Abstract
The in vitro characterization of the catalytic activity of DesVII, the glycosyltransferase involved in the biosynthesis of the macrolide antibiotics methymycin, neomethymycin, narbomycin, and pikromycin in Streptomyces venezuelae, is described. DesVII is unique among glycosyltransferases in that it requires an additional protein component, DesVIII, for activity. Characterization of the metabolites produced by a S. venezuelae mutant lacking desVIII gene confirmed that desVIII is important for the biosynthesis of glycosylated macrolides, but can be replaced by at least one of the homologous genes from other pathways. The addition of recombinant DesVIII protein significantly improves the glycosylation efficiency of DesVII in the in vitro assay. When affinity-tagged DesVII and DesVIII proteins were co-produced in E. coli, they formed a tight (αβ)3 complex that is at least 103-fold more active than DesVII alone. The formation of the DesVII/DesVIII complex requires co-expression of both genes in vivo and cannot be fully achieved by mixing the individual protein components in vitro. The ability of DesVII/DesVIII system to catalyze the reverse reaction with the formation of TDP-desosamine was also demonstrated in a transglycosylation experiment. Taken together, our data suggest that DesVIII assists the folding of DesVII during protein production and remains tightly bound during catalysis. This requirement must be taken into consideration in the design of combinatorial biosynthetic experiments of new glycosylated macrolides.
Glycosyltransferases (GTs) are an important group of enzymes that catalyze the attachment of sugar moieties to acceptor molecules. Although there are over 75,000 putative glycosyltransferases in the Carbohydrate-Active enZymes database (CAZY, www.cazy.org), most of these proteins have never been purified and their catalytic roles have not been verified. Thus far, more than 170 GTs presumably involved in the biosynthesis of microbial secondary metabolites have been identified (1-3). Only a few of them have been characterized in vitro. However, the functions of a number of these GTs have been implicated by gene knockout and/or heterologous expression experiments. One of the first macrolide-related GTs characterized in vitro is OleD, which is responsible for the self-resistance of the oleandomycin producer Streptomyces antibioticus through glucosylation of the antibiotic oleandomycin (4). Among GTs directly involved in the glycosylation of aglycones in macrolide biosynthesis, DesVII was the first one shown to be active in vitro (5). DesVII catalyzes the attachment of TDP-d-desosamine (2) to 10-deoxymethynolide (1) or narbonolide (6) during the biosynthesis of methymycin (4), neomethymycin (5), narbomycin (7), and pikromycin (8) in Streptomyces venezuelae (Scheme 1) (6-8). Our previous in vitro study revealed an unusual requirement for DesVII activity: the need for an additional protein partner, DesVIII (5). Subsequently, two other GTs, EryCIII and AknS, have also been shown to require a DesVIII homolog, EryCII and AknT, respectively, for activityin vitro (9, 10). A similar requirement has now been established in vivo for TylM3/TylM2 and MydC/MycB pairs as well (11).
Scheme 1.
Last steps of the macrolide biosynthesis in Streptomyces venezuelae.
The translated gene sequences for these auxiliary proteins share moderate end-to-end homology with cytochrome P450 enzymes, yet lack the conserved P450 signature motif including an invariant Cys residue that coordinates the heme iron. Although the exact functions of these auxiliary proteins are unclear, the genes coding for DesVIII homologs are only found within the gene clusters encoding the biosynthetic pathway of amino-sugar-bearing metabolites. They typically reside directly upstream from the respective GT gene responsible for the transfer of the amino sugar. Thus, DesVIII and its homologs were initially considered to be necessary for the incorporation of the amino sugar moieties in these metabolites (5). For example, the glycosyltransferases responsible for the attachment of the two amino sugars in spiramycin of Streptomyces ambofaciens, Srm5 and Srm29, were shown to require auxiliary proteins homologous to DesVIII, Srm6 and Srm28, respectively (12). However, the attachment of the third sugar, mycarose, which is devoid of an amino functionality, is catalyzed by glycosyltransferase Srm38 alone.
Interestingly, it has been proposed that AknT, a homolog of DesVIII from the anthracycline biosynthetic pathway, functions as a regulatory subunit that transiently interacts with glycosyltransferase AknS and increases the affinity of AknS for its substrate, thereby facilitating the glycosylation reaction (10). Studies of the EryCIII/EryCII glycosylation system demonstrated that the erythromycin GT (EryCIII) remains fully active in vitro after removal of its auxiliary protein (EryCII) from the pre-incubation mixture (9). These observations contradict the AknS/AknT hypothesis and suggest that EryCII has a chaperone-like function, perhaps facilitating a one-time conformational change that activates EryCIII. Clearly, a better understanding of the exact role of these GT auxiliary proteins is necessary.
In two previous communications, we had reported our preliminary observations of the requirement of DesVIII for DesVII activity (5, 9). Described herein is a full account of the genetic and biochemical characterization of DesVII and DesVIII. We co-expressed the desVII and desVIII genes and found that DesVII and DesVIII form a stable protein complex. Steady-state kinetics comparison of DesVII and the DesVII/DesVIII complex allowed a quantitative evaluation of the activation of DesVII by DesVIII. Our results suggest that DesVIII assists folding of the DesVII polypeptide. However, unlike chaperones, it remains bound to DesVII during catalysis forming a tight DesVII/DesVIII complex. Although the formation of DesVII/DesVIII complex is essential for the catalytic activity, DesVIII is unlikely involved in catalysis directly.
Materials and Methods
General
Enzymes used for the cloning experiments were obtained from Invitrogen (Carlsbad, CA). Antibiotics and biochemicals used in this study were obtained from Sigma-Aldrich (St. Louis, MO) or Fisher Scientific (Pittsburgh, PA) with the exceptions noted below. The growth media components were purchased from BD Diagnostics System (Franklin Lakes, NJ). The 32P labeled nucleotides and the Multiprime DNA Labeling System used for DNA probe labeling in the Southern blot hybridization analysis were obtained from Amersham Biosciences (presently GE Healthcare, Chalfont St. Giles, United Kingdom). Protein concentrations were determined using the Bradford method (13) with bovine serum albumin (BSA) as the standard. The NMR spectra were acquired on a Varian Unity plus 300 or a Varian Inova 500 spectrometer, and chemical shifts are reported in ppm (δ, referenced to the solvent) and coupling constants in hertz (Hz). Flash chromatography was performed on Lagand Chemical silica gel (230-400 mesh) by elution with the specified solvents. Analytical thin-layer chromatography (TLC) was carried out on Polygram Sil G/UV254 plates (0.25 mm, Macherey-Nagel). DNA and protein sequence analysis were done using Vector NTI® Suite Software by InforMax, Inc (presently Invitrogen, Carlsbad, CA).
Plasmids Vectors, and DNA Manipulations
Cosmid pLZ4 (derivative of pNJ1 vector), which harbors the desosamine biosynthetic cluster and part of the polyketide synthase cluster of Streptomyces venezuelae, was constructed in a previous study (6, 14). Cosmid pFD666 was the source of the kanamycin resistance gene (15, 16). Plasmid pKC1139 was used for the conjugal transfer of DNA to S. venezuelae (17), and vector pDHS617 was used in the complementation experiments to introduce exogenous gene(s) into the S. venezuelae KdesVIII mutant strain (18). Cosmid pZHG4 was used as the template for PCR amplification of the tylM3 gene of Streptomyces fradiae (19). Expression vectors of pET series were purchased from Novagen (EMD Chemicals, Gibbstown, NJ). The general methods and protocols for recombinant DNA manipulations are described by Sambrook et al. (20), and those involving Streptomyces strains are described by Hopwood et al. (21) and Kieser et al. (22).
Bacterial Strains
S. venezuelae ATCC 15439, Streptomyces peucetius ATCC 29050, and Streptomyces erythraea ATCC 11635 were acquired from American Type Culture Collection (ATCC, Manassas, VA) as freeze-dried pellets and were revived according to the instructions provided by ATCC. Escherichia coli DH5α was used as a routine cloning host in this study. E. coli BL21(DE3), E. coli BL21 Star(DE3) (Invitrogen) and E. coli Rosetta 2(DE3) (EMD Chemicals) were used as hosts for gene expression as indicated. E. coli S17-1 (23) was employed as the donor strain for conjugal transfer to S. venezuelae.
Cloning of desVII Gene
The desVII gene was amplified by the PCR from cosmid pLZ4 using 5′-GGCCATATGCGCGTCCTGCTGACCTCGTTC-3′ as the forward primer (NdeI restriction site is underlined) and 5′-CGCCTCGAGGTGCCGGGCGTCGGCCGG-3′ as the reverse primer (XhoI restriction site is underlined). The amplified 1280 bp fragment was digested with NdeI and XhoI and ligated into the corresponding sites of pET24b(+) vector. Sequencing of expression plasmid pHC29 revealed that the nucleotide at position 1784 of the desVII gene is a T and not a C as previously reported (see NCBI nucleotide database, sequence AF079762). This result was confirmed by DNA sequencing of the corresponding fragment in the genomic DNA of S. venezuelae. Thus, the sequence of DesVII should be corrected to have a tyrosine (codon TAT) instead of a histidine (codon CAT) at position 104. This is consistent with the conservation of an aromatic amino acid residue, usually tyrosine, at this position in many other homologous glycosyltransferases (see Supporting Information for corrected sequence).
Expression of desVII and Purification of DesVII-C-His6 Protein
Plasmid pHC29 was used to transform E. coli BL21 Star (DE3) for expression of desVII as a C-terminally His6-tagged protein (5). Comparable results were obtained with E. coli BL21 (DE3) as the host (5). The protein DesVII-C-His6 was expressed and purified as previously described (7) with several minor modifications as outlined below. An overnight culture of E. coli BL21 Star (DE3)/pHC29 was used to inoculate LB medium (1 to 100 dilution) containing kanamycin (25 μg/mL). The cultures were incubated with vigorous shaking at 37 °C until the OD600 reading reached approximately 0.6. The temperature was gradually lowered to 16 °C before the culture reached the desired OD600 reading. The cultures were then induced with 0.3 mM isopropyl-β-thiogalactoside (IPTG) and incubation was continued at 16 °C for an additional 16–18 h. The cells were harvested by centrifugation (6,000 g, 4 °C, 10 min) and used as described below.
The protein DesVII-C-His6 was purified using affinity chromatography on Ni-NTA agarose following the purification protocol suggested by the manufacturer (Qiagen, Valencia, CA). All steps were carried out at 4 °C. Briefly, the cell pellet was re-suspended in lysis buffer (50 mM sodium phosphate, 300 mM NaCl, 10% glycerol, 5 mM imidazole, pH 8.0) containing 0.5 mM phenylmethylsulphonyl fluoride (PMSF). Lysozyme was added to a final concentration of 1 mg/mL, and the resulting suspension was incubated on ice for 30 min. The cells were disrupted by sonication in 10 s bursts (36 ×) with 20 s cooling periods between bursts. Cell debris was removed by centrifugation (16,000 g, 4 °C, 30 min). Ni-NTA resin, pre-washed with the lysis buffer, was added to the crude extract, and the mixture was incubated with a slow agitation at 4 °C for 3–4 h. The suspension was loaded onto a column, unbound proteins were removed by washing with lysis buffer containing 15–30 mM imidazole, and bound proteins were eluted with the same buffer containing 250 mM imidazole. The desired fractions were pooled and dialyzed against three changes of 1 L of 50 mM Tris•HCl buffer (pH 8) containing 10% glycerol. The purified DesVII-C-His6 protein was concentrated (precipitated particles removed by centrifugation), aliquotted, flash frozen with liquid nitrogen, and stored at –80 °C. DesVII-C-His6 remains soluble at 100 mg/mL. Typically, approximately 100 mg of DesVII-C-His6 were obtained from a liter of culture.
Glycosyltransferase Activity Assay
A typical assay (50 μL) contained 50 mM Tris•HCl buffer (pH 8), 1–2 mM 10-deoxymethynolide (1, from 100 mM stock in methanol), 1–2 mM TDP-d-desosamine (2), 1–5 μL of DesVII-C-His6 (100 mg/mL, 2 mM), and 5 μL of DesVIII (varied concentrations) (5, 7). The protein portion was substituted with the same amount of buffer in the control reactions. The mixture was incubated at 29 °C. To monitor the progress of the reaction, 20 μL aliquots of the reaction mixture were removed at different time points and extracted with equal volumes of chloroform. The organic extract of each sample was analyzed by TLC, which was developed in a chloroform:methanol:25% NH4OH (90:9.9:0.1) solvent system and stained using a vanillin solution (0.75% vanillin, 1.5% v/v H2SO4, methanol). Alternatively, aliquots of reaction mixture, removed at appropriate time intervals, were quenched with two volumes of cold methanol and the precipitated proteins were removed by centrifugation. Approximately 25 μL of the resulting sample was injected into an HPLC system equipped with the reverse phase analytical C18 column (Microsorb-MV C18, 4.5 × 250 mm, 5 μm, Varian). Compounds were eluted isocratically with 40% acetonitrile in an aqueous buffer (14 mM triethylamine adjusted to pH 2.5 with trifluoroacetic acid) at a flow rate of 1 mL/min. The detector was set at 235 nm. Under these conditions substrate 1 and product 10-deoxymethymycin (3) eluted at approximately 15.0 and 10.5 min, respectively.
Construction of desVIII-disrupted S. venezuelae Mutant (KdesVIII)
The desVIII gene in the genome of S. venezuelae was replaced with the kanamycin resistance marker through double-crossover recombination. To construct the recombination plasmid pDesVIII-K, two fragments flanking the upstream and downstream segments of the desVIII gene were amplified by PCR using cosmid pLZ4 as a template. The following pairs of primers, 5′-CCGGTCTAGACGTGGATCCAGTGGATGC-3′ and 5′-CGCGGAATTCCAGCTACTTCTTCC-3′ (respective engineered restriction sites XbaI and EcoRI are underlined), and 5′-GCGCAAGCTTGGTGTGCATCGGGTAGTTGC-3′ and 5′-CCGGCTGCAGTCAGCAGCTCCTGAGACAC-3′ (respective engineered restriction sites HindIII and PstI are underlined) were used for amplification of the upstream (0.9 kb) and the downstream fragments (1.1 kb), respectively. The fragment (1.38 kb) containing the kanamycin resistance cassette was PCR-amplified from plasmid pFD666 using primers 5′-CGCGTCTAGATACCTACAGCGTGAGC-3′ and 5′-CGCGCTGCAGCCACGAATTAGCC-3′ (respective engineered restriction sites XbaI and PstI are underlined). The PCR products were digested with the corresponding restriction enzymes and the three fragments were ligated into the EcoRI and HindIII sites of pUC119 to give plasmid pDesVIII-d1 in a single reaction. The 3.3 kb insert of pDesVIII-d1 was then removed by digestion with EcoRI and HindIII and cloned into the same sites of pKC1139 to give plasmid pDesVIII-K.
This plasmid was used to transform E. coli S17-1. The resulting strain was used in the subsequent conjugal transfer of pDesVIII-K to S. venezuelae. The conjugal transfer experiments, double-crossover mutant screening, strain propagation, and storage were carried out according to a published procedure (17) with minor modifications (24). Accordingly, a mutant strain S. venezuelae KdesVIII having nucleotides 122–1195 of its desVIII gene (out of 1209 bp) in the chromosomal DNA replaced by the kanamycin resistance marker was generated. The anticipated replacement was confirmed by Southern blot hybridization analysis. The genomic DNA of the wild type S. venezuelae and that of the KdesVIII-92 mutant were isolated (21), digested with BamHI and PstI, separated by agarose gel electrophoresis, and trans-blotted onto a nylon membrane using standard procedures (20). The DNA probes were labeled with 32P-containing nucleotides using a Multiprime DNA Labeling System (Amersham Biosciences, presently GE Healthcare, Chalfont St. Giles, United Kingdom). As expected, when labeled kanamycin resistance gene was used as the probe, a prominent band of 1.4 kb was visible on the autoradiogram for the mutant DNA, but no hybridization with the wild type DNA was observed. In a separate experiment, bands corresponding to the BamHI-PstI fragment of the mutant and the wild type DNA (4.7 kb and 5.5 kb, respectively) were detected when using the labeled 1.0 kb fragment of desR gene located downstream of desVIII, confirming the desired mutation.
Preparation of S. venezuelae KdesVIII Strains Expressing desVII, desVIII, tylM3, eryCII, and dnrQ
Plasmids pDesVII, pDesVIII, pTylM3, pEryCII, and pDnrQ were constructed for the expression of desVII, desVIII, tylM3, eryCII, and dnrQ genes, respectively, in the S. venezuelae KdesVIII mutant. The fragments containing these genes were amplified by PCR using the primers listed in Table 1. These primers were designed to contain restriction sites for EcoRI (underlined in the forward primer) and XbaI (underlined in the reverse primer) for the subsequent cloning. One base in the forward primer for eryCII cloning was modified (T→A, a silent mutation shown in brackets) to eliminate an EcoRI restriction site in the gene. Cosmid pLZ4 was used as the template for the amplification of the desVII and desVIII genes, whereas cosmid pZHG4 was the template for the PCR amplification of the tylM3 fragment. Genomic DNA of S. erythraea and S. peucetius, digested with BamHI, was used as the template for the amplification of eryCII and dnrQ genes, respectively. The PCR products were cloned into the EcoRI-XbaI sites of the vector pDHS617, a derivative of pOJ446 containing an apramycin resistance marker (17). It also contains the promoter sequence pikA of the methymycin/neomethymycin cluster for expression of genes in S. venezuelae (6). Each of the resulting plasmids, pDesVII, pDesVIII, pTylM3, pEryCII, and pDnrQ, was used to transform E. coli S17-1 and then introduced into the KdesVIII-92 mutant via conjugal transfer as described above. Selected colonies of S. venezuelae desVII/KdesVIII, desVIII/KdesVIII, tylM3/KdesVIII, eryCII/KdesVIII, and dnrQ/KdesVIII mutants were propagated on SPA plates (24) containing apramycin (50 μg/mL) and kanamycin (50 μg/mL). For all cases, several strains were selected for each mutant and their macrolide production was similar within each set.
Table 1.
Primers Used for the Amplification of the Heterologous Genesa
| Gene | Size (kb) | Primers (5′ to 3′) | Restriction Sites |
|---|---|---|---|
| desVII | 1.3 | GGCCGAATTCTAAGGAAGGACACGACG | EcoRI |
| GCGCTCTAGATACAGGGGTGAG | XbaI | ||
| desVIII | 1.3 | GGCCGAATTCTGAAGAGATGGCAGAAC | EcoRI |
| GCGCTCTAGACACACGGGACGGTCTGA | XbaI | ||
| tylM3 | 1.3 | GGCCGAATTCTGAGAGGAGAGGCCGTGAACA | EcoRI |
| CGCGTCTAGAGTGCCCTTCTCACT | XbaI | ||
| eryCII | 1.1 | GGCCGAATTCTGAGGAGGGAAT[A]CATGACCAC | EcoRI |
| GCGCTCTAGAGTTACCTCAGAGCTC | XbaI | ||
| dnrQ | 1.4 | GGCCGAATTCTGAGGAGCACGACGATG | EcoRI |
| GCGCTCTAGAGCACCTTCATGGGGTCAC | XbaI | ||
The respective restriction sites are underlined in the sequences of the primers.
Characterization of Metabolite Production by S. venezuelae Strains
The above recombinant strains were grown under conditions described by Zhao et al. (24) based on the procedure of Cane et al., favoring the formation of 12-membered ring macrolides (25). The metabolite production was first analyzed by TLC using conditions described above. Methynolide and derivatives showed a green color after staining with vanillin, whereas neomethynolide derivatives and 10-deoxymethynolide derivatives were stained orange and grey-blue, respectively. To isolate glycosylated compounds produced by S. venezuelae dnrQ/KdesVIII mutant, the crude products from 3 L of a vegetative culture (yellow oil, approximately 120 mg) were first subjected to flash chromatography on silica gel using a gradient of 0–25% methanol in chloroform, and then purified by HPLC on a semi-preparative C18 column (Econosil, 10 × 250 mm, 10 μm) eluted isocratically with 40% acetonitrile in 57 mM ammonium acetate buffer (pH 6.7).
The following compounds were isolated: 10-deoxymethynolide (1, 40 mg), methynolide (9) and neomethynolide (10) (2–5 mg each), methymycin (4) and neomethymycin (5) (3 mg each), pikromycin (8, 4 mg crude). The spectral data of 1, 4, 5 and 8 are identical to those reported. The doubly hydroxylated product, novamethymycin (11, approximately 7 mg) (26), was also purified from the crude extract.
1H NMR (500 MHz, CDCl3) of novamethymycin (11): δ 6.65 (1H, d, J = 16.0, 9-H), 6.31 (1H, d, J = 16.0, 8-H), 4.67 (1H, d, J = 9.0, 11-H), 4.29 (1H, d, J = 7.0, 1′-H), 4.13 (1H, dq, J = 9.0, 6.5, 12-H), 3.61 (1H, d, J = 10.5, 3-H), 3.52 (1H, dqd, J = 11.0, 6.5, 1.5, 5′-H), 3.32 (1H, dd, J = 10.5, 7.0, 2′-H), 2.88–2.80 (2H, m, 3′-H and 2-H), 2.57 (1H, m, 6-H), 2.44 (6H, s, -NMe2), 1.76 (1H, ddd, J = 12.0, 3.5, 1.5, 4′-Heq), 1.67 (1H, bt, J = 13.5, 5a-H), 1.52 (3H, s, 10-Me), 1.44 (1H, m, 5b-H), 1.41 (3H, d, J = 6.5, 2-Me), 1.32 (1H, bq, J = 12.0, 4′-Hax), 1.25 (3H, d, J = 6.5, 5′-Me), 1.20 (1H, m, J = 6.5, 4-H), 1.184 (3H, d, J = 6.5, 12-Me), 1.178 (3H, d, J = 7.0, 6-Me), 1.01 (3H, d, J = 6.5, 4-Me). 13C NMR (125 MHz, CDCl3) of 11: δ 204.1 (C-7), 174.1 (C-1), 148.3 (C-9), 125.4 (C-8), 104.7 (C-1′), 85.5 (C-3), 75.5 (C-10), 74.2 (C-11), 70.0 (C-2′), 68.9 (C-5′), 67.8 (C-12), 65.9 (C-3′), 45.2 (C-6), 44.0 (C-2), 39.8 (NMe2), 33.8 (C-4), 33.7 (C-5), 29.1 (C-4′), 21.07 and 21.04 (5′-Me and 12-Me), 20.2 (10-Me), 17.6 (6-Me), 17.4 (4-Me), 15.9 (2-Me). High resolution FAB-MS of 11: calculated for C25H43NO8 (M+H)+ 486.3067, found 486.3076.
Preparation of DesVIII-C-His6 Protein
Gene desVIII was amplified by PCR using pLZ4 cosmid as the template and the following oligonucleotides as primers: upstream (5′-GCCGCATATGACCGACGACCTGAC-3′, NdeI restriction site is underlined) and downstream (5′-GCGCAAGCTTGGAGCTGCTGAC-3′, HindIII restriction site is underlined). The cloned fragment was ligated into the NdeI-HindIII sites of pET24b(+), and the resulting plasmid, pDesVIII-C-His6, was used to transform E. coli BL21(DE3). An overnight culture of E. coli BL21/pDesVIII-C-His6 was used to inoculate (1 to 50 dilution) LB media (25 μg/mL kanamycin) and grown at 37 °C until the OD600 reading reached 0.5. The production of DesVIII-C-His6 was induced by 1 mM IPTG, and incubation was continued for an additional 5 h at 37 °C. Cells were harvested by centrifugation, and cell pellet was resuspended in the lysis buffer (50 mM sodium phosphate, pH 8, 0.3 M NaCl, 10% glycerol, 5 mM imidazole, 0.4 mM PMSF) in the presence of 1 mg/mL lysozyme. After incubation at 4 °C for 30 min, cells were disrupted by sonication as described above, and the supernatant was removed after centrifugation. The desired protein was entirely in the pellet as determined by SDS-PAGE analysis.
The purification of DesVIII-C-His6 protein under denaturing conditions was first attempted following Qiagen's protocol. Briefly, the pellet was washed with the lysis buffer, and proteins were solubilized in 100 mM sodium phosphate, 10 mM Tris, 8 M urea, pH 8 at room temperature for 1 h. Cell debris was removed by centrifugation, and crude proteins were subjected to the Ni-NTA affinity purification. Unfortunately, DesVIII-C-His6 did not bind to the nickel resin under denaturing conditions.
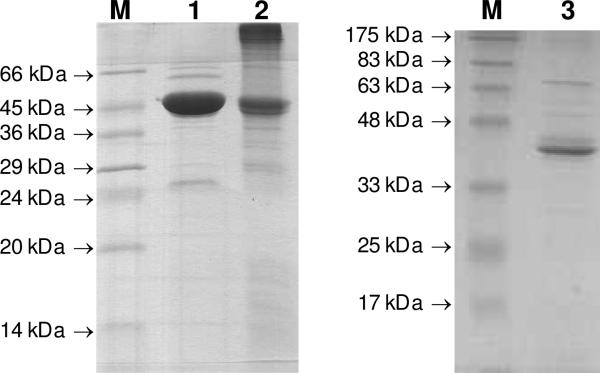
To renature the protein, urea was removed by dialysis against the lysis buffer, and the protein solution was subsequently clarified by centrifugation, and incubated with Ni-NTA resin. After overnight incubation at 4 °C, the mixture was subjected to the eluting protocol as described for the purification of DesVII-C-His6. The collected fractions were dialyzed against 50 mM Tris•HCl buffer (pH 8) containing 10% glycerol, and concentrated until precipitate began to form. Attempts to concentrate further led to a significant protein precipitation. The final concentration of the purified DesVIII-C-His6 was approximately 1 mg/mL (Figure 1, lane 2).
Figure 1.
SDS-PAGE of purified DesVII-C-His6 (lane 1, major band of 47 kDa), DesVIII-CHis6 (lane 2, major band of 44 kDa), and DesVIII-Tev (lane 3, major band of 42 kDa). Lane M shows the molecular weight markers with the indicated weights.
Preparation of MBP-tagged DesVIII-Tev Protein
The preparation of DesVIII-Tev protein was described in brief in a previously report (7), experimental details are provided below. The desVIII gene was amplified by the PCR using pLZ4 cosmid as the template and 5′-GCCGCATATGACCGACGACCTGAC-3′ (NdeI restriction site is underlined) and 5′-GCGCAAGCTTCAGGAGCTGCTGACCG-3′ (HindIII restriction site is underlined) as the forward and reverse primers, respectively. The PCR product was cloned into the NdeI-HindIII sites of the MalE-pET vector (27), and the resulting plasmid pDesVIII-MalEpET was used to transform the expression host E. coli Rosetta 2(DE3). An overnight culture of E. coli Rosetta 2(DE3)/pDesVIII-MalEpET was used to inoculate LB media (1 to 100 dilution, 25 μg/mL kanamycin), and the cells were grown at 37 °C until the OD600 reading reached 0.5– 0.6. The temperature was then lowered to 18 °C, and the production of the protein was induced by 0.3 mM IPTG for 15 h. The cells were harvested by centrifugation. As expected, the overproduced fusion protein, MBP-DesVIII (88.9 kDa), contains the maltose binding protein (MBP) and a His10 tag at its N-terminus (confirmed by Western blot analysis). This region is separated from the DesVIII sequence by the Tev protease cleavage site. The produced MBPDesVIII was soluble.
MBP-DesVIII was subjected to Ni-NTA purification under native conditions following the protocol described for DesVII-C-His6 purification. A protease inhibitor cocktail consisting of a 100 μL aliquot of an aqueous mixture of leupeptin (3 mg), antipain (5 mg), turkey trypsin inhibitor (50 mg), benzamidine-HCl (25 mg), Pefabloc SC (12.5 mg), and aprotinin (2.5 mL) (stored at –20 °C), and a 100 μL aliquot of a mixture of chymostatin (10 mg) and pepstatin A (5 mg) in 2 mL of DMSO (stored at –20 °C) was added to each 100 mL of the lysis buffer used for re-suspension of the cell pellet and during protein elution. Despite using protease inhibitors, there was still a considerable amount of premature cleavage of MBP-DesVIII, most notably at or in the vicinity of the Tev cleavage site.
Preparation of Untagged DesVIII-Tev Protein
To remove the MBP tag from MBP-DesVIII, the recombinant His6-tagged Tev protease was added to MBP-DesVIII to approximately 3% of the total protein concentration. The solution was dialyzed against 1 L of 50 mM sodium phosphate buffer (pH 8) containing 0.1 M NaCl and 10% glycerol, with two changes of buffer overnight to remove imidazole and simultaneously allow for the cleavage of the fusion protein. The complete digestion of MBP-DesVIII was confirmed by the appearance of two major bands, 42.2 kDa for DesVIII-Tev and 46.7 kDa for MBP, on SDS-PAGE. The dialyzed solution was incubated with Ni-NTA resin. Proteins were then eluted with 15 mM imidazole followed by 250 mM imidazole. The fractions containing MBP (250 mM imidazole) were discarded, and the flow-through and 15 mM imidazole fractions containing untagged DesVIII-Tev were combined and dialyzed against the same dialysis buffer as above. While the major protein component was DesVIII-Tev, this sample still contained small amounts of MBP and uncleaved fusion protein that could be detected by Western blot analysis but not by SDS-PAGE. To remove MBP, DesVIII-Tev was concentrated and loaded onto an amylose column. Unbound DesVIII-Tev was eluted with 25 mM Tris•HCl buffer, pH 8, containing 0.1 M NaCl and 10% glycerol, concentrated, and stored in the same buffer (Figure 1, lane 3). The fractions eluted with 20 mM maltose contained mostly MBP. The concentration of DesVIII-Tev was 5.5 mg/mL as estimated by the Bradford assay and is free of MBP and the fusion MBP-DesVIII as assessed by a Western blot. Further purification using size exclusion chromatography was not possible due to protein precipitation on the column.
Activation of DesVII-C-His6 by DesVIII-Tev and Glycosyltransferase Activity Assay
Incubation of DesVII-C-His6 (30 μmol) and DesVIII and DesVIII-Tev (17μmol) was carried out in 1.4 mL of a buffer solution containing 50 mM sodium phosphate, pH 8, 0.3 M NaCl, and 10% glycerol with gentle agitation at 4 °C for 2 h. Ni-NTA resin (0.7 mL) was then added and incubation continued with gentle agitation at 4 °C for an additional 1 h. The proteins were eluted with 1-mL aliquots of the incubation buffer containing 0, 10, 20, and 250 mM imidazole, consecutively. SDS-PAGE analysis of the collected fractions showed the presence of DesVIII-Tev in the 20 and 30 mM imidazole fractions and the “activated DesVII-C-His6” (designated DesVII*) in the 250 mM imidazole fraction. The DesVII* sample was dialyzed against 50 mM Tris•HCl, pH 8, 10% glycerol buffer using a Slide-A-lyzer dialysis cassette (Pierce, a subsidiary of Thermo Fisher Scientific, Waltham, MA), and concentrated using Microcon centrifugal filter YM-30 (Millipore, Billerica, MA). The final concentration of DesVII* was adjusted to approximately 0.2 mM (equivalent to that of the DesVII-C-His6 stock used for the following assay).
Proteins DesVII* and DesVII-C-His6 (as isolated) were also analyzed by HPLC using an Amersham Biosciences AKTA purifier equipped with the reverse phase analytical C4 column (Kromasil C4, 4.5 × 250 mm, 5 μm). The proteins were separated by running a 0–100% linear gradient of buffer A (0.085% trifluoroacetic acid (TFA), 10% water in acetonitrile) in buffer B (0.1% TFA in water) at 1 mL/min over 90 min. The elution profile was monitored at 215 and 280 nm. The proteins were analyzed using liquid chromatography (LC)-electrospray ionization-tandem mass spectrometry (MS/MS). Experiments were performed using a microcapillary LC system (Agilent 1100 series) on-line with an Esquire ion trap mass spectrometer (Bruker, Newark, DE). LC-ESI-MS results: DesVII-C-His6 calcd. 47511 Da, found 47505 Da, DesVII* found 47503 Da.
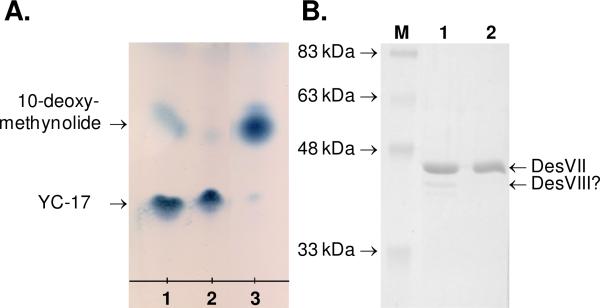
The activity of DesVII* was determined using 2 mM 10-deoxymethynolide (1), 6 mM TDP-d-desosamine (2) in a total volume of 25 μL of 50 mM Tris•HCl buffer, pH 8. The reaction was initiated by the addition of the following protein component to the final concentrations: 1) 24 μM DesVII*; 2) 24 μM DesVII-C-His6, 10 μM DesVIII-Tev; 3) 24 μM DesVII-C-His6. The mixture was incubated at 29 °C for 2 h. Macrolides were extracted with 20 μL of chloroform and analyzed by TLC as described above (Figure 3A).
Figure 3.
A. TLC analysis of glycosyltransferase activity of DesVII* (lane 1), DesVII-C-His6 + DesVIII-Tev (lane 2), and DesVII-C-His6 (lane 3). B. SDS-PAGE analysis of DesVII* (lane 1) and DesVII-C-His6 (lane 2). Lane M shows the molecular weight markers with the indicated weights.
Co-Production of DesVII-C-His6 and DesVIII-N-Stag in E. coli
The desVII gene was sub-cloned into the pET21b(+) vector, which confers ampicillin resistance, for expression as a C-terminally His6-tagged protein. Accordingly, a fragment containing the desVII gene (1.28 kb) was excised from plasmid pHC29 with restriction endonucleases NdeI and XhoI and ligated into corresponding sites of pET21b(+) vector resulting in plasmid pDesVII-pET21. The desVIII gene was amplified by PCR using the pLZ4 cosmid as the template and 5′-GGCCCCATGGTGACCGACGACCTGACG-3′ (NcoI restriction site is underlined) and 5′-GCGCAAGCTTCAGGAGCTGCTGACCG-3′ (HindIII restriction site is underlined) as the forward and reverse primers, respectively. The amplified fragment was cloned into the NcoI-HindIII restriction sites of vector pET29b(+) which confers kanamycin resistance to obtain plasmid pDesVIII-N-Stag. Plasmids pDesVII-pET21 and pDesVIII-N-Stag were used to transform E. coli BL21 Star (DE3). The desired transformants were selected on LB agar plates containing ampicillin (100 μg/mL) and kanamycin (50 μg/mL).
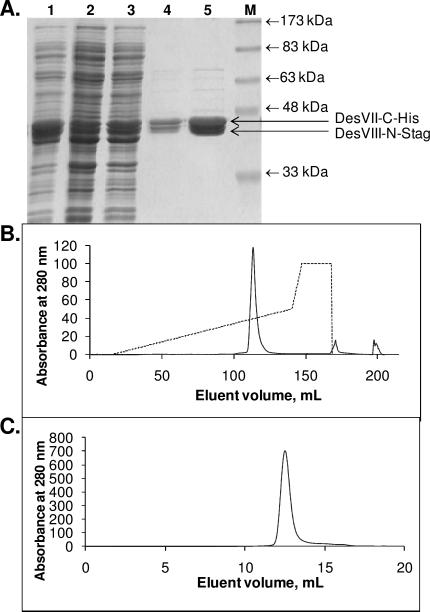
Strain E. coli BL21 Star (DE3)/pDesVII-pET21+pDesVIII-N-Stag was grown as described above for E. coli BL21 Star(DE3)/pHC29 with the exception that both ampicillin and kanamycin were added (60 μg/mL and 25 μg/mL, respectively) and the induction temperature was 18 °C. The expressed proteins, DesVII-C-His6 and DesVIII-N-Stag, were co-purified to near homogeneity using Ni-NTA resin and following the procedure used for the purification of DesVII-C-His6. A protease inhibitor cocktail (as described above) was used during purification. Typically, approximately 120 mg of protein (DesVII-C-His6 and DesVIII-N-Stag, combined) was obtained from 6 L of culture (see Figure 4A). Proteins were concentrated to 50–100 mg/mL, flash frozen with liquid nitrogen, and stored at –80 °C. There was no obvious loss of glycosyltransferase activity upon prolonged storage or several freeze/thaw cycles. Attempts to separate proteins using S-agarose resin were not successful and resulted in co-elution of proteins.
Figure 4.
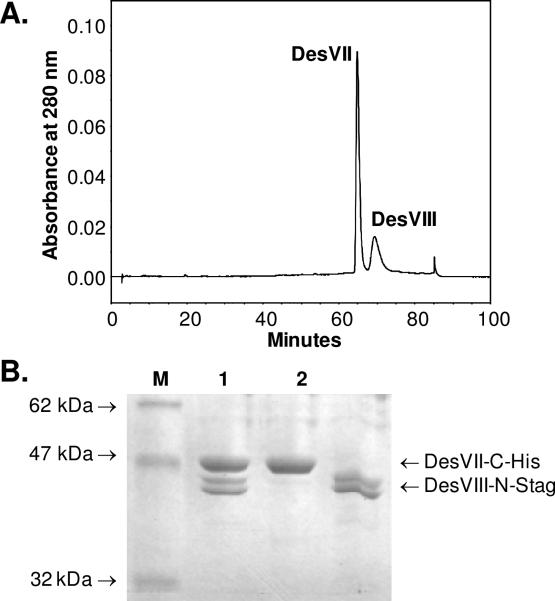
Co-purification of DesVII-C-His6 and DesVIII-N-Stag. A. SDS-PAGE analysis of Ni-NTA purification steps. Lane 1: crude extract, lanes 2–5: fractions eluted with increasing imidazole concentrations, lane M: molecular weight markers with the indicated weights. B. Chromatogram of protein elution (lane 5 in A) from FPLC MonoQ anion-exchange column with the KCl gradient shown as a dashed line. C. Chromatogram of protein elution from FPLC Superdex 200 size-exclusion column.
Elution of DesVII-C-His6 /DesVIII-N-Stag Complex From an Anion Exchange Column
A protein sample of DesVII-C-His6/DesVIII-N-Stag was subjected to separation by anion exchange chromatography using FPLC equipped with a MonoQ HR 10/10 column. Proteins were loaded on the column with buffer A (50 mM Tris•HCl, pH 8, 15% glycerol) at 3 mL/min over 2 column volumes and then eluted with a 0–50% gradient of buffer B (buffer A plus 1 M KCl) in buffer A over 18 column volumes. A single major peak eluted at approximately 350 mM KCl (Figure 4B) was observed (with detector set at 280 nm). It contained both DesVII-C-His6 and DesVIII-N-Stag as determined by SDS-PAGE analysis.
Determination of the Molecular Mass of DesVII-C-His6 /DesVIII-N-Stag Complex Using Size Exclusion Chromatography
A protein sample of DesVII-C-His6/DesVIII-N-Stag was subjected to size exclusion chromatography using FPLC equipped with a Superdex 200 10/300 GL column. Proteins were eluted isocratically using 50 mM sodium phosphate, pH 7, 0.15 M NaCl at 0.4 mL/min. Again, a single major peak, containing both proteins as judged by SDS-PAGE, eluted from the column (Figure 4C). The native molecular mass of the protein complex was determined using the following protein standards (Sigma MW-GF-200, 250 μL each): 1:1 mixture of cytochrome c (12.4 kDa, 4 mg/mL) and β-amylase (200 kDa, 8 mg/mL), 1:1 mixture of carbonic anhydrase (29 kDa, 6 mg/mL) and alcohol dehydrogenase (150 kDa, 10 mg/mL), and bovine serum albumin (BSA, 66 kDa, 10 mg/mL). Blue dextran (2000 kDa, 250 μL of 2 mg/mL) was used to determine the void volume (υ0) of the column. The bed volume of the column (υc) was 24 mL as per the manufacturer's specifications (Amersham Biosciences). Two samples of DesVII-C-His6/DesVIII-N-Stag having concentrations of 7 and 10 mg/mL (250 μL) were examined. The elution volumes (υe, mL) were as follows: blue dextran, 8.5; DesVII-C-His6/ DesVIII-N-Stag, 12.6 ± 0.03; β-amylase, 12.9; alcohol dehydrogenase, 13.8; BSA, 15.0; carbonic anhydrase, 16.9; and cytochrome c, 17.9. The molecular mass (MM) of the two DesVII-CHis6/DesVIII-N-Stag samples deduced from the plot log10(MM) versus (υe–υ0)/(υc–υ0) was found to be 251 kDa and 244 kDa, respectively. Since the calculated molecular mass of the heterodimer of DesVII-C-His6/DesVIII-N-Stag is 92559 Da (DesVII-C-His6: 47511 Da, DesVIII-N-Stag: 45048 Da), these data indicated that DesVII-C-His6/DesVIII-N-Stag likely exists as a trimer (2.6–2.7×) of heterodimers.
HPLC Analysis of Denatured Proteins
Samples of DesVII-C-His6/DesVIII-N-Stag were denatured by the addition of urea to a final concentration of 6–8 M followed by an incubation period at room temperature for at least 30 min. Water was added to these samples to lower the urea concentration to 2 M prior to HPLC analysis. Protein samples (0.1–0.5 mg in 20–100 μL) were injected into an HPLC Microsorb-MV C18 (Varian, 4.6 × 250 mm) column equilibrated with 0.1% trifluoroacetic acid (TFA) in water. After 1 min, proteins were eluted with a 0–70% gradient of 0.085% TFA, 10% water in acetonitrile over 69 min. The flow rate was 1 mL/min and the detector was set at 280 nm. Two major peaks, eluting at 65 min and 69 min (Figure 5A), were collected and solvent was evaporated under reduced pressure. The resulting dry residue was re-suspended in 2 M urea and analyzed by SDS-PAGE (Figure 5B). The peak at 65 min corresponds to DesVII-C-His6 and the peak at 69 min to DesVIII-N-Stag. The area ratio of these two peaks was converted to the mole ratio of proteins based on the calculated molar absorptivity (using Vector NTI software) where one A280 unit corresponds to 0.73 mg/mL of DesVII-C-His6 or 1.71 mg/mL of DesVIII-N-Stag. Results were reproducible regardless of the exact amount of protein injected. To remove any residual proteins from the column, the HPLC column was cleaned by injection of an aliquot (100 μL) of 2 M urea followed by elution with the gradient program described above. This procedure was repeated four times until the baseline was free of protein traces. A ThermoFinnigan LCQ ion trap mass spectrometer was used for ESI-MS analysis of the protein samples separated on HPLC C18 column. The LC-MS analysis was performed by the Analytical Instrumentation Facility Core of the College of Pharmacy, University of Texas at Austin.
Figure 5.
HPLC analysis of the DesVII-C-His6 and DesVIII-N-Stag complex formed upon their co-expression in E. coli. A. HPLC trace of the denatured complex. B. SDS-PAGE of the protein complex prior to HPLC separation (lane 1) and the HPLC fractions corresponding to peaks at 65 and 69 min (lanes 2 and 3, respectively). Lane M shows the molecular weight markers with the indicated weights.
Treatment of DesVII-C-His6/DesVIII-N-Stag with Thrombin
To remove the N-terminal Stag from DesVIII-N-Stag, the DesVII-C-His6/DesVIII-N-Stag complex was treated with thrombin following a protocol provided by the manufacturer (Novagen). The amount of thrombin required for complete cleavage was determined in a series of preliminary small-scale digestion reactions. Briefly, approximately 315 mg of Ni-NTA-purified DesVII-C-His6/DesVIII-N-Stag in 21 mL of 25 mM Tris•HCl (pH 8), 5% glycerol, 4 mM imidazole were treated with 2 mL of 10× thrombin cleavage buffer and 65 units of restriction grade thrombin. The mixture was gently agitated at 4 °C for 13 h. SDS-PAGE analysis showed that cleavage had gone to completion. A small amount of precipitate was removed by centrifugation (4 °C, 30,000 g, 30 min) and 8 mL of Ni-NTA resin was added to the supernatant. The mixture was agitated for 3 h and proteins were eluted with increasing concentrations of imidazole as described earlier. The purified proteins were analyzed by SDS-PAGE, HPLC, and ESI-MS as described above.
Production of DesVIII-N-Stag and Co-Incubation with DesVII-C-His6
Plasmid pDesVIII-N-Stag was used to transform E. coli BL21 Star(DE3). An overnight culture of E. coli BL21 Star (DE3)/pDesVIII-N-Stag was used to inoculate LB media (1 to 100 dilution, 25 μg/mL kanamycin). The cultures were incubated at 37 °C until the OD600 reading reached approximately 0.6 (the temperature was gradually lowered to 20 °C before the culture reached the desired OD600 reading). The cultures were then induced with 0.15 mM IPTG and further incubated at 20 °C for 14 h. The cells were harvested by centrifugation and used in the next step.
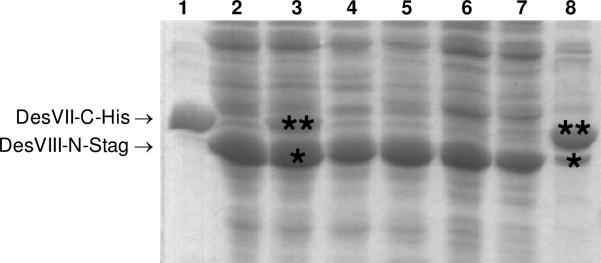
The protein DesVIII-N-Stag was subjected to partial purification using column packed with DEAE CL-6B anion exchange resin (Amersham Biosciences). All steps were carried out at 4 °C. The cell pellet was re-suspended in a buffer consisting of 50 mM Tris•HCl, 15% glycerol (pH 7.5), and protease inhibitor cocktail (added as above). Lysozyme was added to a concentration of 1 mg/mL, and the resulting suspension was incubated on ice for 30 min. The cells were disrupted by sonication in 10 s bursts (36 ×) with a 20-s cooling period between sonication bursts. Cell debris was removed by centrifugation (16,000 g, 4 °C, 30 min). The crude extract was loaded onto the 25 × 140 mm column packed with DEAE CL-6B and equilibrated with buffer A. Proteins were loaded with 30 mL of buffer and then eluted with a gradient of 1 M KCl in the same buffer created with a gradient maker (total volume of 400 mL). Fractions were collected and analyzed by UV spectroscopy (A280), SDS-PAGE, and a western blot using anti-S-tag antibody. Fractions containing DesVIII-N-Stag were pooled, dialyzed against the same buffer, concentrated, and used in the co-incubation experiment. The sample contained approximately 50% of DesVIII-N-Stag proteins as estimated by SDS-PAGE analysis (Figure 7, lane 2).
Figure 7.
Production of DesVII-C-His6 and DesVIII-N-Stag separately followed by co-incubation of proteins leads to reduced protein-protein complex formation as illustrated by SDS-PAGE analysis. DesVII-C-His6 (lane 1) and DesVIII-N-Stag (lane 2) were purified by Ni-NTA chromatography and partially purified by DEAE, respectively. They were incubated together with DesVIII-N-Stag (denoted with *) being in large excess over DesVII-C-His6 (denoted with **) (lane 3), and subjected to purification by Ni-NTA chromatography. Unbound proteins were eluted at low imidazole concentrations (10–30 mM, lanes 4–7). Bound proteins were eluted with 250 mM imidazole (lane 8) and contained an excess of DesVII-C-His6 (**) over DesVIII-N-Stag (*).
DesVII-C-His6 (0.4 mg) was combined with approximately a 2–3 fold excess of partially purified DesVIII-N-Stag in 1.5 mL of 50 mM sodium phosphate, 0.3 M NaCl, 10% glycerol, pH 8.0. The mixture was incubated with gentle agitation at 4 °C for 3 h. Ni-NTA resin (0.2 mL) was added and incubation continued for another 3 h. Proteins were eluted with increasing (10– 250 mM) concentrations of imidazole and fractions were analyzed by SDS-PAGE (Figure 7) and a western blot using anti-His antibody. Proteins eluted with 250 mM imidazole were analyzed by HPLC as described above.
Determination of Kinetic Parameters
A discontinuous HPLC-based assay was used to determine the catalytic activity of DesVII-C-His6/DesVIII-N-Stag and DesVII-C-His6 (as isolated) by monitoring the conversion of 10-deoxymethynolide (1) to 10-deoxymethymycin (3). An assay mixture of 80 μL in 50 mM potassium phosphate buffer, pH 8 contained 10-deoxymethynolide (1) and TDP-d-desosamine (2) with final concentrations listed in Table 2. A stock solution of 10-deoxymethynolide in ethanol was used. The concentration of ethanol in the final assay mixture was kept less than 4% (by volume). Mixtures were pre-incubated at 29 °C. The reaction was initiated by the addition of 1 μL of 12 μM DesVII-C-His6/DesVIII-N-Stag or 3 μL of 1.3 mM DesVII-C-His6 to the final concentrations listed in Table 2. Each reaction was done in duplicate. The reaction mixtures were incubated at 29 °C for a total of 5 min, during which time aliquots of 15 μL were removed every minute and quenched with 35 μL of ice-cold methanol. Precipitated proteins in each sample were removed by centrifugation and an aliquot (27 μL) of the supernatant was subjected to HPLC analysis using an analytical Microsorb-MV C18 column (4.5 × 250 mm, 5 μm, Varian). Isocratic elution using 35% acetonitrile in 14 mM triethylamine (adjusted to pH 2.7 with trifluoroacetic acid) at a rate of 1 mL/min over 20 min gave baseline separation between the substrate (1, 15 min) and the product (3, 12 min). The detector was set at 235 nm. The substrate and product ratios were calculated by integration of the corresponding peaks in the HPLC chromatogram. The rate of the product formation was linear for the duration of the assay. The kinetic parameters were determined by fitting the data to the Michaelis-Menten equation using the GraFit software (Erithacus Software Limited, Horley, Surrey, UK).
Table 2.
Kinetic Parameters for DesVII-C-His/DesVIII-N-Stag and DesVII-C-His. Data for homologous GTs are also shown.
| Assay | [1], mM | [2], mM | [enzyme], μM | kcat, min-1 | KM, mM |
|---|---|---|---|---|---|
| DesVII, variable 1 | 0.37–10.53 | 10.0 | 49 | 0.55 | 5.83 |
| DesVII/DesVIII, variable 1 | 0.07–1.75 | 1.0 | 0.15 | 60.7 | 0.06 |
| DesVII, variable 2 | 9.1 | 2.81–10.18 | 49 | 0.37 | 1.84 |
| DesVII/DesVIII, variable 2 | 3.5 | 0.09–1.46 | 0.15 | 80.0 | 0.16 |
| kcat, min-1 | KM, mM | ||||
| EryCIII*(9) | 1 | 0.050 (A) | |||
| AknS (10) | 0.005 | ND | |||
| AknS + AknT (1:3) (10) | 0.22 | 0.004 (A), 0.288 (D) |
A – acceptor, D – donor, ND – not determined.
Transglycosylation Reaction
The reversibility of the glycosylation reaction was assayed by the discontinuous HPLC assay as described above. A typical assay mixture contained 2 mM of 3, an appropriate amount of TDP, and 1–2 mM of acceptor in 50 mM potassium phosphate buffer, pH 8. The reaction was initiated by the addition of DesVII-C-His6/DesVIII-N-Stag (final concentration of 20 μM) to the assay mixture. The concentration of TDP was 2.3 mM when only the activity of the reverse reaction was measured in the assay, and 0.15–1.5 mM when the transferring capability of the TDP-sugar to another aglycone was assayed. Incubation was carried out at 29 °C for 4, 16, or 24 h. The glyco-swapping activity of the DesVII/DesVIII system shows little dependence on TDP concentration in the range of 0.15–1.5 mM. There was no significant improvement in the yield of the product after prolonged incubation time (> 4 h).
Results and Discussion
DesVII-catalyzed Reaction Requires DesVIII
Activity Assay of Recombinant DesVII-C-His6
As we previously reported (5, 7), the desVII gene was successfully expressed in E. coli as a C-terminal His6-tagged fusion protein and was purified to near homogeneity (Figure 1, lane 1). Despite numerous attempts, the predicted glycosyltransferase activity of the isolated DesVII-C-His6 to catalyze the transfer of desosamine from TDP-d-desosamine (2) to 10-deoxymethynolide (1) to form 10-deoxymethymycin (3) could not be confirmed. This prompted us to consider the possible requirement of an additional protein for DesVII activity. A likely candidate is the protein encoded by the desVIII gene, which is located next to the desVII gene in the desosamine biosynthetic gene cluster, but it is not needed for TDP-d-desosamine biosynthesis (28). Conceivably, the encoded DesVIII protein may instead play a role in the glycosylation and might be the missing component required for DesVII-catalyzed glycosyl transfer reaction.
Disruption of desVIII Gene in the Genome of S. venezuelae
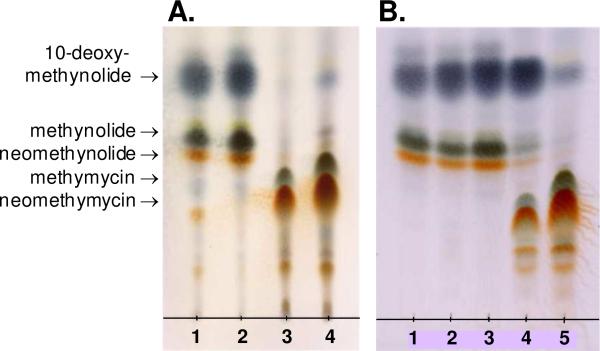
To investigate the role DesVIII plays during macrolide biosynthesis, a S. venezuelae mutant, KdesVIII, was constructed in which the portion of chromosomal DNA containing most of the desVIII gene was replaced with the kanamycin resistance marker. The desired chromosomal mutation in KdesVIII was confirmed using Southern blot hybridization analysis. A comparison of metabolites produced by this mutant and those isolated from the wild-type S. venezuelae by TLC (Figure 2A, lanes 1 and 4) showed that no glycosylated macrolides were generated by the mutant strain. To rule out the polar effect of desVIII disruption on desVII, which is located immediately downstream from desVIII within the same operon (6), an extra copy of the desVII gene on a plasmid was introduced into the KdesVIII mutant. In a separate experiment, the desVIII gene was also introduced and expressed in KdesVIII. TLC analysis showed that the metabolite profile of S. venezuelae desVII/KdesVIII strain (KdesVIII mutant expressing desVII) is identical to that of KdesVIII (Figure 2A, lane 2), whereas the phenotype of the desVIII/KdesVIII strain (KdesVIII mutant expressing desVIII) closely resembles that of the wild type S. venezuelae (Figure 2A, lane 3). It can therefore be concluded that the expression of the downstream genes in KdesVIII strain is not affected by the deletion of the desVIII gene, and the absence of glycosylated products in the metabolite profile of KdesVIII mutant strongly suggests that DesVIII is involved in the attachment of desosamine to the macrolactone aglycones.
Figure 2.
TLC analysis of macrolides produced by different S. venezuelae strains. A. Lane 1: KdesVIII, lane 2: desVII/KdesVIII, lane 3: desVIII/KdesVIII, lane 4: wild type. B. Lane 1: KdesVIII, lane 2: eryCII/KdesVIII, lane 3: tylM3/KdesVIII, lane4: dnrQ/KdesVIII, lane 5: wild type.
Expression and Purification of DesVIII-C-His 6
To unambiguously define the function of DesVIII, it was necessary to purify the protein and assay its activity in vitro. Heterologous expression of desVIII as a C-terminal hexahistidine tagged fusion protein (DesVIII-C-His6) in E. coli resulted in an insoluble protein as reported earlier (5). Changing the expression conditions did not improve the solubility of the expressed protein. Affinity purification under denaturing conditions also failed since DesVIII-C-His6 did not bind to the Ni-NTA resin in the presence of 8 M urea. Fortunately, dialysis of the denatured protein to remove urea followed by purification of the re-folded protein over Ni-NTA resin using non-denaturing protocol yielded soluble but impure DesVIII-C-His6. SDS-PAGE analysis of the collected protein (Figure 1, lane 2) displayed a major band corresponding to DesVIII-C-His6 (43.6 kDa). Western blot analysis confirmed that the corresponding protein has a polyhistidine tag. Attempts to further purify the protein using anion exchange (DEAE) and size-exclusion (Superdex) chromatographic techniques led to protein precipitation. Thus, the partially purified DesVIII-C-His6 was used in our preliminary studies.
The Effect of DesVIII on the DesVII-catalyzed Glycosyl Transfer Reaction
As we reported earlier (5), the addition of partially purified DesVIII-C-His6 to the glycosyltransferase assay and overnight incubation at 29 °C resulted in almost quantitative conversion of aglycone 10-deoxymethynolide (1) to a new product. The control assay in the absence of DesVIII-C-His6 showed barely detectable product formation. A large-scale reaction with DesVII-C-His6 and DesVIII-C-His6 was then carried out and the new product was isolated and subjected to NMR spectroscopy and MS analysis. The spectral data of the glycosylated product are fully consistent with those of 10-deoxymethymycin (3) (29, 30). These results clearly establish that DesVII is indeed the glycosyltransferase, and that the addition of DesVIII is essential to reconstitute the DesVII activity (5).
The Roles of DesVIII and Its Homologs in Glycosyl Transfer Reactions
Homologs of desVIII gene are found in the biosynthetic gene clusters of a number of secondary metabolites. These include eryCII (31% amino acid sequence identity with DesVIII) in the desosamine-containing erythromycin gene cluster of S. erythraea (31, 32), tylM3 (34% sequence identity) in the mycaminose-containing tylosin gene cluster of S. fradiae (33, 34), and dnrQ (35% sequence identity, also known as dnmQ) in the daunosamine-containing daunorubicin gene cluster of S. peucetius (35, 36). The eryCII-encoded product was originally proposed to function as a 3,4-ketoisomerase catalyzing the conversion of a putative 4-keto sugar intermediate to the corresponding 3-ketosugar in the biosynthesis of desosamine (31). However, such a step is not necessary for the formation of daunosamine, and later experiments showed that formation of desosamine also does not involve a ketoisomerization step (28). Interestingly, 3,4-ketoisomerization is a key step during mycaminose biosynthesis, but it is catalyzed by Tyl1a, not TylM3 (37-39). Thus, the biological functions of DesVIII and its homologs, EryCII, TylM3, DnrQ, are different from those originally proposed.
Replacement of desVIII with Its Homologs
Several other GTs involved in the biosynthesis of secondary metabolites were also reported to require an auxiliary protein, e.g., EryCIII/EryCII (9), AknS/AknT (10), and TylM2/TylM3 (11). These results clearly indicate that proteins of DesVIII family play important roles in the sugar attachment process. An interesting question is whether DesVIII and its homologs are interchangeable, because the answer to this question may provide insight into the functions of these proteins. Accordingly, the desVIII homologs, eryCII, tylM3, and dnrQ, were expressed in a S. venezuelae KdesVIII mutant, and the secondary metabolites produced by the resulting strains, eryCII/KdesVIII, tylM3/KdesVIII, and dnrQ/KdesVIII, were analyzed by TLC. As shown in Figure 2B, the phenotypes of two of the mutant strains, eryCII/KdesVIII and tylM3/KdesVIII, are identical to that of the KdesVIII strain, where no glycosylated products were detected. In contrast, dnrQ/KdesVIII is able to produce methymycin (4) and neomethymycin (5), albeit in much lower quantities (approximately 15-fold lower than the wild type strain) (Figure 2B).
The metabolites produced by dnrQ/KdesVIII were purified and characterized. The major product isolated was 10-deoxymethynolide (1), but hydroxylated aglycones, such as methynolide (9) and neomethynolide (10), were also present. In addition, wild type S. venezuelae metabolites methymycin (4), neomethymycin (5), and pikromycin (8) were isolated. Interestingly, the doubly hydroxylated novamethymycin (11) was also purified from the crude extract. The isolation of 11 as minor metabolite of wild type S. venezuelae has been reported previously (26).
Putative Functions of DesVIII and Its Homologs
The above results clearly showed that dnrQ could replace desVIII. The lower yields of glycosylated metabolites from dnrQ/KdesVIII can be attributed to the low expression efficiency of the dnrQ gene in the mutant strain, or a difference between the catalytic properties of DnrQ and DesVIII rendering DnrQ less effective in DesVII activation. In an independent study, Yoon and co-workers expressed dnrQ, eryCII, tylM3, and another homolog of desVIII, oleP1, in a S. venezuelae mutant in which desVIII gene was partially deleted (40). It was found that all of the above genes, with the exception of tylM3, could replace desVIII. In these experiments, the heterologous genes were expressed under the control of a strong ermE promoter, whereas our studies used the native pikromycin promoter pikA. This difference could likely influence the level of protein expression, and consequently, the production of glycosylated compounds.
The results of these complementation experiments shed light on the function of DesVIII and its homologs. As demonstrated here and by Yoon et al., TylM3 cannot functionally replace DesVIII even though the aglycone acceptor (tylactone) and nucleotide-sugar donor (TDP-d-mycaminose) used by TylM2/TylM3 are structurally similar to those [10-deoxymethynolide (1) and TDP-d-desosamine (2)] used by the DesVII/DesVIII system. In contrast, DnrQ (and likely EryCII and OleP1) is a competent DesVIII substitute despite the fact that its aglycone acceptor and nucleotide-sugar donor (anthracyclin and TDP-l-daunosamine) bear little resemblance to those of the methymycin pathway. These observations suggest that DesVIII and its homologs are unlikely to be directly involved in the substrate or product binding, but most probably play roles in the activation of the corresponding glycosyltransferases.
Investigation of DesVIII Role In Vitro
Preparation of Untagged DesVIII-Tev
Being able to separate DesVII and DesVIII after co-incubation would allow us to determine whether DesVII requires DesVIII in every catalytic event or whether DesVII is capable of carrying out multiple turnovers once it is activated by DesVIII. To achieve differential tagging of the two proteins, DesVIII was expressed as a fusion protein with maltose binding protein (MBP) and a His10 tag at its N-terminus as previously reported (7). This fusion protein, MBP-DesVIII (88.9 kDa), was overproduced in E. coli and purified using affinity chromatography. The resulting protein was found to be active in the DesVII-catalyzed glycosyl transfer reaction (7). MBP and the His10 tag were cleaved with Tev protease and untagged protein DesVIII-Tev was concentrated to approximately 5 mg/mL (Figure 1, lane 3). Further concentration led to protein precipitation.
Activated DesVII-C-His6 Appears to be Capable of Catalyzing Multiple Turnovers
Similar to DesVIII-C-His6, the DesVIII-Tev protein greatly enhances the activity of DesVII-C-His6. Most significantly, the “activated” DesVII-C-His6 (DesVII*) remained catalytically active after DesVII-C-His6 and DesVIII-Tev were separated by Ni-NTA resin. As illustrated in Figure 3, the glycosyltransferase activity of DesVII* is significantly higher than that of untreated DesVII-CHis6 (Figure 3A), and is similar to that of DesVII-C-His6 in the presence of DesVIII-Tev. Taken together, these results implicate a “protein chaperon” (or a one-time activator) role for DesVIII in the activation of DesVII for glycosyl transfer. To ensure that DesVII activation does not involve covalent modification, samples of DesVII-C-His6, with and without DesVIII-Tev treatment, were subjected to HPLC and MS analyses. All of the samples tested gave overlapping elution profiles when subjected to HPLC reverse phase C4 chromatography. Their molecular masses determined by ESI-MS were also identical within experimental error. It is thus clear that DesVII activation is not a result of covalent modification, but likely involves a conformational change.
The above conclusion is consistent with a recent study of a closely related desosaminyltransferase, EryCIII, found in the erythromycin biosynthetic pathway where the DesVIII homolog, EryCII, is required for EryCIII in vitro activity (9). It was reported that EryCIII remains active after EryCII is removed. However, close examination of DesVII* by SDS-PAGE analysis revealed the presence of residual DesVIII-Tev in the sample (Figure 3B, line 1) raising the possibility that the observed activation is due to the DesVIII-Tev contaminant. Attempts to further purify DesVII-C-His6 to remove the residual DesVIII-Tev were unsuccessful. Thus, additional investigation was warranted to address whether DesVIII acts as a one-time activator for DesVII or is required all the time for DesVII activation.
DesVII and DesVIII Form a Tight 1:1 Complex When Co-Produced in E. coli
DesVII and DesVIII proteins were co-produced in E. coli as a C-terminally His6-tagged form (DesVII-C-His6) and an N-terminally S-tagged protein (DesVIII-N-Stag), respectively. We anticipated that co-production would allow DesVII-C-His6 to be activated by DesVIII-N-Stag in situ and that the isolation of resulting DesVII* could be achieved by Ni-NTA chromatography. Another advantage of this method is that purification of DesVIII is no longer necessary. DesVIII-N-Stag contains a 16-amino-acid epitope that can be removed by thrombin cleavage, if necessary, to give an untagged DesVIII. The attachment of the S-tag to the N-terminus of DesVIII would enable antibody detection to assess and ensure that the isolated DesVII* sample is free of DesVIII.
Co-Purification of DesVII-C-His6 and DesVIII-N-Stag
Accordingly, desVII and desVIII were cloned and overproduced in E. coli as DesVII-C-His6 and DesVIII-N-Stag proteins. Crude extract containing both proteins was subjected to affinity chromatography using Ni-NTA resin. To our surprise, both proteins remained associated throughout the purification steps and were eluted together with a buffer containing 250 mM imidazole (Figure 4A). Attempts to separate DesVII-C-His6 from DesVIII-N-Stag using anion exchange and size-exclusion chromatography (Figure 4B and 4C) also failed. In all cases, these two proteins were eluted as a single peak and co-purified in approximately equal amounts as estimated by SDS-PAGE. Size-exclusion chromatography showed that the DesVII-C-His6 and DesVIII-N-Stag complex has a molecular mass of approximately 250 kDa.
DesVII-C-His6 and DesVIII-N-Stag Form a Tight 1:1 Complex
To determine the ratio of proteins in the complex, the protein complex was denatured in 8 M urea and the sample was analyzed by HPLC equipped with a reverse phase C18 column. Two proteins eluted at 65 min and 69 min from the column (Figure 5A), and were identified as DesVII-C-His6 and DesVIII-N-Stag, respectively, based on SDS-PAGE analysis (Figure 5B). The ratio of the A280 peak areas (0.72:0.28) for DesVII-C-His6 and DesVIII-N-Stag corresponds to a molar ratio of 1.00 ± 0.10 : 1. Taken together with the estimated native molecular mass, the DesVII-C-His6 and DesVIII-N-Stag complex likely exists as a trimer of heterodimers [i.e., (DesVII•DesVIII)3 or (αβ)3]. It should be noted that DesVIII-N-Stag separated by HPLC displays two bands on SDS-PAGE (Figure 5B, lanes 1 and 3). Yet, MS analysis of the HPLC-purified protein indicated the presence of a single species whose molecular weight is in a good agreement with the calculated value of 45082 Da for DesVIII-N-Stag. Thus, the separation of DesVIII-N-Stag into two bands on SDS-PAGE is likely a result of folding irregularities rather than a truncation of the protein.
S-tag Is Not Important For DesVII-C-His6 :DesVIII-N-Stag Complex Formation
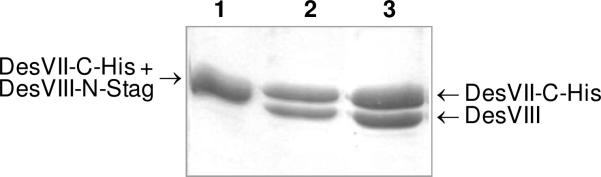
To ensure that the observed complex formation is not an artifact due to the presence of S-tag at the N-terminus of DesVIII, the DesVII-C-His6 and DesVIII-N-Stag complex (Figure 6, lane 1) was subjected to digestion by thrombin to remove the S-tag. Successful removal of the S-tag resulted in the appearance of a new band corresponding to DesVIII on SDS-PAGE (Figure 6, lane 2). DesVII-C-His6 was apparently not affected by thrombin treatment. The resulting mixture was purified by Ni-NTA chromatography, but DesVII-C-His6 and DesVIII again co-eluted (Figure 6, lane 3). HPLC analysis of the protein complex after thrombin cleavage and Ni-NTA purification showed that the ratio of these two proteins (DesVII/DesVIII) remained unchanged (1.05 : 1). The molecular weights of the DesVII-C-His6 and DesVIII fractions collected during HPLC purification were verified by ESI-MS. These findings clearly indicated that tight binding between DesVII and DesVIII is due to strong protein-protein interactions and is independent of the presence of S-tag.
Figure 6.
Removal of S-tag by thrombin cleavage does not affect binding between DesVII-CHis6 and DesVIII. SDS-PAGE analysis shows DesVII-C-His6 and DesVIII-N-Stag co-purified using Ni-NTA chromatography (lane 1, proteins have not been resolved into two bands due to very similar MWs), same sample after treatment with thrombin (lane 2, DesVIII travels faster than DesVIII-N-Stag), and thrombin-treated sample after purification using Ni-NTA resin (lane 3).
Complex Cannot be Reconstituted By Mixing Individually Expressed Proteins
In a separate experiment, DesVIII-N-Stag protein was overproduced in E. coli (in the absence of DesVII-CHis6). Although DesVIII-N-Stag failed to bind to S-protein agarose, it could be partially purified using anion exchange DEAE chromatography (Figure 7, lane 2). Partially purified DesVIII-N-Stag was incubated with DesVII-C-His6 (2-3 fold excess of DesVIII-N-Stag over DesVII-CHis6), and the mixture was then subjected to Ni-NTA chromatography. Interestingly, the proteins that eluted with high imidazole buffer (250 mM) consisted mainly of DesVII-C-His6 with only a small amount (less than 20%, Figure 7, lane 8) of DesVIII-N-Stag present. This finding implies that simultaneous expression of desVII and desVIII is important for efficient DesVII/DesVIII complex formation, which likely occurs during protein translation. This is consistent with two proteins being subunits of a heterosubunit enzyme.
Quantitative Determination of the Activation Effect of DesVIII on the Activity of DesVII
We have shown that the presence of DesVIII has a dramatic effect on the reaction catalyzed by glycosyltransferase DesVII. To quantitatively estimate the effect, we measured and compared the steady state kinetic parameters of the reactions catalyzed by DesVII-C-His6/DesVIII-N-Stag, where the proteins were co-produced, and by DesVII-C-His6 alone. A discontinuous HPLC assay was developed to analyze the extent of conversion of 10-deoxymethynolide (1) to the product 10-deoxymethymycin (3) at appropriate time intervals.
Steady State Kinetics Analysis of the Glycosyl Transfer Reaction
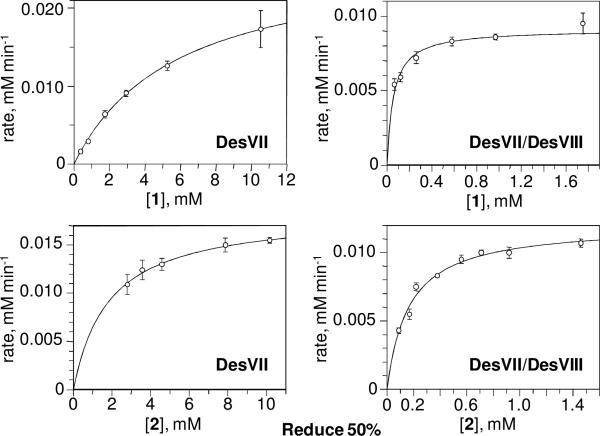
Although product formation by DesVII-C-His6 was previously reported to be most efficient in 50 mM Tris•HCl buffer, pH 9 (5), these assay conditions were not compatible with HPLC analysis. The presence of an unidentified peak, which overlaps with the product peak in chromatogram, makes it difficult to accurately determine the concentration of 3. The assay is further complicated by the necessity to use BSA to stabilize the proteins. We found that using 50 mM potassium phosphate buffer at pH 8 results in only slightly lower conversion (within 20%) but does not require BSA and does not produce extra peaks on the chromatogram. In contrast, using potassium phosphate at pH 7 gave 8- to 10-fold lower conversion to product. Accordingly, pH 8 and 50 mM potassium phosphate buffer were used throughout the kinetic analysis. The kinetic parameters were determined by fitting the data points to the Michaelis-Menten equation (Figure 8) and are listed in Table 2.
Figure 8.
Kinetic data for the activity assay of DesVII-C-His6 and DesVII-C-His6/DesVIII-N-Stag. The assays were performed as described in the text. The lines through the points represent the Michaelis-Menten equation calculated by the fitting program.
It is obvious that the presence of DesVIII dramatically affects KM values for both substrates as well as the value of kcat. There is more than 104-fold overall improvement in the catalytic efficiency (kcat/KM) for the acceptor (10-deoxymethynolide, 1) and more than 2×103-fold improvement for the donor (TDP-d-desosamine, 2), highlighting the DesVIII requirement for the activity of glycosyltransferase DesVII. The fact that KM is affected by the presence of DesVIII indicates that the low activity of DesVII-C-His6 alone is a result of glycosyltransferase being in a different and less active conformation than that of DesVII in DesVII-C-His6/DesVIII-N-Stag. This scenario is different from a situation where lower activity is due to small amount of active species present and the remainder of the protein is inactive in which case KM values would be essentially same.
The 104-fold improvement in the catalytic efficiency of DesVII in the presence of DesVIII could account for the efficient conversion of 1 to 3 by DesVII* observed earlier (Figure 3A, lane 1). Given the kinetic parameters of the reaction, we estimate that full conversion of 1 to 3 could be achieved over the incubation time period of the assay (2 h), even if only less than 2% of DesVII* were DesVIII-bound (existing in the active DesVII-C-His6/DesVIII-Tev form). This minor quantity of DesVIII-Tev would be difficult to detect by SDS-PAGE analysis. Therefore, it is likely that the observed activity of DesVII* is due to contamination with DesVIII-Tev, rather than to the activity of an activated, uncomplexed DesVII.
Interestingly, EryCIII* (EryCIII activated by its DesVIII homolog, EryCII) was reported to have an acceptor KM value (50 μM) similar to that of DesVII/DesVIII (60 μM), but a much lower kcat value (60-fold lower) (9) (Table 2). The AknS/AknT system also has KM values for both substrates similar to those of DesVII/DesVIII. However, the kcat value for AknS is over 270-fold lower than DesVII/DesVIII both in the absence and presence of its DesVIII homolog AknT (10) (Table 2). Perhaps these proteins behave similarly to DesVII/DesVIII, in which case their lower activities could be due to partial reconstitution of each GT complex. In the case of DesVII-CHis6/DesVIII-N-Stag, co-expression is essential for complex formation. Although a 1:3 optimal molar ratio was reported for AknS/AknT when the proteins were co-incubated (10), we found that when DesVII-C-His6 is pre-incubated with more than 3-fold excess of DesVIII-N-Stag, the activity is still lower than that of the co-produced DesVII-C-His6/DesVIII-N-Stag complex (data not shown).
Transglycosylation Catalyzed by DesVII-C-His6 /DesVIII-N-Stag
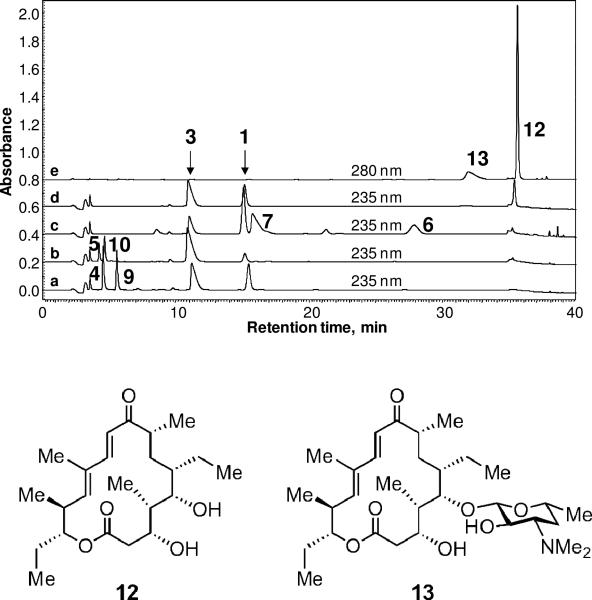
It has been recently demonstrated that a number of biosynthetic GTs can catalyze the reverse reaction, converting the glycosylated natural product to the corresponding aglycone and TDP-sugar in the presence of TDP (41-44). A similar capability was also found for DesVII where formation of aglycone 1 was observed when 3 was incubated with an excess of TDP in the presence of DesVII-C-His6/DesVIII-N-Stag. However, the equilibrium of the reaction strongly favors the glycosylation direction since only less than 10% of 3 was converted to 1 after a 24 h incubation period. To verify that the release of 1 is due to the reverse reaction and not hydrolysis, it is important to show that desosamine can be transferred from 3 to another aglycone acceptor through the intermediacy of 2. Accordingly, the assays were done using catalytic quantities of TDP with different aglycone acceptors added. As shown in Figure 9, methynolide (9) was converted to methymycin (4) (a), neomethynolide (10) was converted to neomethymycin (5) (b), narbonolide (6) was converted to narbomycin (7) (c), and tylactone (12) was converted to desosaminylated tylactone (13) (d, e) under the reaction conditions. These results suggest that 2 is indeed formed and serves as the sugar donor for the second transfer reaction. Interestingly, the conversion from 3 to 1 improved to approximately 30% in the presence of 12, indicating that the addition of an excess acceptor substrate shifts the reaction equilibrium. The reversibility of DesVII/DesVIII and other GTs may be exploited for glycodiversification of secondary metabolites in a combinatorial fashion (2-3, 45-46).
Figure 9.
DesVII/DesVIII-catalyzed transfer of desosamine from the glycosylated macrolide 3 to various aglycone acceptors. HPLC analysis of the reactions is shown. The detector was set at 235 nm, except for the assay with tylactone 12 where absorbance at 280 nm was monitored as well. a) Desosamine is transferred from 3 to 9 with 1 and 4 formed as products; b) 3 + 10 → 1 + 5; c) 3 + 6 → 1 + 7; d) 3 + 12 → 1 + 13 (235 nm, 13 is invisible at this wavelength); d) 3 + 12 → 1 + 13 (280 nm, 1 and 3 are invisible at this wavelength).
Summary
The results reported herein clearly establish that DesVII is the GT responsible for the attachment of TDP-d-desosamine (2) to macrolactones of varied ring sizes. The requirement of DesVIII for DesVII activity is also firmly established. Since purification of functional DesVIII in sufficient quantities proved to be difficult, an expression system for co-production of C-terminally His6-tagged DesVII and N-terminally S-tagged DesVIII was developed. Both proteins, produced in high yields, form a tight complex, which is stable through purification by Ni-NTA affinity, anion exchange and size exclusion chromatography. HPLC analysis of the ureadenatured complex revealed a 1:1 ratio of DesVII and DesVIII. The complex exists as a trimer of heterodimers [(αβ)3] based on the results of size exclusion chromatography. Interestingly, when the proteins are produced separately, co-incubated, and then subjected to Ni-NTA purification, only a small fraction of DesVIII-N-S-tag is bound to DesVII-C-His6. This observation suggests that DesVII and DesVIII need to be co-expressed for proper folding and complex formation. The catalytic efficiency of the DesVII/DesVIII complex (kcat/K [1] = 1012 min-1 mM-1 M, kcat/KM [2] = 500 min-1 mM-1) was determined to be at least 2×103-fold higher than that of DesVII alone (kcat/KM [1] = 0.094 min-1 mM-1, kcat/KM [2] = 0.20 min-1 mM-1). The DesVII/DesVIII complex also catalyzes the reverse reaction and the TDP-d-desosamine product formed can serve as a sugar donor with a different aglycone acceptor. Taken together, these results provide detailed biochemical evidence unraveling the catalytic roles of DesVIII in DesVII-catalyzed glycosyl transfer reaction. It is conceivable that other DesVIII homologs behave similarly in assisting the corresponding GT. The findings presented here are important for further investigation of DesVII/DesVIII catalysis and have a significant impact on the design of new glycosylated macrolide derivatives using a combinatorial approach.
Supplementary Material
Acknowledgment
We thank Dr. Klaus Linse (Protein Microanalysis Facility, Institute for Cellular and Molecular Biology, University of Texas, Austin) for assistance with proteomics experiments, and Drs. Mehdi Moini (Department of Chemistry and Biochemistry, University of Texas, Austin) and Herng-Hsiang Stony Lo (Analytical Instrumentation Facility Core, College of Pharmacy, University of Texas, Austin) for performing mass spectroscopy analysis. This work was supported in part by the National Institutes of Health Grants (GM35906 and GM54346).
Abbreviation
- ATCC
American Type Culture Collection
- BSA
bovine serum albumin
- DEAE
diethylaminoethyl
- DTT
dithiothreitol
- ESI
electrospray ionization
- FPLC
fast protein liquid chromatography
- GT
glycosyltransferase
- HPLC
high performance liquid chromatography
- IPTG
isopropyl β-d-thiogalactoside
- LB
Luria-Bertani
- MBP
maltose binding protein
- NTA
nitrilotriacetic acid
- PAGE
polyacrylamide gel electrophoresis
- PCR
polymerase chain reaction
- PMSF
phenylmethylsulphonyl fluoride
- SDS
sodium dodecyl sulfate
- TDP
thymidine 5′-diphosphate
- TFA
trifluoroacetic acid
- Tris
tris(hydroxymethyl)aminomethane
Footnotes
This work was supported in part by the National Institutes of Health Grants (GM35906 and GM54346).
Supporting Information
Revised sequence of desVII gene. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Thibodeaux CJ, Melançon CE, Liu H.-w. Unusual sugar biosynthesis and natural product glycodiversification. Nature. 2007;446:1008–1016. doi: 10.1038/nature05814. [DOI] [PubMed] [Google Scholar]
- 2.Thibodeaux CJ, Melançon CE, III., Liu H.-w. Natural product sugar biosynthesis and enzymatic glycodiversification. Angew. Chem. Int. Ed. Engl. 2008;47:9814–9859. doi: 10.1002/anie.200801204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erb A, Weiß H, Härle J, Bechtold A. A bacterial glycosyltransferase gene toolbox: generation and applications. Phytochemistry. 2009;70:1812–1821. doi: 10.1016/j.phytochem.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 4.Quiros LM, Carbajo RJ, Braña AF, Salas JA. Glycosylation of macrolide antibiotics. Purification and kinetic studies of a macrolide glycosyltransferase from Streptomyces antibioticus. J. Biol. Chem. 2000;275:11713–11720. doi: 10.1074/jbc.275.16.11713. [DOI] [PubMed] [Google Scholar]
- 5.Borisova SA, Zhao L, Melançon CEI, Kao C-L, Liu H.-w. Characterization of the glycosyltransferase activity of DesVII: analysis of and implications for the biosynthesis of macrolide antibiotics. J. Am. Chem. Soc. 2004;126:6534–6535. doi: 10.1021/ja049967j. [DOI] [PubMed] [Google Scholar]
- 6.Xue Y, Zhao L, Liu H.-w., Sherman DH. A gene cluster for macrolide antibiotic biosynthesis in Streptomyces venezuelae: architecture of metabolic diversity. Proc. Natl. Acad. Sci. USA. 1998;95:12111–12116. doi: 10.1073/pnas.95.21.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borisova SA, Zhang C, Zhang H, Takahashi H, Wong A, Thorson JS, Liu H.-w. Substrate specificity of the macrolide glycosylating enzyme pair DesVII/DesVIII: opportunities, limitations, and mechanistic hypotheses. Angew. Chem. Int. Ed. Engl. 2006;45:2748–2753. doi: 10.1002/anie.200503195. [DOI] [PubMed] [Google Scholar]
- 8.Borisova S, Kim HJ, Pu X, Liu H.-w. Glycosylation of acyclic and cyclic aglycone substrates by macrolide glycosyltransferase DesVII/DesVIII: analysis and implications. Chembiochem. 2008;9:1554–1558. doi: 10.1002/cbic.200800155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan Y, Chung HS, Leimkuhler C, Walsh CT, Kahne D, Walker S. In vitro reconstitution of EryCIII activity for the preparation of unnatural macrolides. J. Am. Chem. Soc. 2005;127:14128–14129. doi: 10.1021/ja053704n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu W, Leimkuhler C, Gatto GJJ, Kruger RG, Oberthur M, Kahne D, Walsh CT. AknT is an activating protein for the glycosyltransferase AknS in l-aminodeoxysugar transfer to the aglycone of aclacinomycin A. Chem. Biol. 2005;12:527–534. doi: 10.1016/j.chembiol.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 11.Melançon CE, Takahashi H, Liu HW. Characterization of tylM3/tylM2 and mydC/mycB pairs required for efficient glycosyltransfer in macrolide antibiotic biosynthesis. J. Am. Chem. Soc. 2004;126:16726–16727. doi: 10.1021/ja043900e. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen HC, Karray F, Lautru S, Gagnat J, Lebrihi A, Huynh TDH, Pernodet J-L. Glycosylation steps during spiramycin biosynthesis in Streptomyces ambofaciens: involvement of three glycosyltransferases and their interplay with two auxiliary proteins. Antimicrob. Agents Chemother. 2010;54:2830–2839. doi: 10.1128/AAC.01602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 14.Zhao L. Ph. D. Thesis. University of Minnesota; Minneapolis, MN: 2000. Biosynthetic studies of d-desosamine and engineered biosynthesis of methymycin/pikromycin analogues carrying modified deoxysugars. [Google Scholar]
- 15.Denis F, Brzezinski R. An improved aminoglycoside resistance gene cassette for use in gram-negative bacteria and Streptomyces. FEMS Microbiol. Lett. 1991;65:261–264. doi: 10.1016/0378-1097(91)90224-x. [DOI] [PubMed] [Google Scholar]
- 16.Denis F, Brzezinski R. A versatile shuttle cosmid vector for use in Escherichia coli and actinomycetes. Gene. 1992;111:115–118. doi: 10.1016/0378-1119(92)90611-r. [DOI] [PubMed] [Google Scholar]
- 17.Bierman M, Logan R, O'Brien K, Seno ET, Rao RN, Schoner BE. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 18.Zhao L, Ahlert J, Xue Y, Thorson JS, Sherman DH, Liu H.-w. Engineering a methymycin/pikromycin-calicheamicin hybrid: construction of two new macrolides carrying a designed sugar moiety. J. Am. Chem. Soc. 1999;121:9881–9882. [Google Scholar]
- 19.Guo Z. Ph. D. Thesis. University of Minnesota; Minneapolis: 1998. Studies of the biosynthesis of the bacterial cell wall and antibiotics deoxysugar components:yersiniose A, mycaminose, and mycarose. [Google Scholar]
- 20.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor Laboratory Press; Woodbury, NY: 1989. [Google Scholar]
- 21.Hopwood DA, Bibb MJ, Chater KF, Kieser T, Bruton CJ, Kieser HM, Lydiate DJ, Smith CP, Ward JM, Schrempf H. Genetic Manipulation of Streptomyces: A Laboratory Manual. The John Innes Foundation; Norwich, UK: 1985. [Google Scholar]
- 22.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. The John Innes Foundation; Norwich, UK: 2000. [Google Scholar]
- 23.Simon R, Priefer V, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology. 1983;1:784–791. [Google Scholar]
- 24.Zhao L, Beyer NJ, Borisova SA, Liu HW. β-Glucosylation as a part of self-resistance mechanism in methymycin/pikromycin producing strain Streptomyces venezuelae. Biochemistry. 2003;42:14794–14804. doi: 10.1021/bi035501m. [DOI] [PubMed] [Google Scholar]
- 25.Cane DE, Lambalot RH, Prabhakaran PC, Ott WR. Macrolide biosynthesis. 7. Incorporation of polyketide chain elongation intermediates into methymycin. J. Am. Chem. Soc. 1993;115:522–526. [Google Scholar]
- 26.Zhang Q, Sherman DH. Isolation and structure determination of novamethymycin, a new bioactive metabolite of the methymycin biosynthetic pathway in Streptomyces venezuelae. J. Nat. Prod. 2001;64:1447–1450. doi: 10.1021/np010146r. [DOI] [PubMed] [Google Scholar]
- 27.Gao P. Ph. D. Thesis. University of Texas; Austin: 2007. Studies of activation, automodification, and transmodification of poly(ADP-ribose) polymerase-1. [Google Scholar]
- 28.Zhao L, Borisova S, Yeung S-M, Liu H.-w. Study of C-4 deoxygenation in the biosynthesis of desosamine: evidence implicating a novel mechanism. J. Am. Chem. Soc. 2001;123:7909–7910. doi: 10.1021/ja010587x. [DOI] [PubMed] [Google Scholar]
- 29.Kinumaki A, Suzuki M. The structure of a new macrolide antibiotic, YC-17. J. Chem. Soc. Chem. Commun. 1972:744–745. [Google Scholar]
- 30.Cane DE, Graziani EI. Methymycin biosynthesis. Isolation of P450 monooxygenase activity in a cell-free system from Streptomyces venezuelae. J. Am. Chem. Soc. 1998;120:2682–2683. [Google Scholar]
- 31.Summers RG, Donadio S, Staver MJ, Wendt-Pienkowski E, Hutchinson CR, Katz L. Sequencing and mutagenesis of genes from the erythromycin biosynthetic gene cluster of Saccharopolyspora erythraea that are involved in l-mycarose and d-desosamine production. Microbiology. 1997;143:3251–3262. doi: 10.1099/00221287-143-10-3251. [DOI] [PubMed] [Google Scholar]
- 32.Salah-Bey K, Doumith M, Michel JM, Haydock S, Cortes J, Leadlay PF, Raynal MC. Targeted gene inactivation for the elucidation of deoxysugar biosynthesis in the erythromycin producer Saccharopolyspora erythraea. Mol. Gen. Genet. 1998;257:542–553. doi: 10.1007/s004380050680. [DOI] [PubMed] [Google Scholar]
- 33.Gandecha AR, Large SL, Cundliffe E. Analysis of four tylosin biosynthetic genes from the tylLM region of the Streptomyces fradiae genome. Gene. 1997;184:197–203. doi: 10.1016/s0378-1119(96)00595-1. [DOI] [PubMed] [Google Scholar]
- 34.Chen H, Guo Z, Liu H.-w. Expression, purification, and characterization of TylM1, an N,N-dimethyltransferase in the biosynthesis of mycaminose. J. Am. Chem. Soc. 1998;120:9951–9952. [Google Scholar]
- 35.Madduri K, Hutchinson CR. Functional characterization and transcriptional analysis of the dnrR1 locus, which controls daunorubicin biosynthesis in Streptomyces peucetius. J. Bacteriol. 1995;177:1208–1215. doi: 10.1128/jb.177.5.1208-1215.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otten SL, Liu X, Ferguson J, Hutchinson CR. Cloning and characterization of the Streptomyces peucetius dnrQS genes encoding a daunosamine biosynthesis enzyme and a glycosyl transferase involved in daunorubicin biosynthesis. J. Bacteriol. 1995;177:6688–6692. doi: 10.1128/jb.177.22.6688-6692.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melançon CE, Yu WL, Liu H.-w. TDP-mycaminose biosynthetic pathway revised and conversion of desosamine pathway to mycaminose pathway with one gene. J. Am. Chem. Soc. 2005;127:12240–12241. doi: 10.1021/ja053835o. [DOI] [PubMed] [Google Scholar]
- 38.Melançon CE, Hong L, White JA, Liu YN, Liu H.-w. Characterization of TDP-4-keto-6-deoxy-D-glucose-3,4-ketoisomerase from the d-mycaminose biosynthetic pathway of Streptomyces fradiae: in vitro activity and substrate specificity studies. Biochemistry. 2007;46:577–590. doi: 10.1021/bi061907y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tello M, Rejzek M, Wilkinson B, Lawson DM, Field RA. Tyl1a, a TDP-6-deoxy-D-xylo-4-hexulose 3,4-isomerase from Streptomyces fradiae: structure prediction, mutagenesis and solvent isotope incorporation experiments to investigate reaction mechanism. Chembiochem. 2008;9:1295–1302. doi: 10.1002/cbic.200800021. [DOI] [PubMed] [Google Scholar]
- 40.Hong JSJ, Park SJ, Parajuli N, Park SR, Koh HS, Jung WS, Choi CY, Yoon YJ. Functional analysis of desVIII homologues involved in glycosylation of macrolide antibiotics by interspecies complementation. Gene. 2007;386:123–130. doi: 10.1016/j.gene.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 41.Minami A, Uchida R, Eguchi T, Kakinuma K. Enzymatic approach to unnatural glycosides with diverse aglycon scaffolds using glycosyltransferase VinC. J. Am. Chem. Soc. 2005;127:6148–6149. doi: 10.1021/ja042848j. [DOI] [PubMed] [Google Scholar]
- 42.Zhang C, Griffith BR, Fu Q, Albermann C, Fu X, Lee IK, Li L, Thorson JS. Exploiting the reversibility of natural product glycosyltransferase-catalyzed reactions. Science. 2006;313:1291–1294. doi: 10.1126/science.1130028. [DOI] [PubMed] [Google Scholar]
- 43.Bode HB, Muller R. Reversible sugar transfer by glycosyltransferases as a tool for natural product (bio)synthesis. Angew. Chem. Int. Ed. Engl. 2007;46:2147–50. doi: 10.1002/anie.200604671. [DOI] [PubMed] [Google Scholar]
- 44.Melançon CE, Thibodeaux CJ, Liu H.-w. Glyco-stripping and glyco-swapping. ACS Chem. Biol. 2006;1:499–504. doi: 10.1021/cb600365q. [DOI] [PubMed] [Google Scholar]
- 45.Griffith BR, Langenhan JM, Thorson JS. Sweetening natural products via glycorandomization. Curr. Opin. Biotechnol. 2005;16:622–630. doi: 10.1016/j.copbio.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 46.Luzhetskyy A, Bechtold A. Features and applications of bacterial glycosyltransferases: current state and prospects. Appl. Microbiol. Biotechnol. 2008;80:945–952. doi: 10.1007/s00253-008-1672-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.