalities and/or cancer, (2) the visual assessment of tissue asymmetry may not be always accurate or consistent due to the inter- and intra-observer variability, and (3) a number of computerized image features have been investigated and proven to be effective in assessing breast tissue patterns or density. Thus, the hypothesis of this study is that a computerized scheme can more accurately and consistently detect and assess the asymmetry of breast tissue patterns or density depicted on bilateral mammograms, and hence be able to identify women with a high-risk of having or developing breast abnormalities, which could help radiologists detect breast cancer at an early stage. To test this hypothesis, we assembled a unique testing image dataset and developed a new computerized scheme to classify these cases into two groups of positive (or “high-risk”) and negative (or “low-risk”) for having or developing breast abnormalities or cancer. A detailed description of the study is reported in the following sections.
II. MATERIALS AND METHODS
2.1. A Testing Image Dataset
Under an Institutional Review Board approved protocol, we have ascertained fully anonymized full-field digital mammography (FFDM) examinations from our clinical facilities and transferred the FFDM images to our research laboratory. All examinations were acquired using Hologic Selenia (Hologic Inc., Bedford, MA) FFDM systems between 2006 and 2008. The established FFDM database includes 6478 images acquired from 1120 women. Among these, 525 were diagnosed with cancer and 595 were benign or negative. In the database, 669 women had only one FFDM examination (namely the “current” examination), 371 had two (one “current” and one “prior”) and 80 had three (one “current” and two “prior”) examinations during 2006 to 2008, respectively. The database was divided into five groups including 1) 477 verified cancer cases (group 1), 2) 48 “interval” cancer cases in which cancers were detected between two periodic screening examinations (group 2), 3) 19 cases that were determined to be “high-risk” cases with surgical removal of the lesions recommended (group 3), 4) 222 cases recalled for a diagnostic workup including biopsy of suspicious findings that were ultimately determined to be benign (group 4), and 5) 354 screening cases rated as negative and not recalled (group 5).
In this study, we selected 200 cases from this established FFDM image database to assemble a testing dataset. The testing dataset includes 100 negative cases and 100 positive cases of having or developing breast abnormalities or cancer. We randomly selected the 100 low-risk cases from group 5 (not recalled screening negative cases) of the FFDM database. The 100 positive cases were selected from the other four groups of the database in which each case depicted “visible” local asymmetrical tissue patterns in the retrospective review (Table 1). In each case, we selected one pair of two FFDM images acquired from the CC view of the left and right breast. Among the 100 positive cases, 60 pairs of bilateral images were selected from “prior” examinations of either the cancer cases (39) or the interval cancer cases (21). All of the “prior” examinations were diagnosed as negative during the original examinations and the cancers were later detected and confirmed at or before the next scheduled annual mammography examinations (typically 12 to 18 months later for the cancer cases and 6 to 9 months later for the interval cancer cases). During the retrospective review of the images acquired from these prior examinations, radiologists did not report any “detectable” masses or other suspected abnormalities. The remainder of the 40 positive cases included 40 pairs of bilateral CC view images acquired from the current examinations of non-cancer cases with suspected lesions identified. Among them, 8 cases were high-risk cases recommended for surgical excision of the lesions and 32 were recalled for additional imaging workup including biopsy but ultimately determined to be benign. In this testing dataset, the majority of cases, namely 61% (122/200) and 7% (13/200), were rated as heterogeneously dense (BIRADS 3) and extremely dense (BIRADS 4), respectively. Figure 1 shows the distribution of breast tissue density as rated by the radiologists during the original image interpretation of these 200 cases.
Table 1.
Distribution of 100 positive cases in the testing dataset
| Number of Cases | Type of Cases | “Current” or “Prior” Examination |
|---|---|---|
| 39 | Cancer | Prior |
| 21 | Interval cancer | Prior |
| 8 | High-risk with surgery excision | Current |
| 32 | Recalled negative | Current |
Figure 1.

2.2. Computation and Assessment of Image Features to Measure Asymmetry of Breast Tissue
A computerized scheme was first applied to compute the gray level histogram of one FFDM image and then adaptively select a threshold based on the valley (the minimum point) between two peaks in the histogram (representing the breast area and direct x-ray exposure background) to segment the breast area depicted on the image. From the segmented breast area, we initially applied the schemes to compute 20 image features. These features have been previously tested to describe or classify breast tissue density or patterns by several research groups [9–15]. In brief, these 20 features can be divided into five groups.
In group 1, we computed the gray level histogram of the segmented breast area. From the histogram, we computed the following five features (Features 1 to 5): (1) the ratio between the number of pixels with the maximum value in the histogram and the total number of pixels depicted inside the segmented breast area, (2) the ratio between the number of pixels with a gray value larger than the average value of the histogram and the total number of pixels depicted inside the segmented breast area, (3) the average change of two adjacent values in the histogram (the local value fluctuation of the histogram), (4) the average value of histogram, and (5) the standard deviation of histogram values.
In group 2, we first computed a map of the local pixel value fluctuation in which a 5×5 square convolution kernel was implemented to scan the segmented breast area. The absolute pixel value differences between the center pixel (i) and each of the other pixels (j) inside the kernel were computed. The maximum difference value computed inside the kernel was used to replace the original digital value of the pixel (i) in the local pixel value fluctuation map. We then computed three features (Features 6 to 8) including (1) the mean (average), (2) the standard deviation, and (3) the skewness recorded on the map.
In group 3, we computed two fractal dimension (texture) based features previously reported by Chang et al. [11]. In brief, each image was first filtered by five Gaussian low-pass filters with varying kernel sizes. After subtraction between the original image and each of the filtered images, we computed the sum of the pixel value in each subtracted image, , Where is a high pass filtered image and its kernel size is obtained by k = 4 × n + 1, n = 1,2,3,4,5. Then the linear regression method is used to fit a straight line in log(G(k)) versus log(k) domain. The first feature (Feature 9) is the slope of the fitted line. The second feature (Feature 10) measures textural “edgeness” (the gradient value of a pixel and its non-overlapping neighborhoods [19]). By computing:
where t is the distance in the number of pixels and g is the gradient. Feature 10 is defined as the slope estimated by fitting a straight line to the log(g (t)) versus log(t)
In group 4, we analyzed the pixel value distributions of the segmented breast areas depicted on the original mammograms and computed four common indices used in the conventional statistical data analysis. These four features (Features 11 to 14) include (1) the mean (average), (2) the standard deviation, (3) the skewness, and (4) the kurtosis of the digital values of all pixels depicted on the segmented breast area.
In group 5, we mimicked the BI-RADS rating method to divide the segmented breast area into four regions based on the pixel value. We computed the maximum pixel value of the breast area and set up three thresholds, namely, 25%, 50%, and 75% of the maximum pixel value. We then computed three features (Features 15 to 17) that represent the ratios between the average pixel value under each threshold level and the average pixel value of the entire breast area (i.e., the ratio of the average value of pixels with value smaller than 50% of the maximum pixel value and the average pixel value of the entire segmented breast area). We also computed the other three ratios between the number of pixels under each threshold and the total number of pixels depicted on the entire segmented breast area, which represent the final three features (Features 18 to 20).
For each testing case, 20 image features were separately computed from two bilateral images. For each feature, the absolute difference (the subtraction) of two values computed from two bilateral images was computed. These bilateral feature differences were used to represent the bilateral mammographic tissue asymmetry. A normalization process was applied for values of each feature. We computed the average value (μ) and standard deviation (σ) of each feature from all 200 testing cases. The feature values were then normalized to values between 0 and 1 using the limit determined by [μ − 2σ, μ + 2σ]. If the value is greater than μ + 2σ, it is assigned to 1. If the value is smaller than μ − 2σ, it is assigned to 0. Thus, the initial feature pool includes 20 normalized features representing the tissue asymmetry of two bilateral images. We then computed and analyzed the performance level of applying each feature to classify the 200 testing cases into two groups of positive and negative of having or developing breast abnormalities or cancer using the ROCKIT program [20]. The classification performance levels when applying for these 20 features were compared.
In addition, we applied each of these 20 features computed from a single mammogram (either left or right breast) to classify the same set of 200 testing cases. For each case, two classification scores are generated (one for each breast). We averaged two classification scores and used it as a new score to represent the likelihood of this case being positive for having or developing breast abnormalities or cancer. These three sets of classification scores were independently analyzed using ROCKIT program. We then compared the classification performance differences between using the bilateral feature differences and three sets of features computed from single mammograms (or their combination).
2.3. Optimization and Evaluation of a Multi-feature based Artificial Neural Network
Although many of the 20 features computed in this study have been applied to assess breast tissue density and patterns in previous studies, some can be redundant. Selection of an “optimal” feature set through the process of discarding redundant features is an important step in developing any data-driven computerized scheme designed for this purpose. In this study, we applied a genetic algorithm (GA) based feature selection protocol to select effective features and build an optimal artificial neural network (ANN). The feed-forward ANN designed in this study included three layers. The first (input) layer included N neurons that connect to N selected features, the second layer included M hidden neurons, and the third (decision) layer included one neuron that generates a likelihood score of a test case being positive of having or developing abnormalities or breast cancer.
A publicly available GA software package [21] was modified and used with a specifically designed binary coding method and a GA fitness function to select an optimal feature set and to determine the ANN structure. Specifically, the GA chromosome used in this study includes 24 genes. The first 20 genes represent one of 20 initially computed features. Using the binary coding method, a value of 1 indicates that the feature represented by the gene in the GA chromosome is selected by the ANN and a value of 0 means that the represented feature is discarded. The last four genes represent the number of hidden neurons. For example, 0110 indicates that six hidden neurons are selected in the ANN. To minimize over-fitting and maintain robustness of the ANN performance, a limited number of training iterations (500), and a large ratio between the momentum (0.9) and learning rate (0.01), was used. The initial GA chromosome was randomly selected by the GA program. Once a GA chromosome was selected, a leave-one-case-out method [22] was used to assess the performance of the ANN-based classifier due to the size limitation of our testing dataset. When using the leave-one-case-out method in this study, 199 cases were used to train the ANN based classifier and the ANN was applied to classify one remaining test case (generating a classification score). This process was repeated 200 times, so that each case in the dataset was used once to test the ANN. The 200 classification scores were generated and connected to the input data of a ROC fitting and analysis program (ROCKIT [20]) that computes the area under the ROC curve (AUC). During the GA optimization step, the initial 100 GA chromosomes in the first generation were randomly generated. After each training cycle, the GA generated a performance score (AUC) for each chromosome. The GA chromosomes that produce higher AUC values have higher probabilities of being selected in generating new chromosomes using the method of crossover and mutation. The GA operation was terminated when it reached a global maximum performance level or a pre-determined number of growth generations (i.e., 100 in this study).
After the optimal ANN based classifier was built, we computed and estimated the performance level when classifying the 200 cases in our dataset into the positive and negative groups. Based on the leave-one-case-out testing method, the ANN-generated classification scores for each testing case over the entire cases of two groups were recorded and tabulated. The performance level (measured by the AUC) was computed using ROCKIT program [20]. In addition, change in the actual sensitivity levels for detecting the different types of cases (i.e., verified cancer cases, high-risk cases recommended for surgery excision, and the other recalled screening cases) as well as the relationships between classification results and several clinical features (i.e., breast tissue BI-RADS density ratings) at a set of different specificity levels were also analyzed and reported.
III. RESULTS
When applying each of the 20 features computed from the difference of two bilateral images to classify the 200 cases in our dataset into two groups of positive and negative of having or developing breast abnormalities or cancer, the average performance level measured by the AUC is 0.590 ranging from 0.485±0.041 to 0.681±0.038 (Figure 2). With the exception of one feature (namely feature 14 – the kurtosis of the digital values of all pixels depicted on the segmented breast area) that yielded an AUC < 0.5, all other 19 features yielded AUC > 0.5, including 8 achieved AUC > 0.6, which is significantly (p < 0.001) higher than the chance (AUC = 0.5), indicating the majority of these features have certain ability to compute and assess the mammographic breast tissue asymmetry and hence identify a fraction of positive cases. Table 2 compares classification performance levels between the use of bilateral feature differences and three sets of features computed from single mammograms including using the features computed from (1) the left breast, (2) the right breast, and (3) the combination (average) of detection results from two breasts. The results indicated that using bilateral feature differences yielded the substantially higher performance including (1) the highest average AUC value, (2) the smallest AUC standard deviation, (3) the largest number of features yielding AUC ≥ 0.6, and (4) the smallest number of features generating AUC ≤ 0.5.
Figure 2.
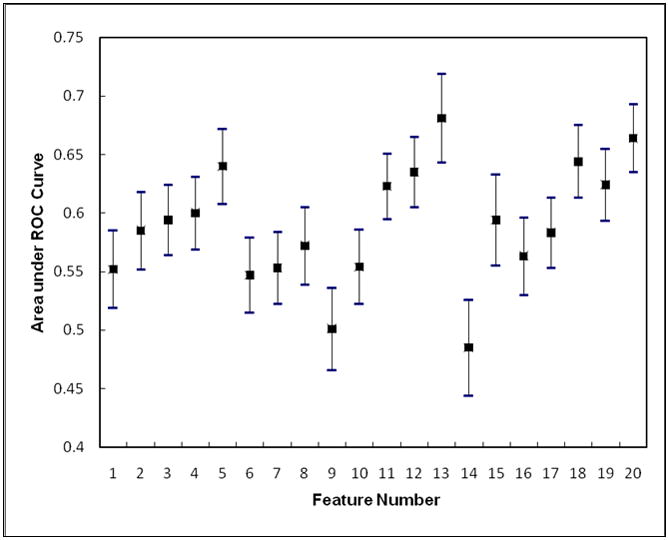
Table 2.
Comparison of classification performance levels (the areas under ROC curves) of using four sets of 20 features computed from (1) the mammograms of left breast, (2) mammograms of the right breast, (3) average of the results from the left and right breasts, and (4) bilateral difference
| Feature type | Average AUC | Standard deviation | Number of AUC ≥ 0.6 | Number of AUC ≤ 0.5 |
|---|---|---|---|---|
| Left breast | 0.495 | 0.059 | 1 | 12 |
| Right breast | 0.505 | 0.058 | 1 | 9 |
| Average of two breasts | 0.505 | 0.066 | 1 | 10 |
| Bilateral differences | 0.590 | 0.051 | 8 | 1 |
The genetic algorithm optimized ANN included six input neurons (represented by six selected image features) and eight hidden neurons. These six features are (1) feature 1 - the ratio between the number of pixels with the maximum value in the grey level histogram and the total number of pixels depicted inside the segmented breast area, (2) feature 4 - the average value of the histogram, (3) feature 8 - the skewness of the pixel values recorded on the map of the local pixel value fluctuation, (4) feature 11 - the mean of the digital values of all pixels depicted on the segmented breast area, (5) feature 12 – the standard deviation of pixel values of the breast area, and (6) feature 13 – the skewness of the pixel values of the breast area. Due to the relatively low correlations among these six features (Table 3), by combining these features, the ANN yielded a significantly improved classification performance with AUC = 0.754±0.024 (p < 0.001). The reduction of the standard deviation value in the computed AUC from a minimum value of 0.033 using each of these six individual features (Figure 2) to 0.024 using the ANN also indicates the reliability improvement of classification performance. Figure 3 shows two fitted ROC curves generated using the ROCKIT program, which represents the best classification performance level using a single feature and the performance level using the ANN.
Table 3.
The correlation coefficients among six features selected to train the ANN based classifier
| Feature 4 | Feature 8 | Feature 11 | Feature 12 | Feature 13 | |
|---|---|---|---|---|---|
| Feature 1 | 0.047 | 0.210 | 0.146 | 0.248 | 0.158 |
| Feature 4 | 0.063 | 0.200 | 0.204 | 0.034 | |
| Feature 8 | 0.164 | 0.145 | 0.205 | ||
| Feature 11 | 0.158 | 0.055 | |||
| Feature 12 | 0.116 |
Figure 3.
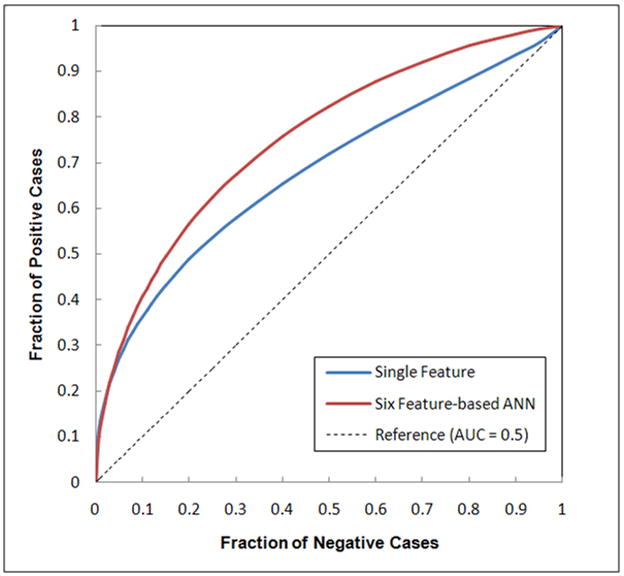
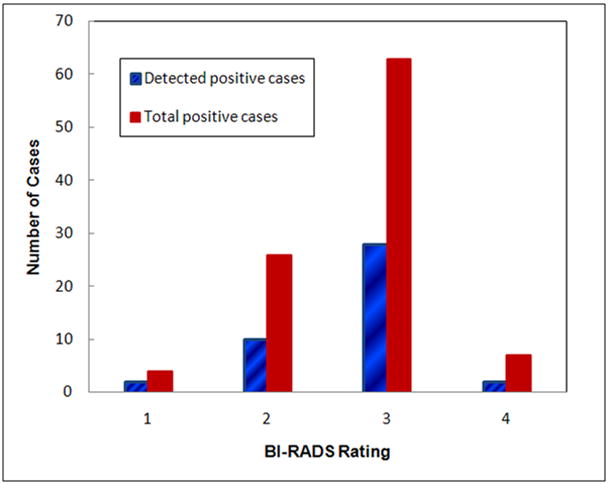
Table 4 summarizes the actual number of high-risk cases that were correctly detected and identified by the ANN at four different specificity levels. The classification sensitivity levels on all 100 positive cases are 42% and 51% at specificity levels of 90% and 80%, respectively. By analyzing the sensitivity levels at four different positive sub-groups (namely, the cancer cases, the interval cancer cases, the high-risk cases with surgery excision, and the recalled and biopsied benign cases), we found that the ANN yielded higher classification sensitivity on actual cancer cases than on suspected but actual benign cases. For example, at 90% and 80% specificity levels, the classification sensitivity levels were 61.9% and 71.4% for the interval cancer case sub-group, while the sensitivities were reduced to 25.0% and 34.4% for the recalled benign case sub-group, respectively. Figure 4 shows and compares the distribution of breast tissue BI-RADS ratings between the 42 ANN-detected high-risk cases at 90% specificity and the total 100 high-risk cases initially selected in the testing dataset. The comparison indicates that unlike the detection of targeted abnormalities (i.e., masses) using the conventional computer-aided detection (CAD) schemes, the classification performance of our computerized scheme was not sensitive to the breast tissue density.
Table 4.
The actual number of positive cases correctly identified (sensitivity levels) at different specificity levels when applying the ANN to classify the dataset with 100 positive and 100 negative cases
| Specificity | 95% | 90% | 85% | 80% |
|---|---|---|---|---|
| Sensitivity on all 100 positive cases | 34 (34.0%) | 42 (42.0%) | 44 (44.0%) | 51 (51.0%) |
| Sensitivity on 39 cancer cases | 14 (35.9%) | 17 (43.6%) | 18 (46.2%) | 20 (51.3%) |
| Sensitivity on 21 interval cancer cases | 12 (57.1%) | 13 (61.9%) | 13 (61.9%) | 15 (71.4%) |
| Sensitivity on 8 high-risk cases (surgery excision) | 2 (25.0%) | 4 (50.0%) | 4 (50.0%) | 5 (62.5%) |
| Sensitivity on 32 recalled negative cases | 6 (18.8%) | 8 (25.0%) | 9 (28.1%) | 11 (34.4%) |
Figure 4.
IV. DISCUSSION
Automatically assessing breast tissue density and detecting suspicious breast abnormalities (i.e., masses) based on the local breast tissue asymmetry has been attracting substantial research interest in developing CAD schemes for mammograms. Previous studies have attempted to detect breast masses by first subtracting the registered bilateral mammograms to identify suspicious abnormalities and then performing statistical analysis of the feature differences computed from the identified regions depicted on two breasts [23–26]. However, due to the difficulty in the registration of two bilateral mammograms, previous CAD yielded substantially high false-positive detection rates [24] and often unreliable results [25]. Therefore, the algorithms for automatically detecting asymmetry of breast tissue density or patterns has not been implemented in commercialized CAD schemes to date. This study is different in which we identified the “global” breast tissue difference (asymmetry) between two bilateral images and not the “local” tissue asymmetry that targets the suspicious abnormalities (i.e., masses). Hence, we avoided possible errors and unreliable assessment results due to the potential misalignment or poor registration between two bilateral images.
In this study, we analyzed the feasibility of using automated methods to detect breast tissue asymmetry between two bilateral mammograms. We first computed 20 image features that have been tested in a number of previous studies by different research groups to evaluate breast tissue density and patterns depicted on mammograms. We then compared the performances of the individual features to classify the cases in our testing dataset into positive and negative of developing or having breast abnormalities or cancer. Although many features can achieve comparable classification performance (Figure 2), we demonstrated that a genetic algorithm optimized multi-feature based ANN yielded the significantly improved classification performance. Unlike previous studies that classified a set of cases with and without known BRCA1/BRCA2 gene mutation based on image feature computation and analysis of the manually selected regions of interest [16–18], our scheme is fully automatic and does not require any human intervention. The scheme is applied to the entire breast areas segmented on the mammograms. Our dataset also included a much diverse composition of cases in particular in the positive group. The cancers were actually detected and confirmed later in 60% (60/100) of the positive cases (6 to 18 months after these “prior” examinations selected in the testing dataset). Since there are no dominant (visually detectable) masses or micro-calcifications depicted on the images of these selected “prior” examinations (in the retrospective review), applying the conventional CAD schemes (including the commercial systems used in the clinical practice to date) to these images is not helpful. Radiologists are likely to interpret or diagnose these examinations as negative with and without using CAD. However, our new computerized scheme can report and flag the suspicious findings of breast tissue asymmetry that relates to the risk of having or developing breast abnormalities or cancer, which may potentially help radiologists detect and identify a fraction of women who have or will rapidly develop breast cancer at least six to 18 months early.
Although previous studies showed that the texture (i.e., fractal dimension or power spectrum) could be used to more accurately describe breast tissue patterns [11] and classify high-risk cases [18], we found that detection and classification of breast tissue asymmetry depicted on bilateral images might be different. Three simple statistic indices of digital value distribution of the pixels depicted on the segmented breast area (namely the mean, standard deviation, and skewness) yielded significantly higher classification performance than the more complicated texture based features (i.e., fractal dimension based features). As a result, the genetic algorithm optimized ANN included these three simple statistic indices and discarded the texture based features. In addition, this study also found that comparing to the use of the breast tissue density related features computed from the single mammograms or the combination of detection results from two images, using bilateral feature differences has significant advantages for this purpose.
Since the ultimate purpose of this study is to develop a new computerized tool or scheme for breast cancer risk stratification to evaluate its potential clinical utility, the underlying concept is different from the conventional CAD schemes that are used as a “second-reader” to assist radiologists in detecting the subtle breast abnormalities that are actually visible or “detectable” on the images [27, 28], In a risk stratification task, any abnormalities or cancers in women would be detected at a later time (and potentially at a later stage) and therefore there may be an important utility for tools that could stratify these women into two groups, those that are at average risk or low-risk and those that are at significantly higher risk of having or developing breast cancer; hence, women determined to be at significantly higher risk should be followed up more closely with other more effective imaging or pathological examinations (i.e., ultrasound, magnetic resonance (MR) imaging, and biopsy) [29]. The risk stratification tool should be developed and assessed based on a “rule in” concept rather than a “rule out” one. Under this paradigm, every additional cancer that would be ultimately detected as a result of the regular mammography practice will likely be detected substantially earlier. To be clinically useful, the performance of the risk stratification tool should be evaluated under high specificity. In this study, we demonstrated the potential of using our computerized scheme as a new risk stratification tool based on the automated detection and analysis of mammographic breast tissue asymmetry. The preliminary testing results showed that among the different sub-groups of positive cases, our scheme yielded higher classification performance in the “prior” examinations of the cancer cases than the recalled benign cases (i.e., 50% and 30% sensitivity at 90% specificity). By flagging warning signs on these examinations, our scheme may help radiologists identify a substantial fraction of the developing cancer cases 6 to 18 months early.
Despite the encouraging results, we recognize that this was a very preliminary study with a number of limitations. First, the size of our testing dataset was limited. Since a risk stratification tool (scheme) targeted on a specific type of high-risk or “positive” cases and it should be operated in the high specificity (i.e., ≥ 90%), the diversity of the negative training cases is very important for increasing the robustness of the scheme. Although we randomly selected 100 negative cases from an established FFDM image database in our group, whether these cases are sufficient to represent or cover a very large pool of negative cases in our medical center (the screening population) remains uninvestigated. Thus, the robustness and generalizability of the reported results need to be further tested in the future studies. Second, the high-risk cases were visually (not randomly) selected from our FFDM database. Each case depicts somewhat local breast tissue asymmetry that was visually “distinguishable” in the retrospective review (with the “truth” already known). Thus, the automated scheme developed in this study can only be applied to detect and identify a fraction of women with mammographic breast tissue density or pattern asymmetry that correlate to a high-risk of having or developing breast abnormalities or cancer at high specificity. Detecting or classifying other types of risk factors for developing breast cancer (i.e., carrying BRCA1/BRCA2 gene mutation) is beyond the scope of this study. Third, since we only used bilateral images of the CC view and investigated a limited number of 20 features, the achieved classification results may not be optimal. In future studies, we will explore more image features and test whether or not, and how, to optimally incorporate breast tissue asymmetry computed from the MLO view to achieve improved classification performance.
In summary, aiming for the breast cancer risk stratification or detecting breast cancer at an early stage, we developed a new automated scheme to detect bilateral mammographic breast tissue asymmetry. We demonstrated that this new scheme was able to detect and identify a fraction of women with high-risk of having or developing breast cancer at high specificity. However, we recognize that this is the first and very preliminary study. Before assessing whether using this tool (scheme) can help radiologists identify more high-risk women and/or detect breast abnormalities or cancer at an early stage in the clinical practice, more work is needed to further improve, optimize, and test the performance and robustness of this type of computerized scheme.
Acknowledgments
This work is supported in part by Grants CA77850 and CA101733 to the University of Pittsburgh from the National Cancer Institute, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wolfe JN. Breast patterns as an index of risk for developing breast cancer. Am J Roentgenol. 1976;126:1130–1139. doi: 10.2214/ajr.126.6.1130. [DOI] [PubMed] [Google Scholar]
- 2.Boyd NF, Martin IJ, Stone J, et al. Mammographic densities as a marker of human breast cancer risk and their use in chemoprevention. Curr Oncol Rep. 2001;3:314–321. doi: 10.1007/s11912-001-0083-7. [DOI] [PubMed] [Google Scholar]
- 3.BI-RADS Breast Imaging Reporting and Data System Breast Imaging Atlas. American College of Radiology; Reston, VA: 2003. [Google Scholar]
- 4.Wolfe JN, Saftlas AF, Salane M. Mammographic parenchymal patterns and quantitative evaluation of mammographic densities: A case-control study. Am J Roentgenol. 1987;148:1087–1092. doi: 10.2214/ajr.148.6.1087. [DOI] [PubMed] [Google Scholar]
- 5.Byng JW, Yaffe MJ, Lockwood LE, et al. Automated analysis of mammographic densities and breast carcinoma risk. Cancer. 1997;80:66–74. doi: 10.1002/(sici)1097-0142(19970701)80:1<66::aid-cncr9>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 6.Brisson J, Diorio C, Masse B. Wolfe’s parenchymal pattern and percentage of the breast with mammographic densities: redundant or complementary classifications? Cancer Epidemiol Biomarkers Prev. 2003;12:728–732. [PubMed] [Google Scholar]
- 7.Boyd NF, Guo H, Martin LJ, et al. Mammographic density and the risk and detection of breast cancer. N Engl J Med. 2007;356:227–236. doi: 10.1056/NEJMoa062790. [DOI] [PubMed] [Google Scholar]
- 8.Berg WA, Campassi C, Langenberg P, Sexton MJ. Breast imaging reporting and data system: Inter- and intra-observer variability in feature analysis and final assessment. Am J Roentgenol. 2000;174:1769–1777. doi: 10.2214/ajr.174.6.1741769. [DOI] [PubMed] [Google Scholar]
- 9.Taboces PG, Correa J, Souto M, et al. Computer-assisted diagnosis: the classification of mammographic breast parenchymal patterns. Phys Med Biol. 1995;40:103–117. doi: 10.1088/0031-9155/40/1/010. [DOI] [PubMed] [Google Scholar]
- 10.Zhou C, Chan HP, Petrick N, et al. Computeized image analysis: estimation of breast density on mammograms. Med Phys. 2001;28:1056–1069. doi: 10.1118/1.1376640. [DOI] [PubMed] [Google Scholar]
- 11.Chang YH, Wang XH, Hardesty LA, et al. Computerized assessment of tissue composition on digitized mammograms. Acad Radiol. 2002;9:898–905. doi: 10.1016/s1076-6332(03)80459-2. [DOI] [PubMed] [Google Scholar]
- 12.Wang XH, Good WF, Chapman BE, et al. Automated assessment of composition of breast tissue revealed on tissue-thickness-corrected mammography. Am J Roentgenol. 2003;180:257–262. doi: 10.2214/ajr.180.1.1800257. [DOI] [PubMed] [Google Scholar]
- 13.Wei J, Chan HP, Helvie MA, et al. Correlation between mammographic density and volumetric fibroglandular tissue estimated on breast MR images. Med Phys. 2004;31:933–942. doi: 10.1118/1.1668512. [DOI] [PubMed] [Google Scholar]
- 14.Li H, Giger ML, Olopade OI, et al. Computerized texture analysis of mammographic parenchymal patterns of digitized mammograms. Acad Radiol. 2005;12:863–873. doi: 10.1016/j.acra.2005.03.069. [DOI] [PubMed] [Google Scholar]
- 15.Glide-Hurst CK, Duric N, Littrup P. A new method for quantitative analysis of mammographic density. Med Phys. 2007;34:4491–4498. doi: 10.1118/1.2789407. [DOI] [PubMed] [Google Scholar]
- 16.Huo Z, Giger ML, Olopade OI, et al. Computerized analysis of digitized mammograms of BRCA1 and BRCA2 gene mutation carriers. Radiology. 2002;225:519–526. doi: 10.1148/radiol.2252010845. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Giger ML, Olopade OI, et al. Fractal analysis of mammographic parenchymal patterns in breast cancer risk assessment. Acad Radiol. 2007;14:513–521. doi: 10.1016/j.acra.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Giger ML, Olopade OI, Chinander MR. Power spectral analysis of mammographic parenchymal patterns for breast cancer risk assessment. J Digital Imaging. 2008;21:145–152. doi: 10.1007/s10278-007-9093-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haralick RM, Shapiro LG. Computer and robot vision. Addison-Wesley; Reading, MA: 1992. [Google Scholar]
- 20.Metz CE. ROCKIT 0.9B Beta Version. University of Chicago; 1998. http://www-radiology.uchicago.edu/krl/ [Google Scholar]
- 21.Kantrowitz M. Prime time freeware for AI, issue 1-1: selected materials from the Carnegie Mellon University, Artificial Intelligence Repository. Prime Time Freeware; Sunnyvale, CA: 1994. [Google Scholar]
- 22.Li Q, Doi K. Reduction of bias and variance for evaluation of computer-aided diagnostic schemes. Med Phys. 2006;33:868–875. doi: 10.1118/1.2179750. [DOI] [PubMed] [Google Scholar]
- 23.Hand W, Semmlow JL, Ackerman LV, Alcorn FS. Computer screening of xeromammograms: A technique for defining suspicious areas of the breast. Comput Biomed Res. 1979;12:445–460. doi: 10.1016/0010-4809(79)90031-4. [DOI] [PubMed] [Google Scholar]
- 24.Yin FF, Giger ML, Doi K, et al. Computerized detection of masses in digital mammograms: Analysis of bilateral subtraction images. Med Phys. 1991;18:955–963. doi: 10.1118/1.596610. [DOI] [PubMed] [Google Scholar]
- 25.Zheng B, Chang YH, Gur D. Computerized detection of masses from digitized mammograms: Comparison of single-image segmentation and bilateral-image subtraction. Acad Radiol. 1995;2:1056–1061. doi: 10.1016/s1076-6332(05)80513-6. [DOI] [PubMed] [Google Scholar]
- 26.Mendez AJ, Tahoces PG, Lado MJ, et al. Computer-aided diagnosis: automatic detection of malignant masses in digitized mammograms. Med Phys. 1998;25:957–964. doi: 10.1118/1.598274. [DOI] [PubMed] [Google Scholar]
- 27.Nishikawa RM. Current status and future directions of computer-aided diagnosis in mammography. Comput Med Imaging Graph. 2007;31:224–235. doi: 10.1016/j.compmedimag.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 28.Fenton JJ, Taplin SH, Carney PA, et al. Influence of computer-aided detection on performance of screening mammography. N Engl J Med. 2007;356:1399–1409. doi: 10.1056/NEJMoa066099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stojadinovic A, Nissan A, Shriver CD, et al. Electrical impedance scanning as a new breast cancer risk stratification tool for young women. J Surg Oncol. 2008;97:112–120. doi: 10.1002/jso.20931. [DOI] [PubMed] [Google Scholar]


