Abstract
The botulinum neurotoxins (BoNTs) are the most potent protein toxins for humans. There are seven serotypes of BoNTs (A-G) based on a lack of cross anti-sera neutralization. BoNTs utilize gangliosides as components of the host receptors for binding and entry into neurons. Members of BoNT/C and BoNT/D serotypes include mosaic toxins that are organized in D/C and C/D toxins. One D/C mosaic toxin, BoNT/D-South Africa (BoNT/D-SA), was not fully neutralized by immunization with BoNT serotype /C or /D, which stimulated this study. Here the crystal structures of the receptor binding domains of BoNT/C, BoNT/D, and BoNT/D-SA are presented. Biochemical and cell binding studies show that BoNT/C and BoNT/D-SA possess unique mechanisms for ganglioside binding. These studies provide new information on how the BoNTs can enter host cells as well as a basis for understanding the immunological diversity of these neurotoxins.
Botulinum neurotoxins (BoNTs) are the most toxic proteins known for humans (1). BoNTs are categorized into 7 serotypes (A-G) based on neutralization characteristics where antiserum to one serotype will neutralize that serotype but not the other six serotypes. BoNTs are synthesized as 150-kDa proteins that possess AB structure-function organization, where AB refer to individual domains within a protein toxin (2, 3). BoNTs are cleaved by bacterial- or host- proteases into the two subunits, light chain (LC, A for the activity domain) and a heavy chain (HC, B for the binding domain) that are linked by a disulfide bond (4). The LC is ~50-kDa and possesses zinc-dependent protease activity (5), while the HC is ~100kDa and contains a receptor binding domain (HCR) and a translocation domain (HCT). HCT is primarily α-helical with a disordered “belt” that wraps around the LC which may serve as a protease chaperone (6). HCR comprises two structural subdomains, an N-terminal jelly-roll motif and a C-terminal β-trefoil conformation.
BoNTs utilize dual receptors for entry into neurons (7). Upon delivery to a cholinergic nerve terminus, BoNT HCRs bind a ganglioside (8), which localizes BoNT to the neuronal plasma membrane. BoNTs then interact with a protein receptor to facilitate internalization. Host proteins that serve as BoNT receptors include the synaptic vesicle proteins, such as Synaptic Vesicle protein 2 (SV2) for BoNT/A, E, and F and Synaptotagmin I/II for BoNT/B and G (9–13). The host protein receptors for BoNT/C and BoNT/D remain to be determined. BoNT serotypes /A, /B, /E, /F, and /G are internalized through synaptic vesicle recycling (7). How BoNT/C and /D enter neurons is not known. Upon acidification, HCT inserts into the synaptic vesicle membrane and translocates LC across the vesicle membrane into the cytosol where the LC (14, 15) dissociates from the HC (16). BoNT LCs are zinc proteases that cleave neuronal soluble N-ethylmaleimide-sensitive-factor attachment protein receptors (SNARE) proteins: BoNT/A and /E cleave SNAP25, BoNT/B, /D, /F, and /G cleave Synaptobrevin-2/ Vesicle Associated Membrane Protein-2 (Syb-2/ VAMP-2). BoNT/C cleaves SNAP25 and Syntaxin 1 (17–20). Thus, BoNTs inhibit synaptic vesicle fusion and subsequent neurotransmitter signaling.
BoNT/C and BoNT/D are not typically associated with natural human infections (21). While BoNT/C does not appear to be toxic to humans following ingestion (22), BoNT/C is toxic for human tissues and cleaves SNAP25 and syntaxin in human neurons (23). BoNT/C was first isolated in 1922 and was shown to be responsible for botulism outbreaks that resulted in mass avian deaths (24–26). Mice deficient in complex ganglioside synthesis are resistant to BoNT/C toxicity in agreement with a direct interaction being observed between BoNT/C and the gangliosides GD1b and GT1b (27). Conversely, BoNT/C is not believed to utilize a synaptic vesicle protein for neuron entry and high affinity binding of BoNT/C to cell lysates is insensitive to proteinase K (27–29). In contrast, BoNT/D does not bind gangliosides, but binds derivatives of phosphatidylethanolamine (27). In addition to the prototype toxins, BoNT/C and BoNT/D mosaics (D/C and C/D) exist that include BoNT/D-South Africa (BoNT/D-SA), a D/C mosaic toxin. Mice immunized with BoNT/C HCR were only partially protected from challenge by BoNT/D-SA, and immunization with BoNT/D HCR did not protect from BoNT/D-SA challenge (30).
To initiate studies to define why vaccination with HCR/C or HCR/D did not provide compete protection against challenge by BoNT/D-SA, the crystal structures of BoNT/C, BoNT/D, and BoNT/D-SA have been solved. This allowed the identification of a unique mechanism for ganglioside binding by BoNT/C and BoNT/D-SA. In addition, we solved the structure of a mutated form of HCR/D-SA, W1252A, which has a decreased affinity for ganglioside binding.
Experimental Procedures
Materials
Escherichia coli codon optimized DNA encoding the HCR domains of BoNT/C-Stockholm (residues 864–1290), BoNT/D-1873 (residues 861–1276), and BoNT/D-South Africa-5995 (residues 867–1285) were synthesized by EZBiolab (Westfield, IN). Chemicals and reagents were obtained from Sigma-Aldrich Co. (St. Louis, MO) and restriction enzymes were purchased from New England Biolabs or Invitrogen (Carlsbad, CA). Sprague–Dawley rat embryonic day 18 cortical neurons were purchased from Brainbits LLC (Springfield, IL) and cultured as described by the supplier. Neuronal cell culture reagents were purchased from Invitrogen.
Expression and Purification of BoNT HCRs
DNA encoding HCRs were subcloned into pET-28a (Novagen) which introduced a 3-FLAG eiptope N-terminal to the HCR. HCRs were purified from E. coli BL-21(DE3) using three sequential steps: Ni2+-nitrilotriacetic acid chromatography, S200-HR gel-filtration chromatography, and DEAE-sephacryl ion exchange chromatography. Typical purifications from a 1-L culture yielded between 5 and 10 mg of HCR. Mutated forms of HCR/C and HCR/D-SA were prepared using a Quikchange site-directed mutagenesis kit (Stratagene).
HCR-Ganglioside Binding Assay
HCR binding to gangliosides was measured in a solid phase assay as previously described (31) Briefly, bovine brain gangliosides (Matreya, LLC) were stored in dimethylsulfoxide (20 mg/ml) at −20°C, diluted into methanol, and applied to non-protein binding 96-well plates (Corning Costar # 3641) at 0.5 or 2.0 μg gangliosides/well. MeOH was evaporated at RT, and the wells were washed three times with phosphate buffered saline (PBS). Plates were blocked with 2% BSA in sodium carbonate buffer (50mM Na2CO3, pH 9.6) for 1 hr. Binding assays were performed in 100 μl of PBS/well for 2 hr at 4°C containing 10 nM of the indicated HC. Bound HCR was detected with α-FLAG M2 monoclonal antibody conjugated to horseradish peroxidase (HRP, diluted 1/10000, Sigma-Aldrich) and detected using TMB-Ultra (Pierce Biochemicals) as the substrate. The reaction was stopped with 1 M H2SO4, and the absorbance at 450nm was determined using a plate reader (Victor 3 V, Perkin-Elmer).
HCR-cell binding assays
Primary rat cortical neurons were plated and cultured in 24-well glass bottom culture dishes pre-coated with poly-D-lysine and laminin (Sigma-Aldrich, St. Louis, MO. and Thermo Fisher Scientific, Wyman, MA). Following 10–14 days, neurons were cooled with two washes in cold PBS and incubated with a dilution series of the indicated HCR in neurobasal media for 1 hr at 4°C. Cells were washed and processed for microscopy. Briefly, fixation in 4% paraformaldehyde in PBS (15 min at RT) was followed by incubation with 150 mM glycine in PBS. Non-specific binding sites were blocked with 10% fetal calf serum, 2.5% fish skin gelatin (Sigma-Aldrich), 0.1% TritonX-100 (Sigma-Aldrich) and 0.05% Tween20 (EMD Chemicals, Gibbstown, NJ) in PBS for 1 hr at room temperature. Cells were incubated overnight with α-FLAG M2 mAb (diluted 1/15000, Sigma-Aldrich) followed by goat α-mouse IgG-488 (diluted 1/500, Invitrogen, Carlsbad, CA) in PBS + for 1 hr at RT. Following washing and fixation, samples were mounted in AF3 anti-fade reagent (Electron Microscopy Sciences, Hatfield, PA). Images were captured on an Eclipse TE2000 inverted microscope (Nikon Instruments Inc. Melville, NY) equipped with a CFI Plan Apo VC 60X oil, NA 1.4-type objective and Cool Snap HQ2 camera (Photometrics, Tuscon, AZ). The average 3×FLAG fluorescence intensity from five random fields was determined and corrected for background fluorescence by subtracting the signal from a minus HCR control sample. The fluorescence signal was analyzed using MetaMorph (Molecular Devices, Sunnyvale, CA). Data were presented using GraphPad Prism (GraphPad Software, Inc. La Jolla, CA), and represent the average of two independent experiments.
Crystallization and Data Collection
Purified HCR proteins were dialyzed and concentrated to 5 mg/ml in buffer containing 20 mM Tris-HCl at (pH 7.6) and 100 mM NaCl. Crystals were produced by vapor diffusion using a hanging drop containing 2 μL protein solution mixed with 2 μL of a well solution consisting of 0.1 M MES (pH 6.5), 12% PEG20K and 0.5 M NaCl for HCR/C; 0.1 M Hepes (pH 7.5), 10% PEG8K and 8% ethylene glycol for HCR/D; 0.1 M MES (pH 6.0), 5% PEG400, 1.8 M ammonium sulphate, and 250 mM NaCl for HCR/D-SA; and 0.1 M Hepes (pH 7.5), 4% PEG400 and 45% ammonium sulfate for the W1252A mutation of HCR/D-SA (HCR/D-SA-W1252A). Diffraction data for HCR/D-SA and HCR/D-SA-W1252A were collected at 100 K at the SBC 19ID beamline, Advanced Photon Source, Argonne National Laboratory, and data for HCR/C and HCR/D were collected using an R-AXIS IV++ with a MicroMax 007 generator at 100 K. HKL2000 (32) was used for data processing. Data collection and processing statistics for all crystals are summarized in Table 1.
Table 1.
Data Collection and Refinement Statistics
| Crystal | HCR/C PDB:3N7K |
HCR/D PDB:3N7J |
HCR/D-SA PDB:3N7L |
HCR/D-SA-W1252A PDB:33N7M |
|---|---|---|---|---|
| Resolution range (Å) | 30-2.5/2.54-2.50 | 30-2.0/2.07-2.0 | 30-2.0/2.03-2.0 | 30-2.6/2.69-2.6 |
| No. total reflections | 171,070 | 486,972 | 246,215 | 100,450 |
| No. unique reflections | 45,896/1,679 | 35,231/2,793 | 41,661/1,610 | 19457/1861 |
| Completeness (%) | 90.9/67.5 | 95.2/77.2 | 97.4/77.7 | 99.0/97.1 |
| Redundancy | 3.7/2.1 | 13.8/7.3 | 5.9/4.4 | 5.2/4.5 |
| I/σ(I) | 8.3/1.9 | 26.8/2.7 | 18.4/2.6 | 15.5/2.5 |
| Unit Cell a, b, c (Å) | 98.2, 77.4, 107.4 | 60.0, 94.2, 95.0 | 57.3, 57.8, 184.2 | 57.4, 57.8, 184.7 |
| β (°) | 116.36 | 90 | 90 | 90 |
| Space Group | P21 | P212121 | P212121 | P212121 |
| Rsymm | 0.105/0.330 | 0.090/0.497 | 0.086/0.434 | 0.098/0.526 |
| Vm (Å3/Da)/solvent content (%) | 3.6/65 | 2.7/52 | 3.0/58 | 3.1/58 |
| Monomers in an asymmetric unit | 2 | 1 | 1 | 1 |
| Refinement | ||||
| Rcrystal/Rfree | 0.192/0.254a | 0.221/0.256 | 0.216/0.257 | 0.222/0.270 |
| Rmsd bond length (Δ)/bond angle (°) | 0.008/1.4 | 0.006/1.3 | 0.006/1.4 | 0.007/1.5 |
| No. protein atoms | 6,919 | 3,335 | 3,412 | 3,403 |
| No. water molecules | 110 | 179 | 169 | 56 |
| No. Sulfate/glycerol atoms | 15/36 | 5/6 | ||
| Average B factor | ||||
| Main-Chain atoms (Å2) | 30.4 | 40.9 | 29.4 | 39.9 |
| Side-chain atoms (Å2) | 30.4 | 43.8 | 32.0 | 40.0 |
| Water molecules (Å2) | 24.1 | 46.7 | 35.4 | 57.1 |
| Sulfate/glycerol (Å2) | 54.6/46.2 | 38.9 | ||
Twin-operator: -h, -k, h+l; Twin-Fraction:=0.113
Without using Twin-Fraction and Twin-operator, Rcrystal/Rfree =0.221/0.276
Structure Determination and Refinement
Structure of HCR/D was solved by the molecular replacement method using MOLREP within the CCP4 (33) program suite and using the structure of the HCR/A (residues 875–1295, PDB code, 3FUO (13)). Structure of HCR/C was solved using HCR/D, the structure of HCR/D-SA solved with HCR/C and HCR/D-SA-W1252A solved with HCR/D-SA. Initial structures obtained from the molecular replacement trials were refined using the program CNS (34). The refinement procedure consisted of rigid body and positional refinements followed by a simulated-annealing protocol. Iterative rounds of positional and temperature factor refinement followed by manual fitting and rebuilding using the graphics program TURBO-FRODO (35) with (2|Fo| – |Fc|) and (|Fo| – |Fc|) difference Fourier maps. At later stages of refinement, water molecules were assigned where electron densities were greater than 3 σ in the (|Fo| – |Fc|) map and situated within 3.3 Å of a potential hydrogen bonding partner. The final models were completed with Rcrystal/Rfree of 0.192/0.254 for HCR/C, 0.221/0.256 for HCR/D, 0.216/0.257 for HCR/D-SA and 0.222/0.270 for HCR/D-SA-W1252A (Table 1).
Results
Amino acid sequence alignment of BoNT HCR/C, HCR/D, and HCR/D-SA
The percent amino acid identities of BoNT/A, BoNT/C and BoNT/D relative to BoNT/D-SA are shown in Table 2. BoNT/D-SA is a mosaic neurotoxin composed of the LC and HCT of BoNT/D and a receptor binding domain (HCR) that resembles BoNT/C as previously described (36). The low identity within the C-terminal subdomain (62%), relative to the identity between the N-terminal subdomains (90%) of the respective HCRs of BoNT/D-SA and BoNT/C indicates genetic drift since the generation of the mosaic gene. The significance of this divergence was defined by Smith and coworkers who reported that vaccination with either HCR/C or HCR/D failed to yield complete protection against challenge by BoNT/D-SA (30). These observations stimulated the characterization of the structure-function properties of the HCRs of BoNT/C, BoNT/D, and BoNT/D-SA.
Table 2.
Amino acid Identity of BoNT/C and BoNT/D with BoNT/D-SA
| % Identity to BoNT/D-SA domainsA | ||||
|---|---|---|---|---|
| Light Chain (1–447) | Translocation (447–866) | HCRN (867–1084) | HCRC (1085–1285) | |
| BoNT/A1 | 33 | 36 | 36 | 31 |
| BoNT/C | 47 | 70 | 90 | 62 |
| BoNT/D | 98 | 95 | 50 | 24 |
Structures of HCR from BoNT/C, BoNT/D, and BoNT/D-SA
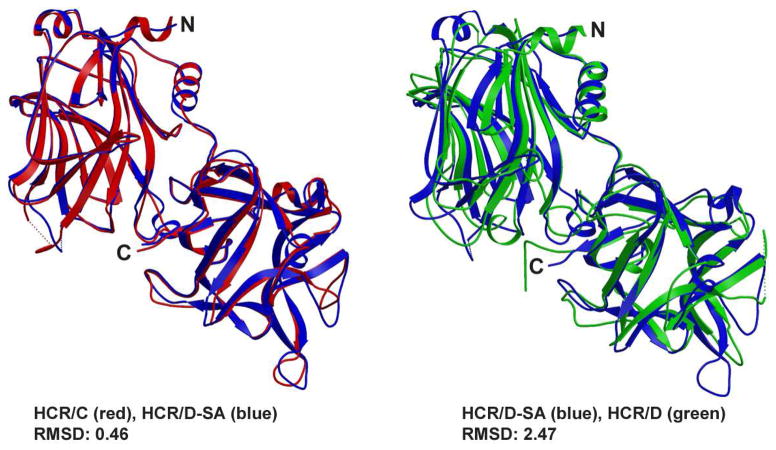
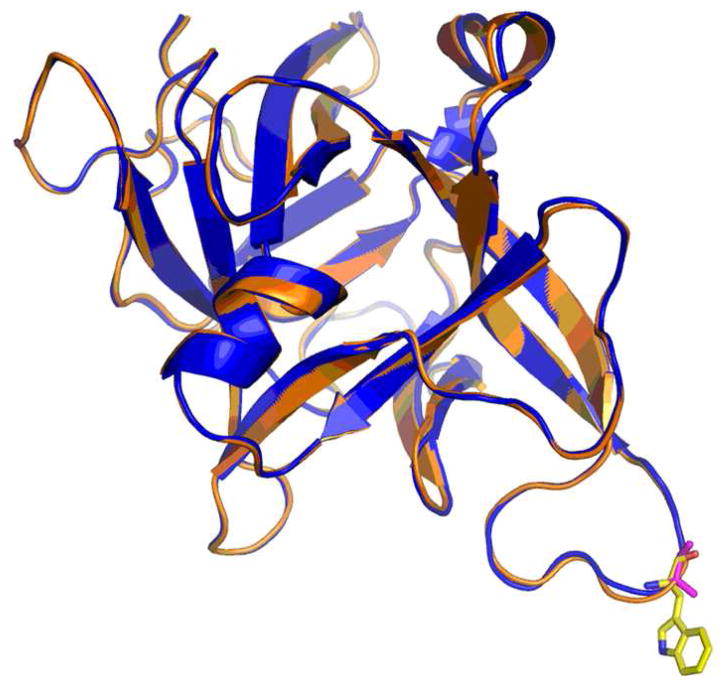
The crystal structures of HCR/C, HCR/D, and HCR/D-SA were solved and observed to have an overall conservation of structure (Figure 1A) among each other as well as among other serotypes of BoNTs (3, 37–39). The HCRs are organized with an N-terminal jelly-roll motif and a C-terminal β-trefoil conformation. Overall, the structures of HCR/D-SA and HCR/D have a root mean square deviation (RMSD) of 2.5 Å and HCR/D-SA and HCR/C have an RMSD of 0.5 Å. The angle between the N- and C- terminal subdomains of HCR/D is more obtuse than that observed between the two subdomains in HCR/C and HCR/D-SA. This change in angel did not appear to be due to changes in crystal packing and likely perturbed the calculated RMSD values for the entire HCR. The calculated RMSD values between the individual subdomains of HCR/D and HCR/D-SA was determined to be 1.5 Å for the N-terminal domain, and 3.6Å for the C-terminal domain. When the RMSD was determined between the C-terminal subdomain of HCR/D and HCR/D-SA excluding loops, the value was found to be 1.8Å, indicating that the majority of structural divergence is in the loop regions of the C-terminal subdomain. The primary divergence of structure within HCR/D-SA and HCR/C was within the loops of the C-terminal subdomain, which includes the ganglioside binding pocket that has been described for several other BoNT serotypes (39).
Figure 1. Crystal structures of HCR/C, HCR/D, and HCR/D-SA.
(A) Shown are overlays of the crystal structure of the HCR/D-SA (blue) with HCR/C (left panel, red) RMSD: 0.46 and HCR/D-SA (blue) with HCR/D (right panel, green) RMSD: 2.47. (B) Structure-based alignments of the C-terminal subdomains of HCR/C, HCR/D, and HCR/D-SA with HCR/A and HCR/B are shown. β strands, α helix, and loops are numbered from residue 1095 of HCR/C. An internal W and F (W1278 and F1280 of HCR/D-SA), highlighted blue, were conserved among the HCRs and provided a point of reference for the alignment. Residues within the ganglioside binding domain of HCR/A are green (E, H, S, W, and Y). Tryptophan within the ganglioside binding loop of HCR/C (W1258) and HCR/D-SA (W1252) are in pink. A W, located at a different position in the GBL of HCR/D (W1238), is pink. Yellow colored residues indicate conserved amino acids within the HCRs that have not been associated ganglioside or receptor binding functions.
Structure-based alignments of HCR/C, HCR/D and HCR/D-SA
The crystal structures were used to construct a structure-based amino acid sequence alignment (Figure 1B), with HCR/A and HCR/B as reference. A C-terminal, buried W (W1278 for HCR/D-SA) and F (F1280 for HCR/D-SA) are structurally conserved among the HCRs and served as a reference point for the alignment. As the main chain of the HCRs proceeds from the buried WF toward the N terminus, the peptide emerges from the interior and assumes a helical conformation. This helix constitutes part of the Ganglioside Binding Pocket (GBP) (Figure 2) observed for BoNT/A, /B, /E, /F, and /G and comprises several conserved residues that define the boundaries of the pocket (40). Y1267 and W1266 of HCR/A are present on the N-terminal side of the helix and contribute to ganglioside binding, while S1264 in the vicinity of the helix and E1203 also participate in ganglioside binding (40). The main chain of the HCRs continue towards the N- terminal domain and form a β-hairpin loop that continues into an anti-parallel-β-sheet that forms the adjacent side of the GBP located opposite W1266 and contains a H which participates in ganglioside binding (H1253 of HCR/A). BoNT/B displays a structurally identical GBP to that observed in BoNT/A. Interestingly, the β-loop formed by the anti-parallel beta-strands which make up one side of the GBP is composed of 10 amino acids rather than a shorter 4 residue loop that is present in HCR/A.
Figure 2. Overlay of the Ganglioside Binding Pocket of HCR/A with the representative residues of HCR/D-SA.
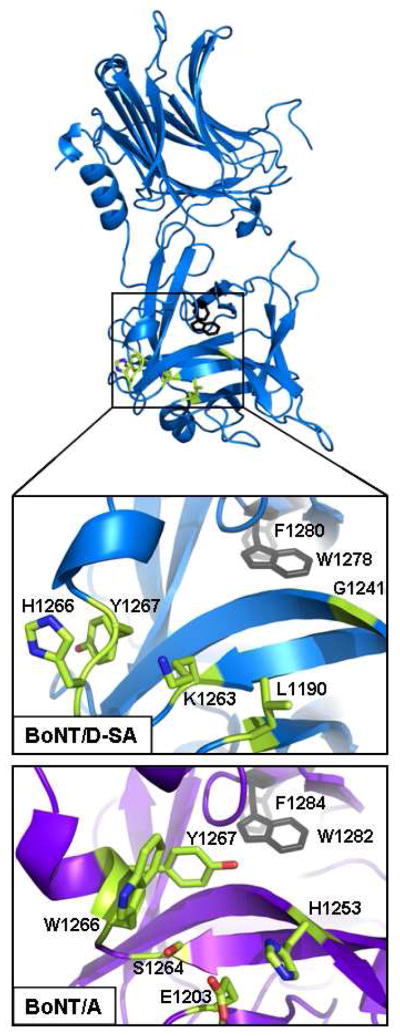
HCR/D-SA (blue) is shown with the following highlighted residues: conserved internal F1280 and W1282 (grey), and corresponding residues that represent the Ganglioside Binding Pocket of HCR/A (green). Ganglioside Binding Pocket of HCR/A (lower) and the corresponding region of HCR/D-SA (upper) were expanded and shown. Residues that contribute to ganglioside binding of HCR/A (E1203, H1253, S1264, W1266 and Y1267) and corresponding residues within HCR/D-SA that align in space are shown in green with nitrogen and oxygen atoms depicted in blue and red respectively. This alignment indicates the major determinant of ganglioside binding, a conserved tryptophan residue present in the ganglioside pocket of BoNT/A, /B, /E, /F, /G and TeNT is absent from the structurally analogous region of HCR/D-SA.
The crystal structures of HCR/C, HCR/D and HCR/D-SA display an overall structural similarity to HCR/A and HCR/B (Figure 1a) that includes the buried WF residues where the main chain proceeds from the interior to an α-helix which is structurally analogous to the GBP of HCR/A. However, within this analogous region neither HCR/D-SA, HCR/D, nor HCR/C contain the conserved W, S, or H, and the phenolic ring of Y is oriented away from the pocket. Proceeding from the GBP, HCR/C, HCR/D, and HCR/D-SA also form β-loops (between β12 and β13 –see Figure 1B) similar in size to HCR/B, and contain a W that was determined to contribute to HCR/C and HCR/D-SA binding to gangliosides, as shown below. This loop will be termed the Ganglioside Binding Loop (GBL) (Figure 3).
Figure 3. Overlay of the Ganglioside Binding Loops of HCR/C, HCR/D, and HCR/D-SA with the representative loops of HCR/A and HCR/B.
HCR/D-SA (blue, upper) is shown with the conserved, buried F1280 and W1282 (black), and the Ganglioside Binding Loop (GBL) W1252 indole ring is also shown at the tip of the loop. The GBL was enlarged, rotated, and shown (lower) aligned with the structurally analogous β-hairpin loops of BoNT/A (purple), /B (orange), /C (red), /D (green). HCR/C and /D-SA loops display a similar overall structural arrangement with the indole ring of the W residue implicated in ganglioside binding (HCR/C (W1258), HCR/D-SA (W1252)) extending away from the HCR molecule. HCR/D, like HCR/C and /D-SA has a tryptophan residue (W1238) in the β-hairpin loop. However, unlike HCR/C and /D-SA, the HCR/D W1238 indole ring does not extend away from the HCR but rather is oriented toward an adjacent β-hairpin loop. BoNT/B has an extended β-hairpin loop like HCR/C, /D, and /D-SA but lacks a tryptophan residue. BoNT/A, in contrast, does not have an extended β-hairpin loop.
Ganglioside binding by HCR/C and HCR/D-SA
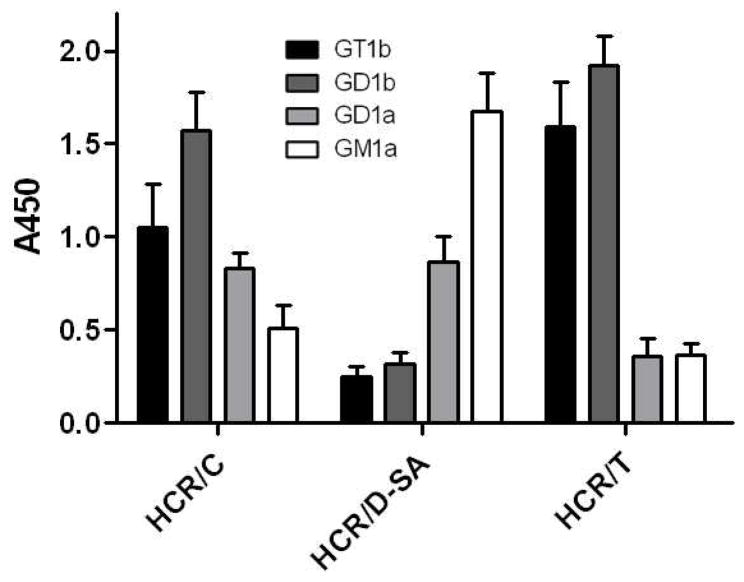
The ability of the HCRs to bind gangliosides was investigated in a solid phase binding assay. Previous studies showed that HCR/C bound GD1b and GT1b (27). Complex gangliosides were immobilized in 96-well plates and 10 nM HCRs were added to each ganglioside well. HCR/TeNT was used as a control to demonstrate effective ganglioside immobilization as HCR/TeNT binds b-series gangliosides with high affinity (41). HCR/C bound GD1b with the highest affinity, followed by GT1b, GD1a, and GM1a, while HCR/D-SA displayed a unique binding preference for GM1a, followed by GD1a with a lower affinity for b-series gangliosides (Figure 4). Thus, HCR/C and HCR/D-SA have unique preferences for gangliosides and the preferred binding of HCR/D-SA to GM1a is a unique property among the BoNT serotypes.
Figure 4. HCR/C and HCR/D-SA binding to complex gangliosides.
Wells (96 well plates) were coated with the indicated ganglioside (0.5 μg per well in 100 μl of MeOH). Plates were dried and 10 nM of HCR/C, HCR/D-SA, or HCR/T was incubated in 100 μl of PBS for 1 hr at 4°C. Wells were washed and bound HCR was detected with anti-FLAG-HRP antibody followed by TMB-Ultra. A450 was determined as the average of three independent experiments.
Role of Tryptophan within the Ganglioside Binding Loop (GBL) in HCR/C and HCR/D-SA binding to gangliosides
Ganglioside binding experiments were conducted to determine the role of the W within the GBL of HCR/C (W1258A) and HCR/D-SA (W1252A) for binding of the HCR to GD1b and GM1a, respectively. Previous studies showed that HCR/C utilized W1258 for cell binding (42). Gangliosides were immobilized on a 96-well plate and HCRs and the mutated HCRs were tested for ganglioside binding affinity. HCR/C bound GD1b in a dose dependent manner, while HCR/C(W1258A) binding to GD1b was reduced (Figure 5). In a similar experiment, HCR/D-SA bound GM1a in a dose dependent manner, while HCR/D-SA(W1252A) binding was reduced (Figure 5). These experiments supported the role of the W within the GBL as contributing to the coordination of ganglioside binding by HCR/C and HCR/D-SA. The crystal structure of the GBL of HCR/D-SA(W1252A) is identical to HCR/D-SA (Figure 6), indicating that the W1252A mutation does not cause structural perturbation to secondary sites on the HCR.
Figure 5. Role of Tryptophan within the ganglioside binding loop in HCR/C and HCR/D-SA binding to complex gangliosides.
Wells (96 well plates) were coated with GD1b (upper panel) or GM1a (lower panel) (0.5 μg (HCR/C) or 2.0 μg (HCR-D-SA) per well in 100 μl of MeOH). Plates were dried and 10 nM of HCR/C or HCR/C (W1258A) (upper panel) or HCR/D-SA or HCR/D-SA (W1252A) (lower panel) was incubated in 100 μl of buffer for 1 hr at 4°C. Bound HCR was detected with an M2 anti-FLAG antibody followed by TMB-Ultra. A450 was determined and plotted as the average of at least duplicate determinations. Background absorbance of HCR bound to well in the absence of ganglioside was subtracted to account for non-specific binding. No binding above background was detected for HCR/D-SA (W1252A) so these values are presented as zero.
Figure 6. Crystal structure of HCR/D-SA(W1252A).
Overlay of the crystal structures of the C-terminal subdomains (residues 1091-1285) of HCR/D-SA(W1252A) (orange) and HCR/D-SA (blue) are shown. The indole ring (yellow) of W1252 of HCR/D-SA and the methyl group (pink) of A1252 of HCR/D-SA(W1252A) are also shown.
Role of the Tryptophan within the GBL in HCR/C and HCR/D-SA binding to primary neurons
W1258 or W1252 within the GBL of HCR/C and HCR/D-SA, respectively, were tested for the ability to coordinate HCR binding to the plasma membrane of neurons by measuring HCRs binding to primary cortical neurons at 4°C. HCR/A did not show detectable binding to neurons at 4°C, consistent with the need for synaptic vesicle fusion to expose the protein receptor (Chen and Barbieri, submitted), SV2, while HCR/T showed a dose-dependent binding to neurons at 4°C consistent with HCR/T utilizing gangliosides present in the outer leaflet of the plasma membrane as receptors for entry into neurons as previously described (31). Both HCR/C and HCR/D-SA bound to neurons in a dose-dependent manner (Figure 7A) with binding observed over the cell body as well as on extended appendages. HCR/D-SA had a lower overall fluorescence intensity relative to HCR/C. In contrast, HCR/D did not show detectable neuron binding above background. Neither HCR/C (W1258A) nor HCR/D-SA (W1252A) bound neurons above background. Figure 7B quantifies the dose-dependent binding pattern of the HCRs. These data indicate HCR/C and HCR/D-SA bind neurons through receptors localized to the plasma membrane and that binding is coordinated by the W within the GBL, while HCR/D did not show detectable binding above that observed for HCR/A.
Figure 7. Binding of HCR/C, HCR/D, and HCR/D-SA to primary cortical neurons.
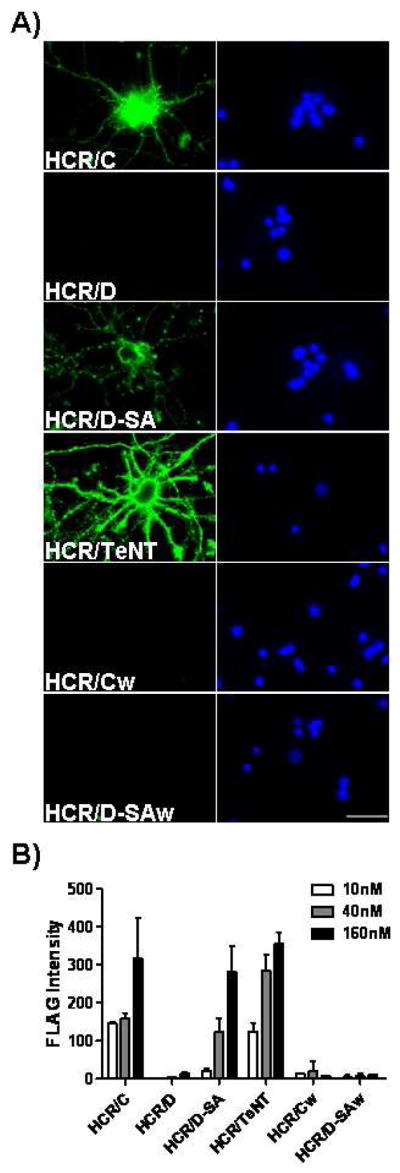
Primary cortical neurons were plated and cultured in 24-well glass bottom culture dishes coated with poly-D-lysine and laminin. Following 10 days of culture, cells were washed and incubated with a dilution series of the indicated HCR in media for 1 hr at 4°C. Bound HCR was identified by immunofluorescence microscopy using mouse anti-Flag mAb followed by goat anti-mouse IgG-488. A) Representative images of the indicated HCR (40 nM) bound to neurons (left panels) and nuclei stained with Hoechst (right panels) are shown (60X magnification). Images were captured at identical exposure times, and analyzed using Image J. Scale Bar = 35μm. B) Quantification of HCR binding to neurons. The values presented are arbitrary fluorescence units and represent the average FLAG fluorescence intensity from five random fields following correction for non-specific fluorescence. The average of two independent experiments is shown.
Discussion
Structures of HCR/C, HCR/D, HCR/D-SA
The crystal structures of HCR/C and HCR/D possess an overall structural homology with other BoNT HCR serotypes, with the N-terminal subdomain adopting a jelly-roll motif and the C-terminal subdomain composed primarily of loops that are organized in a β-trefoil conformation. Nuenket, et al., recently reported crystallization of HCR/D-SA; however, no structural solution was reported (43). In agreement with the primary amino acid sequence alignment, the structure of HCR/D-SA displayed a higher degree of structural similarity to the N terminus of HCR/C than that of HCR/D. There are limited regions of homology within the loops of the C-terminal β-trefoil domain of HCR/D-SA and HCR/C, which is consistent with HCR/C and HCR/D-SA possessing unique ganglioside binding profiles. While the contribution of the GBLs of HCR/C and HCR/D-SA to ganglioside binding is unique, HCR/B shares a β-loop with the GBLs of HCR/C and HCR/D-SA (Figure 1B). The β-loop in HCR/B does not contribute to ganglioside binding and lacks a W; Stevens and coworkers proposed that the β-loop in HCR/B contributes to membrane binding through hydrophobic interactions (38). Conversely, the homologous loop in BoNT/A is short, ~ 4 amino acid residues, which may preclude the loop from inserting into the cell membrane.
Ganglioside Binding Pocket
BoNT/A, /B, /E, /F, and /G contain a ganglioside binding pocket (GBP) (Figure 2) that is conserved by sequence identity (H…SXWY, except serotype G where H is replaced by G and serotype E where H is replaced by K) and structural alignment (44). In HCR/A, H1253 is located on the first β-strand (β12) that makes up the final β-hairpin in the C-terminal domain of the HCR. The peptide continues through the β-hairpin conformation and turns back forming an anti-parallel β-sheet ending in S1264, followed by a short α-helix, which contains W1266 and Y1267 (BoNT/A residue nomenclature). These residues (H1253, S1264, W1266 and Y1267) together with E1203 form a “pocket” for ganglioside binding. The imidazole ring of H and the indole ring of the W are perpendicular to each other within the GBP, while the Y and S form the back wall of the GBP. In a co-crystal structure of BoNT/A with GT1b, the #4 galactose sugar ring of the ganglioside backbone sat parallel to the indole ring of W and interacted through a hydrophobic ring-stacking mechanism. The interaction was further stabilized by contacts with E1203, H1253, and S1264. While the overall main chain organization of the GBP is present in HCR/C, HCR/D, and HCR/D-SA, the residues that contact ganglioside have been substituted precluding ganglioside binding within the GBP of HCR/C and /D-SA. This conclusion is based upon the observation that HCR/C (W1258A) and HCR/D-SA (W1252A) were defective for ganglioside binding (Figure 2). HCR/C, /D and /D-SA lacked H, W, and S, and the Y on the N-terminal side of the α-helix is in a different position relative to HCR/A with the phenolic ring oriented away from the pocket. Moreover, the structure and position of the α-helix which defines the GBP is not conserved in HCR/C and HCR/D-SA relative to HCR/A. Thus, the GBP in HCR/C, HCR/D-SA and HCR/D has a different architecture and slightly different location/ orientation from that of HCR/A.
Ganglioside Binding Loop
Earlier studies showed that BoNT/C toxicity was dependent on complex gangliosides (27, 29). We confirmed the ability of BoNT/C to bind b-series gangliosides and demonstrated that HCR/D-SA bound GM1a. Thus, HCR/C and HCR/D-SA utilize a unique Ganglioside Binding Loop (GBL) (Figure 3) located over 20Å away from the Ganglioside Binding Pocket present in other BoNT serotypes. In the structure-based alignment, HCR/C and HCR/D-SA possess a homologous W within the GBL. Tsukamoto and colleagues reported that mutation of the W1258 to A reduced the ability of the HCR to compete with BoNT/C for synaptosome binding (42); here we show that this W contributes to ganglioside binding.
While each BoNT serotype contains a loop corresponds to the GBL, the loops vary in size and composition. HCR/A has a β-finger (i.e., a two-residue loop) with several hydrophobic residues toward the end of the finger followed by the two consecutive Ns at the tip of the finger which is exposed to solvent. On the other hand, HCR/B has a loop more structurally analogous to HCR/C and HCR/D-SA, but lacking a W. The GBLs of BoNT/C and BoNT/D-SA represent a gain of function of ganglioside binding with a loss of function at the prototypical Ganglioside Binding Pocket.
BoNT/C and BoNT/D-SA have different ganglioside specificities despite structurally similar binding sites. HCR/C interacted with the b-series ganglioside GD1b, while HCR/D-SA preferred binding to the a-series ganglioside, GM1a. The neuronal cell binding experiment supported this observation, since the neuronal plasma membrane is enriched primarily in b-series gangliosides and has lower levels of a-series gangliosides. As a result, HCR/D-SA would be expected to have fewer binding sites on primary cortical neurons than HCR/C or HCR/T. The W within the GBL of HCR/D is located closer to the main body of the HCR (Figure 3), which may interfere with the ability to bind gangliosides (42).
The overlap of the W locations within the GBL of HCR/C and HCR/D-SA indicates that while W is required for ganglioside binding, this residue does not contribute to specificity. Thus, other residues within the GBL may contribute to ganglioside binding specificity. The protein structure of HCR/D-SA (W1258A) is unchanged with respect to the wild-type protein which argues that the W in the GBL contributes directly to ganglioside binding. HCR/C has two Rs flanking either side of the loop (R1251, R1260) and R1253 near the W, which may provide contacts for sialic acid residues in b-series gangliosides. The GBL loop of HCR/D-SA contains an additional D (D1249), which may repel the sialic acid carboxylates of b-series gangliosides. Further studies will be carried out to determine which residues make contact with gangliosides and how gangliosides interact with this novel GBL.
Several other bacterial toxins bind ganglioside receptors, but utilize different mechanisms of binding. For example, the heat-labile enterotoxins of Escherichia coli, as well as cholera toxin also use a tryptophan residue to coordinate sugar binding, as demonstrated by structure and mutagenesis studies (45, 46). However the binding sites are generated by interactions of two subunits of the B-pentamer, as opposed to the beta-trefoil conformation adopted by BoNTs and TeNT. The neuraminidase of influenza virus binds sialic acid by a mechanism lacking a tryptophan residue contact (47). In contrast, the eukaryotic lectins have been described, including ricin toxin, Sambucus nigra agglutinin II, EW29 from the earthworm Lumbricus terrestris, and Hemolytic Lectin CEL-III from Cucumaria echinata (48, 49). These proteins utilize a beta-trefoil domain to bind carbohydrates. These proteins contain multiple carbohydrate binding sites, some of which use tryptophan residues to coordinate sugars while other sites are coordinated using charged residues that can stack with a tyrosine residue. Thus, the botulinum toxins and tetanus toxin are unique prokaryotic proteins that use a beta-trefoil domain and a tryptophan residue to bind gangliosides, which have a similar binding strategy as the eukaryotic lectins.
Structure of HCR/D-SA and vaccine properties
BoNTs are category A select agents because of their extreme potency and duration of paralysis in humans. Though not associated with natural human intoxication, BoNT/C and BoNT/D are toxic in mammalian neuronal tissue (50) and BoNT/C causes paralysis in human neuromuscular preparations (23) and has been implicated as an agent for human therapy (51). In addition, there is interest in developing a multi-serotype vaccine capable of neutralizing all BoNT serotypes. Traditional vaccination strategies use formaldehyde-inactivated BoNT, where toxicity is eliminated, but immunogenicity is retained (30). An alternative vaccine strategy utilizes immunization with recombinant HCRs. The recombinant HCRs can be produced in large quantities and are free of neurotoxin contamination. Mice immunized with a cocktail of HCR domains of the seven prototypical serotypes (HCR/A-G) were resistant to challenge by each neurotoxin (BoNT/A -/G) demonstrating the efficacy of this strategy (52). In addition, anti-sera from mice immunized with the hepta-serotype HCR vaccine blocked binding of HCRs to gangliosides in vitro (41). These studies indicate neutralizing antibodies interfere with receptor recognition. Atassi and colleagues utilized a peptide array and identified immune reactive epitopes adjacent to the ganglioside and protein receptor binding regions of the HCR domain (53, 54). Interestingly, antibodies were not detected that were directed against the GBP, which suggests that the ganglioside pocket does not represent an efficient neutralization site. One caveat to this interpretation is that neutralizing epitopes may be composed of discontinuous segments of the peptide chain that become juxtaposed following protein folding which would preclude detection by peptide array analysis.
Unlike the Ganglioside Binding Pocket, the GBL of BoNT/C and BoNT/D-SA is a β-hairpin loop which protrudes from the HCR. The lack of cross protection observed by mice immunized with HCR/C upon challenge with BoNT/D-SA indicates the neutralizing epitopes are not conserved between these two BoNT subtypes and thus, the β-loop may be a potential site to elicit serotype specific neutralizing antibodies. Consistent with this region contributing to immune stimulation is the recent observation by Fairweather and coworkers, reporting that deletion of the GBL homologous region of HCR/TeNT reduced the capacity to elicit a neutralizing immune response (55). Studies are underway to determine the role of the GBL in eliciting a protective response against botulism.
Acknowledgments
JTB and JJK acknowledge membership and support from National Institutes of Health Regional Center of Excellence for Bio-defense and Emerging Infectious Diseases Research Program, Great Lakes Regional Center of Excellence Award 1-U54-AI-057153. MRB is supported by grant NIH-NINDS NS061763.
We acknowledge the assistance of Amanda Przedpelski for HCR production. We thank the staff at the Advanced Photon Source beamline SBC 19ID for their excellent assistance in data collection.
Abbreviations
- BoNT
botulinum neurotoxin
- HCR
heavy chain receptor binding domain of BoNT
- VAMP2
vesicle-associated membrane protein 2
- SNAP25
synaptosomal-associated protein of 25 kDa
- SNARE
soluble NSF attachment receptors
Footnotes
PDB codes: HCR/C, 3N7K; HCR/D, 3N7J; HCR/D-SA, 3N7L: HCR/D-SA-W1252A, 3N7M
References
- 1.Gill DM. Bacterial toxins: a table of lethal amounts. Microbiol Mol Biol Rev. 1982;46:86–94. doi: 10.1128/mr.46.1.86-94.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lacy DB, Tepp W, Cohen AC, DasGupta BR, Stevens RC. Crystal structure of botulinum neurotoxin type A and implications for toxicity. Nat Struct Mol Biol. 1998;5:898–902. doi: 10.1038/2338. [DOI] [PubMed] [Google Scholar]
- 3.Kumaran D, Eswaramoorthy S, Furey W, Navaza J, Sax M, Swaminathan S. Domain Organization in Clostridium botulinum Neurotoxin Type E Is Unique: Its Implication in Faster Translocation. Journal of Molecular Biology. 2009;386:233–245. doi: 10.1016/j.jmb.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 4.Montecucco C, Schiavo G. Structure and function of tetanus and botulinum neurotoxins. Quarterly Reviews of Biophysics. 1995;28:423–472. doi: 10.1017/s0033583500003292. [DOI] [PubMed] [Google Scholar]
- 5.Montecucco C, Schiavo G. Tetanus and botulism neurotoxins: a new group of zinc proteases. Trends in Biochemical Sciences. 1993;18:324–327. doi: 10.1016/0968-0004(93)90065-u. [DOI] [PubMed] [Google Scholar]
- 6.Brunger AT, Breidenbach MA, Jin R, Fischer A, Santos JS, Montal M. Botulinum Neurotoxin Heavy Chain Belt as an Intramolecular Chaperone for the Light Chain. PLoS Pathog. 2007;3:e113. doi: 10.1371/journal.ppat.0030113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verderio C, Rossetto O, Grumelli C, Frassoni C, Montecucco C, Matteoli M. Entering neurons: botulinum toxins and synaptic vesicle recycling. EMBO reports. 2006;7:995–999. doi: 10.1038/sj.embor.7400796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ochanda JO, Syuto B, Ohishi I, Naiki M, Kubo S. Binding of Clostridium botulinum Neurotoxin to Gangliosides. J Biochem. 1986;100:27–33. doi: 10.1093/oxfordjournals.jbchem.a121702. [DOI] [PubMed] [Google Scholar]
- 9.Dong M, Yeh F, Tepp WH, Dean C, Johnson EA, Janz R, Chapman ER. SV2 Is the Protein Receptor for Botulinum Neurotoxin A. Science. 2006;312:592–596. doi: 10.1126/science.1123654. [DOI] [PubMed] [Google Scholar]
- 10.Dong M, Tepp WH, Liu H, Johnson EA, Chapman ER. Mechanism of botulinum neurotoxin B and G entry into hippocampal neurons. J Cell Biol. 2007;179:1511–1522. doi: 10.1083/jcb.200707184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong M, Richards DA, Goodnough MC, Tepp WH, Johnson EA, Chapman ER. Synaptotagmins I and II mediate entry of botulinum neurotoxin B into cells. J Cell Biol. 2003;162:1293–1303. doi: 10.1083/jcb.200305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong M, Liu H, Tepp WH, Johnson EA, Janz R, Chapman ER. Glycosylated SV2A and SV2B Mediate the Entry of Botulinum Neurotoxin E into Neurons. Mol Biol Cell. 2008;19:5226–5237. doi: 10.1091/mbc.E08-07-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu Z, Chen C, Barbieri JT, Kim JJP, Baldwin MR. Glycosylated SV2 and Gangliosides as Dual Receptors for Botulinum Neurotoxin Serotype F. Biochemistry. 2009;48:5631–5641. doi: 10.1021/bi9002138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montal M. Translocation of botulinum neurotoxin light chain protease by the heavy chain protein-conducting channel. Toxicon. 2009;54:565–569. doi: 10.1016/j.toxicon.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer A, Montal M. Single molecule detection of intermediates during botulinum neurotoxin translocation across membranes. Proceedings of the National Academy of Sciences. 2007;104:10447–10452. doi: 10.1073/pnas.0700046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer A, Montal M. Crucial Role of the Disulfide Bridge between Botulinum Neurotoxin Light and Heavy Chains in Protease Translocation across Membranes. Journal of Biological Chemistry. 2007;282:29604–29611. doi: 10.1074/jbc.M703619200. [DOI] [PubMed] [Google Scholar]
- 17.Schiavo GG, Benfenati F, Poulain B, Rossetto O, de Laureto PP, DasGupta BR, Montecucco C. Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature. 1992;359:832–835. doi: 10.1038/359832a0. [DOI] [PubMed] [Google Scholar]
- 18.Binz T, Blasi J, Yamasaki S, Baumeister A, Link E, Südhof TC, Jahn R, Niemann H. Proteolysis of SNAP-25 by types E and A botulinal neurotoxins. Journal of Biological Chemistry. 1994;269:1617–1620. [PubMed] [Google Scholar]
- 19.Yamasaki S, Baumeister A, Binz T, Blasi J, Link E, Cornille F, Roques B, Fykse EM, Südhof TC, Jahn R. Cleavage of members of the synaptobrevin/VAMP family by types D and F botulinal neurotoxins and tetanus toxin. Journal of Biological Chemistry. 1994;269:12764–12772. [PubMed] [Google Scholar]
- 20.Blasi J, Chapman ER, Yamasaki S, Binz T, Niemann H, Jahn R. Botulinum neurotoxin C1 blocks neurotransmitter release by means of cleaving HPC-1/syntaxin. EMBO J. 1993;12:4821–4828. doi: 10.1002/j.1460-2075.1993.tb06171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oguma K, Yokota K, Hayashi S, Takeshi K, Kumagai M, Itoh N, Tachi N, Chiba S. Infant botulism due to Clostridium botulinum type C toxin. The Lancet. 1990;336:1449–1450. doi: 10.1016/0140-6736(90)93157-k. [DOI] [PubMed] [Google Scholar]
- 22.Gangarosa EJ, Donadio JA, Armstrong RW, Meyer KF, Brachman PS, Dowell VR. Botulism in the United States, 1899–1969. Am J Epidemiol. 1971;93:93–101. doi: 10.1093/oxfordjournals.aje.a121239. [DOI] [PubMed] [Google Scholar]
- 23.Coffield JA, Bakry N, Zhang RD, Carlson J, Gomella LG, Simpson LL. In vitro characterization of botulinum toxin types A, C and D action on human tissues: Combined electrophysiologic, pharmacologic and molecular biologic approaches. Journal of Pharmacology and Experimental Therapeutics. 1997;280:1489–1498. [PubMed] [Google Scholar]
- 24.Neimanis A, Gavier-Widen D, Leighton F, Bollinger T, Rocke T, Morner T. An outbreak of type C botulism in herring gulls (Larus argentatus) in southeastern Sweden. J Wildl Dis. 2007;43:327–336. doi: 10.7589/0090-3558-43.3.327. [DOI] [PubMed] [Google Scholar]
- 25.Davletov B, Bajohrs M, Binz T. Beyond BOTOX: advantages and limitations of individual botulinum neurotoxins. Trends in Neurosciences. 2005;28:446–452. doi: 10.1016/j.tins.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Brand CJ, Schmitt SM, Duncan RM, Cooley TM. An outbreak of type E botulism among common loons (Gavia immer) in Michigan’s upper peninsula. J Wildl Dis. 1988;24:471–476. doi: 10.7589/0090-3558-24.3.471. [DOI] [PubMed] [Google Scholar]
- 27.Tsukamoto K, Kohda T, Mukamoto M, Takeuchi K, Ihara H, Saito M, Kozaki S. Binding of Clostridium botulinum Type C and D Neurotoxins to Ganglioside and Phospholipid. Journal of Biological Chemistry. 2005;280:35164–35171. doi: 10.1074/jbc.M507596200. [DOI] [PubMed] [Google Scholar]
- 28.Baldwin MR, Barbieri JT. Association of Botulinum Neurotoxin Serotypes A and B with Synaptic Vesicle Protein Complexes. Biochemistry. 2007;46:3200–3210. doi: 10.1021/bi602396x. [DOI] [PubMed] [Google Scholar]
- 29.Rummel A, Hafner K, Mahrhold S, Darashchonak N, Holt M, Jahn R, Beermann S, Karnath T, Bigalke H, Binz T. Botulinum neurotoxins C, E and F bind gangliosides via a conserved binding site prior to stimulation-dependent uptake with botulinum neurotoxin F utilising the three isoforms of SV2 as second receptor. J Neurochem. 2009;110:1942–1954. doi: 10.1111/j.1471-4159.2009.06298.x. [DOI] [PubMed] [Google Scholar]
- 30.Webb RP, Smith TJ, Wright PM, Montgomery VA, Meagher MM, Smith LA. Protection with recombinant Clostridium botulinum C1 and D binding domain subunit (Hc) vaccines against C and D neurotoxins. Vaccine. 2007;25:4273–4282. doi: 10.1016/j.vaccine.2007.02.081. [DOI] [PubMed] [Google Scholar]
- 31.Chen C, Fu Z, Kim JJP, Barbieri JT, Baldwin MR. Gangliosides as High Affinity Receptors for Tetanus Neurotoxin. Journal of Biological Chemistry. 2009;284:26569–26577. doi: 10.1074/jbc.M109.027391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minor W, Tomchick D, Otwinowski Z. Strategies for macromolecular synchrotron crystallography. Structure. 2000;8:R105–110. doi: 10.1016/s0969-2126(00)00139-8. [DOI] [PubMed] [Google Scholar]
- 33.CCP4. Acta crystallographica. 1994. pp. 760–763. [Google Scholar]
- 34.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 35.Roussel A, Fontecilla-Camps JC, Cambillau C. CRYStallize: a crystallographic symmetry display and handling subpackage in TOM/FRODO. J Mol Graph. 1990;8:86–88. doi: 10.1016/0263-7855(90)80087-v. [DOI] [PubMed] [Google Scholar]
- 36.Moriishi K, Koura M, Abe N, Fujii N, Fujinaga Y, Inoue K, Ogumad K. Mosaic structures of neurotoxins produced from Clostridium botulinum types C and D organisms. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1996;1307:123–126. doi: 10.1016/0167-4781(96)00006-1. [DOI] [PubMed] [Google Scholar]
- 37.Jin R, Rummel A, Binz T, Brunger AT. Botulinum neurotoxin B recognizes its protein receptor with high affinity and specificity. Nature. 2006;444:1092–1095. doi: 10.1038/nature05387. [DOI] [PubMed] [Google Scholar]
- 38.Chai Q, Arndt JW, Dong M, Tepp WH, Johnson EA, Chapman ER, Stevens RC. Structural basis of cell surface receptor recognition by botulinum neurotoxin B. Nature. 2006;444:1096–1100. doi: 10.1038/nature05411. [DOI] [PubMed] [Google Scholar]
- 39.Stenmark P, Dupuy J, Imamura A, Kiso M, Stevens RC. Crystal structure of botulinum neurotoxin type A in complex with the cell surface co-receptor GT1b-insight into the toxin-neuron interaction. PLoS Pathog. 2008;4:e1000129. doi: 10.1371/journal.ppat.1000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rummel A, Mahrhold S, Bigalke H, Binz T. The HCC-domain of botulinum neurotoxins A and B exhibits a singular ganglioside binding site displaying serotype specific carbohydrate interaction. Molecular Microbiology. 2004;51:631–643. doi: 10.1046/j.1365-2958.2003.03872.x. [DOI] [PubMed] [Google Scholar]
- 41.Chen C, Baldwin MR, Barbieri JT. Molecular Basis for Tetanus Toxin Coreceptor Interactions. Biochemistry. 2008;47:7179–7186. doi: 10.1021/bi800640y. [DOI] [PubMed] [Google Scholar]
- 42.Tsukamoto K, Kozai Y, Ihara H, Kohda T, Mukamoto M, Tsuji T, Kozaki S. Identification of the receptor-binding sites in the carboxyl-terminal half of the heavy chain of botulinum neurotoxin types C and D. Microbial Pathogenesis. 2008;44:484–493. doi: 10.1016/j.micpath.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 43.Nuemket N, Tanaka Y, Tsukamoto K, Tsuji T, Nakamura K, Kozaki S, Yao M, Tanaka I. Preliminary X-ray crystallographic study of the receptor-binding domain of the D/C mosaic neurotoxin from Clostridium botulinum. Acta Crystallogr Sect F Struct Biol Cryst Commun. 66:608–610. doi: 10.1107/S1744309110012182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lacy DB, Stevens RC. Sequence homology and structural analysis of the clostridial neurotoxins. J Mol Biol. 1999;291:1091–1104. doi: 10.1006/jmbi.1999.2945. [DOI] [PubMed] [Google Scholar]
- 45.Merritt EA, Sarfaty S, Akker Fvd, L’Hoir C, Martial JA, Hol WG. Crystal structure of cholera toxin B-pentamer bound to receptor GM1 pentasaccharide. Protein Sci. 1994;3:166–175. doi: 10.1002/pro.5560030202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang RG, Scott DL, Westbrook ML, Nance S, Spangler BD, Shipley GG, Westbrook EM. The three-dimensional crystal structure of cholera toxin. J Mol Biol. 1995;251:563–573. doi: 10.1006/jmbi.1995.0456. [DOI] [PubMed] [Google Scholar]
- 47.Varghese JN, McKimm-Breschkin JL, Caldwell JB, Kortt AA, Colman PM. The structure of the complex between influenza virus neuraminidase and sialic acid, the viral receptor. Proteins. 1992;14:327–332. doi: 10.1002/prot.340140302. [DOI] [PubMed] [Google Scholar]
- 48.Maveyraud L, Niwa H, Guillet V, Svergun DI, Konarev PV, Palmer RA, Peumans WJ, Rouge P, Van Damme EJ, Reynolds CD, Mourey L. Structural basis for sugar recognition, including the Tn carcinoma antigen, by the lectin SNA-II from Sambucus nigra. Proteins. 2009;75:89–103. doi: 10.1002/prot.22222. [DOI] [PubMed] [Google Scholar]
- 49.Hatakeyama T, Unno H, Kouzuma Y, Uchida T, Eto S, Hidemura H, Kato N, Yonekura M, Kusunoki M. C-type lectin-like carbohydrate recognition of the hemolytic lectin CEL-III containing ricin-type -trefoil folds. J Biol Chem. 2007;282:37826–37835. doi: 10.1074/jbc.M705604200. [DOI] [PubMed] [Google Scholar]
- 50.Kalandakanond S, Coffield JA. Cleavage of intracellular substrates of botulinum toxins A, C, and D in a mammalian target tissue. J Pharmacol Exp Ther. 2001;296:749–755. [PubMed] [Google Scholar]
- 51.Eleopra R, Tugnoli V, Rossetto O, Montecucco C, De Grandis D. Botulinum neurotoxin serotype C: a novel effective botulinum toxin therapy in human. Neuroscience Letters. 1997;224:91–94. doi: 10.1016/s0304-3940(97)13448-6. [DOI] [PubMed] [Google Scholar]
- 52.Baldwin MR, Tepp WH, Przedpelski A, Pier CL, Bradshaw M, Johnson EA, Barbieri JT. Subunit vaccine against the seven serotypes of botulism. Infect Immun. 2008;76:1314–1318. doi: 10.1128/IAI.01025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dolimbek BZ, Steward LE, Aoki KR, Atassi MZ. Immune recognition of botulinum neurotoxin B: antibody-binding regions on the heavy chain of the toxin. Mol Immunol. 2008;45:910–924. doi: 10.1016/j.molimm.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 54.Dolimbek BZ, Aoki KR, Steward LE, Jankovic J, Atassi MZ. Mapping of the regions on the heavy chain of botulinum neurotoxin A (BoNT/A) recognized by antibodies of cervical dystonia patients with immunoresistance to BoNT/A. Mol Immunol. 2007;44:1029–1041. doi: 10.1016/j.molimm.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 55.Qazi O, Sesardic D, Tierney R, Soderback Z, Crane D, Bolgiano B, Fairweather N. Reduction of the ganglioside binding activity of the tetanus toxin HC fragment destroys immunogenicity: implications for development of novel tetanus vaccines. Infect Immun. 2006;74:4884–4891. doi: 10.1128/IAI.00500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]