Abstract
Conantokins are naturally-occurring small peptide antagonists of ion flow through NMDA/glycine activated-N-methyl-D-aspartate receptor (NMDAR) ion channels. One member of the conantokin family, conantokin (con)-G, a 17-residue peptide, is selective for NMDARs containing the NR2B subunit, whereas the homologous peptides, con-T and con-R, show broader selectivity for NR2 subunits. In this study, con-G, con-R, and con-T variants were chemically synthesized and employed to investigate their subunit selectivities as inhibitors of agonist-evoked ion currents in HEK-293 cells expressing various combinations of NMDAR subunits that contain NR1a or NR1b combined with NR2A or NR2B. Using truncation and point mutants, as well as chimeric conantokins, we determined that the N-terminus of con-G contains all the determinants for NR2B selectivity. With this information, a large number of conantokin variants were synthesized and used to establish minimal sequence determinants for selectivity. Tyr at position 5 broadens the NR2 selectivity, and recovery of NR2B selectivity in Tyr5 peptides was achieved by incorporating Ala or Gly at position 8. NR2B selectivity in con-R can be conferred through deletion of the Ala at position 10, thereby shifting the γ-carboxyglutamate (Gla) at position 11 to position 10, where a Gla naturally occurs in con-G and con-T. The nature of the amino acid at position 6 is also linked to subunit selectivity. Our studies suggest that the molecular determinants of conantokins that dictate NMDAR subunit selectivity are housed in specific residues of the N-termini of these peptides. Thus, it is possible to engineer desired NMDAR functional properties into conantokin-based peptides.
Keywords: conantokins, NMDAR subtypes, NMDAR antagonists, electrophysiology, ion channels
INTRODUCTION
Conantokins are small naturally-occurring peptides that specifically antagonize ion flow through NMDA/glycine-activated N-methyl-D-aspartate receptor (NMDAR) ion channels. Several conantokins have been identified in the venoms of snails of the genus Conus (McIntosh et al., 1984, Haack et al., 1990, White et al., 2000, Jimenez et al., 2002, Teichert et al., 2007, Gowd et al., 2008, Twede et al., 2009), all of which contain multiple copies of the amino acid, γ-carboxyglutamate (Gla or γ), as well as other co/post-translationally modified amino acids. The conantokins, which are not ion channel blockers, but act as competitive inhibitors with glutamate/NMDA (Donevan and McCabe, 2000, Klein et al., 2001b), are effective in animal models of neuropathies, e.g., pain (Xiao et al., 2008), epilepsy (Xi et al., 2002), protection against neuronal apoptosis after ischemic stroke (Williams et al., 2002), and opiate addiction (Wei et al., 2006).
NMDARs are ligand-gated neuroreceptors, coagonized by glutamate (or NMDA)/glycine. D-serine is also a potent coagonist with glutamate, and, in fact, has been proposed to be the physiological ligand (Wolosker, 2006). These receptors are constituted from two heterodimers of distinct NR subunits (Furukawa et al., 2005). One of the requisite subunits is NR1, the ubiquitously expressed glycine-binding component of the NMDAR (Grimwood et al., 1995). One of the 8 splice variants of NR1 (NR1a–h) associates with one or more of the more restrictively expressed independent gene products encoding the glutamate-binding NR2 subunit, viz., NR2A, NR2B, NR2C, and/or NR2D (Laube et al., 1997). The non-glutamate-binding NR3 subunit can also form glycine excitatory receptors with NR1 (Chatterton et al., 2002). Much interest has been shown in one of the conantokins, conantokin-G (con-G) from Conus geographus, because it specifically antagonizes NMDARs containing the NR2B subunit. Since NR2B is temporally and regionally regulated in the brain, and is not present in all NMDARs, specific targeting of receptors containing this subunit will only affect subsets of NMDARs (Ma and Hargreaves, 2000), thus reducing generalized side-effects on the total NMDAR, as is the case with nonselective NMDAR blockers. This rationale has been borne out in clinical trials, in which the NR2B-specific agent, CP-101,606, was demonstrated to be well-tolerated in patients with mild-moderate tramautic brain injury or hemorrhagic stroke (Merchant et al., 1999). Thus, selective inhibition of NMDARs containing NR2B is an emerging therapeutic goal, and NR2B-selective agents, including con-G (CGX-1007), have been examined in clinical trials of pain.
Because of the importance of possessing a repertoire of NR2B-selective allosteric inhibitors of NMDAR ion channels for use as both drug templates and pharmacological tools, we examined three conantokin peptides, viz., the NR2B-selective inhibitor, con-G, and the nonselective inhibitors con-T and con-R. In order to determine the molecular determinants of these peptides that lead to selectivity for NR2B-containing NMDARs, we designed and synthesized new peptide variants based on the parent peptides, con-G, con-T, and con-R′. These variant peptides were then tested for their inhibitory effects on current flow through ion channels of HEK293 cells expressing NMDARs of varying subunit combinations (NR1a/2A, NR1a/2B, NR1b/2A, and NR1b/2B). The results of this study are summarized herein.
EXPERIMENTAL PROCEDURES
Peptide synthesis
The amino acid sequences of the conantokins relevant to this study are provided in Table 1. One of these peptides, con-R′, consists of residues 1-17 of the 27-residue con-R parent. This truncated variant was employed because previous data demonstrated that no differences existed in cation binding properties and NMDAR antagonist activities between WT con-R and con-R′ (Blandl et al., 2000, Klein et al., 2003). Solid phase peptide synthesis was employed using an Applied Biosystems (Foster City, CA) Model 433A peptide synthesizer. All reagents and methods have been described previously (Prorok et al., 1996).
Table 1.
Sequences of synthetic parent conantokins and their variant peptidesa
| con-G | GEγγL5QγNQγ10LIRγK15SN(NH2) |
|---|---|
| con-G[1–11] | GEγγLQγNQγL |
| con-G[Q6A] | GEγγLAγNQγLIRγKSN(NH2) |
| con-G[γ7A] | GEγγLQANQγLIRγKSN(NH2) |
| con-G[γ7K] | GEγγLQKNQγLIRγKSN(NH2) |
| con-G[N8M] | GEγγLQγMQγLIRγKSN(NH2) |
| con-G[Q9A] | GEγγLQγNAγLIRγKSN(NH2) |
| con-G[L5Y/N8A] | GEγγYQγAQγLIRγKSN(NH2) |
| con-G[Q6A/Q9A] | GEγγLAγNAγLIRγKSN(NH2) |
| con-T | GEγγY5QKMLγ10NLRγA15EVKKN20A(NH2) |
| con-T[Y5L] | GEγγLQKMLγNLRγAEVKKNA(NH2) |
| con-T[K7A] | GEγγYQAMLγNLRγAEVKKNA(NH2) |
| con-T[K7γ] | GEγγYQγMLγNLRγAEVKKNA(NH2) |
| con-T[M8A] | GEγγYQKALγNLRγAEVKKNA(NH2) |
| con-T[M8G] | GEγγYQKGLγNLRγAEVKKNA(NH2) |
| con-T[M8N] | GEγγYQKNLγNLRγAEVKKNA(NH2) |
| con-T[L9A] | GEγγYQKMAγNLRγAEVKKNA(NH2) |
| con-T[M8A/+A10] | GEγγYQKALAγNLRγAEVKKNA(NH2) |
| con-G/T | GEγγLQγNQγNLRγAEVKKNA(NH2) |
| con-R′: | GEγγV5AKMAA10γLARγ15NI(NH2) |
| con-R′ [A6Q] | GEγγVQKMAAγLARγNI(NH2) |
| con-R′ [K7A] | GEγγVAAMAAγLARγNI(NH2) |
| con-R′ [M8A] | GEγγVAKAAAγLARγNI(NH2) |
| con-R′ [M8N] | GEγγVAKNAAγLARγNI(NH2) |
| con-R′ [A6Q/A9Q] | GEγγVQKMQAγLARγNI(NH2) |
| con-R′ [ΔA10] | GEγγVAKMA AγLARγNI(NH2) |
| con-R′ [V5L/ΔA10] | GEγγLAKNA AγLARγNI(NH2) |
| con-R′ [V5Y/ΔA10] | GEγγYAKNA AγLARγNI(NH2) |
| con-G/Rχ | GEγγLQγNQγLARγNI(NH2) |
All mutated or deleted amino acids are underlined and placed in bold font.
Expression of recombinant (r) NMDAR subtypes
Plasmid pEGFP-N1, encoding a red-shifted variant of wild-type (WT) green fluorescent protein (GFP), was purchased from Clontech Laboratories (San Diego, CA). cDNAs encoding rat NR1a, NR1b, NR2A, and NR2B (subcloned into pRC/CMV, pRK7, or pRK5) were provided by Dr. David Lynch (University of Pennsylvania). HEK293 cells (ATCC), grown to ∼50% confluency on 60 mm poly-D-lysine-coated dishes, were transiently transfected with recombinant NR subunits using calcium phosphate precipitation (Sheng et al., 2007). Cotransfections with pEGFP-N1 were used for positive selection of transfected cells. Plasmid ratios for diheteromeric combinations were 1:3:6 for GFP:NR1a/b:NR2A/B, respectively. Following transfection, the cells were maintained in fresh medium containing 500 μM ketamine to prevent excitotoxic cell death from glutamate present in the culture medium. After 48 hr post-transfection, the cells were plated in 35 mm poly-D-lysine-coated dishes for electrophysiological recordings.
Electrophysiology
Electrophysiological patch clamp whole cell recordings were performed at room temperature as described (Sheng et al., 2007). The cells were bathed in extracellular solution consisting of 140 mM NaCl, 3 mM KCl, 2 mM CaCl2, 10 mM Na-Hepes, and 20 mM dextrose, pH 7.35. Glycine (10 μM) was also present to saturate its site on the NMDAR. Recording pipettes, constructed from borosilicate glass (Drummond Scientific, Broomall, PA), with a resistance of 2–4 MΩ, were back-filled with a solution containing 140 mM CsF, 1 mM CaCl2, 2 mM MgCl2, 10 mM EGTA, 10 mM Cs-Hepes, 2 mM tetraethylammonium chloride, and 4 mM Na2ATP, pH 7.35.
The GFP fluorescence in cells was visualized using a Nikon Eclipse TE200 microscope (Fryer) with a fluorescence detection system. The application of agonist solutions was accomplished at 2.5 ml/min via a rapid solution changer (RSC-200; Biologic Science Instruments, Knoxville, TN) equipped with a 9-barrel straight-head (exchange time, 15 msec). Whole-cell currents were recorded, amplified, digitized, and data acquired as previously published (Sheng et al., 2007). All measurements were performed at room temperature with cells voltage-clamped at −70 mV, pH 7.35. The responses in a particular cell were normalized to maximum current evoked with 100 μM NMDA/10 μM glycine in the absence of peptide.
Each data point is an average of at least three measurements collected from at least three cells. Current measurements of the acquired data were made using pCLAMP (Axon) and Origin (Microcal, Northampton, MA) software. The data were averaged and are presented as the mean ± SEM. Statistical analyses were performed using the two-tailed Student’s t-test, and repetitive measures of analysis of variance (Sigma Stat, Jandel Scientific, San Rafael, CA; and Origin software). Significance was assigned at P< 0.05.
RESULTS
As previously described (Sheng et al., 2007), the affinity of NMDA is similar across the four NR subunit combinations examined, viz., NR1a/NR2A, NR1a/NR2B, NR1b/NR2A, NR1b/NR2B. In addition, for the interpretation of the data that follow, it can be assumed that the rates of inhibition onset and offset, as well as the steady-state levels of inhibition, are ascribable to the conantokins alone, and are not complicated by differential subunit fast responses to NMDA. We chose a concentration of 100 μM NMDA, wherein approximately 85–90% saturation of the NMDA response was achieved, for all subsequent experiments.
C-terminal residues of conantokins do not affect NR2B selectivity
The first peptide examined was an 11 amino acid, C-terminal truncation variant of con-G, viz., con-G[1–11] (all peptide sequences are provided in Table 1). N-terminal truncations were not considered, since the first 4 residues are essential for function of the conantokins (Blandl et al., 2001, Warder et al., 2001). Fig. 1 illustrates the comparison of functional effects of different NMDAR complexes with parent con-G and con-G[1–11], as well as with an example full-length con-G variant, con-G[Q6A]. After removal of 6 amino acids from C-terminus of con-G, it is seen that con-G[1–11] does not potently antagonize NMDAR, and the percentage inhibition is less than 60%, even when a high concentration (60 μM) of con-G[1–11] utilized. This is explained by the fast off-rate of inhibition during the wash-out process (Table 2). Nonetheless, con-G[1–11] remains NR2B-selective, suggesting that while the C-terminus of con-G affects its potency, it does not affect its NR2B selectivity, and residues in N-terminus strongly contribute to the bias of con-G for NR2B-containing receptors.
Fig. 1.
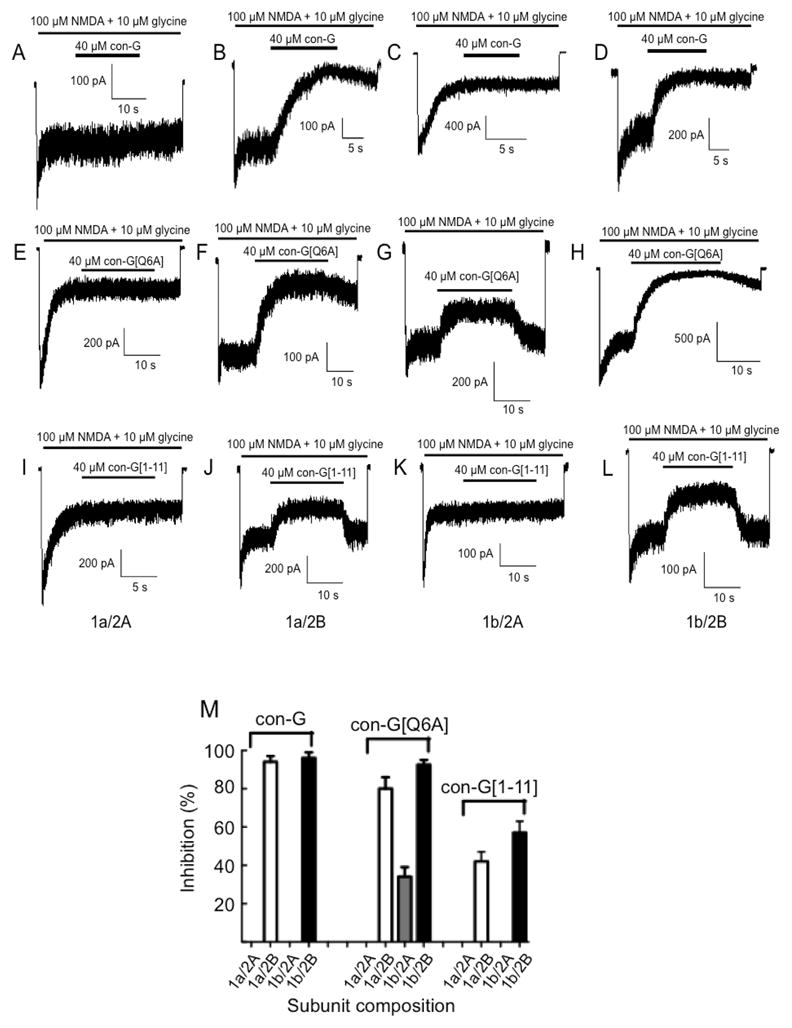
Inhibition of NMDA/glycine-induced ion flow through recombinant NMDAR channels by con-G, con-G[Q6A], con-G[1–11]. Peptides (40 μM) were applied as indicated by the lower horizontal line accompanying each trace, while NMDA (100 μM)/glycine (10 μM) was applied throughout each experiment (as denoted by the upper line). The recombinant NMDARs consisted of the subunit combinations, NR1a/NR2A, NR1a/NR2B, NR1b/NR2A, and NR1b/NR2B. (A–D) con-G; (E–H) con-G[1–11]; (I–L) con-G[Q6A]. (M) Comparison of maximal inhibition by con-G, con-G[Q6A], and con-G[1–11] of steady state whole cell currents in HEK-293 cells transfected with the four different NMDAR subunit complexes. Voltage-clamped recordings were obtained at −70 mV, pH 7.35, 25o C.
Table 2.
Inhibition by synthetic conantokin variants toward NMDAR subunit complexes
| Variant | % maximal inhibition | |||
|---|---|---|---|---|
| 1a/2A | 1a/2B | 1b/2A | 1b/2B | |
| con-Ga | 0 | 94 ± 3 | 0 | 96 ± 3 |
| con-G[1–11] | 0 | 42 ± 5 | 0 | 57 ± 6 |
| con-G[Q6A] | 0 | 80 ± 6 | 34 ± 5 | 92 ± 3 |
| con-G[γ7A] | 0 | 90 ± 4 | 0 | 95 ± 3 |
| con-G[γ7K] | 0 | 82 ± 5 | 0 | 94 ±2 |
| con-G[N8M] | 0 | 73 ± 5 | 0 | 90 ± 2 |
| con-G[Q9A] | 0 | 78 ± 5 | 0 | 92 ± 2 |
| con-G[L5Y/N8A] | 0 | 90 ± 3 | 0 | 91 ± 3 |
| con-G[Q6A/Q9A] | 0 | 70 ± 6 | 0 | 92 ± 4 |
| con-Ta | 90 ± 4 | 92 ± 4 | 93 ± 2 | 97 ± 2 |
| con-T[Y5L] | 80 ± 6 | 85 ± 3 | 86 ± 7 | 90 ± 6 |
| con-T[K7A] | 93 ± 2 | 85 ± 5 | 91 ± 3 | 94 ± 2 |
| con-T[K7γ] | 78 ± 4 | 86 ± 5 | 82 ± 6 | 90 ± 2 |
| con-T[M8A]a | 0 | 73 ± 8 | 0 | 81 ± 5 |
| con-T[M8G] | 0 | 80 ± 5 | 0 | 84 ± 4 |
| con-T[M8N]a | 48 ± 6 | 82 ± 5 | 56 ± 7 | 90 ± 4 |
| con-T[L9A] | 93 ± 2 | 92 ± 5 | 91 ± 3 | 95 ± 2 |
| con-T[M8A/+A10] | 0 | 63 ± 4 | 0 | 67 ± 5 |
| con-G/T | 0 | 80 ± 5 | 0 | 86 ± 4 |
| con-R′a | 93 ± 2 | 94 ± 2 | 94 ± 3 | 97 ± 2 |
| con-R′ [A6Q] | 50 ± 7 | 84 ± 5 | 72 ± 6 | 90 ± 4 |
| con-R′ [K7A] | 93 ± 3 | 91 ± 3 | 93 ± 4 | 95 ± 2 |
| con-R′ [M8A] | 0 | 85 ± 3 | 0 | 88 ± 4 |
| con-R′ [M8N] | 45 ± 6 | 53 ± 5 | 60 ± 7 | 66 ± 5 |
| con-R′ [A6Q/A9Q] | 0 | 70 ± 6 | 25 ± 5 | 79 ± 5 |
| con-R′ [ΔA10] | 0 | 55 ± 6 | 0 | 65 ± 5 |
| con-R′ [V5L/ΔA10] | 0 | 69 ± 4 | 0 | 71 ± 3 |
| con-R′ [V5Y/ΔA10] | 82 ± 6 | 87 ± 5 | 84 ± 4 | 90 ± 3 |
| con-G/R′ | 0 | 30 ± 4 | 0 | 35 ± 5 |
Data from a(Sheng et al., 2007).
Sequence determinants that reside in the N-terminus of conntokins control NR2B selectivity
Two chimeric peptides, con-G/T and con-G/R′, were constructed using the 10 residue N-terminus of con-G to replace the corresponding regions in con-T and con-R′ respectively (Table 1). Both chimeric peptides were found to be NR2B-selective with different degrees of inhibition at 40 μM concentrations of peptides (Table 2). The parent, con-T, inhibits all four of the NMDAR subunit complexes potently. The inhibitory profile of con-G/T resembles that of con-G in that no inhibition is observed with NR2A-containing NMDAR complexes, and this was confirmed by employing concentrations of peptides up to 100 μM. On the other hand, NR2B inhibition by con-G/T remains potent. Con-G/R′ behaves similarly to con-G/T with respect to subunit responses, albeit with reduced potency (<40% maximal inhibition) compared to con-G/T (>80% maximal inhibition).
Point mutations in the N-termini of conantokins reveal loci that govern NR2B selectivity
The above results strongly suggest that the sequence determinants that control conantokin selectivity reside in the N-terminal regions of these peptides. Examination of the N-termini of con-G, con-T, and con-R′ (Table 1) reveals that there are four amino acid differences between con-G and con-T, at sequence positions 5, 7, 8, and 9, and five such differences between con-G and con-R′, at sequence positions 5–9, in addition to an Ala insertion in con-R′ at position 10. It has been previously recognized (Klein et al., 2001a) that replacement of Leu5 in con-G with Tyr, the counterpart residue in con-T, yields a peptide (con-G[L5Y]) that is able to antagonize all four NR1/2 complexes. However, the reciprocal replacements in con-T and con-R′, viz., con-T[Y5L] (Table 2) and con-R′ [V5L] (Klein et al., 2001a), fail to alter the selectivity of the resultant variants, which remains the same as that of their parent peptides, as does con-G[L5V] (Klein et al., 2001a). These data suggest that while a Tyr at position 5 in con-G is sufficient for producing a selectivity switch in con-G, the results with the con-T and con-R′ variants, as well as with con-G[L5V], reveal that Leu5 is not solely responsible for the NR2B-selective properties of con-G, nor do the counterpart Tyr5 and Val5 residues in con-T and con-R′ influence the nonselectivity in these peptides.
Substitution of Gln6 of con-G with an Ala, the residue that occurs in con-R′, leads to a partial selectivity change (Table 2, Fig. 1). This variant, viz., con-G[Q6A], shows a strong inhibitory preference for NR2B-containing subunits (Table 3, Figure 1), demonstrating that Gln6 in con-G is a key residue for conferring NR2B selectivity to this peptide. The tendency of con-R′ [A6Q] towards increased NR2B selectivity (Table 2) strengthens this conclusion. In addition, while con-G[Q6A] shows no inhibitory effects toward NR1a/NR2A, even at concentrations of up to 100 μM con-G[Q6A], there is a weak inhibitory effect toward NR1b/NR2A (Table 2, Figure 1). This discovery of a residue at position 6 that has the potential of functionally discriminating between NR1a and NR1b is a novel finding of this work.
An obvious candidate for the selectivity switch is the amino acid at position 7. In con-G this locus is occupied by a Gla, but in con-T and con-R′, is occupied by a Lys. Despite the charge reversal associated with the Gla (−2) to Lys (+1) replacement, our data using con-G[γ7K], con-G[γ7A], con-T[K7γ], con-T[K7A], and con-R′ [K7A] (Table 2) demonstrate that this striking difference between the sequences of NR2B-selective con-G and the more nonselective con-T and con-R′ is without consequence to the NMDAR subunit preferences of these peptides. A similar argument can be made for position 8 residues, since con-G[N8M], con-T[M8N], and con-R′ [M8N] do not change their selectivity profiles from that of their parent peptides. However, one peptide, con-T[M8N], has a slight bias for inhibiting NR2B-containing receptors (Sheng et al., 2007). A prominent feature that emerges from sequence comparisons of con-G, con-T, and con-R′ with the NR2B-selective Conus parius conantokins, con-Pr1, and con-Pr2, is the presence of a Tyr5 in both of the latter peptides, and a Gly and Ala in position 8 in con-Pr1 and con-Pr2, respectively. In a previous publication (Sheng et al., 2007), we reported that a con-T-based peptide with selectivity for the NR2B receptor subtype could be obtained through replacement of Met8 with Ala. To test the hypothesis that the presence of a Gly, as well as an Ala residue in position 8, can confer NR2B subunit selectivity on a Tyr5-containing conantokin, we obtained functional data for con-T[M8G] (Table 2). Like the Ala variant, this peptide is also highly selective for NR2B-containing receptors. This propensity was likewise observed for con-R′ [M8A]. Hence, as with the L5Y substitution in con-G, a single amino acid replacement in con-T or con-R′ appears to be sufficient for a transformation between subunit selective and nonselective modes of NMDAR inhibition. Confirmation of this supposition was provided by the inhibitory profile of con-G[L5Y/N8A], in which the nonselective activity of con-G[L5Y] reverted to that of a NR2B-selective peptide by virtue of the presence of an Ala at position 8.
We constructed another variant, con-G[Q9A] (Table 2), thus inserting the Ala9 of con-R′ into con-G. However, this change did not generate a NR2A-inhibitory peptide, as occurred for the Ala substitution of Gln6 in con-G. Incorporation of the Ala6xxAla9 pattern in con-G, as occurs in con-R′, also failed to confer NR2A-inhibitory activity on the resultant peptide, as evidenced by the strict NR2B selectivity displayed by con-G[Q6A/Q9A]. However, the converse replacements in con-R′ resulted in abrogation of the NR1a/2A activity of the resultant variant, con-R′ [A6Q/A9Q], suggesting that the presence of Gln residues at positions 6 and 9 are partially coupled with respect to NMDAR activity and can confer a NR2B-selective bias in the context of an otherwise non-selective conantokin.
Insertion and deletion mutations in conantokins alter NMDAR selectivity
Lastly, a salient difference between con-R′ and con-G and con-T involves the insertion, in the former, of an Ala at position 10. We found that con-R′ was converted into a NR2B-selective variant through deletion of this residue, in the peptide, con-R′ [ΔA10]. Similarly, con-R′ [V5L/ΔA10] displayed NR2B selectivity (Table 2). On the other hand, con-R′ [V5Y/ΔA10] remains nonselective. Hence, the coupling of an aliphatic residue at position 5 with the deletion of Ala10 produces a con-R′-based peptide that displays inhibitory activity only at NR1a/2B- and NR1b/2B-containing NMDAR complexes. However, the Ala10 deletion cannot compensate for the presence of aromatic Tyr in sequence position 5. Conversely, the insertion of an Ala10 is insufficient for the recovery of NR2A activity in the NR2B-selective variant con-T[M8A], as highlighted by the functional data for con-T[M8A/+A10] (Table 2). This is consistent with the NR2B-selective inhibitory activity noted for con-R′ [M8A], which contains the same Ala8xxGla11 pattern as con-T[M8A/∇A10].
DISCUSSION
Conantokins are potent and selective peptide antagonists of NMDARs, and their subunit selectivities have been widely investigated (Donevan and McCabe, 2000, Klein et al., 2001a, Sheng et al., 2007). To this point, no universal conclusions have been forwarded regarding the molecular basis for the NMDAR subunit selectivities of the conantokins. Con-G, and three recently characterized conantokin peptides, viz., con-Pr1, con-Pr2, and con-Pr3 (Teichert et al., 2007), are the only reported peptides in the conantokin family that are either strongly or weakly selective for NR2B- versus NR2A-containing NMDARs.
Our goal in this project was to establish the minimum number of mutations that would suffice to convert con-G to a nonselective NMDAR inhibitor, and allow con-T and con-R′ to become NR2B-selective inhibitors. We built on previous work in which it was demonstrated that the replacement of Leu5 of con-G with a Tyr, the analogous residue in con-T, was sufficient to impart nonselective functionality to the con-G peptide (Klein et al., 2001a). However, the nonselective NMDAR inhibitory activities of con-T and con-R′ were unaffected by the converse substitutions, that yielded con-T[Y5L] and con-R′ [V5L]. These findings suggest that, unlike the case for con-G, the amino acid at position 5 in both con-T and con-R′ does not govern the selectivity of conantokin peptides, but is coupled to another residue or residues within these peptides for this purpose. Such a cooperative effect could facilitate con-T and con-R′ accommodation within the conantokin sites of action in NR2A- and NR2B-containing receptors alike.
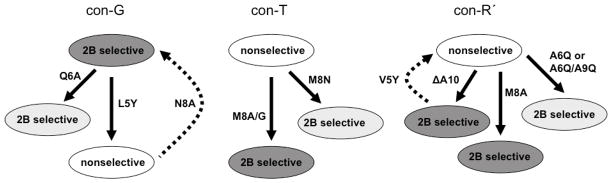
Figure 2 represents a summary of the primary sequence changes that can impart a selectivity interchange (or modify selectivity) in the three conantokins examined. Among the three conantokins investigated, a single amino acid modification can exert a selectivity switch; an L5Y substitution in con-G, an M8A substitution in con-T and con-R′, and an A10 deletion in con-R′. As a general conclusion, it appears that subunit selectivity is highly sensitive to the nature of the amino acid at sequence position 8. For both con-T and con-R′, the presence of an Ala at this position is adequate for NR2B selectivity. Furthermore, the presence of Ala8 overrides the conferral of nonselectivity that attends the L5Y substitution in con-G. The Ala10 deletion in con-R′, which shifts the Gla at position 11 to position 10 as occurs in con-G and con-T, elicits a selectivity switch in con-R′, but its insertion in con-T[M8A] cannot reverse the NR2B-selective nature of this peptide (Table 2). Hence, the con-R′ nonselectivity conferred by the presence of A10 may be acting in synergy with sequence elements of con-R′ that are not present in con-T or con-T[M8A]. An obvious candidate for this coupling is the aliphatic Val5, which is homologous to the Leu5 in con-G. However, other residues must also be invoked in accounting for the NR2B-selectivity of con-G and con-R′ [ΔA10], since con-T[Y5L] retains the nonselectivity of the parent peptide. While a V5Y substitution in con-R′ [ΔA10] reverts the peptide to that of a more nonselective nature, the basis for this may simply reside in the high sequence identity between the N-terminal segments of con-T and con-R′ [V5Y/ΔA10] (8 out of the first 10 amino acids). The nature of the residue at position 6 also influences the subunit preferences of the conantokins, as the presence of a Gln6 in both con-G and con-R′ increases the bias of each peptide towards NR2B-containing subunits.
Fig. 2.
A summary of the mutations in con-G, con-T, and con-R′ that alter their selectivity profiles with respect to NR2A- and NR2B-containing NMDARs.
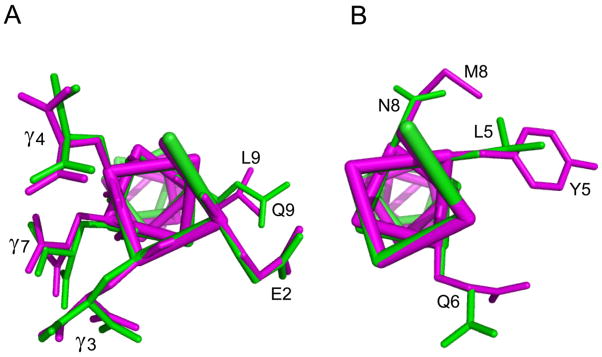
Currently, no structural data exist regarding the NMDAR-bound conformations of the conantokins, or the locations of the conantokin binding sites in the receptors. Thus, we focused herein on molecular features of the ligand that led to NR2B-selectivity. In this regard, insights on the selectivity determinants in the conantokins can be proposed upon comparisons of the crystal structures of Ca2+-complexed con-G and con-T[K7γ] (Cnudde et al., 2007). Both peptides form antiparallel dimers, with the dimerization interface mediated by interstrand bridging between Ca2+ and Gla residues 3, 7, 10, and 14. These structures are physiologically relevant, insofar as physiological Ca2+ concentrations (ca., 2 mM) are high enough to allow for significant population of the dimeric state, and electrophysiological results have revealed robust NMDAR activity of the con-G dimer (Dai et al., 2007). Furthermore, because con-T[K7γ] exhibits the same nonselective NMDAR activity as con-T, this leaves only 3 sequence positions, namely 5, 8, and 9, that can be responsible for the switch between selective and nonselective modes of NMDAR inhibition. These residues may be involved in direct interactions with their cognate receptor binding sites or may act indirectly by affecting the orientation(s) of key binding residues. As shown in Fig. 3A, similar side chain dispositions occur for Glu2 and Gla residues 3, 7, and 4. These amino acids are common to both peptides. The side-chains of Gln9 of con-G and Leu9 of con-T likewise adopt similar geometries and, despite differences in hydrophobic character, their functions with NMDARs are inconsequential to selectivity since an Ala9 substitution in either of these peptides does not alter their NMDAR subunit preferences (Table 2). Unlike other identical sequence elements within the N-termini of con-G and con-T[K7γ], the side-chain of Gln6 adopts a dramatically different conformation between the two peptides, with the bond between the γ- and carbonyl carbons rotated by about 90° between the two peptides. This suggests a mechanism wherein the conantokin function sites in NR2A- and NR2B-containing receptors can accommodate the Gln6 of con-T[K7γ], but only NR2B-receptors can allow for this side-chain in con-G. The acquisition of some NR2A activity in con-G upon a Q6A substitution is consistent with this interpretation, and may occur because the smaller Ala side-chain can access functional sites complementary to position 6 in both NR2A and NR2B receptor types.
Fig. 3.
Comparison of the three-dimensional structures of con-G (green) and con-T[K7γ] (magenta) modeled from X-ray crystallographic data of these peptides (Cnudde et al., 2007). The overlays depict residues in the N-terminal regions of con-G and con-T[K7γ] that have (A) similar and (B) distinct side-chain geometries and conformations.
From examination of Fig. 3B, it is likely that a Met at position 8 confers NR2A activity on con-T (and con-R′). The abrogation of the NR2A inhibitory activity of con-T by replacement of Met8 by Gly or Ala, and the NR2B bias displayed by con-T[M8N], are compatible with this conclusion. Con-G[N8M] still retains NR2A activity since it may be the case that the side-chain orientation of Met8 in the con-T[Y5L] is affected by those residues not shared with con-G[N8M], in a manner that allows activity with NR2A-containing receptors. In con-R′, which, like con-G, contains an aliphatic residue at position 5, this role can be fulfilled by the Ala insert at position 10, since deletion of this residue is sufficient to confer NR2B selectivity to the resultant variant.
Lastly, the structural model presented in Fig. 3B highlights the differences in length and volume that occurs between the Leu and Tyr at position 5 of con-G and con-T con-T[K7γ]. Previously acquired structure-activity data (Prorok and Castellino, 2001) have revealed that a sizable hydrophobic residue is required at this position, since severely diminished NMDAR inhibition results from Ala5 substitutions in con-G, con-T and con-R′. However, as highlighted in this and other studies (Klein et al., 2001a), NMDAR selectivity does not solely rely on the nature of the residue at this position, since con-R′ (which contains a Val at position 5) and con-T[Y5L] are nonselective NMDAR inhibitors. The results presented herein strongly suggest that the presence of a Tyr at position 5, in tandem with a bulky hydrophobic or non-charged amino acid at position 8, provides for inhibitory activity at NR2A- and NR2B-containing NMDAR subunit combinations. The amino acid at sequence position 6 and the presence or absence of an Ala at position 10, also contribute to selectivity.
The work described herein reinforces the possibility of making simple modifications of one or two residues in order to engineer different subtype selectivity profiles into the known conantokins. Currently, all trials, in vitro or in vivo, for use of NR2B-selective conantokins employ con-G. Having other NR2B-selective mutations available would allow other conantokins to be employed with other advantageous properties incorporated, e.g., blood-brain barrier permeability.
Acknowledgments
We thank Ms. Brigid E. Brown for assistance with peptide synthesis and Dr. Min Wang for providing the structures in Figure 3.
This work was supported by grant HL019982 (to F.J.C.) from the NIH.
Abbreviations
- NMDAR
N-methyl-D-aspartate receptor
- NR1
N-methyl-D-aspartate receptor subunit 1, with its 8 splice variants (a–h)
- NR2
N-methyl-D-aspartate receptor subunit 2, with its 4 gene products (A–D)
- NR3
N-methyl-D-aspartate receptor subunit 3, with its two variants, A and B
- con
conantokin
- HEK
human embryonic kidney
- γ
γ-carboxyglutamic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blandl T, Warder SE, Prorok M, Castellino FJ. Structure-function relationships of the NMDA receptor antagonist peptide, conantokin-R. FEBS Lett. 2000;470:139–146. doi: 10.1016/s0014-5793(00)01309-0. [DOI] [PubMed] [Google Scholar]
- Blandl T, Zajicek J, Prorok M, Castellino FJ. Sequence requirements for the N-methyl-D-aspartate receptor antagonist activity of conantokin-R. J Biol Chem. 2001;276:7391–7396. doi: 10.1074/jbc.M006648200. [DOI] [PubMed] [Google Scholar]
- Chatterton JE, Awobuluyi M, Premkumar LS, Takahashi H, Talantova M, Shin Y, Cui JK, Tu SC, Kevin ASK, Nakanishi N, Tong G, Lipton SA, Zhang DX. Excitatory glycine receptors containing the NR3 family of NMDA receptor subunits. Nature. 2002;415:793–798. doi: 10.1038/nature715. [DOI] [PubMed] [Google Scholar]
- Cnudde SE, Prorok M, Dai Q, Castellino FJ, Geiger JH. The crystal structures of the calcium-bound con-G and con-T[K7gamma] dimeric peptides demonstrate a metal-dependent helix-forming motif. J Am Chem Soc. 2007;129:1586–1593. doi: 10.1021/ja065722q. [DOI] [PubMed] [Google Scholar]
- Dai Q, Sheng Z, Geiger JH, Castellino FJ, Prorok M. Helix-helix interactions between homo- and heterodimeric gamma-carboxyglutamate-containing conantokin peptides and their derivatives. J Biol Chem. 2007;282:12641–12649. doi: 10.1074/jbc.M609087200. [DOI] [PubMed] [Google Scholar]
- Donevan SD, McCabe RT. Conantokin G is an NR2B-selective competitive antagonist of N-methyl-D-aspartate receptors. Mol Pharmacol. 2000;58:614–623. doi: 10.1124/mol.58.3.614. [DOI] [PubMed] [Google Scholar]
- Furukawa H, Singh SK, Mancusso R, Gouaux E. Subunit arrangement and function in NMDA receptors. Nature. 2005;438:185–192. doi: 10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
- Gowd KH, Twede V, Watkins M, Krishnan KS, Teichert RW, Bulaj G, Olivera BM. Conantokin-P, an unusual conantokin with a long disulfide loop. Toxicon. 2008;52:203–213. doi: 10.1016/j.toxicon.2008.04.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimwood S, Le Bourdelles B, Whiting PJ. Recombinant human NMDA homomeric NR1 receptors expressed in mammalian cells form a high-affinity glycine antagonist binding site. J Neurochem. 1995;64:525–530. doi: 10.1046/j.1471-4159.1995.64020525.x. [DOI] [PubMed] [Google Scholar]
- Haack JA, Rivier J, Parks TN, Mena EE, Cruz LJ, Olivera BM. Conantokin-T. A γ-carboxyglutamate-containing peptide with N-methyl-D-aspartate antagonist activity. J Biol Chem. 1990;265:6025–6029. [PubMed] [Google Scholar]
- Jimenez EC, Donevan S, Walker C, Zhou LM, Nielsen J, Cruz LJ, Armstrong H, White HS, Olivera BM. Conantokin-L, a new NMDA receptor antagonist: determinants for anticonvulsant potency. Epilepsy Res. 2002;51:73–80. doi: 10.1016/s0920-1211(02)00101-8. [DOI] [PubMed] [Google Scholar]
- Klein RC, Prorok M, Castellino FJ. Direct binding properties of conantokins to native N-methyl-D-aspartate receptors. J Pept Res. 2003;61:307–317. doi: 10.1034/j.1399-3011.2003.00059.x. [DOI] [PubMed] [Google Scholar]
- Klein RC, Prorok M, Galdzicki Z, Castellino FJ. The amino acid residue at sequence position 5 in the conantokin peptides partially governs subunit-selective antagonism of recombinant N-methyl-D-aspartate receptors. J Biol Chem. 2001a;276:26860–26867. doi: 10.1074/jbc.M102428200. [DOI] [PubMed] [Google Scholar]
- Klein RC, Warder SE, Galdzicki Z, Castellino FJ, Prorok M. Kinetic and mechanistic characterization of NMDA receptor antagonism by replacement and truncation variants of the conantokin peptides. Neuropharmacology. 2001b;41:801–810. doi: 10.1016/s0028-3908(01)00119-8. [DOI] [PubMed] [Google Scholar]
- Laube B, Hirai H, Sturgess M, Betz H, Kuhse J. Molecular determinants of agonist discrimination by NMDA receptor subunits: Analysis of the glutamate binding site on the NR2B subunit. Neuron. 1997;18:493–503. doi: 10.1016/s0896-6273(00)81249-0. [DOI] [PubMed] [Google Scholar]
- Ma Q, Hargreaves RJ. Localization of N-methyl-D-aspartate NR2B subunits on primary sensory neurons that give rise to small-caliber sciatic nerve fibers in rats. Neuroscience. 2000;101:699–707. doi: 10.1016/s0306-4522(00)00419-x. [DOI] [PubMed] [Google Scholar]
- McIntosh J, Olivera BM, Cruz L, Gray W. γ-Carboxyglutamate in a neuroactive toxin. J Biol Chem. 1984;259:14343–14346. [PubMed] [Google Scholar]
- Merchant RE, Bullock MR, Carmack CA, Shah AK, Wilner KD, Ko G, Williams SA. A double-blind, placebo-controlled study of the safety, tolerability and pharmacokinetics of CP-101,606 in patients with a mild or moderate traumatic brain injury. Ann NY Acad Sci. 1999;890:42–50. doi: 10.1111/j.1749-6632.1999.tb07979.x. [DOI] [PubMed] [Google Scholar]
- Prorok M, Castellino FJ. Structure-function relationships of the NMDA receptor antagonist conantokin peptides. Curr Drug Targets. 2001;2:313–322. doi: 10.2174/1389450013348542. [DOI] [PubMed] [Google Scholar]
- Prorok M, Warder SE, Blandl T, Castellino FJ. Calcium binding properties of synthetic γ-carboxyglutamic acid containing marine cone snail “sleeper” peptides, conantokin-G and conantokin-T. Biochemistry. 1996;35:16528–16534. doi: 10.1021/bi9621122. [DOI] [PubMed] [Google Scholar]
- Sheng Z, Dai Q, Prorok M, Castellino FJ. Subtype-selective antagonism of N-methyl-D-aspartate receptor ion channels by synthetic conantokin peptides. Neuropharmacology. 2007;53:145–156. doi: 10.1016/j.neuropharm.2007.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichert RW, Jimenez EC, Twede V, Watkins M, Hollmann M, Bulaj G, Olivera BM. Novel conantokins from conus parius venom are specific antagonists of NMDA receptors. J Biol Chem. 2007;282:36905–36913. doi: 10.1074/jbc.M706611200. [DOI] [PubMed] [Google Scholar]
- Twede V, Teichert R, Walker C, Gruszczynski P, Kazmierkiewicz R, Bulaj G, Olivera B. Conantokin-Br from Conus brettinghami and selectivity determinants for the NR2D subunit of the NMDA receptor. Biochemistry. 2009;48:4063–4073. doi: 10.1021/bi802259a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warder SE, Blandl T, Klein RC, Castellino FJ, Prorok M. Amino acid determinants for NMDA receptor inhibition by conantokin-T. J Neurochem. 2001;77:812–822. doi: 10.1046/j.1471-4159.2001.00281.x. [DOI] [PubMed] [Google Scholar]
- Wei J, Dong M, Xiao C, Jiang F, Castellino FJ, Prorok M, Dai Q. Conantokins and variants derived from cone snail venom inhibit naloxone-induced withdrawal jumping in morphine-dependent mice. Neurosci Lett. 2006;405:137–141. doi: 10.1016/j.neulet.2006.06.040. [DOI] [PubMed] [Google Scholar]
- White HS, McCabe RY, Armstrong H, Donevan SD, Cruz LJ, Abogadie FC, Torres J, Rivier JE, Paarmann I, Hollmann M, Olivera BM. In vitro and in vivo characterization of conantokin-R, a selective NMDA receptor antagonist isolated from the venom of the fish-hunting snail Conus radiatus. J Pharmacol Exp Ther. 2000;292:425–432. [PubMed] [Google Scholar]
- Williams AJ, Dave JR, Lu XM, Ling G, Tortella FC. Selective NR2B NMDA receptor antagonists are protective against staurosporine-induced apoptosis. Eur J Pharmacol. 2002;452:135–136. doi: 10.1016/s0014-2999(02)02327-0. [DOI] [PubMed] [Google Scholar]
- Wolosker H. D-serine regulation of NMDA receptor activity. Sci STKE. 2006;2006:41. doi: 10.1126/stke.3562006pe41. [DOI] [PubMed] [Google Scholar]
- Xi G, Hua Y, Keep RF, Younger JG, Hoff JT. Brain edema after intracerebral hemorrhage: the effects of systemic complement depletion. Acta Neurochir Suppl. 2002;81:253–256. doi: 10.1007/978-3-7091-6738-0_66. [DOI] [PubMed] [Google Scholar]
- Xiao C, Huang Y, Dong M, Hu J, Hou S, Castellino FJ, Prorok M, Dai Q. NR2B-selective conantokin peptide inhibitors of the NMDA receptor display enhanced antinociceptive properties compared to non-selective conantokins. Neuropeptides. 2008;42:601–609. doi: 10.1016/j.npep.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]