Abstract
Essential for the normal functioning of a cell is the maintenance of genomic integrity. Failure in this process is often catastrophic for the organism, leading to cell death or mis-proliferation. Central to genomic integrity is the faithful replication of DNA during S phase. The GINS complex has recently come to light as a critical player in DNA replication through stabilization of MCM2-7 and Cdc45 as a member of the CMG complex which is likely responsible for the processivity of helicase activity during S phase. The GINS complex is made up of 4 members in a 1:1:1:1 ratio: Psf1, Psf2, Psf3, And Sld5. Here we present the first analysis of the function of the Sld5 subunit in a multicellular organism. We show that Drosophila Sld5 interacts with Psf1, Psf2, and Mcm10 and that mutations in Sld5 lead to M and S phase delays with chromosomes exhibiting hallmarks of genomic instability.
Keywords: GINS, SLD5, CMG Complex, DNA Replication, Elongation
INTRODUCTION
The maintenance of genomic stability is essential for the normal growth and development of organisms. Genomic instability has been linked to the development of cancers and other disease states [1,2]. Central to the maintenance of genomic stability is the high fidelity replication of DNA during S phase [2,3,4]. The GINS complex has been identified as an essential DNA replication factor that along with Cdc45 and Mcm2-7 (the CMG complex) constitutes the core of the Replisome Progression Complex (RPC) [5]. The CMG complex has been shown to be required for the initiation of DNA replication and the subsequent progression of the replication fork [6]. The absence of the CMG complex during DNA replication results in stalling of the replication fork presumably due to the decrease in the helicase activity of Mcm2-7 [5,6,7]. The GINS complex likely plays a role in stabilization of the interaction between Cdc45 and the Mcm2-7 hexamer and, as such, is important for maintaining the CMG complex during elongation[8] [6].
The GINS complex is an evolutionarily conserved heterotetrameric complex that consists of: Sld5, Psf1, Psf2, and Psf3. Crystal structures of the human GINS complex reveal a structure resembling an elongated spindle with a central pore [9]. Within the GINS complex Sld5 interacts with Psf1 via N terminal alpha helical domains and with Psf2 through a combination of alpha helical and beta sheet domains in the C terminal half of the protein [9].
The Sld5 subunit of the GINS complex was first identified in the budding yeast S. cerevisiae as synthetically lethal with a mutation in dpb11, a protein required for the loading of DNA polymerase epsilon [10,11]. Although a number of biochemical, tissue culture, and gene expression studies have characterized the GINS complex, no studies have addressed the function of Sld5 within the context of a multicellular metazoan [6,12,13,14,15,16,17]. Using mutant alleles of Sld5 and tissue specific RNAi in Drosophila we show that Sld5 is required for normal cell cycle progression and the maintenance of genomic integrity.
MATERIALS AND METHODS
Fly husbandry / stocks
All fly stocks were maintained on Drosophila K12 media (US Biological # D9600-07B) at room temperature. Drosophila strains used for this study were acquired from the Bloomington Fly stock center, the Exelixis Drosophila Stock Collection at Harvard Medical School, and the Vienna Drosophila RNAi Center. These strains consisted of: wt (Flybase ID: FBst0006326, ), w; Sld5c010719/TM3,Sb,Ser (derived from Flybase ID:FBst1005902), yw; Sld5A462/TM3,Sb,Ser (Flybase ID:FBst0016124), w; Df(3R)BSC140/ TM6B (deletion spanning Sld5 and Dak1, Flybase ID: FBst0009500), w; Df(3R)BSC751/TM6C, Sb (Deletion spanning Dak1 but not Sld5, Flybase ID: FBst0026849), w; P{GD8365}v43588 (Gal4 inducible Sld5 RNAi, Flybase ID: FBst0465151), P{w[+mW.hs]=GawB}167Y, w (Gal4 expression in 3rd instar brain, Flybase ID: FBst0003741), and y,w; D/TM3, Sb, GFP (3rd chromosome GFP balancer, Flybase ID: FBst0005195).
Transgenic rescue flies were generated through germline transformation by Best Gene, Inc. (Chino Hills, CA). The Sld5 P Element rescue construct consisted of the pTWM backbone with an insert containing the genomic Sld5 region (+2000bp upstream: Sld5 coding region: stop codon removed for fusion to myc tag in pTWM). A germline transformant was selected with a non-lethal insert in the 2nd chromosome and used to generate fly line; apXa T(2:3)/ p[Sld5+]; TM3, Sb through standard genetic crosses.
Yeast 2 hybrid
Yeast manipulation and growth were conducted using standard protocols as in [18] and found in manufacture's protocols (Clonetech, Matchmaker™ Yeast two-hybrid system). Yeast strain AH109 (Clonetech) was used as the reporter strain. Plasmids used were pGBKTetT7 and pGADT7 (Clontech) except that both were converted to the Gateway™ cloning system (Invitrogen) by insertion of Gateway™ cassette into the MCS. In addition the KanR gene in pGBKT7 was disrupted by insertion of TetR gene to facilitate use in the Gateway™ system. Entry clones generated for the genes outlined figure 2 were constructed using DNA that was PCR amplified from an early embryo cDNA library. All entry clones were sequence verified prior to LR reactions with two-hybrid plasmids. Resulting two-hybrid clones were then sequence verified to ensure proper reading frame was maintained and no ectopic mutations were introduced.
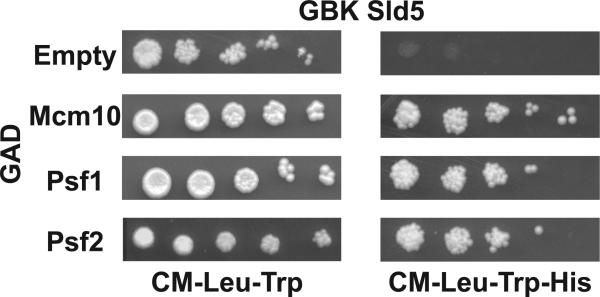
Figure 2. Drosophila Sld5 interacts with GINS partners and Mcm10.
Yeast two-hybrid analysis showing that Sld5 interacts with Drosophila Psf1, Psf2, and Mcm10. Bait and prey constructs are as indicated and interactions were detected by the activation of the HIS3 reporter gene and 5X serial dilutions onto media lacking histidine (right panel).
Arrest point
Sld5A462/TM3, Sb,GFP and Sld5c010719/TM3, Sb,GFP flies respectively were isolated in a collection chamber and fed yeast paste for 24 hours. After removing the yeast paste a fresh grape plate with a film of yeast paste spread over its surface was introduced for the flies to deposit their embryos. Embryos were collected for 4-5hours. The embryos were examined under an Olympus SZX7 Dissecting Stereo Microscope with X-Cite Series Q Epifluorescence illumination for GFP and separated based on their phenotype. Non-glowing flies were homozygous for the Sld5 mutation and glowing embryos were isolated as heterozygous controls. Growth of each group was monitored and development of homozygous mutants was compared to heterozygous controls.
Brain Squash for M phase index
3rd instar wandering larva were harvested and dissected in 1XPBS pH 7.2 with 1% PEG 8000. Removed larval brains were transferred to hypotonic solution (Sodium Citrate 0.5%) and incubated for 10 min. Brains were then fixed in Acetic acid : Methanol : Water 11:11:2 for 30 seconds. Brains were then transferred to cleaned microscope slide and overlaid with a siliconized coverslip. The microscope slide and coverslip were sandwiched between filter paper and an additional microscope slide. This was then placed in a machinist vise and pressure was applied using a torque wrench to 15Nm. Following 2 min incubation at this pressure, slide and coverslip were removed and lowered into liquid nitrogen. Once equilibrated slide and coverslip were removed and the coverslip popped off and slide washed gently with 100% EtOH, allowed to air dry and mounted with 7ul of Vectashield™ with DAPI. Mitotic index determination were performed on these squash preparations by selecting 10 random well populated fields of view for each brain squash using a 20X objective. Total nuclei were counted for each field and this was divided by the total number of mitotic figures observed in each field to generate the fraction of cells in mitosis. Statistical analysis was performed using the Minitab™ software package.
EdU incorporation assays
S-phase detection in Drosophila neural tissue was ascertained using the Click-It® reaction kit from Invitrogen (Cat. # C10337). Brains were dissected in fresh Grace's unsupplemented cell culture medium. An equal volume of 200μM EdU solution in DMSO was added to the well and brains from each strain were incubated for 30 minutes in the dark at room temperature. Following the incubation the liquid was removed from each well and the brains were rinsed three times with 1X PBS. Brains were fixed in fresh 3.7% Formaldehyde in PBS incubating at room temperature for 15 minutes in the dark. The liquid was then removed and the brains were rinsed two times with 1X PBS. Brains were then permeabilized using 0.1% Triton X100 in PBS for 15 minutes at room temperature in the dark. The liquid was removed and the brains were rinsed two times with 1X PBS. Brains were then incubated in the Click-It® reaction cocktail as per the manufacturer's instructions for 30 minutes. The brains were rinsed two times with the reaction rinse buffer provided by the manufacturer. After removing the rinse buffer Hoescht 33342 was prepared as per the manufacturer's instructions for nuclear visualization for 10 minutes. The Hoescht solution was then removed and the brains were washed four times with 1X PBS. Brains were then mounted on Poly-lysine coated slides with Vectashield™. S phase indices were performed on these tissues
S phase indices
Tissues were dissected and incubated with EdU as above. The brains were then transferred to a well containing 0.5% Sodium Citrate Solution to allow the brains to swell for 10 minutes. The brains were then placed in an 11:11:2 Acetic Acid, Methanol and Water solution for 30 seconds. The brains were immediately transferred to a slide containing a dot (~5μl of 1X PBS) and a Sigmacote® coverslip was placed on top of the brain. The brain was then squashed as for mitotic indices above. The slide was then gently dipped into liquid nitrogen. The coverslip was then popped off and the slide was washed with 3% BSA in 1X PBS. A 0.5% PBT-X solution was added for 20 minutes followed by two washes in 3% BSA in 1X PBS. The Click-It Reaction Cocktail was applied and the tissue incubated for 30 minutes in the dark followed by two washes with 3% BSA in 1X PBS. A 1X Hoescht 33342 (5μg/mL) solution in 1X PBS was added and the tissue incubated for 20 minutes in the dark followed by two 1X PBS washes. PBS was removed and replaced by 7ul of Vectashield™ with DAPI followed by a coverslip. Calculation of the S phase index was performed as for M phase indices except that EdU positive cells were counted. Statistical analysis was performed using the Minitab™ software package.
RESULTS
Identification of Sld5 mutant alleles
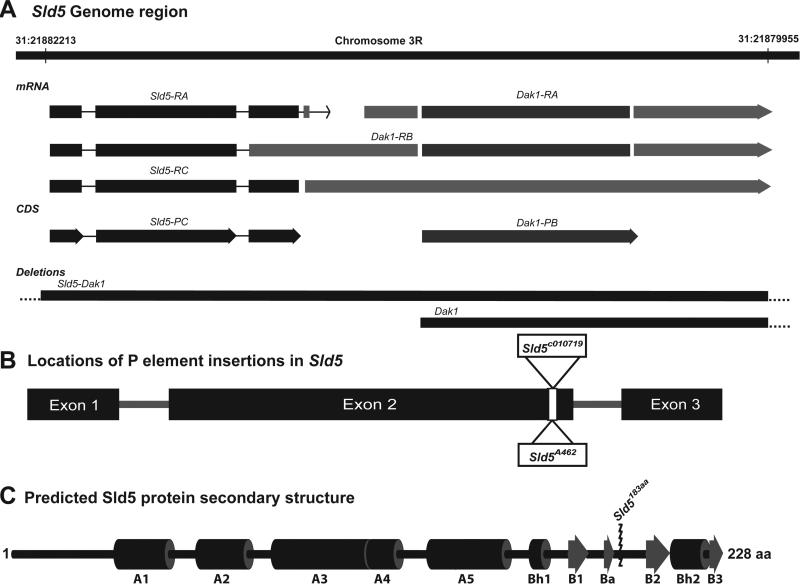
The Sld5 gene is found on chromosome 3R in the Drosophila genome and is located in close proximity to Dak1 (Figure 1A). Transcript models for this region suggest that Sld5 and Dak1 may be part of a multi-cistronic transcript [19] (Figure 1A). Two independent, recessive lethal, P element insertion lines were identified for the Sld5 coding region: Sld5c010719 and Sld5A462 (Figure 1B). Careful analysis of the sequence flanking these independent P element insertions reveals that both are inserted at exactly the same location in the second exon of Sld5 [19]. Disruption of the coding sequence at this location results in small linker sequence from the P element, VNSSMYVS, followed by a stop codon. In total 45 amino acid residues are removed from the C terminal of the native Sld5 protein (Figure 1C). Sequence alignments of Drosophila Sld5 with Human Sld5, combined with the published crystal structure of human Sld5 [9], reveals that the truncation alleles in Drosophila remove a portion of Sld5 that is critical for interaction with Psf2 and the stability of the GINS complex (Figure 1C).
Figure 1. Sld5 Gene Region, P-element insertion, and Protein layout.
A. Sld5 gene is located on chromosome 3R and may be part of a multicistronic mRNA that also contains Dak1. Two different deletion lines were used to show that Sld5 mutations were specific to Sld5 and not also defective for Dak1. B. Map of the Sld5 gene showing insertion site of the two independent P element lines used in this study. C. Map of the predicted 1° protein structure of Sld5 based on alignment with the human protein [9,22,23] comprised of a N terminal “A” domain comprised mainly of alpha helices and a C terminal “B” domain with 4 beta sheet regions. The insertion of the respective P elements truncates the protein removing critical residues in the B domain required for stability of the GINS complex [12].
Homozygous individuals for both Sld5 mutant strains arrested at the late embryo/first instar larval stages. The lethality of these homozygous lines was rescued by the introduction of a Sld5 transgene driven by a native promoter (see materials and methods). In order to rule out the possibility that phenotypes observed in these mutants was due to disruption of the adjacent Dak1 gene each of the Sld5 mutant line was crossed to two separate lines: one containing a deletion spanning both Dak1 and Sld5 and another that just spanned Dak1 (Figure 1A). Complementation of the Sld5 lesions only occurred when crossed to the Dak1 deficiency line, but did not occur when crossed to the Sld5-Dak1 deficiency line. When taken together the results of these crosses point to the fact that the phenotypes observed in these mutant lines are due to defects in Sld5 and not the proximal Dak1 gene.
Drosophila Sld5 interactions
As a member of the GINS complex, Sld5 make contacts with both Psf1 and Psf2 [9]. In order to investigate whether Drosophila Sld5 also contacts Drosophila Psf1 and Psf2 a two-hybrid approach was taken. Sld5 was found to interact by two-hybrid with both Psf1 and Psf2 (Figure 2). In addition Sld5 was found to interact with Drosophila Mcm10 (Figure 2) which has been previous identified as co-purifying with the CMG complex [17]. One-hybrid activity was not observed for the GAD fusion proteins tested (data not shown).
Mitotic defects
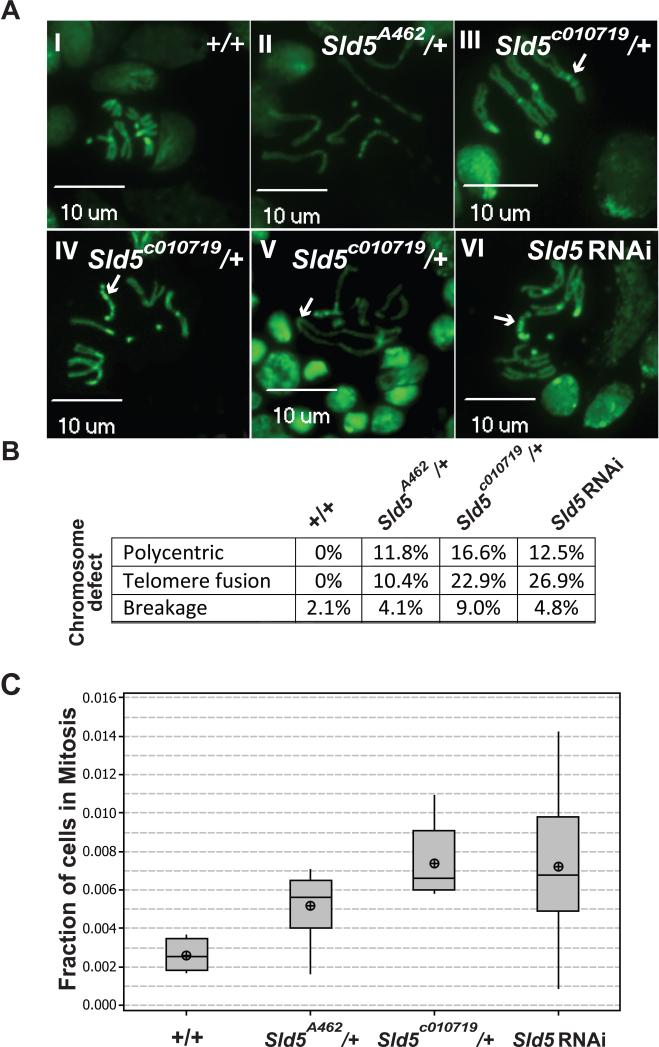
Wandering 3rd instar larval brains are the tissue of choice for examining mitosis in Drosophila. Brain tissue from wild-type, tissue from larva heterozygous for the respective Sld5 mutant alleles, and tissue from a UAS:: Sld5 RNAi line driven by a brain specific Gal4 expression were dissected, squashed, and visualized for mitotic figures. While wild-type mitotic figures appeared normal, numerous types of mitotic defects were observed in the Sld5 mutant lines and the RNAi depletion line (Figure 3A). All of the metaphase chromosomes observed in the Sld5 defective backgrounds were characterized by defects in condensation as evidenced by longer than normal chromosomes. In addition a variety of mitotic aberrations were observed including: polycentric chromosomes (Figure 3A, III, IV, VI), chromosome breakage (Figure 3A, II, V), and telomere fusions (Figure 3A, V). Quantitation of the types of mitotic defects observed suggests that similar frequency of defects is observed in all Sld5 defective lines tested (Figure 3B). Mitotic index measurements for these tissues demonstrated significant M phase delays in the mutant and RNAi larval brains compared to wild-type: Sld5A462/+ had 98% more cells in mitosis (p=0.002), Sld5c0107192/+ had 182% more (p<0.000), and Sld5 RNAi had 175% more cells in mitosis compared to wild-type (p=0.007)(Figure 3C).
Figure 3. Observed Mitotic Defects and M-Phase Delay in wandering 3rd instar brains.
A. Mitotic figures: I) Representative wild type mitotic figure. II-VI) Representative mitotic defects from both Sld5 mutant alleles. II) Condensation defects, III & IV & V1) polycentric chromosomes (arrows), V) telomere fusions (arrow). B. Table of types of the frequency of defects observed in the different genotypes and the RNAi knockdown (n>100 for each). C. Box plot of Mitotic index for the genotypes indicated showing M phase delays in both Sld5 mutants and in RNAi treated.
S phase
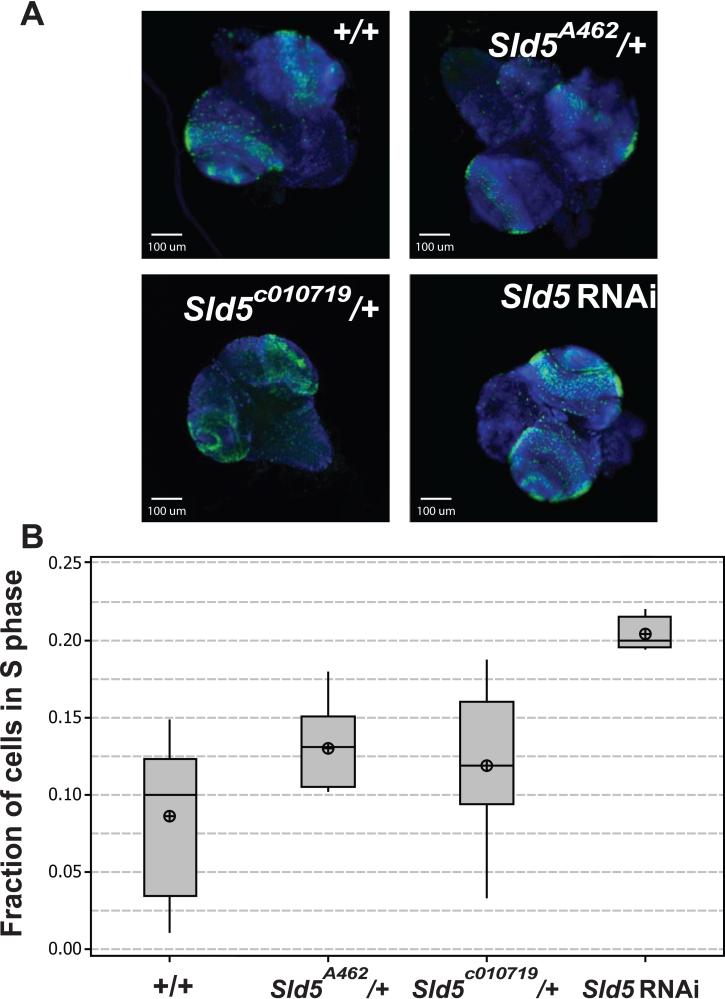
The GINS complex is required for efficient DNA synthesis during S phase [12]. In order to test for S phase defects in the Sld5 defective lines EdU incorporation assays were performed on dissected 3rd instar larval brains from the same genotypes as for mitotic indices above (Figure 4A). Larvae heterozygous for the Sld5 alleles had S phase delays as compared to wild-type (Figure 4B). Sld5A462/+ larvae had significantly more cells (51%) in S phase compared to wild-type (p=0.034), while, not statistically significant, Sld5c0107192/+ larvae had 38% more cells in S phase compared to wild-type (p=0.142). When Sld5 was depleted by RNAi, larval brains had significantly more cells in S phase than wild-type with 138% more (p<0.000).
Figure 4. Measurement of S phase indices using EdU incorporation.
A. Micrographs of whole dissected 3rd instar brains showing DNA (blue) and EdU incorporation (green). B. Box plots of S phase indices for the genotypes and RNAi knockdown as indicated. Both the mutant alleles show slight S phase delays while the RNAi treatment shows severe S phase delay.
DISCUSSION
Here we present the first phenotypic characterization of defects associated with a mutation in Sld5 in a mutlicellular organism. Two independent lines, each with a P element insertion removing a portion of the critical B domain responsible for interaction with Psf2 and overall stability of the GINS complex [12, were analyzed for cell cycle defects and mitotic chromosome abnormalities. The analysis of tissue specific RNAi lines corroborates the findings in the mutant lines and suggests that the dosage of Sld5 appears to be critical for proper cell cycle progression and the maintenance of genomic stability as larvae tissues from heterozygous individuals demonstrate M and S phase delays as well as chromosome aberrations. This is consistent with reports from mouse studies, where haploinsufficiency of PSF1 leads to impaired stem-cell proliferation in response to haematopoietic stress {Ueno, 2009 #551]. Moreover, our observations of chromosome abnormalities in mutant and RNAi backgrounds is consistent with assertions that the GINS complex may play a separable role in chromosome biology [20]. It is clear, however, that questions remain as the chromosome defects we have observed may in fact be the result of defects in S phase manifesting themselves in M phase.
Another intriguing possibility arises when one considers that the GINS complex is both required for the efficient helicase activity of Mcm2-7 [8] and is a phosphorylation target for the ATR and ATM protein kinases, two key mediators of DNA damage response and the intra S checkpoint [21]. Human Psf2 was identified as a phosphorylation target for the ATR and ATM protein kinases [21]and careful examination of Drosophila Psf2 reveals several possible phosphorylation sites as well. Recently it has been shown in untransformed Human Dermal Fibroblasts that depletion of Psf1 and Psf2 by siRNA leads to genomic instability and activation of Chk2. These studies along with our observations suggest a possible model whereby a number of replication origins fire without GINS complex present. The resulting replication forks are defective to the extent that the helicase activity of Mcm2-7 is decreased. Moreover, the regulation of these forks by the intra S checkpoint is compromised due to the absence of the GINS complex. This testable model would lead to our observed delay in S phase as well as well as the observation of mitotic chromosome defects detected in the Sld5 mutant backgrounds.
CONCLUSION
What is clear is that, although the GINS complex is a late addition to the lexicon of DNA replication proteins, it is quickly becoming apparent that it plays a central role in coordinating DNA replication with cell cycle checkpoints and is essential for the maintenance of genomic integrity. Here we present findings that show that within the context of a mutlicellular organism, defects in Sld5 lead to cell cycle delays in both S and M phase. In addition we show that Sld5 is required for the maintenance of chromosome integrity. These findings set the ground work for further investigation in Drosophila to delineate the role of the GINS complex with respect to DNA replication, checkpoint control, and the critical maintenance of genomic integrity.
ACKNOWLEDGEMENTS
We would like to thank Michael Reubens for plasmid construction, the East Carolina University Core Imaging and Core Genomics Facilities, East Carolina University for start-up funds and the NIH (1R15GM093328-01) awarded to T.W.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
- Drosophila Sld5 interacts with Psf1, PPsf2, and Mcm10
- Haploinsufficiency of Sld5 leads to M phase delay and genomic instability
- Sld5 is also required for normal S phase progression.
REFERENCES
- 1.Bogliolo M, Cabre O, Callen E, Castillo V, Creus A, Marcos R, Surralles J, Bogliolo M, Cabre O, Callen E, Castillo V, Creus A, Marcos R, Surralles J. The Fanconi anaemia genome stability and tumour suppressor network. Mutagenesis. 2002;17:529–538. doi: 10.1093/mutage/17.6.529. [DOI] [PubMed] [Google Scholar]
- 2.Umar A, Kunkel TA, Umar A, Kunkel TA. DNA-replication fidelity, mismatch repair and genome instability in cancer cells. European Journal of Biochemistry. 1996;238:297–307. doi: 10.1111/j.1432-1033.1996.0297z.x. [DOI] [PubMed] [Google Scholar]
- 3.Koepp DM. The replication stress response and the ubiquitin system: a new link in maintaining genomic integrity. Cell Div. 2010;5:8. doi: 10.1186/1747-1028-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atkinson J, McGlynn P. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 2009;37:3475–3492. doi: 10.1093/nar/gkp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aparicio T, Ibarra A, Mendez J, Aparicio T, Ibarra A, Mendez J. Cdc45-MCM-GINS, a new power player for DNA replication. Cell Division. 2006;1:18. doi: 10.1186/1747-1028-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aparicio T, Guillou E, Coloma J, Montoya G, Méndez J. The human GINS complex associates with Cdc45 and MCM and is essential for DNA replication. Nucleic Acids Res. 2009;37:2087–2095. doi: 10.1093/nar/gkp065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Labib K, Gambus A, Labib K, Gambus A. A key role for the GINS complex at DNA replication forks. Trends in Cell Biology. 2007;17:271–278. doi: 10.1016/j.tcb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang YP, Wang G, Bermudez V, Hurwitz J, Chen XS. Crystal structure of the GINS complex and functional insights into its role in DNA replication. Proc Natl Acad Sci U S A. 2007;104:12685–12690. doi: 10.1073/pnas.0705558104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamimura Y, Masumoto H, Sugino A, Araki H. Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol Cell Biol. 1998;18:6102–6109. doi: 10.1128/mcb.18.10.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Araki H, Leem SH, Phongdara A, Sugino A. Dpb11, which interacts with DNA polymerase II(epsilon) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc Natl Acad Sci U S A. 1995;92:11791–11795. doi: 10.1073/pnas.92.25.11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacNeill SA. Structure and function of the GINS complex, a key component of the eukaryotic replisome. Biochem J. 2010;425:489–500. doi: 10.1042/BJ20091531. [DOI] [PubMed] [Google Scholar]
- 13.Han Y, Ueno M, Nagahama Y, Takakura N. Identification and characterization of stem cell-specific transcription of PSF1 in spermatogenesis. Biochem Biophys Res Commun. 2009;380:609–613. doi: 10.1016/j.bbrc.2009.01.133. [DOI] [PubMed] [Google Scholar]
- 14.Kamada K, Kubota Y, Arata T, Shindo Y, Hanaoka F. Structure of the human GINS complex and its assembly and functional interface in replication initiation. Nat Struct Mol Biol. 2007;14:388–396. doi: 10.1038/nsmb1231. [DOI] [PubMed] [Google Scholar]
- 15.De Falco M, Ferrari E, De Felice M, Rossi M, Hübscher U, Pisani FM. The human GINS complex binds to and specifically stimulates human DNA polymerase alpha-primase. EMBO Rep. 2007;8:99–103. doi: 10.1038/sj.embor.7400870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walter BE, Henry JJ. Embryonic expression of pre-initiation DNA replication factors in Xenopus laevis. Gene Expr Patterns. 2004;5:81–89. doi: 10.1016/j.modgep.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Moyer SE, Lewis PW, Botchan MR, Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christensen TW, Tye BK. Drosophila MCM10 interacts with members of the prereplication complex and is required for proper chromosome condensation. Mol Biol Cell. 2003;14:2206–2215. doi: 10.1091/mbc.E02-11-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, Zhang H, FlyBase C. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang HK, Bailis JM, Leverson JD, Gomez EB, Forsburg SL, Hunter T. Suppressors of Bir1p (Survivin) identify roles for the chromosomal passenger protein Pic1p (INCENP) and the replication initiation factor Psf2p in chromosome segregation. Mol Cell Biol. 2005;25:9000–9015. doi: 10.1128/MCB.25.20.9000-9015.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 22.Boskovic J, Coloma J, Aparicio T, Zhou M, Robinson CV, Mendez J, Montoya G. Molecular architecture of the human GINS complex. EMBO Rep. 2007;8:678–684. doi: 10.1038/sj.embor.7401002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi JM, Lim HS, Kim JJ, Song OK, Cho Y. Crystal structure of the human GINS complex. Genes & Development. 2007;21:1316–1321. doi: 10.1101/gad.1548107. [DOI] [PMC free article] [PubMed] [Google Scholar]