Abstract
MicroRNAs (miRNAs) have emerged as an essential regulatory component in plants. Many of the known miRNAs are evolutionarily conserved across diverse plant species and function in the regulatory control of fundamentally important biological processes such as developmental timing, patterning, and response to environmental changes. Expression of miRNAs in plants involves transcription from MIRNA loci by RNA polymerase II (pol II), multi-step processing of the primary transcripts by the DICER-LIKE1 (DCL1) complex, and formation of effector complexes consisting of mature miRNAs and ARGONAUTE (AGO) family proteins. In this short review, we present the most recent advances in our understanding of the molecular machinery as well as the regulatory mechanisms involved in the expression of miRNAs in plants.
Keywords: miRNA expression, Transcriptional regulation, Transcription factors, Pri-miRNA processing, Arabidopsis thaliana
1. Introduction
Small RNAs of 21- to 24-nucleotide (nt) in size have emerged as an important regulatory component expressed by most eukaryotic genomes (reviewed in [1] and [2]). Studies over the past decade have firmly established the important role of small RNAs in the control of gene expression, epigenetic modification of the genome, and defense against viruses [1, 2]. In general, these small RNA molecules arise from transcripts that form either bimolecular or intra-molecular double-stranded RNA (dsRNA) precursors. Processing of dsRNA precursors by the evolutionarily conserved RNA silencing machinery gives rise to mature small RNAs that function in diverse cellular processes. Based on their distinct biogenesis mechanisms, endogenous small RNAs are classified into two broad categories, microRNAs (miRNAs) that arise from characteristic imperfect hairpin-like precursors, and small interfering RNAs (siRNAs) that arise typically from bimolecular dsRNA precursors. miRNAs, which are the focus of this review series, are a class of endogenous small RNAs that negatively regulate the expression of protein-coding mRNAs through sequence-specific interactions, typically leading to target mRNA cleavage or translational repression in plants [3, 4]. Some miRNAs in plants target non-protein-coding RNA transcripts derived from defined genetic loci known as TRANS-ACTING SIRNAs (TAS). The miRNA-guided cleavage of TAS transcripts initiates the production of trans-acting siRNAs (ta-siRNAs) from the cleaved transcripts, which in turn function to negatively regulate their protein-coding mRNA targets [1]. Significantly, many of the known miRNAs in plants are evolutionarily conserved and function in fundamentally important biological processes such as developmental timing, patterning, and response to environmental changes [3]. This short review will focus on miRNA expression and its regulation in plants, with an emphasis on the most recent advances in our understanding of the molecular machinery and regulatory mechanisms involved in the expression of miRNAs in plants.
2. Transcription of MIRNA Genes and Its Regulation
Mature miRNAs are expressed typically as a class of 21-nt endogenous small RNAs in plants. Expression data from RNA blot assays, high-throughput sequencing analysis, and in situ hybridization have collectively shown that the abundance of mature miRNAs in cells is tightly regulated. For instance, the abundance of mature miRNAs varies greatly among different miRNAs [3], which may result from differences in the transcriptional activity among MIRNA genes. Moreover, the same miRNA can be found in different abundance among tissue types or developmental stages, indicating the spatially and temporally regulated expression patterns of plant miRNAs [3]. Such regulation may take place at both the transcriptional and post-transcriptional levels, as will be discussed below in more detail.
2.1 Plant MIRNA genes are transcribed by RNA polymerase II
miRNAs in plants arise from defined genetic loci known as MIRNA genes. Most plant MIRNA genes exist as independent transcription units and locate typically in the genomic regions that are not occupied by protein-coding genes, previously known as intergenic regions (IGR). To date, nearly 200 and over 400 MIRNA loci have been identified in the genomes of Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa), respectively [5]. Several lines of evidence indicate that plant MIRNA genes are transcribed by RNA polymerase II (pol II). First, the primary transcripts of plant miRNAs so far characterized contain a 5′cap and a 3′ poly (A) tail that are characteristic of a pol II transcript. Secondly, sequence analysis of MIRNA loci in Arabidopsis thaliana identified core promoter elements including TATA box and transcription initiator (INR) elements that are typically found in pol II genes [6, 7]. A key feature of a primary miRNA transcript, termed pri-miRNA, is its capability of forming a characteristic hairpin-like imperfect stem-loop structure. In most cases, a pri-miRNA folds back to form a single hairpin-like structure that gives rise to only one unique mature miRNA species. However, “polycistronic” MIRNA loci have recently been reported in plants, where a single pri-miRNA can form two or more hairpins, each containing a distinct mature miRNA species [8]. In addition, while the vast majority of plant miRNAs appear to be derived from independent transcription units defined by distinct MIRNA loci, deviations from this paradigm have been observed. For example, intronic miRNAs can be embedded in intronic sequences of some protein-coding genes and can therefore be produced as by products of host gene expression [9]. Certain transposable elements (TEs) may also give rise to miRNA and siRNA dual-coding transcripts, as have been observed in both Arabidopsis and rice [10], although the functional significance of these miRNAs remains to be investigated.
2.2 Transcriptional regulation of miRNA expression in plants
Just as do their protein-coding gene counterparts, transcriptional regulation for miRNA gene expression involves cis-regulatory elements and trans-acting factors. Since a large set of plant miRNAs are known to target mRNAs encoding diverse developmentally important transcription factor (TF) family members [3], many of which are specific to the plant lineage [11], an interesting question is whether some of these TFs are also involved in the regulation of miRNA expression. Taking advantage of the MIRNA loci whose transcription start sites (TSS) have been experimentally mapped, several groups have taken an informatics approach to search for cis-regulatory elements that are enriched or over-represented in the putative promoter region of MIRNA genes [12–14]. In addition to the core promoter elements such as the TATA-box, sequence motifs present in the promoters of protein-coding genes are also found in the putative promoters of MIRNA genes. Intriguingly, binding motifs for several transcription factors including LEAFY (LFY), AUXIN RESPONSE FACTORS (ARFs), and AtMYC2 (a basic helix-loop-helix [bHLH] type transcription factor) were found to be over-represented in Arabidopsis MIRNA loci when compared with their presence in the promoters of protein-coding genes [12]. The finding of ARF-binding motifs in MIRNA promoters is particularly interesting because mRNAs for multiple ARF family members are known to be targeted, either directly or indirectly, by members of several conserved plant miRNA families including miR160, miR167, and miR390 [3] (Fig. 1A). These observations could therefore suggest a possible ARF-mediated negative feedback regulatory loop for miRNA expression in plants [12] (Fig. 1B), although the functionality of these computationally identified cis-elements remains to be experimentally validated.
Figure 1. Transcriptional regulation of miRNA expression in Arabidopsis thaliana.
A. Certain ARF family members are regulated by miRNAs, either directly (miR160 and miR167) or indirectly (miR390). B. A possible negative feedback regulatory loop involving miR160, miR167 and certain ARF family members. Numerous copies of ARF recognition motif (TGTCTC) are found in the putative promoter regions of MIR160b, MIR167a, MIR167b, and miR167d [12]. C. SPL-mediated cross talk between miR156 and miR172. miR156 targets mRNAs for 10 members of the SPL family of transcription factors including SPL9 and SPL10, both of which promote the expression of MIR156a [18]. miR156 also regulates the expression of miR172 through SPL9 and SPL10 which function redundantly in promoting the transcription of MR172b [18]. TOE1 and TOE2, two of the six AP2 family transcription factors that are known to be the regulatory targets of miR172, also appear to positively regulate the expression of MIR172b [18], although evidence for a direct interaction between these AP2-like TFs and the MIR172 promoter is currently lacking. D. Putative SPL7 recognition sequences are found in the promoter regions of several low-copper-inducible MIRNAs. Each vertical light blue bar represents the presence of a putative SPL7 recognition sequence motif (GTAC) [26], as revealed by a computational scan within the putative promoter regions for each of the MIRNAs. Up to 1,000bp of the the putative promoter sequence, arbitrarily defined as the sequence upstream of a curated miRNA hairpin precursor [5] was included in the analysis. Note that while the putative promoter regions were drawn to scale, the miRNA hairpin precursor regions (black boxes) were not.
Further insights into the transcriptional regulation of miRNA expression in plants came from analyses on the cross talk between miR156 and miR172. In Arabidopsis, the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) TFs are regulated by miR156 [3]. The interactions between miR156 and mRNAs of several SPL family members are involved in the regulation of developmental timing, including the transitions from juvenile to adult and from the vegetative to the reproductive phase [15, 16]. Members of the APETALA2 (AP2)-like family of TFs are regulated by miR172 and involved in regulation of flowering time and floral organ development [3, 17]. Interestingly, miR156 and miR172 exhibit a complementary temporal expression pattern in Arabidopsis: while miR156 expression declines during the transition from the juvenile to the adult phase, the expression of miR172 increases [18]. This complementary expression pattern between miR156 and miR172 has also been observed in maize [19]. A recent report from the Poethig laboratory has provided an interesting mechanistic link between these observations [18]. SPL9 and SPL10, two of the 10 SPL family TFs that are targeted by miR156, promote the expression of miR172 by directly activating its transcription, which is consistent with the presence of multiple copies of putative SPL binding elements in the MIR172b promoter [18]. Moreover, SPL9 and SPL10 appear to also promote the expression of miR156 through transcription activation, thereby forming a negative feedback loop that regulates their own expression [18] (Fig. 1C). This SPL-mediated regulatory module allows a sequential action of miR156 and miR172, which contributes to the regulatory network that governs developmental timing [18, 20].
Several plant miRNAs have been shown to respond to environmental cues, such as limited nutrients supply, cold, or light signals [3, 13, 21, 22]. Expression of Arabidopsis miR398, for example, is induced in response to copper limitation [23, 24]. miR398 targets a cytosolic CSD1 and a chloroplastic CSD2, two closely related Cu/Zn superoxide dismutases, as well as COX5b-1, a subunit of the mitochondrial cytochrome c oxidase [3, 23]. Significantly, miR398-mediated down-regulation of its target gene expression under copper limitation likely serves as an important mechanism for plants to mobilize copper from non-essential to essential copper proteins, such as plastocyanin that is indispensable for photosynthesis [24]. Similarly, the expression of miR397, miR408, and miR857-- the other three Arabidopsis miRNAs that are known to target mRNAs for copper-containing proteins, also appears to be inducible under low cooper conditions, thereby contributing to the proposed mechanism for copper economy [25](Fig. 1D). Further analysis of miR398 expression has identified SPL7 as an essential TF that activates miR398 transcription under low copper conditions [26]. The Arabidopsis SPL7 is a homolog of the Chlamydomonas reinhardtii protein known as Copper response regulator1 (Crr1), a TF that is required for switching between plastocyanin and cytochrome C6 in response to low copper conditions. Both in vitro and in vivo evidence indicates that SPL7 functions to activate miR398 expression through its interaction with the multiple GTAC motifs found in the promoters of both MIR398b and MIR398c, all within a 300 bp region upstream of the TSS [26] (Fig. 1D). The fact that multiple GTAC motifs are also present in the promoter region of miR397, miR408, and miR857, respectively (Fig. 1D), and that a loss-of-function mutation in SPL7 impaired the copper deficiency-induced expression of these miRNAs strongly indicate that SPL7 plays a central regulatory role in miRNA-mediated copper homeostasis [26].
3. Processing of MIRNA Transcripts and Its Regulation
Central to miRNA biogenesis is the recognition and stepwise processing of pri-miRNAs by the conserved miRNA pathway machinery, leading to the accurate excision and release of functional mature miRNAs. Although the precise mechanism remains to be elucidated for routing a pri-miRNA into the miRNA pathway instead of into the translation machinery, the framework for miRNA biogenesis in plants has emerged, mainly based on genetic and biochemical studies in Arabidopsis [3, 4].
3.1 The nuclear cap-binding complex plays a role in pri-miRNA processing
The pri-miRNAs in plants possess a 5′ cap and a 3′ poly(A) tail [3]. Some of them are also known to contain one or more introns that undergo canonical splicing [3]. Processing of the pri-miRNAs is therefore believed to take place shortly after a nascent transcript emerges from the pol II complex. The early events presumably involve at least some of the machinery responsible for capping, splicing, and polyadenylation of protein-coding transcripts. Indeed, recent reports from several groups have shown the involvement of the nuclear cap-binding complex (CBC) in pri-miRNA processing [27–29]. CBC is a heterodimer consisting of two protein subunits (CBP80 and CBP20). The Arabidopsis CBP80 is also known as ABSCISCIC ACID (ABA) HYPERSENSITIVE1 (ABH1) since it was isolated in a forward genetic screen for mutants that are hypersensitive to the plant hormone ABA. Loss-of-function mutations in ABH1/CBP80 and CBP20 resulted in accumulation of unspliced transcripts to elevated levels as observed through transcriptome profiling using whole-genome tiling array technology, which was indicative of defective intron splicing, thereby confirming the speculated role of Arabidopsis CBC in promoting efficient intron splicing [29]. Significantly, both the cbp80/abh1 and cbp20 mutants were found also to accumulate elevated levels of pri-miRNAs, whereas the levels of mature miRNA accumulation were substantially reduced [27–29]. These observations have therefore established dual roles of the Arabidopsis CBC in pre-mRNA splicing and pri-miRNA processing.
Intriguingly, both the cbp80/abh1 and cbp20 mutants exhibited pleiotropic abnormalities including the “serrated leaves” that strikingly resemble the phenotype observed in the serrate (se) mutant [30]. SE encodes a C2H2-type zinc-finger domain-containing protein which has recently been shown to be required for pri-miRNA processing [3, 30–32] (and see below). A tantalizing question to ask would be whether SE also plays a role in pre-mRNA splicing. Indeed, whole-genome tiling array-based transcriptome profiling detected the accumulation of higher levels of unspliced transcripts for a subset of genes that partially overlap with those observed in the cbp80/abh1 and cbp20 mutants [29]. Of note, analysis of intron-containing pri-miRNAs suggest that the pri-miRNA processing function of CBC and SE appears to be independent from their role in pre-mRNA splicing [29]. These observations therefore have not only identified a novel role for SE in pre-mRNA splicing, but also provided hints for potential mechanisms by which the CBC and SE function in the two distinct processes. One possibility is that SE may serve as a common mediator for facilitating the interactions between the CBC and components of the spliceosome, in the case of pre-mRNA splicing, and also between the CBC and the pri-miRNA processing machinery, as has been proposed by Laubinger and colleagues [29](Fig. 2A). However, evidence for a direct interaction between CBC and SE is currently lacking, and other modes of action for SE in pri-miRNA processing remain possible (see below).
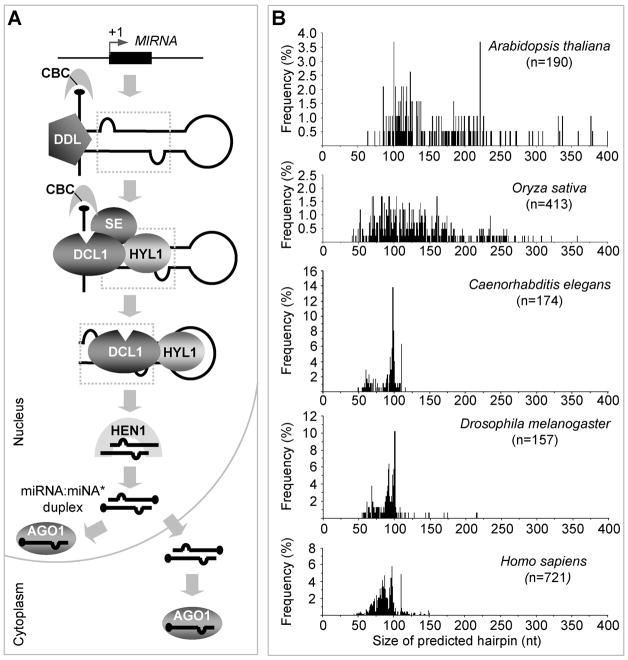
Figure 2. miRNA biogenesis in plants.
A. Multi-step processing of pri-miRNA and miRNA maturation in a plant cell. Expression of miRNAs in plants begins with the pol II transcription of a MIRNA locus that is typically located in a genomic region not occupied by protein-coding genes, previously known as intergenic regions (IGR). The primary transcripts of a miRNA (pri-miRNA), which bear a 5′cap and a 3′ poly (A) tail, are capable of forming a characteristic hairpin-like secondary structure. CBC, a heterodimer consisting of CBP80 and CBP20 subunits of the nuclear cap-binding complex has been shown to play a role in pri-miRNA processing, likely through its direct interaction with a nascent transcript. DDL, a forkhead-associated domain-containing protein that has been shown to physically interact with pri-miRNA in vitro, likely function to stabilize the hairpin structure and help recruit DCL1 to its substrate. DCL1 and two other factors, HYL1, a dsRNA-binding protein, and SE, a C2H2-type zinc-finger domain-containing protein physically interact with each other and form a processing complex (DCL1 complex) in subnuclear bodies. The stepwise processing of a pri-miRNA by the DCL1 complex ultimately gives rise to a miRNA: miRNA* duplex that is subsequently recognized and end-methylated by HEN1. Loading of the miRNA strand into an AGO protein to form a functional miRISC marks the end of miRNA biogenesis. B. Size distribution of predicted hairpin precursors for plant and animal miRNAs. For each of the five model organisms (Arabidopsis thaliana, Oryza sativa, Caenorhabditis elegans, Drosophila melanogaster, and Homo sapiens), the size distribution of miRNA hairpin precursors was analyzed by plotting the size (as curated in miRBase [5]; release 14) versus its frequency of occurrence.
3.2 Processing of pri-miRNA transcripts by the DICER-LIKE1complex
Genetic analysis has identified multiple components that function in the miRNA biogenesis pathway. DICER-LIKE1 (DCL1), one of the four Arabidopsis DCLs belonging to the multi-domain ribonuclease III (RNase III)-like protein family, is a key enzyme in miRNA biogenesis [3]. The DICER (DCR) or DCL family proteins contain a helicase domain at the N-terminus, a PAZ domain in the middle, and the dual RNase III domains as well as a dsRNA-binding motif (dsRBM) at the C-terminus. Structural analysis of a Giardia intestinalis DCR has revealed that DCR itself functions as a molecular ruler that measures and cleaves dsRNA from the helical end, the length of the small RNAs being determined by the distance between the PAZ domain that binds the end of dsRNA and the catalytic RNase III domains [33]. HYPONASTIC LEAVES1 (HYL1), a dsRNA-binding protein, and SE (as described above) are also required for pri-miRNA processing [3, 30–32]. Importantly, DCL1, HYL1, and SE physically interact with each other and co-localize in the same subnuclear bodies (termed the dicing bodies or D-bodies) that also contain pri-miRNAs [34–36]. The speculated roles of HYL1 and SE in miRNA biogenesis include stabilizing the pri-miRNA structure and facilitating the correct positioning of DCL1 on its substrates, thereby ensuring accurate processing [31, 32, 37, 38] (Fig. 2A). In a recent biochemical analysis, a recombinant Arabidopsis DCL1 alone was shown to be capable of processing either pri-miRNA or pre-miRNA into 21-nt small RNAs in vitro. However, correctly processed miRNAs accounted for only a small portion of the small RNA population [39]. Addition of either a recombinant HYL1 or SE into the reactions substantially increased the rate and accuracy of DCL1 processing. Further improvement in DCL1 processing accuracy was achieved when both HYL1 and SE were present in the reactions, which resulted in a remarkable, nearly 80% processing accuracy [39]. The suboptimal pri-miRNA processing accuracy achieved by the DCL1-HYL1-SE complex (DCL1 complex) in vitro could be an indication that certain components required for more efficient and accurate DCL1 processing are still missing. DAWDLE (DDL), a forkhead-associated domain-containing protein could be one such component [40, 41]. Loss-of-function mutations in Arabidopsis DDL led to reduced levels of pri-miRNAs as well as mature miRNAs, although such an effect is not specific to miRNAs since the accumulation of endogenous siRNAs was also reduced. In addition, DDL was shown to bind a pri-miRNA and interact with DCL1 in vitro, supporting its speculated role of facilitating the functional interactions between DCL1 and pri-miRNAs [41] (Fig. 2A).
The precise molecular mechanism by which the DCL1 complex recognizes and acts on a pri-miRNA to accurately produce a distinct miRNA in plants is not well understood. In animal systems, the pri-miRNAs undergo a two-step processing catalyzed by two different RNase III enzyme complexes. First, the Drosha complex in the nucleus cleaves pri-miRNAs at the loop distal site of the stem (usually about 11-nt away from the junction of single-stranded RNA [ssRNA] and dsRNA) and releases the “stem-loop” miRNA precursors (pre-miRNAs) [42]. The pre-miRNAs are then exported to the cytoplasm, where the DCR complex cleaves at the loop proximal site to release a small RNA duplex of approximately 22-nt long. One strand will become a mature miRNA, and the other becomes the non-functional passenger strand called a miRNA* (the small RNA residing on the opposite side of the miRNA in a hairpin precursor) [42]. The miRNA biogenesis in plants differs from their animal counterparts in several aspects. First, plant genomes are not known to encode Drosha orthologs. Instead, the DCL1 complex in the nucleus is believed to be responsible for the processing of both pri-miRNAs and pre-miRNAs. Secondly, the hairpin portion of the plant pri-miRNAs is, in general, much longer and more variable in size compared with the animal miRNAs (Fig. 2B) [3]. In searching for structural determinants of pri-miRNA processing in plants, several recent studies have provided interesting mechanistic insights on DCL1-mediated miRNA biogenesis in Arabidopsis [43–46]. Using a range of elegantly designed approaches, structure-function analyses of pri-miRNAs derived from miR167a [45], miR171a [45], miR172a [44, 46], and miR390a [43] revealed that an approximately 15-nt stem segment next to the loop-distal end of the miRNA:miRNA* duplex was an essential element for accurate processing. This segment likely serves as a docking site for the DCL1 complex to make the first, loop-distal cleavage in a canonical “base-to-loop” processing mode. Interestingly, pri-miRNAs harboring unusually long stems appear to have adopted a non-canonical, “loop-to-base” processing mode, as have been shown for miR319 and miR159 in which up to four, instead of two, cleavage events are needed to release the miRNA:miRNA* duplex [47, 48].
3.3 End modification and maturation of miRNAs
The multi-step processing of a pri-miRNA by the DCL1 complex ultimately releases a small RNA duplex consisting of a miRNA and a miRNA* (Fig. 2A). As a hallmark of the DCR/DCL cleavage, each small RNA in the miRNA:miRNA* duplex contains a 2-nt overhang at the 3′-end. In plants, the miRNA:miRNA* duplex is recognized by, and serves as a substrate for, HUA ENHANCER1 (HEN1), the S-adenosylmethionine-dependent small RNA methyltransferase that deposits a methyl group to the 2′-hydroxyl group of the 3′terminal nucleotide in each of the small RNA strands [3, 49]. This 2′-O-methylation on the 3′-terminal nucleotide is common to all known classes of silencing small RNAs in plants and is believed to stabilize the small RNAs by protecting them from other type of cellular activities such as oligouridylation [3]. Arabidopsis hen1 mutants are impaired in mature miRNA accumulation and exhibit pleiotropic developmental abnormalities reminiscent of phenotypes for certain hypomorphic dcl1 alleles [3, 50]. These observations strongly indicate the HEN1-mediated 2′-O-methylation as a crucial step in plant miRNA biogenesis (Fig. 2A).
The final step of miRNA maturation involves selective incorporation of the miRNA strand into an ARGONAUTE (AGO)-containing effector complex, known as the RNA-induced silencing complex (RISC). In most cases, the miRNA strand of a miRNA:miRNA* duplex is selectively loaded onto an AGO complex and serves as a guide for its sequence-specific interaction with target transcripts, whereas the miRNA* strand is excluded from the AGO complex and thought to be short-lived in vivo. Among the ten AGO family proteins in Arabidopsis, AGO1 is apparently the most important family member for the miRNA pathway, as evidenced by both genetic and biochemical data [3, 51] (Fig. 2A). In plants, the fate of each small RNA strand in a miRNA:miRNA* duplex is likely to be determined by multiple factors. First, functional small RNA duplexes often exhibit asymmetrical thermostability between the two ends. In many cases, the small RNA with its 5′ end located at the less stable end of the duplex is preferentially selected to form a RISC [3]. This “strand asymmetry” rule originally observed from animal small RNAs also plays an important role in the formation of miRNA-associated RISCs in plants [52]. Moreover, plant AGO proteins show distinct preference for small RNAs with a different 5′-terminal nucleotide [53–55]. Analysis of small RNAs from distinct immunoprecipitated Arabidopsis AGO complexes have revealed the preferential association of AGO1 with small RNAs bearing a 5′-terminal uridine (5′U), AGO2 and AGO4 with small RNAs bearing a 5′-terminal adenosine (5′A), and AGO5 with small RNAs bearing a 5′-terminal cytosine (5′C), respectively [53–55]. Since the majority of plant miRNAs so far identified have a 5′U, as is the case for animal miRNAs [3, 5], the above observations explain nicely for AGO1 being the major AGO family member that functions in miRNA pathway in plants [51, 56].
So where in the plant cell do mature miRNAs reside? Because many plant miRNAs have been shown to guide cleavage of their targets mRNAs, presumably in the cytoplasm [3], and recent genetic and biochemical studies have implicated plant miRNAs in translational repression [57, 58], it is widely believed that plant miRNAs function in the cytoplasm. Although a cellular fractionation-based study in Arabidopsis has shown that a majority of miRNAs accumulate in the cytoplasm [59], it is currently unclear if plant miRNAs function exclusively in the cytoplasm. Limited cellular localization data indicate that HEN1 and AGO1 are found in both the cell nucleus and cytoplasm [34], which is consistent with the role of these proteins in multiple small RNA pathways [60]. Co-localization with HYL1 in the subnuclear D-bodies has also been observed for HEN1 and AGO1, respectively [34], suggesting that dicing, end modification, and formation of miRNA-containing effector complexes (miRISCs) could be intimately coupled steps in the nucleus during plant miRNA biogenesis. Interestingly, HYL1 has recently been shown to plays a role in directing the strand selection during the formation of miRNA-AGO1 complex, likely through a mechanism similar to that of the Drosophila R2D2 [52]. The fact that single-stranded, mature miRNAs also exist in the nucleus [59], and that HYL1 and AGO1 interact in the nuclear D-bodies [34] could indicate that at least some of the plant miRNAs may incorporate into AGO1 and form functional miRISCs in the nucleus. In fact, indirect evidence suggests that the miR162-directed cleavage of DCL1 mRNA likely takes place in the nucleus, before the intron 14 is spliced out from the DCL1 transcripts [59].
3.4 Post-transcriptional regulation of miRNA expression in plants
Once a pri-miRNA is made, the birth of a mature miRNA could be, in principle, controlled at multiple levels. Efficient and accurate processing of a pri-miRNA requires the presence of key miRNA pathway components in the same cell. The miRNA expression domain could therefore be affected by the spatiotemporal expression pattern of proteins that are involved in pri-miRNA processing and miRNA maturation. Although this has not been extensively examined, existing data support such a possibility. For instance, Arabidopsis SE has been shown to exhibit distinct spatial expression patterns during development [61]. SE mRNA was present in the shoot and root meristems as well as in the adaxial side of cotyledons in early stage embryos, but the expression level was reduced in the later stages and was absent from mature embryos [61]. Distinct spatial expression patterns for SE were also observed in developing Arabidopsis leaves and floral organs [61]. Although the role of SE is not limited in pri-miRNA processing, as discussed above, it is reasonable to expect that the expression pattern of SE will affect miRNA expression. The distinct spatial expression pattern of AGO7, one of the ten Arabidopsis AGO family members that is known to function in miR390-directed TAS3 ta-siRNA biogenesis [62, 63] serves as another example. AGO7 expression was shown to be highly restricted to the vasculature and pith regions beneath the shoot apical meristem (SAM) and the adaxial-most cell layers of developing leaf primordia. The localized expression of AGO7, in conjunction with that of TAS3 helps establish the activity domain of miR390, thereby contributing to the TAS3 ta-siRNA-mediated leaf patterning [54, 64].
The mRNAs of Arabidopsis DCL1 and AGO1 are known miRNA targets, suggesting a negative feedback regulatory mechanism for DCL1 and AGO1 expression [3]. The negative feedback regulatory circuits involving miR162-directed cleavage of DCL1 mRNA [65] and miRNA168-directed cleavage of AGO1 mRNA [66, 67], respectively, are believed to play an important role in miRNA homeostasis, since similar regulatory mechanisms appear to be deeply conserved among diverse plant species [68]. The maintenance of AGO1 homeostasis in Arabidopsis has been analyzed in depth, revealing differential roles played by the MIR168a and MIR168b loci [69], as well as an additional layer of regulation involving AGO1 silencing mediated by AGO1-derived siRNAs [70]. The Arabidopsis AGO2, which has been shown to preferentially bind to 21-nt small RNAs with a 5′A [53–55], is also known to be regulated by a miRNA (miR403) [71], although a role of AGO2 in miRNA pathway has yet to be established.
4. Conclusions
In this short review, we have summarized some of the most recent studies that have advanced our knowledge on miRNA expression and its regulation in plants. The essential regulatory role of plant miRNAs has been well established genetically, as manifested not only by the embryonic lethal phenotypes of the null dcl1 and ago1 mutants, but also by the developmental defects associated with perturbation in the expression of a single miRNA [3]. However, deciphering the miRNA-mediated regulatory networks that govern plant development or response to environmental cues remains a challenging task for plant biologists. A major challenge for genetic dissection of miRNA-mediated regulation in plants has been the presence of multi-member miRNA families in which more than one MIRNA locus would give rise to identical or near-identical mature miRNAs, which is often the case for more evolutionarily conserved miRNAs [3]. However, emerging evidence appears to suggest that such a potential genetic redundancy does necessarily prevent informative locus-specific genetic analysis, as has been illustrated for loci encoding miR160 [72], miR164 [73], and miR168 [69], respectively. One could therefore expect that future analysis on locus-specific miRNA expression may lead to identification of important cis-regulatory elements as well as trans-acting factors that have shaped regulatory diversification among paralogous MIRNA loci. One of the most intriguing findings we have discussed here is the “cross talk” between miR156 and miR172 [18] (Fig. 1C), for it provides a proof-of-concept for the speculated “regulation of a regulator (miRNA) by a regulator (TF) that is regulated by another regulator (miRNA)” in a miRNA-mediated regulatory cascade that governs developmental timing in plants.
With the excellent genomic and genetic tools available for the model plant Arabidopsis, along with the recently established in vitro pri-miRNA processing system [39], as well as other biochemical tools coupled with the power of next-generation DNA sequencing technology, we anticipate the closing of many of the existing gaps in our current understanding of miRNA expression and its control mechanisms in plants within the next few years to come.
Acknowledgments
We thank Nathan Collie and Chris Rock for critically reading the manuscript. We apologize to colleagues whose work we could not cover due to space limitations. Research in the authors’ laboratory is currently supported by a grant from the National Institutes of Health (to Z.X.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet. 2007;8:884–96. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- 2.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their Regulatory Roles in Plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 4.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–87. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 5.Griffiths-Jones S, Saini HK, Dongen SV, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–8. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC. Expression of Arabidopsis MIRNA genes. Plant Physiol. 2005;138:2145–54. doi: 10.1104/pp.105.062943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou X, Ruan J, Wang G, Zhang W. Characterization and identification of microRNA core promoters in four model species. PLoS Comput Biol. 2007;3:e37. doi: 10.1371/journal.pcbi.0030037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merchan F, Boualem A, Crespi M, Frugier F. Plant polycistronic precursors containing non-homologous microRNAs target transcripts encoding functionally related proteins. Genome Biol. 2009;10:R136. doi: 10.1186/gb-2009-10-12-r136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–25. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piriyapongsa J, Jordan IK. Dual coding of siRNAs and miRNAs by plant transposable elements. RNA. 2008;14:814–21. doi: 10.1261/rna.916708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu G. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–10. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 12.Megraw M, Baev V, Rusinov V, Jensen ST, Kalantidis K, Hatzigeorgiou AG. MicroRNA promoter element discovery in Arabidopsis. RNA. 2006;12:1612–9. doi: 10.1261/rna.130506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou X, Wang G, Sutoh K, Zhu JK, Zhang W. Identification of cold-inducible microRNAs in plants by transcriptome analysis. Biochim Biophys Acta. 2008;1779:780–8. doi: 10.1016/j.bbagrm.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Zhou X, Wang G, Zhang W. UV-B responsive microRNA genes in Arabidopsis thaliana. Mol Syst Biol. 2007;3:103. doi: 10.1038/msb4100143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev Cell. 2005;8:517–27. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 16.Wu G, Poethig RS. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133:3539–47. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chuck G, Meeley R, Irish E, Sakai H, Hake S. The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat Genet. 2007;39:1517–21. doi: 10.1038/ng.2007.20. [DOI] [PubMed] [Google Scholar]
- 18.Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138:750–9. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuck G, Cigan AM, Saeteurn K, Hake S. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet. 2007;39:544–9. doi: 10.1038/ng2001. [DOI] [PubMed] [Google Scholar]
- 20.Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138:738–49. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 21.Jia X, Ren L, Chen QJ, Li R, Tang G. UV-B-responsive microRNAs in Populus tremula. J Plant Physiol. 2009;166:2046–57. doi: 10.1016/j.jplph.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 22.Sire C, Moreno AB, Garcia-Chapa M, Lopez-Moya JJ, San Segundo B. Diurnal oscillation in the accumulation of Arabidopsis microRNAs, miR167, miR168, miR171 and miR398. FEBS Lett. 2009;583:1039–44. doi: 10.1016/j.febslet.2009.02.024. [DOI] [PubMed] [Google Scholar]
- 23.Sunkar R, Kapoor A, Zhu JK. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006;18:2051–65. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamasaki H, Abdel-Ghany SE, Cohu CM, Kobayashi Y, Shikanai T, Pilon M. Regulation of copper homeostasis by micro-RNA in Arabidopsis. J Biol Chem. 2007;282:16369–78. doi: 10.1074/jbc.M700138200. [DOI] [PubMed] [Google Scholar]
- 25.Abdel-Ghany SE, Pilon M. MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J Biol Chem. 2008;283:15932–45. doi: 10.1074/jbc.M801406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamasaki H, Hayashi M, Fukazawa M, Kobayashi Y, Shikanai T. SQUAMOSA Promoter Binding Protein-Like7 Is a Central Regulator for Copper Homeostasis in Arabidopsis. Plant Cell. 2009;21:347–61. doi: 10.1105/tpc.108.060137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gregory BD, O’Malley RC, Lister R, Urich MA, Tonti-Filippini J, Chen H, Millar AH, Ecker JR. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell. 2008;14:854–66. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 28.Kim S, Yang JY, Xu J, Jang IC, Prigge MJ, Chua NH. Two cap-binding proteins CBP20 and CBP80 are involved in processing primary MicroRNAs. Plant Cell Physiol. 2008;49:1634–44. doi: 10.1093/pcp/pcn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laubinger S, Sachsenberg T, Zeller G, Busch W, Lohmann JU, Rätsch G, Weigel D. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2008;105:8795–800. doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grigg SP, Canales C, Hay A, Tsiantis M. SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature. 2005;437:1022–6. doi: 10.1038/nature04052. [DOI] [PubMed] [Google Scholar]
- 31.Lobbes D, Rallapalli G, Schmidt DD, Martin C, Clarke J. SERRATE: a new player on the plant microRNA scene. EMBO Rep. 2006;7:1052–8. doi: 10.1038/sj.embor.7400806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang L, Liu Z, Lu F, Dong A, Huang H. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 2006;47:841–50. doi: 10.1111/j.1365-313X.2006.02835.x. [DOI] [PubMed] [Google Scholar]
- 33.Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–8. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 34.Fang Y, Spector DL. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol. 2007;17:818–23. doi: 10.1016/j.cub.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujioka Y, Utsumi M, Ohba Y, Watanabe Y. Location of a possible miRNA processing site in SmD3/SmB nuclear bodies in Arabidopsis. Plant Cell Physiol. 2007;48:1243–53. doi: 10.1093/pcp/pcm099. [DOI] [PubMed] [Google Scholar]
- 36.Song L, Han MH, Lesicka J, Fedoroff N. Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc Natl Acad Sci U S A. 2007;104:5437–42. doi: 10.1073/pnas.0701061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurihara Y, Takashi Y, Watanabe Y. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA. 2006;12:206–12. doi: 10.1261/rna.2146906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tagami Y, Motose H, Watanabe Y. A dominant mutation in DCL1 suppresses the hyl1 mutant phenotype by promoting the processing of miRNA. RNA. 2009;15:450–8. doi: 10.1261/rna.1297109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong Z, Han MH, Fedoroff N. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci U S A. 2008;105:9970–5. doi: 10.1073/pnas.0803356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morris ER, Chevalier D, Walker JC. DAWDLE, a forkhead-associated domain gene, regulates multiple aspects of plant development. Plant Physiol. 2006;141:932–41. doi: 10.1104/pp.106.076893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu B, Bi L, Zheng B, Ji L, Chevalier D, Agarwal M, Ramachandran V, Li W, Lagrange T, Walker JC, Chen X. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci U S A. 2008;105:10073–8. doi: 10.1073/pnas.0804218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 43.Cuperus JT, Montgomery TA, Fahlgren N, Burke RT, Townsend T, Sullivan CM, Carrington JC. Identification of MIR390a precursor processing-defective mutants in Arabidopsis by direct genome sequencing. Proc Natl Acad Sci U S A. 2009;107:466–71. doi: 10.1073/pnas.0913203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mateos JL, Bologna NG, Chorostecki U, Palatnik JF. Identification of MicroRNA Processing Determinants by Random Mutagenesis of Arabidopsis MIR172a Precursor. Curr Biol. 2010;20:49–54. doi: 10.1016/j.cub.2009.10.072. [DOI] [PubMed] [Google Scholar]
- 45.Song L, Axtell MJ, Fedoroff NV. RNA Secondary Structural Determinants of miRNA Precursor Processing in Arabidopsis. Curr Biol. 2010;20:37–41. doi: 10.1016/j.cub.2009.10.076. [DOI] [PubMed] [Google Scholar]
- 46.Werner S, Wollmann H, Schneeberger K, Weigel D. Structure Determinants for Accurate Processing of miR172a in Arabidopsis thaliana. Curr Biol. 2010;20:42–8. doi: 10.1016/j.cub.2009.10.073. [DOI] [PubMed] [Google Scholar]
- 47.Addo-Quaye C, Snyder JA, Park YB, Li YF, Sunkar R, Axtell MJ. Sliced microRNA targets and precise loop-first processing of MIR319 hairpins revealed by analysis of the Physcomitrella patens degradome. RNA. 2009;15:2112–21. doi: 10.1261/rna.1774909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bologna NG, Mateos JL, Bresso EG, Palatnik JF. A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J. 2009;28:3646–56. doi: 10.1038/emboj.2009.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang Y, Ji L, Huang Q, Vassylyev DG, Chen X, Ma JB. Structural insights into mechanisms of the small RNA methyltransferase HEN1. Nature. 2009;461:823–7. doi: 10.1038/nature08433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schauer SE, Jacobsen SE, Meinke DW, Ray A. DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci. 2002;7:487–91. doi: 10.1016/s1360-1385(02)02355-5. [DOI] [PubMed] [Google Scholar]
- 51.Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008;13:350–8. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 52.Eamens AL, Smith NA, Curtin SJ, Wang MB, Waterhouse PM. The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA. 2009;15:2219–35. doi: 10.1261/rna.1646909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, Chen S, Hannon GJ, Qi Y. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–27. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–41. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 55.Takeda A, Iwasaki S, Watanabe T, Utsumi M, Watanabe Y. The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 2008;49:493–500. doi: 10.1093/pcp/pcn043. [DOI] [PubMed] [Google Scholar]
- 56.Wu L, Zhang Q, Zhou H, Ni F, Wu X, Qi Y. Rice MicroRNA effector complexes and targets. Plant Cell. 2009;21:3421–35. doi: 10.1105/tpc.109.070938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–90. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 58.Lanet E, Delannoy E, Sormani R, Floris M, Brodersen P, Crete P, Voinnet O, Robaglia C. Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell. 2009;21:1762–8. doi: 10.1105/tpc.108.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS. Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci U S A. 2005;102:3691–6. doi: 10.1073/pnas.0405570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 2006;20:759–71. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- 61.Prigge MJ, Wagner DR. The Arabidopsis SERRATE gene encodes a zinc-finger protein required for normal shoot development. Plant Cell. 2001;13:1263–79. doi: 10.1105/tpc.13.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Alexander AL, Carrington JC. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol. 2006;16:939–44. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- 63.Garcia D, Collier SA, Byrne ME, Martienssen RA. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr Biol. 2006;16:933–8. doi: 10.1016/j.cub.2006.03.064. [DOI] [PubMed] [Google Scholar]
- 64.Chitwood DH, Nogueira FT, Howell MD, Montgomery TA, Carrington JC, Timmermans MC. Pattern formation via small RNA mobility. Genes Dev. 2009;23:549–54. doi: 10.1101/gad.1770009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xie Z, Kasschau KD, Carrington JC. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr Biol. 2003;13:784–9. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- 66.Vaucheret H, Mallory AC, Bartel DP. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol Cell. 2006;22:129–36. doi: 10.1016/j.molcel.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vazquez F, Gasciolli V, Crete P, Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol. 2004;14:346–51. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 68.Axtell MJ, Snyder JA, Bartel DP. Common Functions for Diverse Small RNAs of Land Plants. Plant Cell. 2007 doi: 10.1105/tpc.107.051706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vaucheret H. AGO1 homeostasis involves differential production of 21-nt and 22-nt miR168 species by MIR168a and MIR168b. PLoS ONE. 2009;4:e6442. doi: 10.1371/journal.pone.0006442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mallory AC, Vaucheret H. ARGONAUTE 1 homeostasis invokes the coordinate action of the microRNA and siRNA pathways. EMBO Rep. 2009;10:521–6. doi: 10.1038/embor.2009.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during transacting siRNA biogenesis in plants. Cell. 2005;121:207–21. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 72.Liu X, Huang J, Wang Y, Khanna K, Xie Z, Owen HA, Zhao D. floral organs in carpels, a loss-of-function mutation uncovers the role of the MicroRNA160a gene in organogenesis and the mechanism of its expression regulation in Arabidopsis. The Plant Journal. 2010 doi: 10.1111/j.1365-313X.2010.04164.x. in press. [DOI] [PubMed] [Google Scholar]
- 73.Baker CC, Sieber P, Wellmer F, Meyerowitz EM. The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr Biol. 2005;15:303–15. doi: 10.1016/j.cub.2005.02.017. [DOI] [PubMed] [Google Scholar]