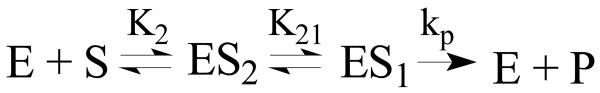Abstract
The first and rate-limiting step of the kynurenine pathway, in which tryptophan (Trp) is converted to N-formylkynurenine is catalyzed by two heme-containing proteins, Indoleamine 2,3-dioxygenase (IDO) and Tryptophan 2,3-dioxygenase (TDO). In mammals, TDO is found exclusively in liver tissue, IDO is found ubiquitously in all tissues. IDO has become increasingly popular in pharmaceutical research as it was found to be involved in many physiological situations, including immune escape of cancer. More importantly, small-molecule inhibitors of IDO are currently utilized in cancer therapy. One of the main concerns for the design of human IDO (hIDO) inhibitors is that they should be selective enough to avoid inhibition of TDO.
In this work we have used a combination of classical molecular dynamics (MD) and hybrid quantum-classical (QM/MM) methodologies to establish the structural basis that determine the differences in a) the interactions of TDO and IDO with small ligands (CO/O2) and b) the substrate stereo-specificity in hIDO and TDO. Our results indicate that the differences in small ligand bound structures of IDO and TDO arise from slight differences in the structure of the bound substrate complex. The results also show that substrate stereo-specificity of TDO is achieved by the perfect fit of L-Trp, but not D-Trp, which exhibits weaker interactions with the protein matrix-. For hIDO, the presence of multiple stable binding conformations for L/D-Trp reveal the presence of a large and dynamic active site. Taken together, our data allow determination of key interactions useful for the future design of more potent hIDO-selective inhibitors.
Keywords: TDO, IDO, Molecular Dynamics, Structure, Affinity, oxygen, CO, dioxygenase, inhibitors
Tryptophan and Indoleamine 2,3-dioxygenase (TDO and IDO respectively) are two heme containing enzymes that catalyze the conversion of L-Tryptophan to N-Formyl kynurenine (NFK), the first and rate limiting step of L-Trp catabolism through the kynurenine pathway.1 TDO has been found in mammals, insects and also in bacteria. In mammals it is found mainly in the liver where it is responsible for Trp catabolism.2 Available TDO crystal structures are that of Xanthomonas campestris (xcTDO, PDBid: 2NW8)3 and Ralstonia metallidurans (rmTDO, PDBid: 2NOX)4, and in both cases the enzyme was found to crystallize in a tetrameric form with several intersubunit contacts. xcTDO was crystallized in complex with L-Trp, showing several tight contacts between them, the most relevant being the interaction of the indoleamine group of L-Trp with His55, the amino group of L-Trp with the heme A propionate and the carboxylate grop of L-Trp with Arg117, while the aromatic ring is held in place by Phe51, Leu120 and Tyr113.
IDO is a monomeric enzyme, only present in mammals. The crystal structure of the human enzyme (hIDO) (PDB id 2D0T)5 shows that it shares significant structural similarity with xcTDO. The reported structure lacks the L-Trp substrate and instead Phenyl-Imidazol (PI) occupies the active site.
Additionally, structure-based sequence alignment of xcTDO and hIDO reveals that the key residues involved in L-Trp binding as found in xcTDO are present in the topologically equivalent positions in hIDO, such as Arg117 (Arg231 in hIDO), Phe51 (Phe163 in hIDO) and Leu120 (Leu234 in hIDO). Strikingly, His55 in xcTDO, which is hydrogen bonded to the indoleamine group of L-Trp and is responsible for proper positioning of the substrate, is missing in hIDO and replaced by Ser167.
In the past few years, there has been an increasing interest in IDO reaction and inhibition mechanisms, due to its implication in immune response and cancer,6 since inhibition of hIDO has been show to prevent T-cell activation7,8 and its overexpression in cancer cells has been linked to immune escape.9,10 Moreover, in-vivo experiments show that combination of chemotherapeutic agent with hIDO inhibitor significantly promotes tumour regression.6 In this context, an important question arises as to how to inhibit hIDO without interfering normal TDO function.
An interesting comparison between TDO and hIDO concerns the reported differences in substrate selectivity towards L and D enantiomers of Trp. In human TDO (hTDO), while L and D-Trp have similar affinities as indicated by the reported Km values, the kcat is 10 times lower for D-Trp.11 Interestingly, xcTDO has ≈140 times higher affinity for L-Trp and is completely inactive towards D-Trp. On the other hand, hIDO displays a similar kcat for the two substrates, but D-Trp has 173 folds higher Km12. The structural reasons for these differences are unknown, deciphering the key elements that determine the enantioselectivity of hIDO and TDO has potential implications for structure based hIDO-selective inhibitor design.
Ligand binding and oxygen reactivity characteristics of hIDO and TDO are distinctive: while hIDO is able to form a stable oxy complex regardless of Trp presence, for TDO it was believed that the enzyme was unable to bind O2 in the absence of L-Trp.1,13 However, recently Batabyal et al. showed the existence of a transient oxy complex of hTDO in absence of L-Trp, which rapidly autooxidizes to the ferric enzyme, in sharp contrast to hIDO where the process is much slower.14 CO has been widely used as a probe for the electrostatic potential of the distal pocket of heme proteins. Specifically, the νC-O and νFe-C frequencies allow sensing the environment of the bound ligand. Corresponding results for the CO complexes of hTDO and hIDO show that whereas hIDO displays an increase in νFe-C frequency (and a decrease in νC-O frequency) upon L-Trp binding, indicating a polar interaction between CO and L-Trp, the opposite trend is found in hTDO11, suggesting that L-Trp could be located in different conformations in the two enzymes leading to the different CO environments.11 The differences in the structures of the O2 and CO complexes and in the reactivities of the O2 complexes point to subtle differences between the two enzymes which may be key elements determining their designated biological function.
In this work we have used a combination of Molecular Dynamics (MD) simulations and hybrid quantum mechanics – molecular mechanics (QM-MM) calculations to compare the structure of TDO and hIDO with small ligands (CO/O2). We have also used a combination of Docking and MD simulations to analyze the comparative binding of L and D-Trp isomers in both proteins. The results obtained allow understanding of important differences between hIDO and TDO and their interactions with their substrates, with a potential impact on understanding their catalytic mechanism and the design of hIDO-selective inhibitors.
Computational Methods
TDO Starting structures
Starting from the crystal structure of ferrous xcTDO with Trp (PDB id 2NW8)3, a one-subunit model of TDO was constructed (Figure S1). The crystal structure presents two subunits, and in each subunit approximately the first 15 aminoacids form a short helix that penetrates the structure of the opposite subunit. With this in mind, the starting structure was constructed by removing from subunit A residues Glu19 to Ser35, and adding residues Arg21 to Ser 35 of subunit B. (residues 19 and 20 of subunit B are missing in the crystal structure). This structure was used for all the MD and QM-MM simulations.
IDO starting structures
For IDO, the starting structure corresponds to the crystallized hIDO monomeric structure (PDBid 2D0T)5. Since in the corresponding structure the loop formed by residues 361 to 379 is missing, it will not be considered in the models. In order to obtain the oxy/CO bound or free hIDO D/L-Trp complexes we removed the PI ligand and built the oxy/CO or free structure in-silico. After a small relaxation of the structure, we performed a short 5 ns MD simulation for both oxy and deoxy states and selected snapshots for flexible docking of the ligands.
Docking simulations
To obtain structures of the protein substrate complexes, we docked D/L-Trp in hIDO, using the Autodock program.15 We performed 100 flexible ligand docking runs for a set of 10 selected IDO structures (obtained from the hIDO-substrate-free Molecular Dynamics simulations, 5 oxy structures and 5 deoxy structures) using a 0.25A resolved grid, centered on the protein active site. The resulting structures were analyzed and clustered into groups of possible complex conformations which were subsequently used for the MD simulations. As a positive control we docked L-Trp into the TDO X-ray structure, and obtained 100% structures corresponding to the experimental complex with similar docking energies as those obtained for L/D-Trp hIDO complexes. This procedure was also applied in order to obtain the D-Trp xcTDO complex. For this case we docked the D-Trp in the crystal xcTDO structure.
Classical Molecular Dynamics Simulations
The starting structures, for each complex, were immersed in a pre-equilibrated octahedral box of TIP3P water molecules. The standard protonation state at physiological pH was assigned to ionizable residues. Special attention was paid to the protonation states of histidines, which were assigned on the basis of the hydrogen bonding patterns with neighboring residues. All simulations were performed at 300 K and pressure of 1 bar using Berendsen thermostat and barostat16. Periodic boundary conditions and Ewald sums (grid spacing of 1 Å) were used to treat long range electrostatic interactions. The SHAKE algorithm was used to keep bonds involving hydrogen atoms at their equilibrium length. A 1 fs time step for the integration of Newton's equations was used. The Amber ff99SB force field17 was used for all residues but the heme, whose parameters were developed and thoroughly tested by our group in previous works18,19. All simulations were performed with the PMEMD module of the AMBER9 package.20 Equilibration consisted of an energy minimization of the initial structures, followed by a slow heating up to the desired temperature. For each structure 10ns MD production runs were performed. Frames were collected at 2ps intervals, which were subsequently used to analyse the trajectories.
Ligand Binding Energies
From the production MD simulations the binding free energy as well as its contributions were computed for each structure using the MM-GBSA (Molecular Mechanics, Generalized-Born and Solvent Accesibility) approach21 implemented in AMBER. Computed contributions consist of:
Where ΔEGAS corresponds to the ligand binding energy in the gas phase (i.e. in vacuum), and ΔEELEC, ΔEVDW, ΔEINT are the corresponding electrostatic, van der Waals and internal (bonds, angles and dihedrals) contributions to the energy. Moreover, the solvation free energy (ΔGSV) contribution to the binding was estimated using the Generalized Born implicit solvation method.22 The sum of ΔEGAS and ΔGSV provides an estimation the binding free energy ΔGB.
Free energy Profiles
The free energy profiles for the interconversion between the two conformations for the substrate-bound hIDO were calculated using the Umbrella Sampling method.23 Based on analysis of the equilibrium simulations, for both cases (L-Trp and D-Trp), the distance between the Cε2 of Trp and the Cα of Ser167 was chosen as the reaction coordinate (RC), for the biased potential. The RC was divided in ten windows, covering a distance of 4.5 Å and for each window, 2ns MD simulations were performed, using a 10 kcal/mol. Å2 force constant for the harmonic restraint potential.
QM-MM calculations
QM-MM calculations were performed for the O2 and CO bound protein complexes of TDO and IDO with and without L-Trp. The initial structures for the QM-MM calculations were obtained from the MD runs of the corresponding complexes. Selected snapshots for each structure were chosen and cooled down slowly to 0 K. Starting form these frozen structures full hybrid QM-MM geometry optimizations were performed using a conjugate gradient algorithm, at the DFT level with the SIESTA code using our own QM-MM implementation.18,24,25 For all atoms, basis sets of double beta plus polarization quality were employed. All calculations were performed using the generalized gradient approximation functional proposed by Perdew, Burke, and Ernzerhof.26 Only residues located less then 10 Å apart from the heme reactive center were allowed to move freely in the QM-MM runs. The iron porphyrinate, the distal ligand and the imidazol of the proximal histidine were selected as the quantum subsystem. The rest of the protein unit, together with water molecules, was treated classically. The interface between the QM and MM portions of the system was treated by the scaled position link atom method. The SIESTA code showed an excellent performance for medium and large systems, and also proved to be appropriate for biomolecules, and specifically for heme models. Further technical details about the QM-MM implementation can be found elsewhere.25
O2 binding energies (ΔEO2) were calculated as:
where Eprot-O2 is the energy of the oxy protein, Eprot is the energy of the deoxy protein and EO2 is the energy of the isolated oxygen molecule.
Results
The results section is organized as follows. First, we analyze the CO and O2 adducts of both IDO and TDO in the substrate free (SF) and L-Trp protein complexes, combining MD and QM/MM simulations. Then, we show the MD results on xcTDO oxy complexes with L and D Trp, and continue by analyzing the IDO complexes with L and D-Trp by means of flexible docking calculations and MD simulations. Finally, all results are analyzed in a comparative way.
Small ligand binding
CO vs O2 binding
Both TDO and IDO need to bind oxygen in order to catalyze dioxygenation. Interestingly, and as mentioned in the introduction, they behave differently towards it. Additionally, CO binding reflects differences in the active site interactions. In order to get a better understanding of how the active site structure and dynamics modulates small ligand binding, we have performed 5ns MD simulations of CO bound xcTDO and hIDO in the presence or absence of Trp. Additionally, MD simulations of the TDO and IDO oxy complexes in the absence of Trp were also performed. Finally, based on selected CO/O2 bound structures hybrid QM-MM calculations were performed to analyze the electronic structure in each case.
xcTDO
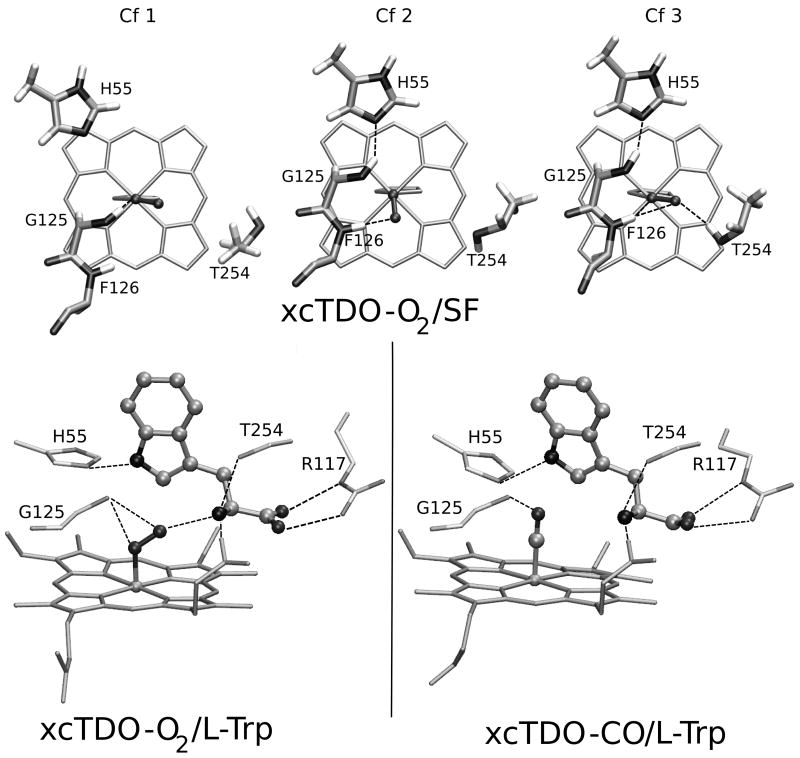
MD simulations of SF xcTDO CO or O2 complexes show that for CO a unique conformation is observed, in which the CO is forming an hydrogen bond (HB) interaction with the backbone NH group of Phe126, and due to the absence of Trp, Gly125 NH rotates in order to establish an HB with His55. Interestingly when oxygen is bound three conformations are observed depending on the possible interactions of the bound O2 with the distal cavity of the protein. The first conformation (Cf1) involves an HB interaction between the backbone NH of Gly125 and the O2. The second conformation is similar to the one obtained for the SF complex with CO, in which the O2 interacts with Phe126 NH and Gly125 NH with His55. Furthermore, a third conformation (Cf3) appears, in which not only the Phe126 interacts with the O2 but also the side chain of Thr254 rotates and forms a HB with the bound O2. As will be shown below, the presence of multiple conformations significantly impacts in the stability of the oxy TDO complex. The QM-MM optimized structures for all three obtained conformations are shown in Figure 1a.
Figure 1.
QM-MM optimized structures of xcTDO active site. Top pannel corresponds to the three conformations found in SF TDO-O2 complex. Lower panel corresponds to the L-Trp-bound xcTDO-O2 (left) and -CO complex (right), respectively.
The MD simulations of the Trp bound CO and oxy states also illuminate differences between the CO and O2 interactions with the Trp-ligand, especially concerning the HB interaction between bound O2/CO with the NH3+ group of L-Trp. This is reflected in the mean value and standard deviation of the N(NH3+)-O(CO-O2) distance which resulted (2.79 ± 0.12) Å for O2 and (3.77 ± 0.37) Å for CO, indicating the loss of this important HB for the CO complex. The origin of this difference is due to the more negatively charged and bent O2 ligand, allowing a proper HB to be established. Relevant structural parameters for QM-MM optimized structures of O2/CO-bound complexes are shown in Table SI. Comparing complex structures as shown in Figure 1, it can be observed that while in the oxy complex the O2 is in a bent conformation pointing to the NH3+ group, in the CO complex the ligand is positioned perpendicular to the heme plane, as found normally in heme-CO adducts.
To address the issue of oxygen binding to SF or Trp bound TDO, we have computed the O2 binding energies of the above described complexes using a QM-MM scheme. As shown by previous works from our group27,28 the presence of multiple HB patterns in the distal site affect significantly the affinity for O2, and therefore all conformations should be considered. As shown in Table I, and consistently with the HB patterns, the higher affinity in the substrate free protein is obtained for Cf3. However, although Cf1 and Cf2 display slightly lower affinities they show a moderate O2 binding energy. These results confirm that TDO is able to bind O2 in the absence of Trp, as indicated by recent experimental data.14
Table I.
O2 binding energies of hIDO and xcTDO with and without L-Trp. Values are given in kcal/mol. SF stands for substrate-free enzyme.
| hIDO | xcTDO | |||||
|---|---|---|---|---|---|---|
| L-Trp | SF | L-Trp | SF | |||
| Cf1 | Cf2 | Cf1 | Cf2 | Cf3 | ||
| 32.4 | 34.5 | 34.2 | 30.5 | 27.6 | 29.8 | 35.3 |
hIDO
The substrate free hIDO shows that bound CO is stabilized in the active site by a hydrogen bond interaction with the backbone NH group of Gly265. A similar situation is observed in the SF hIDO in complex with O2, in which the O2 is hydrogen bonded to Ala264-NH and more weakly to Gly265-NH. In contrast to what is observed for TDO, only one configuration of the distal site has been obtained. This HB pattern is also reflected in the oxygen affinity calculations (shown in Table I), which indicate that IDO is able to bind tightly O2 with and/or without Trp.
For hIDO in the presence of Trp, as will be shown below, two different conformations (Cf1 and Cf2) were obtained in the docking protocol and were shown to be stable in the MD simulations. Thus, both Cf1 and Cf2 obtained structures were simulated with CO and O2. For Cf1, and as observed for xcTDO, the HB interaction between the NH3+ group of Trp and the CO is not established during the simulation, highlighting again the differences between the two ligands. Although the indoleamine group of Trp stays close to the CO (the average distance between the O atom of CO and the H atom of the NH indolic group is 3.05 ± 0.29 Å), the perpendicular position of the CO and the Trp respect to the heme plane do not allow a proper orientation for an HB interaction. In Cf2, on the other hand, where the Trp indole group lies almost parallel to the heme plane, an HB between Trp-NH and the ligand is found. Additionally, both CO and O2 ligands are stabilized in the pocket by an HB interaction with the NH of Ala 264. Relevant geometrical parameters of these structures can be found in Table SII of the Suplementary Information.
Taken together the results of the ligand complexes allow interpretation of previous Resonance Raman (RR) results on CO bound enzymes with and without Trp, showing that the CO probe molecule is subjected to different environments in each case.11 As shown by the simulations, binding of L-Trp to xcTDO does not result in new HB to the ligand, since the Trp-NH3+ does not interact with the bound CO. However, the Trp aromatic region is very close to the CO probably creating a more hydrophobic environment compared to the SF enzyme, and in agreement with the above mentioned RR observations. The change from polar to hydrophobic ligand environment upon Trp binding in xcTDO is also consistent with the behaviour towards oxygen. In the SF xcTDO, oxygen is able to form a stable complex with multiple HB, and moderate to high binding energy. However probably due to the easy accession of solvent molecules, the bound oxygen rapidly oxidizes. Binding of Trp significantly blocks water access to the oxygen protecting the enzyme from autooxidation, as shown experimentally.
For hIDO the increase in polarity revealed by the CO frequencies indicates that L-Trp is strongly HB to the bound CO. These results can be interpreted considering the population of Cf2, showing a strong and direct HB with the bound CO ligand.
L/D Trp binding to oxy xcTDO
We start by analyzing MD simulations of L and D oxy TDO complexes. In both simulations, the backbone RMSD remains under 2 Å (See Figure S2), and both structures equilibrate after the first nanosecond, then being the first nanosecond excluded from the analysis. Visual inspection of the TDO-L-Trp simulation indicates that the interactions that stabilize the substrate, in the original X-ray structure are mostly conserved during the timescale of the simulation. Firstly, L-Trp carboxylate group forms strong electrostatic interactions with Arg117 side chain, and Tyr113-OH group. In addition the NH3+ group of L-Trp interacts with Thr254-OH and with the heme carboxylate A. The apolar region of L-Trp is fixed by the aromatic cluster formed by Phe51, Phe116 and Tyr113. Finally, His55 establishes a strong HB with the indoleamine group of Trp. (See Figure 2). All these interactions fix the L-Trp in a rigid and unique position.
Figure 2.
The averaged structures of xcTDO-O2 with D- and L-Trp obtained from the MD simulation. Panel a corresponds to the L-Trp-bound xcTDO-O2, panel b shows superimposed structures of L and D-Trp complexes, depicted in black and green, respectively, with only one heme displayed for simplicity.
The D-Trp-oxy xcTDO structure was obtained in-silico by means of flexible docking calculations, as explained in the Computational Methods section. These calculations lead to a single structure, very similar to the complex with L-Trp. Visual inspection of the molecular dynamics simulation of the xcTDO complex with D-Trp shows that, even though the most important interactions between Trp and the protein residues are similar when compared to the L-Trp complex, there are some subtle differences that warrant careful analysis. Table II shows the results of a systematic, comparative HB analysis for L and D-Trp. These results indicate that the main interactions that stabilize Trp in the active site are slightly weakened in the D-Trp case, as judged by the decreased population of the HB. The biggest difference concerns the interaction between the NH3+ group of Trp and the Thr254-OG that is almost lost in the D-Trp complex. This indicates that Thr254 may play a key role in regulating the difference between L and D-Trp affinity of xcTDO.
Table II.
HB analysis of the MD simulation s results of L and D-Trp bound xcTDO-O2. Values correspond to the percentage of time in which the HB are formed. For this analysis, distances shorter than 3.5 Å and angles greater than 120 degrees were considered as HB interactions.
| xcTDO | |||
|---|---|---|---|
| Donor / Acceptor Pair | L-Trp | D-Trp | |
| Trp NH3+ | Op (O2) | < 1 | 8 |
| Ot (O2) | 95 | 65 | |
| T254 OH | 96 | 15 | |
| Trp COO- | R 117 Nε | 100 | 100 |
| R117NH1 | 100 | 100 | |
| T254 NH | 99 | 89 | |
| Y113 OH | 100 | 100 | |
| Trp NE1 | H55 Nε2 | 99 | 96 |
The comparison between the calculated average structures obtained in the MD simulations for the L and D-Trp complexes shows that both structures remain very similar during the simulation, both with an average backbone RMSD of under 1 Å (1.18 Å considering all the heavy atoms). However, a subtle but significant difference in the position of Trp side chain is evident (shown in Figure 2), impacting the Trp interaction with His55. While in the L-Trp structure the His55- Nε - Trp-Hε1 distance and His55-Nε - Trp-Hε1 – TrpNε1 angle mean values are 1.931 Å and 153.8 degrees respectively, in the D-Trp this values are 2.005 Å and 145.9 deg. This indicates that although in both cases the HB is established, for D-Trp the interaction is slightly weaker than for L-Trp.
In order to further compare L and D-Trp binding, we have computed the binding energy for the two ligands in oxy xcTDO, using the MM-GBSA approximation described in the Computational Methods section. This approach, although it might lead to overestimated absolute binding free energy values (since for example ligand and protein entropy losses are not considered) allows generally obtaining accurate relative trends for different ligands or mutant proteins. Consistently with the above observed HB analysis, the results presented in Table III show that the binding energy for D-Trp is 5.6 kcal/mol lower than for L-Trp, and, the reduction in binding energy is almost due to electrostatic interactions.
Table III.
Binding Free Energies (kcal/mol) and experimental Michaelis constant (Km) (mM) for xC-TDO and hIDO.
| Electrostatic | van der Waals | Total Gas | GB Solvation | Total | Km | ||
|---|---|---|---|---|---|---|---|
| xC-TDO | |||||||
| L-Trp | -136.9 | -28.9 | -165.7 | 114.0 | -51.7 | 0.1143 | |
| D-Trp | -132.3 | -28.9 | -161.2 | 115.1 | -46.1 | 163 | |
| hIDO | |||||||
| L-Trp | Cf1 | -104.4 | -27.6 | -132.0 | 107.8 | -24.2 | 0.01512 |
| Cf2 | -109.5 | -26.6 | -136.0 | 106.4 | -29.6 | ||
| D-Trp | Cf1 | -85.1 | -27.4 | -112.5 | 92.4 | -20.1 | 2.612 |
| Cf2 | -79.1 | -26.9 | -106.0 | 85.8 | -20.2 | ||
L/D Trp binding to oxy hIDO
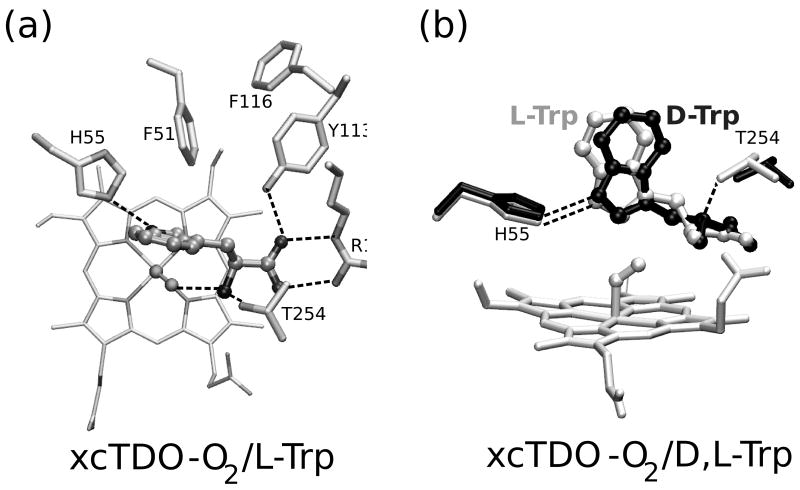
Since no X-Ray structures of the IDO D/L-Trp complexes are available, in order to obtain putative D/L-Trp bound hIDO structures we used a combined MD-Docking approach. Starting from ten equilibrated structures, obtained from a 5ns long oxy and free IDO MD simulation we performed 100 flexible L/D-Trp docking runs for each. For L-Trp, the analysis of the resulting structures showed two major clusters of the complex. The first conformation (L-Cf1) appeared in 44% cases with docking scoring energies between 7.0-8.2 kcal/mol. The structure is equivalent to the xcTDO L-Trp bound structure and similar to previous structures obtained by the docking methodology.29 The indole ring lies in a plane perpendicular to the heme, with the indole C2-C3 bond close to the dioxygen molecule, ideal for dioxygenase chemistry, and corresponds to the reactive structure as described previously by our group.30,31 The Trp carboxylate forms a salt bridge with the side chain of Arg231, while the Trp-NH3+ group interacts with the heme propionate A carboxylate and the Ot of the bound O2. Additionally, the aromatic Trp region forms hydrophobic interactions with Phe163 and Phe226.
A second conformation (L-Cf2) appeared in 35% of the docking simulations with binding energies between 6.0 and 7.0 kcal/mol. In this conformation, the Trp is further away from the active site iron, and the indolic ring lies almost parallel to the heme plane (ppositely to cf1). The Trp-NH points directly towards the O2 ligand and the C2-C3 bond is farther away from the O2. Additionally, while the Trp-NH3+ interaction with heme propionate A is conserved, the Trp carboxylate forms HB interactions with the Ser 235 hydroxyl side chain, and several backbone amides (Gly 236, Gly 261, Gly 262), losing the interaction with Arg231.
Starting from these two structures we performed a 10ns long MD simulation. The average structures obtained from these simulations are presented in Figure 3. In the simulation corresponding to the Cf1 conformation, all above mentioned Trp-protein interactions are conserved, with the exception of the Trp-NH3+ HB to O2 that shows dynamical fluctuations. During the simulation, the NH3+ moves closer to the heme propionate, and also rotates to interact with the carbonyl groups of Gly261 and Gly 262. This Trp-backbone movement allows strengthening the HB between the O2 and the Trp-NH indolic group.
Figure 3.
The average structures of D- and L-Trp-bound hIDO-O2 obtained from the MD simulation. Panels a and b correspond to the L-Trp complexes and show the Cf1 and Cf2 conformations, respectively. Panels c and d correspond to the D-Trp complexes in the Cf1 and Cf2 conformations, respectively.
On the other hand, the Cf2 MD simulation shows some interesting changes in the Trp interactions with the protein matrix. During the simulation, a rotation of the Cα-Cβ Trp bond occurs, causing the interactions of the Trp backbone groups to change from the above described ones to a structure where the Trp carboxylate strongly interacts with Arg 231 (as in Cf1). The amino group, which initially interacts with the heme propionate, after the rotation, forms an HB to Ser263-OH group and Gly261 backbone carbonyl groups. A detailed analysis of HB interactions is presented in Table IV below.
Table IV.
HB analysis of the MD simulation results of L- and D-Trp-bound hIDO-O2. The values correspond to the percentage of time in which the H Bonds are established. For this analysis, distances shorter than 3.5 Å and angles greater than 120 degrees were considered as HB interactions. In all cases, the last 6 ns were used except for D-Trp Cf1, where only the first 4ns of the simulation, before the conformational transition, was analyzed.
| hIDO | |||||
|---|---|---|---|---|---|
| Donor / Acceptor Pair | L-Trp [Cf1] |
L-Trp [Cf2] |
D-Trp [Cf1] |
D-Trp [Cf2] |
|
| Trp NH3+ | G 262 O | 49 | 15 | 64 | 84 |
| G 261 O | - | 18 | 28 | 44 | |
| S 263 OH | 3 | 74 | 63 | 4 | |
| Trp COO- | R231NH1 | 90 | 100 | - | - |
| R231NH2 | - | 64 | 54 | - | |
| R 231 Nε | 100 | - | 49 | - | |
| S 235 OH | - | 64 | 4 | 25 | |
| G261N | - | 4 | 23 | 70 | |
| Trp NE1 | Op (O2) | 42 | 80 | 23 | |
| Ot (O2) | 14 | 95 | 38 | 99 | |
From the global protein perspective our results show that Cf1 and Cf2 average structures display an overall backbone RMSD with the X-ray structure below 2 Å, showing only minor structural changes due to substrate binding. The Cα-RMSD vs. residue plot shows that most of the protein fluctuates less than 1.5 Å (Figure S4), with only some regions showing a higher deviation from the starting structure. Considering side chains movements due to Trp binding, in both Cf1 and Cf2, Arg231 moves away from the solvent to interact with the Trp carboxylate. A difference is also observed in the relative position of heme group and the distal helix with respect to the proximal histidine helix. While in Cf2 the heme is in the same position as in the starting structures and the distance between the proximal and distal helices (measured as the proximal His CA to distal Phe163 CA distance) increases from 13.2 to 14.6 Å, in Cf1 Trp is further inside the active site and moves the helices further apart to 15.6 Å. This pushes the CD side of the heme slightly down. In TDO, an induced fit mechanism was proposed for Trp binding, due to movement of the αJK and αDE and Arg231 side chain movement.3 The side chain, heme and relative helix movement upon Trp binding also suggest a possible induced fit mechanism for Trp binding in IDO.
For D-Trp the docking results are slightly different than those obtained for L-Trp. The most populated conformation (50% Docking simulations, with docking scoring energies of about 7.0 kcal/mol) resembles the L-Trp Cf2 conformation, and will therefore be referred to as D-Trp Cf2. It displays the Trp aromatic ring located between Phe226, and Phe163 with the Trp-NH pointing towards the bound oxygen. However, the Trp carboxylate does not interact with Arg231, but with the amide groups of Gly261 and Gly236. In contrast to the L-Trp Cf2 conformation, the NH3+ group interaction with Ser263-OH group was almost absent in D-Trp Cf2. Another possible conformation, obtained with the docking protocol with only minor population contribution (ca 5%) shows the D-Trp ligand further inside the active site, similar to L-Trp Cf1, in which the Trp-NH3+ group is pointing towards O2 ligand and the Trp carboxylate is interacting with Arg231. Additionally, the hydroxyl group of Ser263 also helps to locate the Trp in the reactive position interacting with the NH3+ group of Trp, oppositely to what is observed in L-Trp Cf1, where Ser263 only interacts with the heme propionate. This conformation will therefore be referred to as D-Trp Cf1.
Starting from these two initial IDO/D-Trp structures we have also performed a 10ns MD simulation. Interestingly, while D-Trp Cf2 remains in the initial conformation during the timescale of the simulation, the D-Trp Cf1 suffers a transition after 4ns that locates the Trp closer to Cf2. The transition involves initially the loss of the Trp- NH3+ interactions with O2 and Ser263, and then the loss of the salt bridge between the Trp carboxylate and Arg231. The transition clearly suggests a small barrier for D-Trp between both conformations.
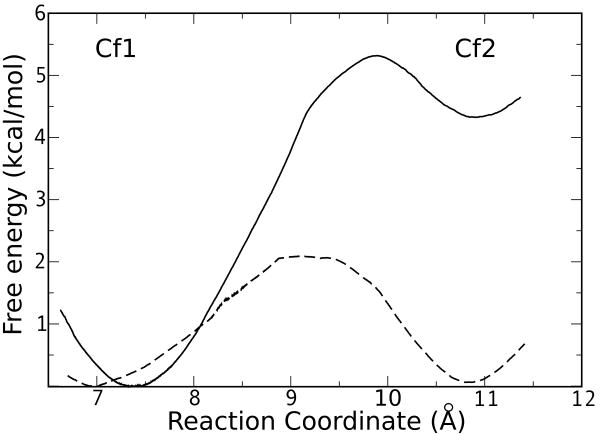
In order to determine the relative populations of Cf1 and Cf2 for L and D-Trp, and the free energy barrier separating the two conformers, we have computed the free energy profile for the interconversion process by means of Umbrella Sampling (US) simulations. As shown in Figure 4, the results indicate that while for L-Trp the most populated conformation corresponds to Cf1, with a free energy 4.3 kcal/mol lower than Cf2, and the Cf2 to Cf1 transition displays a small barrier of 1 kcal/mol. For D-Trp the two conformations dysplay similar free energies, separated by a small barrier of 2 kcal/mol, hence the Cf1-Cf2 transition for D-Trp is completely accessible at room temperature, in agreement with the MD results shown above.
Figure 4.
Free energy profile for the interconversion between Cf1 and Cf2. The results for L and D-Trp are depicted in solid and dashed lines, respectively.
As for TDO, we estimated the D/L-Trp binding energies and their contributions using the MM-GBSA explained in the Computational Methods section. The estimated binding energy for D-Trp in IDO is between 4 and 10 kcal/mol less favourable than for L-Trp in any of the two observed conformations (Table III). The difference in binding energies of L/D Trp in IDO shows the same trend as the experimental Km values showing a clear preference for L-Trp.32 As in xcTDO the difference is mainly due to electrostatic interactions. The most important difference between the complexes with L and D-Trp in IDO is the loss of the Trp interaction with the side chain of Arg231, which is replaced by the much weaker interactions with different amide groups of the protein backbone.
In summary, the results for IDO and TDO indicate that while in TDO a single conformation is found for L and D Trp that binds tightly to the protein, due to strong electrostatic interactions, in IDO the Trp ligand may adopt different conformations. The multiplicity of conformations observed for hIDO reflects a significantly larger active site, when compared with TDO. The larger and more flexible active site is probably the cause of hIDO substrate diversity.
Discussion
The aim of the present work was to a) compare the structure of TDO and IDO with small ligands (CO/O2) and b) understand the structural basis for comparative binding of L and D-Trp isomers in IDO and TDO. Using a combination of MD and QM/MM methodologies our results shed light into these key issues allowing for a better molecular based interpretation of the ongoing active research in this field.2,3,5,6,11,14,31,33,34
Small ligand binding and Trp ligand interactions
The analysis of small ligand binding (O2/CO) show that in the substrate free enzymes both ligands establish HB interactions with the protein in both hIDO and xcTDO. The HB analysis and O2 binding energy show that both proteins should be able to form an oxy complex in the absence and presence of substrate. The ligand binding results also show that the presence of Trp significantly alters the bound ligand environment, and new interactions are established between the bound O2/CO and the Trp substrate. Our CO/O2 binding results are consistent with the previously obtained RR data, validating the obtained ligand and substrate bound structures and allowing a structural interpretation of the RR results as described above. RR results indicate that in TDO no HB interactions are observed between the CO and bound Trp as shown by the MD simulations. In IDO, on the other hand RR data suggest formation of a strong HB between bound CO and Trp.11 The existence of Cf2 for IDO, in which the indoleamine group of Trp strongly interacts with CO, explains this issue. It should be noted that, although Trp bound in Cf1 does not form an HB interaction with bound CO, this conformation must be considered since it corresponds to the reactive conformation, which occurs in the oxy complex.30 A point of attention should be made however when extrapolating CO RR data to oxy structures, since the charge density and geometry of the two ligands are different and may in some cases lead to different conformations, as shown by the data presented here.
Substrate stereoselectivity in TDO and hIDO
Enzyme kinetic studies show that both hIDO and xcTDO show a clear preference for L-Trp over D-Trp binding and they also show clear differences in reactivity. Our data show that for xcTDO both L/D-Trp bind in a very similar way, but the active site is overall better suited for the L isomer. The comparison of the two complex structures and the HB analysis shows that D-Trp position is shifted as compared to the L-Trp structure, and consequently the most important protein-substrate HB interactions (those with the Trp-backbone and His55 Trp-NH) are weakened. It seems that xcTDO structure is perfectly fitted for L-Trp binding. D-trp is able to bind but fits poorly, and although the same interactions are formed between xcTDO and L-Trp as those with D-Trp, the dynamical fluctuations for the D-isomer are larger making the overall binding energy and therefore the affinity lower. Thus stereoselectivity in TDO arises from a subtle dynamic effect as revealed by the MD data. The results also clearly show that the Trp substrate binding in xcTDO is mainly of electrostatic nature highlighting the importance of the Trp charged backbone. Overall, the data yields the expected tendency in the preference for the L-isomer, but the difference cannot be attributed to one particular interaction, but to the whole active site.
For hIDO the picture is more complex due to the existence of multiple conformations for both D/L isomers. The first significant difference concerns the stability of the two conformations. For L-Trp Cf1 is about 4 kcal/mol more stable than Cf2, while for D-Trp both conformations show the same free energy. Since the Cf1 is considered to be the reactive conformation30, the predominance of Cf1 in the L-Trp-protein complex accounts for the enhanced enzymatic efficiency for L-Trp, compared to D-Trp. Also, the thermodynamic analysis shows that in hIDO Trp binding is mainly driven by electrostatic interactions, but the contribution from van der Waals interactions is slightly more significant than that in TDO.
Implications of multiple hIDO-Trp conformations
One of the most relevant results obtained in this work corresponds to the presence and structure of two different oxy-hIDO L-Trp conformations. Comparative analysis of both structures with xcTDO shows that Cf1 corresponds to the equivalent oxy xcTDO L-Trp structure, which was shown to be the reactive structure in our previous work.30,31 The coexistence of both structures is strongly suggested by their similar binding free energy, their high population in the docking results, the stability of each of them during the simulation, the low free energy barriers for the interconversion between both structures, and their consistence with the observed RR data. Looking at both conformations with respect to the global protein structure, and considering that Cf1 is the reactive structure, the interesting question arises of the role played by Cf2. Although a precise role for it cannot be derived from our data, the following picture seems possible. Cf2 would correspond to a first docking site of the Trp substrate on its way towards the enzyme reactive site corresponding to Cf1. This view is supported by the increased helix displacement and heme distortion observed for free hIDO, hIDO-Cf2, hIDO-Cf1 direction of events. In the same line of thought, a Cf2-like structure could represent a transient docking site for NFK in its way out.
The effect of these two conformations on the overall enzyme function is difficult to predict but a preliminary analysis relating the experimental kinetic data with the observed structural features can be performed and compared with those for xcTDO. According to enzyme kinetics mechanism35 two consecutive substrate binding sites as observed for hIDO are described by Scheme 1.
Scheme 1.
Schematic illustration of the hIDO reaction. ES2 and ES1 represent the protein-L-Trp complexes in Cf2 and Cf1 conformations, respectively. K2 is the L-Trp dissociation equilibrium constant of ES2, K21 is the equilibrium constant between ES2 and ES1 and kp is the rate constant of the chemical reaction.
According to Scheme 1 the Michaelis-Menten parameter Km is defined in the following equation, highlighting the role played by the two conformations in the observed Km value.35
In this sense, the Km is inversely proportional to the affinity to the ES2 site (characterized by the dissociation constant K2), and can be further reduced if the equilibrium is displaced towards ES1 (high K21).
The equations derived from Scheme 1 in the context of the computed values allow obtaining an explanation of the different observed Km values determined for L and D-Trp in hIDO. The results for the Free Energy profile calculated with US for hIDO indicate that, while in the L-Trp-enzyme complex the Cf1 is favoured by 4.3 kcal/mol, in the D-Trp complex both conformations are equally populated. Therefore, the 173-fold lower Km for L-Trp compared to D-Trp can at least be partially explained due to strongly shifted equilibrium towards the Cf1 conformation, which results in a significantly higher K21 constant for L-Trp.
Size and flexibility of IDO active site and its relevance for the design of selective inhibitors
Our results also have potential implications for the design of selective hIDO inhibitors. Since inhibition of hIDO has been shown as therapeutic strategy for treating immune disorders and cancer, the search for synthetic hIDO inhibitors has drawn considerable interest. One of the main concerns for the design of hIDO inhibitors is that they should be selective enough to avoid inhibition of hTDO and interference with normal Trp catabolism. In this context substrate analogues may have low perspectives. Interestingly, our results show that the xcTDO (and presumably hTDO) active site is much smaller. The almost perfect fit of L-Trp inside xcTDO, suggest that Trp analogues with bulky substituents such as N-Methyl Trp may show unexpectedly high selectivity towards hIDO compared to TDO. Furthermore, the extension of the hIDO active site, revealed by the presence of multiple Trp binding sites in a dynamic equilibrium, suggests the possibility of binding larger molecules as compared with TDO. In this context, our results may explain how the complex exiguamine-A, which has 2-3 times the size of a Trp is a nanomolar inhibitor of hIDO. Finally the binding energy contribution analysis shows that TDO and IDO active sites have significant electrostatic nature, suggesting the use of amphiphilic drugs as potential inhibitors.
Conclusions
In summary, our results show that in xcTDO stereoselectivity towards L-Trp arises due to less favorable fitting of D-Trp in the active site, with the consequence of weakening of electrostatic interactions. For hIDO our results show that the active site is much bigger and flexible which results in the presence of multiple conformations of the L-Trp complex. For the D isomer multiple binding sites are revealed with weaker binding energies. The results also show that Trp binding in both enzymes is electrostatically driven, highlighting the importance of Trp backbone. Finally, the study of the substrate-free oxy complex show that both enzymes stabilize the bound O2 with HB interactions, and that the presence of the substrate significantly alters bound oxygen properties.
Supplementary Material
Acknowledgments
This work was partially supported by grants from NIH (GM086482) to SRY and from Universidad de Buenos Aires 08-X625 to MAM and 08-X074 to DAE, ANPCYT 07-1650 to MAM and 06-25667 to DAE, Conicet PIP 01207 and Guggenheim Foundation grant awarded to DAE. DAE and MAM are members of CONICET, LC holds a CONICET PhD fellowship. MA acknowledges NSF-CHE-0755022 grant for the international REU experience in Argentina. Computer power was provided by the Centro de computacion de alto rendimiento (CECAR) at the FCEN-UBA and Teragrid (TG-MCA 05S010).
References
- 1.Sono M, Roach MP, Coulter ED, Dawson JH. Heme-Containing Oxygenases. Chem Rev. 1996;96(7):2841–2888. doi: 10.1021/cr9500500. [DOI] [PubMed] [Google Scholar]
- 2.Takikawa O. Biochemical and medical aspects of the indoleamine 2,3-dioxygenase-initiated L-tryptophan metabolism. Biochem Biophys Res Commun. 2005;338(1):12–19. doi: 10.1016/j.bbrc.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 3.Forouhar F, Anderson JLR, Mowat CG, Vorobiev SM, Hussain A, Abashidze M, Bruckmann C, Thackray SJ, Seetharaman J, Tucker T, Xiao R, Ma LC, Zhao L, Acton TB, Montelione GT, Chapman SK, Tong L. Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase. Proc Natl Acad Sci U S A. 2007;104(2):473–478. doi: 10.1073/pnas.0610007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Kang SA, Mukherjee T, Bale S, Crane BR, Begley TP, Ealick SE. Crystal structure and mechanism of tryptophan 2,3-dioxygenase, a heme enzyme involved in tryptophan catabolism and in quinolinate biosynthesis. Biochemistry. 2007;46(1):145–155. doi: 10.1021/bi0620095. [DOI] [PubMed] [Google Scholar]
- 5.Sugimoto H, Oda Si, Otsuki T, Hino T, Yoshida T, Shiro Y. Crystal structure of human indoleamine 2,3-dioxygenase: catalytic mechanism of O2 incorporation by a heme-containing dioxygenase. Proc Natl Acad Sci U S A. 2006;103(8):2611–2616. doi: 10.1073/pnas.0508996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muller AJ, DuHadaway JB, Donover PS, Sutanto-Ward E, Prendergast GC. Inhibition of indoleamine 2,3-dioxygenase, an immunoregulatory target of the cancer suppression gene Bin1, potentiates cancer chemotherapy. Nature medicine. 2005;11(3):312–319. doi: 10.1038/nm1196. [DOI] [PubMed] [Google Scholar]
- 7.Munn DH, Shafizadeh E, Attwood JT, Bondarev I, Pashine A, Mellor AL. Inhibition of T Cell Proliferation by Macrophage Tryptophan Catabolism. The Journal of Experimental Medicine. 1999;189(9):1363–1372. doi: 10.1084/jem.189.9.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grohmann U, Fallarino F, Puccetti P. Tolerance, DCs and tryptophan: much ado about IDO. Trends in Immunology. 2003;24(5):242–248. doi: 10.1016/s1471-4906(03)00072-3. [DOI] [PubMed] [Google Scholar]
- 9.Friberg M, Jennings R, Alsarraj M, Dessureault S, Cantor A, Extermann M, Mellor AL, Munn DH, Antonia SJ. Indoleamine 2,3-dioxygenase contributes to tumor cell evasion of T cell-mediated rejection. Int J Cancer. 2002;101(2):151–155. doi: 10.1002/ijc.10645. [DOI] [PubMed] [Google Scholar]
- 10.Uyttenhove C, Pilotte L, Theate I, Stroobant V, Colau D, Parmentier N, Boon T, Van den Eynde BJ. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat Med. 2003;9(10):1269–1274. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- 11.Batabyal D, Yeh SR. Human Tryptophan Dioxygenase: A Comparison to Indoleamine 2,3-Dioxygenase. J Am Chem Soc. 2007;129(50):15690–15701. doi: 10.1021/ja076186k. [DOI] [PubMed] [Google Scholar]
- 12.Lu C, Lin Y, Yeh SR. Inhibitory Substrate Binding Site of Human Indoleamine 2,3-Dioxygenase. J Am Chem Soc. 2009;131(36):12866–12867. doi: 10.1021/ja9029768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishimura Y, Nozaki M, Hayaishi O, Nakamura T, Tamura M, Yamazaki I. The Oxygenated Form of l-Tryptophan 2,3-Dioxygenase as Reaction Intermediate. J Biol Chem. 1970;245(14):3593–3602. [PubMed] [Google Scholar]
- 14.Batabyal D, Yeh SR. Substrate-Protein Interaction in Human Tryptophan Dioxygenase: The Critical Role of H76. J Am Chem Soc. 2009;131(9):3260–3270. doi: 10.1021/ja807969a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 2005;19(14):1639–1662. [Google Scholar]
- 16.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81(8):3684–3690. [Google Scholar]
- 17.Wang J, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J Comput Chem. 2000;21(12):1049–1074. [Google Scholar]
- 18.Marti MA, Crespo A, Capece L, Boechi L, Bikiel DE, Scherlis DA, Estrin DA. Dioxygen affinity in heme proteins investigated by computer simulation. J Inorg Biochem. 2006;100(4):761–770. doi: 10.1016/j.jinorgbio.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 19.Bidon-Chanal A, Martí MA, Estrin DA, Luque FJ. Exploring the nitric oxide detoxification mechanism of mycobacterium tuberculosis truncated haemoglobin N. NATO Science for Peace and Security Series A: Chemistry and Biology. 2009:33–47. [Google Scholar]
- 20.Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham TE, DeBolt S, Ferguson D, Seibel G, Kollman P. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and. ComputPhysComm. 1995;91(1):1–41. [Google Scholar]
- 21.Lee MR, Duan Y, Kollman PA. Use of MM-PB/SA in estimating the free energies of proteins: Application to native, intermediates, and unfolded villin headpiece. Proteins: Struct Func Bioinf. 2000;39(4):309–316. [PubMed] [Google Scholar]
- 22.Still WC, Tempczyk A, Hawley RC, Hendrickson T. Semianalytical treatment of solvation for molecular mechanics and dynamics. J Am Chem Soc. 1990;112(16):6127–6129. [Google Scholar]
- 23.Marti MA, Capece L, Bidon-Chanal A, Crespo A, Guallar V, Luque FJ, Estrin DA. Nitric Oxide reactivity with Globins as investigated through computer simulation. Methods in enzymology. 2008;437:477–498. doi: 10.1016/S0076-6879(07)37024-9. [DOI] [PubMed] [Google Scholar]
- 24.Bikiel DE, Boechi L, Capece L, Crespo A, De Biase PM, Di Lella S, González Lebrero MC, Martí MA, Nadra AD, Perissinotti LL, Scherlis DA, Estrin DA. Modeling heme proteins using atomistic simulations. Phys Chem Chem Phys. 2006;8(48):5611–5628. doi: 10.1039/b611741b. [DOI] [PubMed] [Google Scholar]
- 25.Crespo A, Scherlis DA, Marti MA, Ordejon P, Roitberg AE, Estrin DA. A DFT-Based QM-MM Approach Designed for the Treatment of Large Molecular Systems: Application to Chorismate Mutase. J Phys Chem B. 2003;107(49):13728–13736. [Google Scholar]
- 26.Perdew JP, Burke K, Ernzerhof M. Generalized Gradient Approximation Made Simple. Phys Rev Lett. 1996;77(18):3865. doi: 10.1103/PhysRevLett.77.3865. [DOI] [PubMed] [Google Scholar]
- 27.Martí MA, Capece L, Bikiel DE, Falcone B, Estrin DA. Oxygen affinity controlled by dynamical distal conformations: The soybean leghemoglobin and the Paramecium caudatum hemoglobin cases. Proteins: Structure, Function, and Bioinformatics. 2007;68:480–487. doi: 10.1002/prot.21454. [DOI] [PubMed] [Google Scholar]
- 28.Martí MA, Bikiel DE, Crespo A, Nardini M, Bolognesi M, Estrin DA. Two distinct heme distal site states define Cerebratulus lacteus mini-hemoglobin oxygen affinity. Proteins: Structure, Function, and Bioinformatics. 2006;62:641–648. doi: 10.1002/prot.20822. [DOI] [PubMed] [Google Scholar]
- 29.Macchiarulo A, Nuti R, Bellocchi D, Camaioni E, Pellicciari R. Molecular docking and spatial coarse graining simulations as tools to investigate substrate recognition, enhancer. BBA - Proteins and Proteomics. 2007;1774(8):1058–1068. doi: 10.1016/j.bbapap.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 30.Capece L, Lewis-Ballester A, Batabyal D, Di Russo N, Yeh SR, Estrin DA, Marti MA. The first step in the dioxygenation reaction carried out by Tryptophan dioxygenase and Indoleamine 2,3- dioxygenase as revealed by QM-MM studies. J Biol Inorg Chem. 2010 doi: 10.1007/s00775-010-0646-x. Published Online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis-Ballester A, Batabyal D, Egawa T, Lu C, Lin Y, Marti MA, Capece L, Estrin DA, Yeh SR. Evidence for a Ferryl Intermediate in Heme-based Dioxygenases: Mechanistic Implications. Proc Natl Acad Sci U S A. 2009;106(41):17371–17376. doi: 10.1073/pnas.0906655106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takikawa O, Kuroiwa T, Yamazaki F, Kido R. Mechanism of interferon-gamma action. Characterization of indoleamine 2,3-dioxygenase in cultured human cells induced by interferon-gamma and evaluation of the enzyme-mediated tryptophan degradation in its anticellular activity. J Biol Chem. 1988;263(4):2041–2048. [PubMed] [Google Scholar]
- 33.Chauhan N, Basran J, Efimov I, Svistunenko DA, Seward HE, Moody PCE, Raven EL. The Role of Serine 167 in Human Indoleamine 2,3-Dioxygenase: A Comparison with Tryptophan 2,3-Dioxygenase. Biochemistry. 2008;47(16):4761–4769. doi: 10.1021/bi702405a. [DOI] [PubMed] [Google Scholar]
- 34.Chauhan N, Thackray SJ, Rafice SA, Eaton G, Lee M, Efimov I, Basran J, Jenkins PR, Mowat CG, Chapman SK, Raven EL. Reassessment of the Reaction Mechanism in the Heme Dioxygenases. J Am Chem Soc. 2009;131(12):4186–4187. doi: 10.1021/ja808326g. [DOI] [PubMed] [Google Scholar]
- 35.Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. New York: W.H. Freeman and Company; 1999. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.