Abstract
Many rodent species act as reservoir hosts of zoonotic cutaneous leishmaniasis in endemic areas. In the present study a simple and reliable assay based on nested PCR was developed for the detection and identification of Leishmania parasites from rodent skin samples. We designed Leishmania-specific primers that successfully amplified ITS regions of Leishmania major, Leishmania gerbilli and Leishmania turanica using nested PCR. Out of 95 field collected Rhombomys opimus, 21 were positive by microscopic examination and 48 by nested PCR. The percentage of gerbils infected with L. major, L. gerbilli and L. turanica was 3.2%, 1.1% and 27.4%, respectively. In 15.8% of the rodents, we found mixed natural infections by L. major and L. turanica, 1.1% by L. major and L. gerbilli, and 2.1% by the three species. We concluded that this method is simple and reliable for detecting and identifying Leishmania species circulating in rodent populations.
Keywords: Leishmania major, Leishmania gerbilli, Leishmania turanica, Rodent, Rhombomys opimus, Cutaneous leishmaniasis, Nested PCR, Iran
1. Introduction
Cutaneous leishmaniasis due to Leishmania major (CLM) is an increasing disease and a public health problem in many regions of the Old World. L. major is widely distributed in various rodent populations in arid and savannah regions (Gramiccia and Gradoni, 2005). Rodents belonging to the subfamily Gerbillinae are the main reservoir hosts of cutaneous leishmaniasis in Iran and other countries where CLM is endemic (Dubrovsky, 1979; Strelkova, 1996; Yaghoobi-Ershadi et al., 1996). Gerbils are the most abundant mammals in natural ecosystems of Old World deserts (Dubrovsky, 1979). Many rodent species act as reservoir hosts of CLM: Rhombomys opimus (great gerbil) in Central Asia, Northern Afghanistan and Iran; Meriones libycus (Libyan jird) in the Arabian Peninsula, Central Asia and Iran; Meriones hurrianae (Indian desert jird) in India and Iran; Psammomys obesus (fat sand rat) and Meriones crassus in Northern Africa and Middle East; Ratus ratus and Arvicanthis niloticus in Sudan (Abdalla et al., 2003) and Tatera spp. in subsaharan Africa and Iran (Gramiccia and Gradoni, 2005). R. opimus (Cricetidae: Gerbillinae) is the principal reservoir host of L. major over the vast territory of the Turan lowland (west and south Kazakhstan and Central Asia with adjacent parts in Afghanistan and Iran), Mongolia, and apparently, in some provinces of China. In the Turan lowland, naturally infected R. opimus were found in more than 200 places from where they were investigated and showed a higher infection rate than any other mammal (other rodents, insectivores, carnivores) investigated (Dubrovsky, 1979). All the proven sand fly vectors of CLM belong to the subgenus Phlebotomus, including Phlebotomus papatasi, the principal vector, and related species Phlebotomus salehi and Phlebotomus duboscqi. Well-described stable zoonotic cutaneous leishmaniasis (ZCL) foci are associated with L. major and P. in North Africa and the Middle East, and with R. in central Asia, Afghanistan and Iran (Gramiccia and Gradoni, 2005; Yaghoobi-Ershadi et al., 2003; Parvizi et al., 2005).
The geographic distribution and role of rodents as reservoir hosts of CLM in Iran are well known and R. opimus is considered the most important in Central and North East Iran (Yaghoobi-Ershadi and Javadian, 1996; Yaghoobi-Ershadi et al., 1996). The main foci of CLM in Iran are located in Esfahan Province, Central Iran, where the current study was carried out. Only single infection of L. major or Leishmania turanica has been detected and identified from rodents in Iran yet (Yaghoobi-Ershadi et al., 1996, 2004; Mohebali et al., 2004; Parvizi et al., 2008; Hajjaran et al., 2009). Recently, L. major, L. turanica and Leishmania gerbilli sensu lato were detected in sand flies in the northeast and centre of Iran (Parvizi and Ready, 2008).
One of the major obstacles for the control and understanding of this neglected disease is the detection and identification of Leishmania parasites in animal reservoirs. L. major infection is usually accompanied by non-pathogenic (sub-clinical) L. turanic or L. gerbilli (Strelkova, 1996; Strelkova et al., 2001) which are microscopy-confounding. Traditional techniques (direct examination and culture) commonly used for diagnosing leishmaniasis do not differentiate Leishmania species (Ben-Ismail et al., 1992; Shahbazi et al., 2008). Gold-standard isoenzyme characterization requires large scale parasite culture without contamination (Evans, 1989) and also it is possible that infections of Leishmania parasites will be missed due to disparate growth rates of different parasites in blood agar cultures (Ibrahim et al., 1994; Abdalla et al., 2003).
Nested PCR provides a rapid, sensitive, and specific alternative to traditional techniques. Moreover, diagnosis of Leishmania infection and species identification is done simultaneously. Here, we have developed a simple and reliable method to detect and identify Leishmania species from skin samples from naturally infected great gerbils using a nested PCR method and identified the parasites circulating in these animal reservoirs.
2. Materials and methods
2.1. Gerbil collection
The investigation was conducted over a period of 24 months from October 2006 to October 2008 in three rural districts (Borkhar, Sejzi and Badrood) of Esfahan Province, Central Iran where CLM is endemic. Active colonies of gerbils were identified and caught using 20–45 Sherman traps baited with cucumber. The trapped gerbils were transferred to the animal house facility at the Esfahan Health Research and Training Center, Institute of Public Health, Esfahan, Iran, and maintained for parasitological and molecular testing. The captured rodents were identified by morphological characters (Etemad, 1978) and only the great gerbils, R. opimus, were selected for the study.
2.2. Direct parasitological test
In the laboratory, the rodents were anaesthetized using intramuscular Ketamine hydrochloride (60 mg/kg) and Xylazine (5 mg/kg). Regardless of the presence of lesions, impression smears were prepared from the ear lobes of the animals (Edrissian et al., 1982) and stained by Giemsa. Samples were examined under the light microscope (1000×) for detection of Leishmania amastigotes.
After preparing direct smears, ear lobe samples were removed from anesthetized rodents and transferred to 500 μl of cold PBS (pH 7.4), thoroughly disrupted by grinding with a pestle and kept at −20 °C until use. The animals were nursed to recovery after these procedures. Animal procedures were approved by the Ethics Committee of Tehran University of Medical Sciences, Tehran, Iran.
2.3. Polymerase chain reaction (PCR) assay
2.3.1. Deoxyribonucleic acid (DNA) extraction
Genomic DNA was extracted and purified using a conventional phenol–chloroform protocol (Sambrook and Russel, 2001) with slight modification. Briefly, a 100 μl of each disrupted tissue sample was transferred to a 1.5 ml microtube containing 200 μl of lysis buffer (100 mM Tris–HCl, pH 8; 10 mM EDTA, pH 8; 1% SDS; 100 mM NaCl; 2% Triton X-100) containing proteinase K (100 mg/ml), vortexed, and incubated at 56 °C for 1 h. Three hundred microliters of phenol–chloroform (1:1) were added, vortexed and centrifuged and chloroform extraction was performed again. An equal volume of isopropanol and 1/10 volume of 3 M sodium acetate (pH 5.2) were added to the supernatant and centrifuged for 15 min at 5000g and the precipitant washed with 70% ethanol and centrifuged for 5 min at 800g. The pellet was air dried and resuspended in 20 μl of distilled water and stored at −20 °C until use.
2.3.2. Primers design
To design universal primers for detecting and identifying the common Leishmania species found in gerbils (L. major, L. gerbilli and L. turanica) we focused on ribosomal DNA using GenBank sequences of internal transcribed spacer ITS2 (Table 1). The sequences from each species were aligned and compared using DNASIS software (Hitachi Software Engineering Co. Tokyo). The external primers, Leish out F (5′-AAA CTC CTC TCT GGT GCT TGC-3′) and Leish out R (5′-AAA CAA AGG TTG TCG GGG G-3′), and internal primers, Leish in F (5′-AAT TCA ACT TCG CGT TGG CC-3′) and Leish in R (5′-CCT CTC TTT TTT CTC TGT GC-3′) were selected to distinguish among the parasite species in a nested PCR system. Predicted PCR products are shown in Table 1.
Table 1.
Expected size (bp) of products following first round and second round PCR using GenBank sequences of internal transcribed spacer ITS2.
| Leishmania species | ITS2 products with external primers | Predicted size of ITS2 products with internal primers | Genbank accession numbers of L. major, L. turanica and L. gerbilli |
|---|---|---|---|
| L. major | 483 | 245 or 233* | FJ753394, FJ753393, FJ753392, FJ753391, DQ300195, AJ786166, AJ786165, AJ786164, AJ786163, AY260965, AJ300481, AJ272383, AJ000310 |
| L. gerbilli | 441 | 206 | AJ300486 |
| L. turanica | 399 | 141 | AJ272382, AJ272381, AJ272380, AJ272379, AJ272378, AJ000309, AJ000308, AJ000307 |
This pattern was not observed in our standard strains or in samples from the gerbils.
2.3.3. PCR conditions
PCR amplification was carried out in an Applied Biosystems thermocycler. The initial PCR contained 0.6 μM of each forward (Leish out F) and reverse (Leish out R) external primers, 12.5 μl Taq DNA polymerase, 2X Master Mix Red (Amplicon, Denmark) and sterile distilled water to a final volume of 25 μl. An initial denaturation step at 95 °C for 5 min was followed by 30 cycles of: denaturation at 94 °C for 30 s, annealing at 60 °C for 45 s and extension at 72 °C for 1 min, with a final extension step of 72 °C for 5 min. The first round reactions were performed in duplicate with 1 and ½ μl volumes of DNA template. The second-round (nested) PCR was performed in a final volume of 20 μl containing 1 μl of a 1:10 dilution in distilled water of the first-round PCR product as template, 0.3 μM of each forward (Leish in F) and reverse (Leish in R) internal primers, 10 μl of Taq DNA polymerase and 2X Master mix Red. The reactions were cycled under the following conditions: 95 °C for 2 min, 25 cycles of 94 °C for 15 s, 62 °C for 30 s, 72 °C for 45 s followed by 72 °C for 5 min. PCR products were separated by 1.5% (w/v) agarose gel electrophoresis in TBE buffer (0.09 mM Tris, 0.09 mM boric acid and 20 mM EDTA, pH 8.3), visualized with ethidium bromide (0.5 μg/ml) and photographed.
Reference strains L. major (MRHO/IR/75/ER), L. gerbilli (MRHO/CN/60/GERBILLI) and L. turanica (MRHO/SU/1983/MARZ-051) were used as positive controls; samples of normal dermal tissues of naive gerbil (bred under laboratory conditions, without any exposure to Leishmania) and/or distilled water were used as negative controls. The PCR product of the negative control of the first-round PCR was used as the negative control in the second round and also the PCR product of the positive control of the first-round PCR was used as positive control in the second round. Necessary precautions such as adding mineral oil on the master mix, using filter pipette tips, sterilizing equipments with 10% sodium hypochlorite solution to avoid cross-contamination were taken.
3. Results
In the present study a simple and reliable assay based on nested PCR was developed for the detection and identification of Leishmania species involved in rodent leishmaniasis. The designed Leishmania-specific external and internal primers of the nested PCR successfully amplified the ITS2 region of the reference strains L. major, L. gerbilli and L. turanica, generating a single major product with a distinct size for each species (Fig. 1). The primers could detect and identify all three species by fragment size polymorphisms in both rounds of PCR but the sensitivity of the second round was augmented significantly (Table 2). The nested PCR assay tested on normal skin tissues of a naive gerbil and standard Leishmania strains showed no false negatives or false positives, respectively.
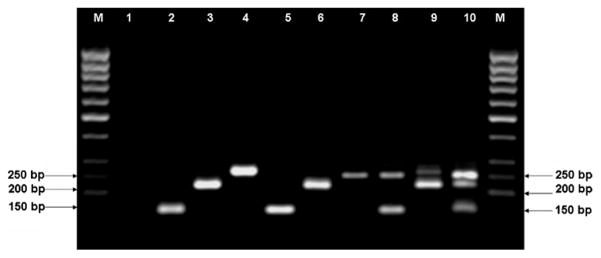
Fig. 1.
Agarose (1.5%) gel electrophoresis of nested PCR products. M, 50 bp ladder (Fermentas); lane 1, negative control (distilled water); lane 2, L. turanica; lane 3, L. gerbilli; lane 4, L. major; lanes 2–4 are reference strains; lane 5, L. turanica; lane 6, L. gerbilli; lane 7, L. major; lane 8, mixed infection of L. major and L. turanica; lane 9, mixed infection of L. major and L. gerbilli; lane 10, mixed infection of L. major, L. gerbilli and L. turanica. Lanes 5–10 are from skin samples of Rhombomys opimus rodents (field samples).
Table 2.
Detection of Leishmania parasites from rodent skin samples by direct examination, first-round PCR and nested PCR.
| Diagnosis | No. of positive samples/no. of examined samples | % Positive |
|---|---|---|
| Clinical examination | ||
| Direct examination | 21/95 | 22.1 |
| First-round PCR for samples positive by direct examination | 12/21 | 57.1 |
| First-round PCR for samples negative by direct examination | 7/74 | 9.5 |
| First-round PCR for all samples | 19/95 | 20 |
| Nested PCR for samples positive by direct examination | 21/21 | 100 |
| Nested PCR for samples negative by direct examination | 27/74 | 36.5 |
| Nested PCR for all samples | 48/95 | 50.5 |
Specimens from 95 R. opimus, captured from CLM foci, were examined by two diagnostic techniques, direct (microscopic) examination and nested PCR. Out of 95 specimens, 21 specimens were positive by microscopic examination and 19 were positive by first-round PCR using external primers. Importantly, 48 specimens were positive after further testing with nested PCR (Table 2). Furthermore, compared to only 12 (57.1%) samples detected by the first-round PCR (of the 21 specimens positive by direct examination), all 21 (100%) specimens were positive by nested PCR. Out of 74 specimens negative by direct examination, 9.5% were positive by first-round PCR and 36.5% by nested PCR (Table 2). Out the 95 investigated gerbils, 3.2% were infected with L. major, 1.1% with L. gerbilli and 27.4% with L. turanica. We also found mixed natural infections with L. major and L. turanica in 15.8% of the rodents; L. major and L. gerbilli in 1.1% and 2.1% were infected with all three species. 94.6 of the infected gerbils showed no cutaneous leishmaniasis lesion on their ear lobes.
4. Discussion
Traditional techniques (direct examination and culture) commonly used for diagnosing leishmaniasis do not differentiate Leishmania species and their sensitivity is also less than molecular techniques (Ben-Ismail et al., 1992; Faber et al., 2003; Shahbazi et al., 2008). It is possible that infections of Leishmania parasites will be missed due to disparate growth rates of different parasites in blood agar cultures (Ibrahim et al., 1994). Nested PCR provides a rapid, sensitive, and specific alternative to traditional techniques.
Moreover, diagnosis of Leishmania infection and species identification is done simultaneously. In this study a nested PCR method was successfully developed and applied for the detection and identification of rodent Leishmania infections commonly find in Iran and central Asia. Our results indicate that this method is sensitive and specifically distinguishes among L. major, L. gerbilli and L. turanica, the three most common parasites of R. opimus, the principal reservoir of L. major in Iran. The designed Leishmania-specific primers tested on DNA from pure standard strains of L. major, L. gerbilli and L. turanica generated a single major product different in size for each of the three parasite species. The size of these ITS2 ribosomal gene products corresponded to those predicted from GenBank sequence data. The second round PCR was much more sensitive than the first round. Both first and second primer pairs have been designed to identify all tested Leishmania species, therefore it is expected having more positive samples in the second round PCR than the first one.
Detection of Leishmania infection in over 50% (Table 2) of skin samples from field collected rodents attests to the sensitivity of the developed nested PCR. The specificity, sensitivity and rapid unambiguous distinction of mixed infections by nested PCR justifies its use as a diagnostic test for the three investigated Leishmania species.
The results of the current study show that L. major, L. gerbilli and L. turanica are circulating in R. opimus populations from Central Iran. It seems that there is some variation among L. major strains. The three parasite species L. major, L. gerbilli and L. turanica have been identified in naturally infected gerbils from Turkmenistan, Uzbekistan and Kazakhstan (Strelkova, 1996; Strelkova et al., 2001). L. turanica was reported as the dominant species in R. opimus populations in hypoendemic, mesoendemic and hyperendemic foci of ZCL in Turkmenistan and Uzbekistan (Strelkova et al., 2001). In vast territories of Central Asia, mixed infections of wild rodents with L. major and L. turanica are typical (Strelkova, 1996). In our study areas, L. turanica was the dominant species in R. opimus populations. We rarely found R. opimus infected with L. major or L. gerbilli alone and in most cases L. major infection was accompanied by L. turanica. In addition, this is the first report to our knowledge of a mixed natural co-infection with L. major, L. turanica and L. gerbilli in wild R. opimus populations from Iran or elsewhere.
In most of the previous studies conducted in Iran, only L. major has been isolated from great gerbils and characterized by using isoenzymes or DNA-based molecular techniques (Yaghoobi-Ershadi et al., 2003; Mohebali et al., 2004; Parvizi et al., 2008; Rassi et al., 2008). There are rare reports of L. turanica infection in rodents in Iran (Mohebali et al., 2004; Yaghoobi-Ershadi et al., 2004; Hajjaran et al., 2009). Identification of Leishmania species have mostly been based on culture and usually only one species of Leishmania is detected and identified by each examination (Yaghoobi-Ershadi et al., 2003; Mohebali et al., 2004; Hatam et al., 2005; Hajjaran et al., 2009), As the method presented in this study is not based on culture, it is simpler as well as more sensitive for detecting and identifying Leishmania parasites from skin samples in rodents. Gerbils infected by L. major are important in the transmission cycle of CLM (Gramiccia and Gradoni, 2005; Strelkova, 1996; Yaghoobi-Ershadi et al., 2003) and the distribution of L. major, the causative agent of ZCL in Central Asia, has been found to coincident with that of R. opimus (Strelkova, 1996). Therefore, it is important to accurately assess the rate of L. major infection in R. opimus and other important reservoirs.
In CLM foci where L. major, L. gerbilli and L. turanica circulate in the reservoir population of R. opimus and where L. major–L. turanica co-infections are common, the nested PCR method described in the current study is an reliable assay to distinguish the three species of Leishmania and improve our estimates of reservoir infection rates with L. major, the human infecting species.
Acknowledgments
This research was supported by Research deputy of Tehran University of Medical Sciences, Project No. 3025. We are very grateful to Dr. P. Parvizi, Pasteur Institute of Iran, who provides us with the standard strains of L. turanica and L. gerbilli and also Mr. D. Iravani and Ms. M. Vaziri, Pasteur Institute of Iran for their technical assistance in this project. We also thank Dr. M. Jeddi-Tehrani, Avicenna Research Institute, ACECR, for his valuable comment on the molecular experiments.
Sincere thanks are also extended to head and staff of Esfahan Research and Training Center, Institute of Public Health Research for their assistance in the project. We are also grateful to Mr. E. Eskandar and Ms. A. Mirmohammadi, Center for Research and Training in Skin Diseases and Leprosy for their kind helps.
References
- Abdalla NM, Eldosh AA, Abdulgani AM, Yusif BE, Magzoub MM. Typing and Characterization of leishmania sub-clinical isolates from Nuba Mountain, West of Sudan. Infection Genetic and Evolution. 2003;2 (4):277. [Google Scholar]
- Ben-Ismail R, Smith DF, Ready PD, Ayadi A, Gramiccia M, Ben-Osman A, Ben-Rachid MS. Sporadic cutaneous leishmaniasis in north Tunisia: identification of the causative agent as Leishmania infantum by the use of a diagnostic deoxyribonucleic acid prob. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1992;86:508–510. doi: 10.1016/0035-9203(92)90087-s. [DOI] [PubMed] [Google Scholar]
- Dubrovsky Y. WHO Traveling Seminar on Leishmaniasis Control. Ministry of Health; Moscow: 1979. Biology of Great Gerbil – The Principal Carrier of the Great of Zoonotic Cutaneous Leishmaniasis. [Google Scholar]
- Edrissian Gh, Zovein Z, Nadim A. A simple technique for presentation of smears from the ear of Rhombomys opimus for the detection of leishmanial infection. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1982;76:706–707. doi: 10.1016/0035-9203(82)90255-3. [DOI] [PubMed] [Google Scholar]
- Etemad E. Mammals of Iran Rodents and Their Identification Keys. Vol. 1. National society of Guardian ship of Natural Resources and Human Environment; Tehran: 1978. [Google Scholar]
- Evans D. UNDP World Bank/WHO Special Programme for Research and Training in Tropical Diseases. World Health Organization; Geneva: 1989. [Google Scholar]
- Faber WR, Oskam L, Van Gool T, Kroon NCM, Knegt-Junk KJ, Hofwegen H, Van Der Wal AC, Kager PA. Value of diagnostic techniques for cutaneous leishmaniasis. Journal of the American Academy of Dermatology. 2003;49 (1):70–74. doi: 10.1067/mjd.2003.492. [DOI] [PubMed] [Google Scholar]
- Gramiccia M, Gradoni L. The current status of zoonotic leishmaniases and approaches to disease control. International Journal for Parasitology. 2005;35:1169–1180. doi: 10.1016/j.ijpara.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Hajjaran H, Mohebali M, Alimoradi S, Abaei MR, Edrissian GhH. Isolation and characterization of pathogenic Leishmania turanica from Nesokia indica (Rodentia, Muridae) by PCR-RFLP and ITS1 sequencing in Iran. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2009;103:1177–1179. doi: 10.1016/j.trstmh.2008.08.016. [DOI] [PubMed] [Google Scholar]
- Hatam GR, Riyad M, Bichichi M, Hejazi SH, Guessous-Idrissi N, Ardehali S. Isoenzyme characterization of Iranian Leishmania isolates from cutaneous leishmaniasis. Iranian Journal of Science and Technology, Transaction A. 2005;29:65–70. [Google Scholar]
- Ibrahim ME, Smith AJ, Ali MH, Barker DC, Kharazmi A. The polymerase chain reaction can reveal the occurrence of naturally mixed infections with Leishmania parasites. Acta Tropica. 1994;57:327–332. doi: 10.1016/0001-706x(94)90078-7. [DOI] [PubMed] [Google Scholar]
- Mohebali M, Javadian E, Yaghoobi-Ershadi MR, Akhavan AA, Hajjaran H, Abaei MR. Characterization of leishmania infection in rodents from endemic areas of Islamic Republic of Iran. Eastern Mediterranean Health Journal. 2004;10:591–599. [PubMed] [Google Scholar]
- Parvizi P, Mauricio I, Aransay AM, Miles MA, Ready PD. First detection of Leishmania major in peridomestic Phlebotomus papatasi from Isfahan province, Iran: comparison of nested PCR of nuclear ITS ribosomal DNA and semi-nested PCR of minicircle kinetoplast DNA. Acta Tropica. 2005;93:75–83. doi: 10.1016/j.actatropica.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Parvizi P, Ready PD. Nested PCRs and sequencing of nuclear ITS-rDNA fragments detects three Leishmania species of gerbils in sandflies from Iranian foci of zoonotic cutaneous leishmaniasis. Tropical Medicine and International Health. 2008;13:1159–1171. doi: 10.1111/j.1365-3156.2008.02121.x. [DOI] [PubMed] [Google Scholar]
- Parvizi P, Moradi Gh, Akbari Gh, Farahmand M, Ready PD, Piazak N, Assmar M, Amirkhani A. PCR detection and sequencing of parasite ITS-rDNA gene from reservoir host of zoonotic cutaneous leishmaniasis in central Iran. Parasitology Research. 2008;103:1289–1295. doi: 10.1007/s00436-008-1124-z. [DOI] [PubMed] [Google Scholar]
- Rassi Y, Abai MR, Javadian E, Rafizadeh S, Imamian H, Mohebali M, Fateh M, Hajjaran H, Ismaili M. Molecular study on vectors and reservoir hosts of zoonotic cutaneous leishmaniasis in Central of Iran. Bulletin de la Societe de Pathologie Exotique. 2008;101:425–428. [PubMed] [Google Scholar]
- Sambrook J, Russel DW. Molecular Cloning: A Laboratory Manual. 3. Cold Spring Harbor Cold Spring Harbor Laboratory Press; New York: 2001. [Google Scholar]
- Shahbazi F, Shahabi S, Kazemi B, Mohebali M, Abadi AR, Zare Z. Evaluation of PCR assay in diagnosis and identification of cutaneous leishmaniasis: a comparison with the parasitological methods. Parasitology Research. 2008;103:1159–1162. doi: 10.1007/s00436-008-1111-4. [DOI] [PubMed] [Google Scholar]
- Strelkova MV. Progress in studies on Central Asian foci of zoonotic cutaneous leishmaniasis a review. Folia Parasitologica. 1996;43:1–6. [PubMed] [Google Scholar]
- Strelkova MV, Eliseev LN, Ponirovsky EN, Dergacheva TL, Annacharyeva DK, Erokhin PI, Evans DA. Mixed leishmanal infection in Rhombomys opimus: a key to the persistence of Leishmanha major from one transmission season to the next. Annals of Tropical Medicine and Parasitology. 2001;95:811–819. doi: 10.1080/00034980120111154. [DOI] [PubMed] [Google Scholar]
- Yaghoobi-Ershadi MR, Javadian E. Epidemiological study of reservoir hosts in an endemic area of zoonotic cutaneous leishmaniasis in Iran. Bulletin of the World Health Organization. 1996;74:587–590. [PMC free article] [PubMed] [Google Scholar]
- Yaghoobi-Ershadi MR, Akhavan AA, Mohebali M. Meriones libycus and Rhombomys opimus (Rodentia:gerbillidae) are the main reservoir hosts in a new focus of zoonotic cutaneous leishmaniasis in Iran. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1996;90:503–504. doi: 10.1016/s0035-9203(96)90295-3. [DOI] [PubMed] [Google Scholar]
- Yaghoobi-Ershadi MR, Akhavan AA, Zahraei-Ramazani AR, Abai MR, Ebrahimi B, Vafaei-Nezhad R, Hanafi-Bojd AA, Jafari R. Epidemiological study in a new focus of cutaneous leishmaniasis in the Islamic Republic of Iran. Eastern Mediterranean Health Journal. 2003;9:816–826. [PubMed] [Google Scholar]
- Yaghoobi-Ershadi MR, Akhavan AA, Zahraei-Ramazani AR, Abai MR, Ebrahimi B, Vafaei-Nezhad R, Hanafi-Bojd AA, Jafari R. Letter to editor. Eastern Mediterranean Health Journal. 2004;10:1. [PubMed] [Google Scholar]