Abstract
Simian virus 40 (SV40) large T antigen (TAg) is a multifunctional oncoprotein essential for productive viral infection and for cellular transformation. We have used microarray analysis to examine the global changes in cellular gene expression induced by wild-type T antigen (TAgwt) and TAg-mutants in mouse embryo fibroblasts (MEFs). The expression profile of approximately 800 cellular genes was altered by TAgwt and a truncated TAg (TAgN136), including many genes that influence cell cycle, DNA-replication, transcription, chromatin structure and DNA repair. Unexpectedly, we found a significant number of immune response genes upregulated by TAgwt including many interferon stimulated genes (ISGs) such as ISG56, OAS, Rsad2, Ifi27 and Mx1. Additionally, we also observed activation of STAT1 by TAgwt. Our genetic studies using several TAg mutants reveal an unexplored function of TAg and indicate that the LXCXE motif and p53 binding are required for the upregulation of ISGs.
Keywords: SV40, T antigen, gene expression, immune response, interferon
INTRODUCTION
Simian virus 40 (SV40) is a small DNA virus that under permissive conditions replicates in cells leading to the propagation of the viral progeny. However, under nonpermissive conditions it is oncogenic in animals and capable of inducing transformation of multiple cell lines in cell culture systems. To perform these functions SV40 encodes two major transforming proteins, the large T antigen (T antigen) and the small t antigen (t antigen). The small t antigen targets the cellular protein phosphatase PP2A and is required for transformation under some conditions. On the other hand, the expression of large T antigen alone is sufficient for SV40's oncogenic potential in most cases (Ahuja, Saenz-Robles, and Pipas, 2005).
The large T antigen (TAg) is a 708 amino acid protein that contains several independently folding domains/regions. These domains/regions interact with host cellular proteins and thus provide an excellent tool to understand the different cellular processes affected by viral infection. Three regions of TAg are essential to elicit cellular transformation (Ahuja, Saenz-Robles, and Pipas, 2005; Ali and DeCaprio, 2001). The LXCXE motif and the J domain reside in the N-terminus of TAg, and are involved in the inactivation of Rb family functions. The LXCXE motif mediates binding to the Rb family members (pRb, p107 and p130) (DeCaprio et al., 1988). The J domain, via binding to the chaperone hsc70, stimulates the ATPase activity of hsc70, and the resulting energy is used to release E2F from Rb (Sullivan, Gilbert, and Pipas, 2001). This release leads to the upregulation of E2F transactivation activity and subsequent progression of cells into S phase. The C- terminus of TAg is essential for inactivation of the tumor suppressor p53 (Kierstead and Tevethia, 1993; Pipas and Levine, 2001), as binding of p53 by TAg prevents its degradation by Mdm2 and results in p53 stabilization. Therefore, high levels of p53 are found in TAg transformed cells (Oren, Maltzman, and Levine, 1981).
Infection of cells by viruses often results in a potent and dramatic shift in the transcriptional activity of host cellular genes. These changes reflect the strategies developed by both host and pathogen to facilitate their own survival. For example, many viruses induce the expression of host cell genes involved in DNA replication and cell cycle to enhance its own replication. On the other hand, the induction of interferon (IFN) and IFN-stimulated genes (ISGs) is the hallmark of the host response to create an antiviral state. As a countermeasure to this response, many viruses encode proteins that interfere with the IFN induction as well as the products of the ISGs (Randall and Goodbourn, 2008). An understanding of the host-pathogen relationship requires the study of complex biological processes which result in altered global gene expression.
Previously we used a mouse cDNA array consisting of ~8000 genes to compare gene expression induced by wild-type TAg in different systems (Cantalupo et al., 2009). Moreover, we have also reported differential gene expression induced by wild-type TAg and mutants in the mouse intestine (Rathi et al., 2009). These studies demonstrated that wild-type TAg regulates a large number of known E2F-responsive genes in both systems. Our studies in intestine showed that these genes require the LXCXE motif and J domain to be regulated by TAg (Rathi et al., 2009; Rathi, Saenz Robles, and Pipas, 2007). In addition, we found sets of genes that were uniquely regulated by TAg in one system but not in the other. For example, TAg downregulates several components of the cytochrome P450 pathway in intestine, but this pathway is not altered by TAg in mouse embryo fibroblasts (MEFs) (Cantalupo et al., 2009; Saenz-Robles et al., 2007b). Similarly, we noted that some immune response genes were regulated by TAg in MEFs, but not in intestine (Cantalupo et al., 2009). However, the limited gene coverage of the array used in these studies coupled with a lack of accompanying genetics prevented us from discerning if those genes were independently regulated or if they were responding to the activation of an immune response pathway. In the current study we examine TAg's effects on gene expression in MEFs, a well characterized cell culture system. In this case, we have used mouse whole genome arrays to examine global changes in cellular gene expression in MEFs induced by the presence of wild-type T antigen (TAgwt) and TAg-mutants (Fig.1A) The mutants used for this study are TAgN136 (expresses the first 136 amino acids of T antigen), TAg3213 (mutation in the LXCXE motif) and TAgD44N (mutation in the J domain). The transformation potential of these mutants has been studied in several different systems. In particular, TAgN136 is unable to transform MEFs while TAg3213 and TAgD44N show reduced efficiency in transformation (Hahn et al., 2002; Markovics et al., 2005; Stubdal et al., 1997; Thompson et al., 1990). The current study reveals an unexplored function of TAg in regulation of immune response genes.
Fig. 1.
(A) Domain maps of SV40 TAgwt and mutants. J domain (J), Rb-protein (pRb, p130 and p107) binding motif (LXCXE), nuclear localization signal (NLS), origin binding domain (OBD), Zn domain (Zn), ATPase domain (ATPase), and host range domain (HR) are labeled on the linear representation of TAg. Grey circle represents cellular proteins interacting with TAg or mutants. (B) Global patterns of cellular gene expression in MEFs expressing wild-type or mutant T antigens. Genes 3 fold up or down with present call were selected for this analysis (see materials and methods for details).
RESULTS
Global patterns of cellular gene expression in MEFs expressing wild-type or mutant T antigens
We analyzed the RNA profile from two day post-confluent MEFs stably expressing wild-type or mutant T antigens. Protein levels of the wild-type or mutant TAgs in MEFs were assessed by western blot analysis (data not shown) and only those clones were selected for the study which were expressing similar levels of protein. The Affymetrix mouse whole genome chip, which consists of 21,635 unique genes, was used for microarray analysis. Genes showing three or more than three fold upregulation or downregulation in comparison to normal MEFs were selected for gross analysis of gene expression (Fig.1B). Using these criteria, we found that MEFs expressing TAgwt upregulates 446 genes and downregulates 558 genes in comparison to normal MEFs. MEFs expressing a truncated TAg (TAgN136) upregulated 593 genes and downregulated 512 genes. Noticeably, TAgN136 upregulated more genes than wild-type TAg. Mutation of the LXCXE motif (TAg3213) or the J domain (TAgD44N) resulted in a significant reduction in the number of genes upregulated by TAgwt and TAgN136, suggesting the importance of these domains in gene regulation. MEFs expressing TAg3213 upregulated119 genes and downregulated 399 genes, while MEFs expressing TAgD44N upregulated 117 genes and downregulated 470 genes. Thus, the number of genes downregulated by TAg was not affected significantly by these mutations. These results differ from our previous studies in intestine, where nearly all gene regulation required the TAg J domain and LXCXE motif (Rathi et al., 2009).
TAg regulates E2F-dependent gene expression and blocks p53-dependent targets
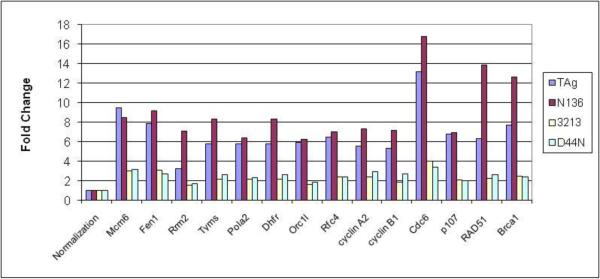
The LXCXE motif and J domain are required for the complete inactivation of Rb-family members and thus for the induction of E2F target genes (Srinivasan et al., 1997; Stubdal et al., 1997; Zalvide, Stubdal, and DeCaprio, 1998). Our previous study has shown that mice expressing TAgN136 in the villus enterocytes regulated the same set of E2F-target genes as TAgwt while TAg3213 and TAgD44N did not regulate a significant number of genes in this system (Rathi et al., 2009). Consistent with our studies in enterocytes (Rathi et al., 2009; Saenz-Robles et al., 2007a) we found a profound upregulation of E2F target genes in MEFs expressing TAgwt and TAgN136. This regulation was significantly reduced in MEFs expressing TAg3213 and TAgD44N (Fig. 2). We confirmed the expression levels of several E2F-target genes (BRCA1, RRM2, DHFR, B-myb, TS and Cyclin E) by semiquantitative RT-PCR analysis (data not shown).
Fig. 2.
TAg regulates gene expression by inactivating RB-family proteins. TAgwt and TAgN136 show significant upregulation while TAg3213 and TAgD44N show intermediate upregulation of E2F target genes. Fold change of the specified genes were determined by microarray analysis for the respective category: TAgwt (TAg), TAgN136 (N136), TAg3213 (3213) and TAgD44N (D44N). “Normalization” represents fold change of 1 (not regulated) in each category.
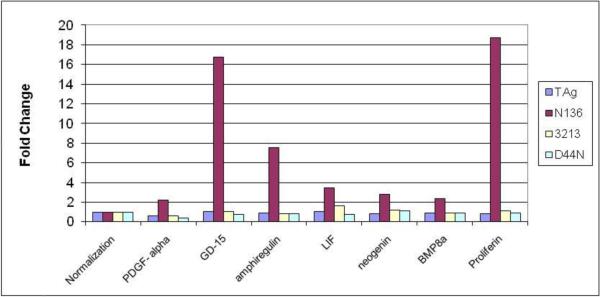
TAgwt binds to the tumor suppressor p53 and blocks p53 dependent transcription (Jiang et al., 1993; Lane and Crawford, 1979; Linzer and Levine, 1979). Consistent with the microarray data p53 target genes were upregulated in cell lines expressing TAgN136 while these genes were not altered in MEFs expressing TAg3213, TAgD44N, or TAgwt (Fig.3). We confirmed the expression levels of some of the p53-target genes (Lrdd, p21, NOXA, Mdm2, Cyclin G1, Fas, PERP, Pten and Rprm) by semiquantative RT-PCR analysis (data not shown). Interestingly, we also found a number of growth factors upregulated only by TAgN136 (Fig.4) and confirmed the expression levels of some of these genes (GD15, LIF, BMP8a, PDGF-α, amphiregulin and proliferin) by semiquantitative RT-PCR analysis (data not shown). To our knowledge, these genes are not known to be regulated by p53. The basis for their regulation by TAgN136 and the biological significance of their expression is under investigation.
Fig. 3.
TAgN136 upregulates p53 target genes. Fold change of the specified genes were determined by microarray analysis for the respective category: TAgwt (TAg), TAgN136 (N136), TAg3213 (3213) and TAgD44N (D44N). “Normalization” represents fold change of 1 (not regulated) in each category.
Fig. 4.
Upregulation of growth factors by TAgN136. Fold change of the specified genes were determined by microarray analysis for the respective category: TAgwt (TAg), TAgN136 (N136), TAg3213 (3213) and TAgD44N (D44N). “Normalization” represents fold change of 1 (not regulated) in each category.
TAg regulates expression of immune response genes
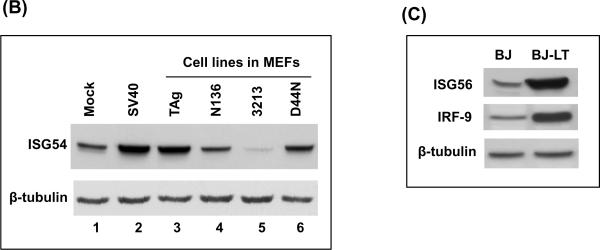
Many immune response genes were upregulated in MEFs stably expressing TAgwt as well as in cell abortively infected with SV40 virions (Table 1 and Fig.5). Many of these genes are known interferon-stimulated genes (ISGs). The products of ISGs are induced in response to virus infection and act on infected as well as uninfected cells to activate a global antiviral state (Randall and Goodbourn, 2008; Sarkar and Sen, 2004). These ISGs are upregulated both in MEFs infected with SV40 virus as well as MEFs stably expressing TAg. We confirmed the expression levels of some of the ISGs such as Ifi44, Ifi27, Oas2, GTPase2, Rsad2, Mx1, Oasl2, and IRF7 by semiquantitative RT-PCR (Fig. 5A). We found upregulation of these genes by MEFs expressing TAgwt and TAgD44N (lanes 3–4 and 9–10) but not by TAgN136 and TAg3213 (lanes 5–6 and 7–8). We also determined the protein levels of ISG54 in MEFs abortively infected with SV40 and MEFs stably expressing TAg or its mutants (Fig. 5B). We found that basal level of expression of ISG54 in normal MEFs (lane 1) was increased in MEFs infected with SV40 (lane 2) and MEFs stably expressing TAgwt or TAgD44N (lane 3 and 6). In order to confirm that induction of these immune response genes by TAg is not restricted to MEFs, we checked the protein levels of ISG56 and IRF9 in human fibroblast (BJ cells) expressing a vector control or large TAg (BJ-LT) (Fig. 5C). The levels of ISG56 and IRF9 were significantly increased in BJ-LT cells relative to BJ cells (Fig. 5C)
Table-1.
Immune response genes regulated by SV40, TAg and mutants.
| Specific Function Biological Process Description | Entrez Gene ID | Gene Symbol | SV40 | TAg | N136 | 3213 | D44N | ||
|---|---|---|---|---|---|---|---|---|---|
| Transcription | Interferon type I biosynthetic process | interferon regulatory factor 7 | 54123 | Irf7 | 29.1 | 4.2 | 1.0 | 0.8 | 5.7 |
| interferon dependent positive acting transcription factor 3 gamma | 16391 | Irf9 | 3.7 | 2.8 | 0.9 | 0.7 | 2.4 | ||
| JAK/Stat signaling | signal transducer and activator of transcription 1 | 20846 | Stat1 | 5.8 | 3.1 | 1.5 | 0.5 | 2.0 | |
| signal transducer and activator of transcription 2 | 20847 | Stat2 | 7.5 | 1.9 | 1.0 | 0.5 | 1.5 | ||
| Cytokine and chemokine signaling | N-myc (and STAT) interactor | 64685 | Nmi | 3.5 | 4.3 | 1.4 | 1.2 | 2.3 | |
| PML body | promyelocytic leukemia | 18854 | Pml | 1.8 | 2.5 | 3.1 | 1.4 | 1.9 | |
| nuclear antigen Sp100 | 20684 | Sp100 | 4.5 | 7.2 | 1.1 | 1.1 | 2.2 | ||
|
| |||||||||
| RNA binding | Interferon-induced | 2'-5' oligoadenylate synthetase 1C | 114643 | Oas1c | 2.5 | 2.4 | 2.2 | 1.5 | 2.5 |
| 2'-5' oligoadenylate synthetase-like 1 | 231655 | Oasl | 16.0 | 2.7 | 1.2 | 1.1 | 3.5 | ||
| 2'-5' oligoadenylate synthetase 1A | 246730 | Oas1a | 13.8 | 18.9 | 4.0 | 1.3 | 15.1 | ||
| 2'-5' oligoadenylate synthetase 2 | 246728 | Oas2 | 9.5 | 9.1 | 2.1 | 1.2 | 4.2 | ||
| 2'-5' oligoadenylate synthetase-like 2 | 23962 | Oasl2 | 82.4 | 16.3 | 0.6 | 0.4 | 7.1 | ||
|
| |||||||||
| Binding to other proteins | Interferon-induced | interferon-induced protein with tetratricopeptide repeats 1 | 15957 | Ifit1/ISG56 | 55.7 | 7.7 | 2.0 | 0.5 | 4.6 |
| interferon-induced protein with tetratricopeptide repeats 2 | 15958 | Ifit2 / ISG54 | 4.3 | 2.4 | 1.7 | 0.9 | 2.1 | ||
| interferon-induced protein with tetratricopeptide repeats 3 | 15959 | Ifit3 / ISG49 | 13.5 | 4.3 | 1.1 | 0.4 | 3.0 | ||
| interferon activated gene 202B | 26388 | Ifi202b | 7.7 | 3.3 | 1.2 | 0.8 | 2.3 | ||
| interferon activated gene 203 | 15950 | Ifi203 | 10.8 | 2.7 | 1.2 | 1.0 | 2.0 | ||
| interferon activated gene 204 | 15951 | Ifi204 | 8.3 | 3.2 | 0.9 | 1.1 | 1.8 | ||
|
| |||||||||
| NC* | Interferon-induced | interferon-induced protein 35 | 70110 | Ifi35 | 3.6 | 3.1 | 1.8 | 1.0 | 2.1 |
| interferon-induced protein 44 | 99899 | Ifi44 | 49.8 | 17.0 | 1.1 | 0.5 | 6.5 | ||
| interferon, alpha-inducible protein 27 | 76933 | Ifi27 | 21.6 | 21.5 | 3.4 | 1.6 | 6.5 | ||
| interferon gamma inducible protein 47 | 15953 | Ifi47 | 9.0 | 4.2 | 0.9 | 0.9 | 1.9 | ||
|
| |||||||||
| Helicase | Interferon-induced | interferon induced with helicase C domain 1 | 71586 | Ifih1 | 22.6 | 8.9 | 3.0 | 0.6 | 4.7 |
| Interferon type I biosynthetic process | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 | 230073 | Ddx58 / RIG-I | 5.5 | 4.2 | 1.2 | 0.9 | 1.6 | |
|
| |||||||||
| Catalytic activity | Virus response | radical S-adenosyl methionine domain containing 2 | 58185 | Rsad2 | 37.9 | 12.4 | 1.5 | 1.2 | 6.7 |
|
| |||||||||
| GTPase | Interferon-induced | myxovirus (influenza virus) resistance 1 | 17857 | Mx1 | 11.4 | 4.1 | 1.1 | 0.9 | 1.8 |
| myxovirus (influenza virus) resistance 2 | 17858 | Mx2 | 8.0 | 3.2 | 0.9 | 1.0 | 2.5 | ||
| GTPase, very large interferon inducible 1 | 74558 | Gvin1 | 3.4 | 2.5 | 1.7 | 0.5 | 1.1 | ||
| interferon inducible GTPase 1 | 60440 | Iigp1 | 23.2 | 6.8 | 0.3 | 0.3 | 2.8 | ||
| interferon inducible GTPase 2 | 54396 | Iigp2 | 10.1 | 8.5 | 1.0 | 0.6 | 4.4 | ||
| interferon gamma induced GTPase | 16145 | Igtp | 15.0 | 8.1 | 1.7 | 0.8 | 4.0 | ||
|
| |||||||||
| Protein Kinase | Interferon-induced | eukaryotic translation initiation factor 2-alpha kinase 2 | 19106 | Eif2ak2 / PKR | 3.1 | 2.3 | 1.1 | 0.9 | 2.0 |
|
| |||||||||
| ubiquitin like/protein binding | Interferon-induced | interferon, alpha-inducible protein | 100038882 | Isg15 | 33.2 | 9.8 | 1.7 | 0.4 | 4.1 |
|
| |||||||||
| Exonulcease | Interferon-induced | interferon-stimulated protein | 57444 | Isg20 | 3.8 | 1.9 | 1.1 | 1.6 | 1.8 |
|
| |||||||||
| Adenosine Deaminase | Interferon-induced | adenosine deaminase, RNA-specific | 56417 | Adar | 5.2 | 5.0 | 1.8 | 0.9 | 2.5 |
|
| |||||||||
| Receptor | Response to microbial infections | toll-like receptor 3 | 142980 | Tlr3 | 3.4 | 3.9 | 1.8 | 1.1 | 2.6 |
| Cell adhesion | lectin, galactose binding, soluble 9 | 16859 | Lgals9 | 3.5 | 3.9 | 0.9 | 1.1 | 2.4 | |
| Cell proliferation | CD274 antigen | 60533 | Cd274 | 2.4 | 2.6 | 1.3 | 1.3 | 1.2 | |
| Interferon-induced | RIKEN cDNA 5830458K16 gene | 67775 | Rtp4 | 28.3 | 7.4 | 1.6 | 0.4 | 4.3 | |
|
| |||||||||
| Endopeptidase | Antigen presentation | proteosome (prosome, macropain) subunit, beta type 8 (large multifunctional peptidase 7) | 16913 | Psmb8 | 10.6 | 5.5 | 1.5 | 0.4 | 2.6 |
| proteosome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) | 16912 | Psmb9 | 4.1 | 6.3 | 3.6 | 0.6 | 2.4 | ||
|
| |||||||||
| MHC class II presentation | Antigen presentation | Ia-associated invariant chain | 16149 | Cd74 | 1.1 | 2.1 | 0.8 | 1.9 | 1.4 |
|
| |||||||||
| MHC class I presentation | Antigen presentation | histocompatibility 2, K1, K region | 14972 | H2-K1 | 6.1 | 4.5 | 2.0 | 1.6 | 3.4 |
| histocompatibility 2, D region locus 1 | 14964 | H2-D1 | 3.5 | 4.2 | 3.0 | 1.5 | 2.5 | ||
| histocompatibility 2, K1, K region | 100044874 | LOC1000 44874 | 6.9 | 5.2 | 2.0 | 1.8 | 3.9 | ||
| histocompatibility 2, T region locus 23 | 15040 | H2-T23 | 5.1 | 3.6 | 3.0 | 0.9 | 2.0 | ||
| histocompatibility 2, D region locus 1 | 14980 | H2-L | 2.4 | 2.2 | 1.6 | 0.9 | 1.5 | ||
|
| |||||||||
| GTP binding | Interferon-induced | guanylate nucleotide binding protein 2 | 14469 | Gbp2 | 11.7 | 2.8 | 1.8 | 0.5 | 1.6 |
| macrophage activation 2 like | 100702 | Mpa2l | 18.3 | 8.5 | 0.8 | 0.6 | 3.1 | ||
|
| |||||||||
| transferase activity | protein localization | poly (ADP-ribose) polymerase family, member 9 | 80285 | Parp9 | 5.3 | 4.0 | 1.2 | 0.5 | 2.3 |
| Virus response | zinc finger CCCH type, antiviral 1 | 78781 | Zc3hav1 | 3.8 | 2.8 | 1.0 | 1.2 | 2.1 | |
|
| |||||||||
| cyclic-nucleotide phosphodiesterase activity | IFN-induced | SAM domain and HD domain, 1 | 56045 | Samhd1 | 2.5 | 2.3 | 1.4 | 1.8 | 2.0 |
|
| |||||||||
| dTTP biosynthesis | LPS response | thymidylate kinase family LPS-inducible member | 22169 | Tyki | 39.6 | 8.5 | 0.7 | 0.8 | 4.6 |
| placenta-specific 8 | 231507 | Plac8 | 11.9 | 2.0 | 0.1 | 0.5 | 0.3 | ||
|
| |||||||||
| phospholipid binding | defense response | lymphocyte antigen 6 complex, locus E | 17069 | Ly6e | 3.5 | 2.3 | 1.1 | 1.7 | 1.4 |
|
| |||||||||
| Ubiquitin ligase | Interferon-induced | deltex 3-like (Drosophila) | 209200 | Dtx3l | 6.5 | 3.7 | 1.3 | 0.7 | 2.6 |
|
| |||||||||
| Peptidase | Interferon-induced | ubiquitin specific peptidase 18 | 24110 | Usp18 | 49.0 | 20.4 | 1.8 | 0.7 | 9.7 |
|
| |||||||||
| Protein/nucleic acid binding | Interferon-induced | tripartite motif protein 34 | 94094 | Trim34 | 3.4 | 2.2 | 1.2 | 0.8 | 0.9 |
List of selected immune response genes showing differential regulation (in fold change) by SV40, TAg and mutants compared to wild-type MEFs.
NC* = Not classified.
Fig. 5.
Upregulation of interferon-stimulated genes (ISGs) by TAg. (A) Transcript levels of ISGs were analyzed in normal MEFs and MEFs expressing TAgwt (TAg), TAgN136 (N136), TAg3213 (3213) and TAgD44N (D44N). Two independent clones of each cell line were used for this experiment. cDNAs were reverse-transcribed from equal amounts of total RNA and subjected to PCR using specific primers. Transcript level of alcohol dehydrogenase 5 (Adh5) was used as a loading control. (B) MEFs were infected with SV40 (lane 2) or stably expressing TAg or its mutants (lane 3–6) were used. Whole-cell extracts from these cells were subjected to immunoblot for ISG54. Mock (lane 1) represent normal MEFs. β-tubulin was used as a loading control. (C) Whole cell extract from BJ cells (human fibroblast) expressing empty vector or TAg were subjected to immunoblots for ISG56 and IRF-9. β-tubulin was used as a loading control.
Interferons are not made by MEFs expressing TAg
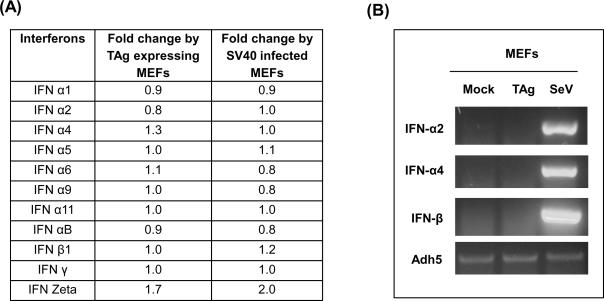
While many IFN-stimulated genes were upregulated in TAg expressing MEFs, the levels of IFN-α and -β mRNAs were not altered (Fig. 6A). Next, we tested the expression levels of IFN-α2, IFN-α4 and IFN- β in TAg expressing MEFs by semiquantitative RT-PCR analysis (Fig. 6B). MEFs infected with Sendai virus were used as a positive control and these cells showed a robust increase in IFN-α and -β mRNA levels. However, there was no increase in IFN mRNA levels in TAg expressing cells. This suggests that ISG induction is not due to the production of IFNs.
Fig. 6.
Interferon mRNAs are not increased in MEFs expressing TAg. (A) The table represents the fold change of the interferon genes, determined by microarray analysis in MEFs expressing TAg or infected with SV40 virions. (B) Transcript levels of Interferon α2, α4 and β1 were analyzed in normal MEFs and MEFs expressing TAgwt (TAg). MEFs infected with Sendai Virus were used as a positive control. cDNAs were reverse-transcribed from equal amounts of total RNA and subjected to PCR using specific primers. Transcript level of Alcohol dehydrogenase 5 (Adh5) was used as a loading control.
Activation of STAT1 by SV40 TAg
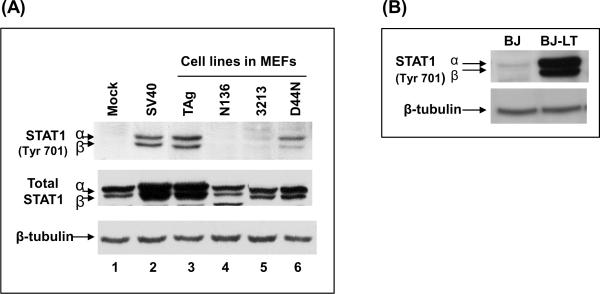
Stat1 plays a major role in both IFN-α/β and IFN-γ induced signaling pathways. Upon exposure to IFNs, Stat1 is activated by tyrosine phosphorylation by tyrosine kinases such as JAK1 (Darnell, Kerr, and Stark, 1994; Goodbourn, Didcock, and Randall, 2000). STAT1 is also an ISG whose transcription is upregulated by IFNs. Stat1 mRNA levels were increased in MEFs expressing TAg (see Table 1). To test for Stat1 activation, we analyzed the phosphorylation status of Stat1 by western blot analysis using whole cell extracts from MEFs stably expressing TAgwt or mutants as well as MEFs abortively infected with SV40 virions (Fig. 7A). We found phosphorylation of Stat1 at Tyr701 in MEFs infected with SV40 (lane 2) as well as MEFs expressing TAgwt (lane 3) or TAgD44N (lane 6). However, Stat1 were not phosphorylated in MEFs stably expressing TAgN136 (lane 4) or TAg3213 (lane 5). In addition, the steady-state levels of Stat1 protein was increased in MEFs infected with SV40 or stably expressing TAgwt. Stat1 was also phosphorylated at Tyr701 in BJ cells expressing TAgwt (Fig. 7B). Thus, T antigen-induced STAT1 phosphorylation followed the same genetics as ISG induction.
Fig. 7.
Activation of STAT1 by TAg. (A) MEFs infected with SV40 (lane 2) or stably expressing TAg or its mutants were used (lane 3–6). Whole-cell extracts from these cells were subjected to immunoblots for phospho-specific (Tyr701) STAT1 and total STAT1. Mock (lane 1) represent normal MEFs. β-tubulin was used as a loading control. (B) Whole cell extract from BJ cells (human fibroblast) expressing empty vector (BJ) or TAg (BJ-LT) were subjected to immunoblots for phospho-specific (Tyr701) STAT-1. β-tubulin was used as a loading control.
Regulation of immune response genes by TAg requires interaction with p53
TAg directly binds to the DNA-binding surface of p53 thus blocking its ability to bind promoters and to regulate gene expression (Bargonetti et al., 1993; Jiang et al., 1993). As a consequence, complex formation between TAg and p53 interferes with the expression of p53-regulated genes involved in several biological processes such as cell cycle (p21, cyclin G1), DNA repair (GADD45), apoptosis (Bax) and signal transduction (IGF-BP3). We found that MEFs expressing TAgN136, a truncation mutant unable to bind p53, did not show induction of immune response genes (Fig. 5A). To determine if p53 binding to TAg is required for ISG regulation, we used TAg mutant Patch-1 (Ahuja et al., 2009). Mutations comprising patch-1 specifically alter the p53-TAg interface and thus fail to bind and inactivate p53 (Ahuja et al., 2009). Figure 8 shows that Patch-1 is defective for the upregulation of ISGs. We conclude that T antigen binding and perhaps stabilization of p53 is required for ISG induction.
Fig. 8.
Regulation of immune response genes by TAg requires interaction with p53. Transcript levels of Interferon-stimulated genes including Oasl2, Mx1 and IRF-7 were analyzed in normal MEFs and MEFs expressing TAgwt (TAg), TAgN136 (N136) or p53 binding defective mutant, Patch-1 (P1). cDNAs were reverse-transcribed from equal amounts of total RNA and subjected to PCR using specific primers. Transcript level of Alcohol dehydrogenase 5 (Adh5) was used as a loading control.
DISCUSSION
SV40 is thought to induce transformation in part by acting on key transcriptional regulators and thereby altering cellular gene expression. Thus, SV40 TAg antagonizes the ability of Rb proteins to repress E2F-dependent gene expression leading to the expression of genes required for cell cycle entry and progression while simultaneously blocking p53-dependent transcription and consequently inhibiting apoptosis. Consistent with this view, we previously reported that TAg upregulates E2F-dependent genes in both primary MEFs and transgenic murine enterocytes and that p53-dependent transcription was not induced in either of these systems (7). However, these studies were limited by the use of a Agilent mouse cDNA array which only included 8462 genes of the estimated 21,000 mouse genes.
We have now extended these studies by the use of mouse whole-genome arrays and by including three key mutants (TAgN136, TAg3213 and TAgD44N) in addition to TAgwt in the analysis. TAg3213 and TAgD44N are defective for Rb protein inactivation but retain the ability to bind p53, while TAgN136 is defective for p53 interaction but retains the ability to inhibit the Rb proteins. In addition, to eliminate genes whose regulation may be altered as a consequence of cell line establishment, we have analyzed gene expression in MEFs abortively infected with SV40.
TAg induces E2F-dependent transcription and blocks expression of p53- target genes
Studies of transgenic enterocytes expressing wild-type or mutant TAg demonstrated that nearly all TAg-dependent gene regulation in these cells can be explained by the inactivation of the Rb proteins and resultant upregulation of E2F-dependent gene transcription. Neither wild-type nor the mutant TAg's affected the expression of p53-regulated genes, consistent with the lack of p53 expression in this cell type (Markovics et al., 2005).
In this report we examined the consequences of wild-type and mutant TAg's in MEFs. Consistent with our studies in enterocytes we found that TAg induction of E2F-dependent transcription depends on both a functional J domain and Rb-binding LXCXE motif, and that the first 136 amino acids of TAg, that contains both of these elements is capable of upregulating these genes. In contrast to enterocytes, MEFs upregulated p53-dependent genes in response to TAgN136. However, p53-dependent genes were not induced, or in some cases were repressed, by TAgwt and by TAg3213 and TAgD44N, each of which retains the ability to bind p53.
Induction of immune response genes by TAg requires the LXCXE motif and p53 binding
Previously we noticed that TAg regulated a set of immune response genes in MEFs and that these same genes were not regulated by TAg in transgenic enterocytes (7). The use of the whole genome mouse array in this study clearly shows the upregulation of a large number of immune response genes by SV40 T antigen (Table 1). These genes represent two major classes: (1) Interferon-stimulated genes (ISGs), such as the OAS family, MX1, ISG56, ISG54, ISG15, Cig5, GTPase, P200 gene family and PKR; and, (2) genes involved in interferon induction and signaling such as IRF-7, IRF-9, RIG-1, STAT1 and STAT2. This collection of genes was upregulated in both MEFs abortively infected with SV40 as well as in MEFs stably expressing TAg indicating that their altered regulation is not an artifact arising from cell line establishment.
The genetic data suggests that the immune response genes are regulated by a common mechanism. All of these genes are regulated by TAgwt and by TAgD44N but not by TAgN136or TAg3213. This suggests a requirement for both the LXCXE motif and a TAg function or functions carboxy-terminal to amino acid 136 The inability of TAg3213 to regulate the immune response genes suggests that TAg binding to Rb family members plays a role in this effect. However, the inactivation of Rb proteins by TAg is thought to be J domain dependent so the observation that TAgD44N is capable of regulating these genes argues against this possibility. Perhaps the LXCXE motif can also act on Rb proteins in a J-domain-independent manner. Alternatively, the LXCXE motif may target cellular proteins other than Rb and these unknown targets may play a role in regulating the immune response genes.
TAgN136 is fully able to inactivate Rb family members, yet it is unable to induce ISGs. This indicates that the regulation of immune response genes requires one or more activities residing in the carboxy-terminal region of TAg. One candidate activity is the ability of TAg to bind p53 and we found that a p53 binding defective mutant of TAg (Patch-1) is unable to upregulate ISGs (Fig. 8). This indicates that the TAg-p53 interaction is necessary for ISG induction. At present we can not distinguish between a “loss of function” model, in which p53 normally functions to repress ISG induction and TAg blocks this p53 action, or a “gain-of-function” model in which the TAg-p53 complex is actively involved in ISG expression. Bocchetta et al have shown that TAg-p53 complexes are required to activate the insulin-like growth factor-I promoter (Bocchetta et al., 2008). Consistent with a “gain of function” hypothesis, enterocytes, which lack p53 expression, do not show induction of immune response genes (Cantalupo et al., 2009; Rathi et al., 2009). In either case, binding of TAg to p53 alone is not sufficient for ISG induction as TAg3213 can bind to p53 but is unable to induce the immune response genes. Collectively, our mutant analysis suggests cooperation between LXCXE motif of TAg and binding with p53 in the regulation of immune response genes.
TAg activates STAT-1 in the absence of interferon production
Interestingly, TAg is capable of inducing the downstream interferon pathway without affecting the levels of IFN-α or IFN-β. We found that TAg induces STAT1 Tyr701 phosphorylation suggesting that TAg can activate the interferon signaling pathway independent of interferon production. One possibility is p53-dependent activation of STAT1 by c-Abl1 tyrosine kinase instead of the classical JAK-STAT pathway as reported by Youlyouz-Marfak et al (Youlyouz-Marfak et al., 2008). Furthermore, TAg mutants capable of inducing ISG expression also induce STAT-1 phosphorylation while mutants defective for ISG induction do not induce STAT-1 phosphorylation. This suggests that the primary mechanism by which TAg induces ISG's is through STAT-1 signaling.
In conclusion, we have shown that SV40-transformed MEFs have activated the interferon pathway in the absence of interferon production. The biological consequences of this activation remain unclear. However, one practical consequence of this observation is in the common use of TAg in immortalization of MEFs obtained from knockout mice. Our observation clearly demonstrates that this practice should be critically evaluated while testing functional consequences of gene knockouts, especially in the studies involving genes that modulate innate immune signaling pathways.
Several herpesviruses, such as Epstein–Barr virus, herpes simplex virus, Kaposi's sarcoma-associated herpesvirus and HCMV have been shown to activate IFN-responsive genes, such as MxA and OAS (Browne et al., 2001; Mossman et al., 2001; Poole et al., 2002; Ruvolo et al., 2003; Zhu et al., 1998). However, in these cases the induction occurs in the context of a productive infection and the effects of the ISG's are later mitigated by other viral functions. One interesting question that arises is how TAg expressing MEFs are able to survive in the presence of high levels of ISGs which are known to create growth inhibitory environment. Future studies are needed to identify the cellular target(s) on which TAg acts to elicit ISG expression and to explore the connection between ISGs and transformation by TAg.
MATERIALS AND METHODS
Isolation of primary fibroblasts, cell culture conditions, and establishment of cell lines
Mouse embryo fibroblasts (MEFs) were harvested from 13.5-day-old FVB embryos as described previously (Markovics et al., 2005) and grown in DMEM (Mediatech, cat# 10-013-CV) supplemented with 10% heat-inactivated fetal bovine serum (Hyclone, cat# SH30071.03), 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen, cat# 15140-122). MEFs were grown at 37°C in 5% CO2. Stable MEF cell lines were made with pRSVB-neo plasmid that expresses a Geneticin (G418) resistance gene and a cDNA or genomic version of the wild-type or mutant T antigens (N136, 3213 (E107K, E108K) and 5110 (D44N)) under the control of the Rous sarcoma virus (RSV) promoter (Srinivasan et al., 1997). Primary cultures of MEFs were transfected with above plasmids using Lipofectamine reagent (Invitrogen), according to the manufacturer's instructions. Upon selection in culture with 0.4 mg/ml of G418 (Invitrogen, cat# 11811-031), transformed colonies (foci) were selected and individually grown and several independent cell lines were established.
SV40 infection
For abortive SV40 infections, confluent MEFs were infected with SV40 at a multiplicity of infection 10 pfu/cell (M.O.I = 10) as described (Tremblay, Sachsenmeier, and Pipas, 2001). Cells were incubated for an additional 3 to 4 days before the cells were harvested with trypsin. The cell pellet was washed three times with cold PBS/EDTA and frozen at −80°C for protein and RNA extraction.
RNA extraction
Wild-type MEFs and MEFs expressing TAgwt and TAg mutants were grown to confluence and fed with fresh medium. After 2 days, the cells were harvested and the cell pellets were washed with cold PBS/EDTA three times. Two independent wild-type MEFs, three independent MEFs expressing TAgwt and mutants were used to prepare RNA. Three independent mock and SV40 infections were conducted. Total RNA from transfected or infected MEFs was isolated using the RNeasy kit according to the manufacture's protocol (Qiagen Inc.). Genomic DNA contamination was eliminated by digesting the RNA with RNase-free DNase. The yield (at absorbance A260) and purity (A260/A280 ratio) of each RNA sample were determined by using a spectrophotometer (Eppendorf).
Microarrays
Total RNA was sent to the Genomics and Proteomics Core Laboratories (University of Pittsburgh) for hybridization to the Mouse 430 2.0 whole genome array (Affymetrix) which contains 45,101 probesets representing 21,635 unique genes. CEL files for each array were converted into RMA expression values using BRB-Array Tools (Rich Simon, National Cancer Institute, http://linus.nci.nih.gov/BRB-ArrayTools.html). An average fold change ratio (Experimental/Control) and a one-sample T-test was calculated for each probe set. The total number of genes which were selected showed three fold up or downregulation in an individual experimental class (TAgwt / TAgN136/ TAg3213/ TAgD44N). We applied additional criteria of present and absent calls provided by Affymetrix data files. To be included for consideration, an upregulated gene needs to be present in all the replicates of an experimental class and a downregulated gene needs to be present in all the replicates of the control class (wild-type MEFs). Microarray data submitted to GEO, under the accession number: GSE20620
Reverse transcription PCR (RT-PCR) analysis
cDNA synthesis from 1 μg of total RNA was performed using Superscript II Reverse Transcriptase (Invitrogen). PCR was performed with equal amounts of cDNA using GoTAq polymerase (Promega) for 25 cycles with specific primers for the different transcripts. Amplification with primers for the Alcohol dehydrogenase 5 (Adh5) transcript was used as a normalizing control. PCR reaction products were resolved through a 2% agarose gel in 1× TAE and stained with GelStar (Cambrex Bio Science). The cDNA was amplified with PCR using primers specific for each gene as shown in Table 2. To ensure that these reactions were within the linear range of the assay, we optimized the number of cycles required to obtain non-saturated signals. Exponential amplifications of PCR products were obtained as follows: 2 min. at 94°C; a series of 25 cycles at 94°C for 30 sec., variable annealing temperatures for 30 sec., and 72°C for 30 sec., and a final extension step of 5 min. at 72°C. The annealing temperatures and product sizes for each gene are described in Table 2. The products were resolved on 2% agarose gels and stained with GelStar (BioWhittaker Molecular Applications).
Table-2.
Primers used in this study.
| Gene | Forward (F) or Reverse (R) | Primer Sequence (5' to 3') | Product size (bp) | Annealing temp. (°C) | Extention time (sec) |
|---|---|---|---|---|---|
| Adh | F | TGCACCACCAACTGCTTAG | 152 | 58 | 30 |
| R | GATGCAGGGATGATGTTC | ||||
| Ifi44 | F | GAGAGAACAGGGAATGAAGAAGGC | 134 | 52.4 | 30 |
| R | CCAACAGAATTGCGATTGGTCC | ||||
| Ifi27 | F | CCATAGCAGCCAAGATGATGTCTG | 121 | 55 | 30 |
| R | GCATTTGTTGATGTGGAGAGTCC | ||||
| Oas2 | F | AAAACCAACCGCTCCCAGTTCGTC | 488 | 57.1 | 30 |
| R | GCAATGTCAAAGTCATCTGTGCC | ||||
| GTPase2 | F | CTTCCACCTGCTTGTTCTTTGG | 266 | 55.4 | 30 |
| R | TCACAGTTTCCTCAGTGCTGGG | ||||
| Rsad2 | F | CAATCACACCCAGCAGCAGTTAG | 209 | 54.4 | 30 |
| R | AGCGATGCCTCAGAACACAGTG | ||||
| Mx1 | F | CAGCACCTGAAAGCCTACTACCAG | 135 | 53.6 | 30 |
| R | GGTGTCCTGTAAAAGCTGAAGCATC | ||||
| Oasl2 | F | TTACAGAACAGCCAGAGCTATACGG | 548 | 56.1 | 40 |
| R | CAAGGGAGATAGATTTACGTCCACG | ||||
| IRF7 | F | ACACCATCTACCTGGGTTTTG | 243 | 54 | 60 |
| R | TTGGGATTCTGAGTCAAGGC | ||||
| IFN-α2 | F | CATCTGCTGCTTGGAATACAACC | 197 | 56.7 | 30 |
| R | GGGGCTGTGTTTCTTCTCTCTCAG | ||||
| IFN-α4 | F | TCAATGACCTCAAAGCCTGTGTG | 211 | 56.6 | 30 |
| R | CACTCCTCCTCACTCAGTCTT | ||||
| IFN-β | F | AAGAGTTACACTGCCTTTGCCATC | 138 | 53.2 | 30 |
| R | AAACACTGTCTGCTGGTGGAGTTC |
Immunoblot analysis
Lysates were prepared from 2-day post-confluent cells in lysis buffer [50 mM HEPES pH 7.9, 400 mM KCl, 0.5 mM EDTA, 0.1% NP40, 10% glycerol, 1 mM DTT, 0.5 mM Na3VO4, 0.5 mM NaF, 1 μg/ml pepstatin and a protease inhibitor tablet (Roche)] for 30 min on ice, then centrifuging the lysate for 10 min at 4°C. Protein concentration was determined by Bradford assay (Bio-Rad). Appropriate dilutions of the following primary antibodies were used: anti-STAT1 (Cell signaling); anti-Phospho-STAT1 (Tyr701) (Cell signaling), anti-ISG54 (Thermo Fischer); anti-ISG56 (gift from Dr. Saumendra N. Sarkar), anti-IRF-9 (C-20; Santa Cruz), anti-β-tubulin-HRP (Santa Cruz). Goat anti-mouse A2554 and goat anti-rabbit A0545 (Sigma) were used as secondary antibodies.
ACKNOWLEGEMENTS
We thank Dr. William C. Hahn (Department of Medical Oncology, Dana-Farber Cancer Institute) for providing BJ and BJ-LT cells. This work was supported by National Institutes of Health grant CA40586 to J.M.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahuja D, Rathi AV, Greer AE, Chen XS, Pipas JM. A structure-guided mutational analysis of simian virus 40 large T antigen: identification of surface residues required for viral replication and transformation. J Virol. 2009;83(17):8781–8. [Google Scholar]
- Ahuja D, Saenz-Robles MT, Pipas JM. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene. 2005;24(52):7729–45. doi: 10.1038/sj.onc.1209046. [DOI] [PubMed] [Google Scholar]
- Ali SH, DeCaprio JA. Cellular transformation by SV40 large T antigen: interaction with host proteins. Semin Cancer Biol. 2001;11(1):15–23. doi: 10.1006/scbi.2000.0342. [DOI] [PubMed] [Google Scholar]
- Bargonetti J, Manfredi JJ, Chen X, Marshak DR, Prives C. A proteolytic fragment from the central region of p53 has marked sequence-specific DNA-binding activity when generated from wild-type but not from oncogenic mutant p53 protein. Genes Dev. 1993;7(12B):2565–74. doi: 10.1101/gad.7.12b.2565. [DOI] [PubMed] [Google Scholar]
- Bocchetta M, Eliasz S, De Marco MA, Rudzinski J, Zhang L, Carbone M. The SV40 large T antigen-p53 complexes bind and activate the insulin-like growth factor-I promoter stimulating cell growth. Cancer Res. 2008;68(4):1022–9. doi: 10.1158/0008-5472.CAN-07-5203. [DOI] [PubMed] [Google Scholar]
- Browne EP, Wing B, Coleman D, Shenk T. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J Virol. 2001;75(24):12319–30. doi: 10.1128/JVI.75.24.12319-12330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantalupo PG, Saenz-Robles MT, Rathi AV, Beerman RW, Patterson WH, Whitehead RH, Pipas JM. Cell-type specific regulation of gene expression by simian virus 40 T antigens. Virology. 2009;386(1):183–91. doi: 10.1016/j.virol.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JE, Jr., Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264(5164):1415–21. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- DeCaprio JA, Ludlow JW, Figge J, Shew JY, Huang CM, Lee WH, Marsilio E, Paucha E, Livingston DM. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54(2):275–83. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- Goodbourn S, Didcock L, Randall RE. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J Gen Virol. 2000;81(Pt 10):2341–64. doi: 10.1099/0022-1317-81-10-2341. [DOI] [PubMed] [Google Scholar]
- Hahn WC, Dessain SK, Brooks MW, King JE, Elenbaas B, Sabatini DM, DeCaprio JA, Weinberg RA. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol Cell Biol. 2002;22(7):2111–23. doi: 10.1128/MCB.22.7.2111-2123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Srinivasan A, Lozano G, Robbins PD. SV40 T antigen abrogates p53-mediated transcriptional activity. Oncogene. 1993;8(10):2805–12. [PubMed] [Google Scholar]
- Kierstead TD, Tevethia MJ. Association of p53 binding and immortalization of primary C57BL/6 mouse embryo fibroblasts by using simian virus 40 T-antigen mutants bearing internal overlapping deletion mutations. J Virol. 1993;67(4):1817–29. doi: 10.1128/jvi.67.4.1817-1829.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278(5701):261–3. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- Linzer DI, Levine AJ. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17(1):43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- Markovics JA, Carroll PA, Robles MT, Pope H, Coopersmith CM, Pipas JM. Intestinal dysplasia induced by simian virus 40 T antigen is independent of p53. J Virol. 2005;79(12):7492–502. doi: 10.1128/JVI.79.12.7492-7502.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossman KL, Macgregor PF, Rozmus JJ, Goryachev AB, Edwards AM, Smiley JR. Herpes simplex virus triggers and then disarms a host antiviral response. J Virol. 2001;75(2):750–8. doi: 10.1128/JVI.75.2.750-758.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren M, Maltzman W, Levine AJ. Post-translational regulation of the 54K cellular tumor antigen in normal and transformed cells. Mol Cell Biol. 1981;1(2):101–10. doi: 10.1128/mcb.1.2.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipas JM, Levine AJ. Role of T antigen interactions with p53 in tumorigenesis. Semin Cancer Biol. 2001;11(1):23–30. doi: 10.1006/scbi.2000.0343. [DOI] [PubMed] [Google Scholar]
- Poole LJ, Yu Y, Kim PS, Zheng QZ, Pevsner J, Hayward GS. Altered patterns of cellular gene expression in dermal microvascular endothelial cells infected with Kaposi's sarcoma-associated herpesvirus. J Virol. 2002;76(7):3395–420. doi: 10.1128/JVI.76.7.3395-3420.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89(Pt 1):1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Rathi AV, Saenz Robles MT, Cantalupo PG, Whitehead RH, Pipas JM. Simian virus 40 T-antigen-mediated gene regulation in enterocytes is controlled primarily by the Rb-E2F pathway. J Virol. 2009;83(18):9521–31. doi: 10.1128/JVI.00583-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathi AV, Saenz Robles MT, Pipas JM. Enterocyte proliferation and intestinal hyperplasia induced by simian virus 40 T antigen require a functional J domain. J Virol. 2007;81(17):9481–9. doi: 10.1128/JVI.00922-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvolo V, Navarro L, Sample CE, David M, Sung S, Swaminathan S. The Epstein-Barr virus SM protein induces STAT1 and interferon-stimulated gene expression. J Virol. 2003;77(6):3690–701. doi: 10.1128/JVI.77.6.3690-3701.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz-Robles MT, Markovics JA, Chong JL, Opavsky R, Whitehead RH, Leone G, Pipas JM. Intestinal hyperplasia induced by simian virus 40 large tumor antigen requires E2F2. J Virol. 2007a;81(23):13191–9. doi: 10.1128/JVI.01658-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz-Robles MT, Toma D, Cantalupo P, Zhou J, Gong H, Edwards C, Pipas JM, Xie W. Repression of intestinal drug metabolizing enzymes by the SV40 large T antigen. Oncogene. 2007b;26(35):5124–31. doi: 10.1038/sj.onc.1210310. [DOI] [PubMed] [Google Scholar]
- Sarkar SN, Sen GC. Novel functions of proteins encoded by viral stress-inducible genes. Pharmacol Ther. 2004;103(3):245–59. doi: 10.1016/j.pharmthera.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Srinivasan A, McClellan AJ, Vartikar J, Marks I, Cantalupo P, Li Y, Whyte P, Rundell K, Brodsky JL, Pipas JM. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol Cell Biol. 1997;17(8):4761–73. doi: 10.1128/mcb.17.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubdal H, Zalvide J, Campbell KS, Schweitzer C, Roberts TM, DeCaprio JA. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol Cell Biol. 1997;17(9):4979–90. doi: 10.1128/mcb.17.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CS, Gilbert SP, Pipas JM. ATP-dependent simian virus 40 T-antigen-Hsc70 complex formation. J Virol. 2001;75(4):1601–10. doi: 10.1128/JVI.75.4.1601-1610.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DL, Kalderon D, Smith AE, Tevethia MJ. Dissociation of Rb-binding and anchorage-independent growth from immortalization and tumorigenicity using SV40 mutants producing N-terminally truncated large T antigens. Virology. 1990;178(1):15–34. doi: 10.1016/0042-6822(90)90375-2. [DOI] [PubMed] [Google Scholar]
- Tremblay JD, Sachsenmeier KF, Pipas JM. Propagation of wild-type and mutant SV40. Methods Mol Biol. 2001;165:1–7. doi: 10.1385/1-59259-117-5:1. [DOI] [PubMed] [Google Scholar]
- Youlyouz-Marfak I, Gachard N, Le Clorennec C, Najjar I, Baran-Marszak F, Reminieras L, May E, Bornkamm GW, Fagard R, Feuillard J. Identification of a novel p53-dependent activation pathway of STAT1 by antitumour genotoxic agents. Cell Death Differ. 2008;15(2):376–85. doi: 10.1038/sj.cdd.4402270. [DOI] [PubMed] [Google Scholar]
- Zalvide J, Stubdal H, DeCaprio JA. The J domain of simian virus 40 large T antigen is required to functionally inactivate RB family proteins. Mol Cell Biol. 1998;18(3):1408–15. doi: 10.1128/mcb.18.3.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Cong JP, Mamtora G, Gingeras T, Shenk T. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc Natl Acad Sci U S A. 1998;95(24):14470–5. doi: 10.1073/pnas.95.24.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]