Abstract
Heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) is a RNA binding protein that plays important role in the biogenesis of mRNA, such as alternative splicing and mRNA stability. We have previously demonstrated that hnRNP A1 has diminished protein levels and shows cytoplasmic accumulation in senescent human diploid fibroblasts. Recent reports showed that p38 MAP kinase (p38 MAPK), a member of the MAP kinase family is necessary and sufficient for the cytoplasmic accumulation of hnRNP A1 by stress stimuli such as osmotic shock. p38 MAP kinase has been shown to be involved in cell proliferation and the induction of senescence in response to extracellular stimuli. However, the relationship between hnRNP A1 and p38 MAPK and the roles of hnRNP A1 in cellular senescence have not yet been elucidated. Here we show that hnRNP A1 forms a complex with phospho-p38 MAPK in vivo. Inhibition of p38 MAPK activity with SB203580 elevated hnRNP A1 protein levels and prohibited the cytoplasmic accumulation of the protein, but not hnRNP A2, in senescent cells. The phosphorylation level of hnRNP A1 was elevated in senescent cells. Reduction of hnRNP A1 and A2 levels by siRNA transfection induced a senescence-like morphology and elevated the level of F-actin, a marker of senescence. These results suggest that the expression levels and subcellular distribution of hnRNP A1 are regulated in a p38 MAPK-dependent manner, probably via its phosphorylation. Our results also suggest that hnRNP A2 in addition to hnRNP A1 may play a role in establishing the senescence phenotype.
Keywords: senescence, fibroblasts, IMR-90 cells, HS74 cells, hnRNP A1, A2, ras, p38 MAP kinase, siRNA, F-actin, SB203580
Introduction
The property of cells to respond to a variety of environmental changes is essential for their homeostasis, metabolism and survival. Intracellular signaling transduction pathways are involved in transduction of the extracellular stress signals to downstream target genes. Protein kinase families are known to play a central role in these processes.
The mitogen-activated protein kinase (MAPK) pathway consists of three protein kinases, MAPK, MAPK kinase (MAPKK) and MAPKK kinase (MAPKK-K). MAPKK-Ks activate specific MAPKKs by phosphorylation and the MAPKKs in turn activate distinct MAPKs by phosphorylation.1–3 p38 MAP kinase (p38 MAPK) is one of the factors that plays a critical role in the process. p38 MAPK is activated by multiple environmental stresses and inflammatory cytokines2,4 through dual phosphorylation on its threonine/tyrosine residues in the T-loop. The activated p38 MAPK signaling pathway then activates the downstream transcription factors, such as ATF2 and MEF-2C.5–8 p38 MAPK is activated by Ras proto-oncogene family members that encode small GTP-binding proteins and this process is mediated by MKK3 and MKK6.9 Interestingly, high levels of Ras expression induces p38-mediated premature senescence, or the early onset of replicative senescence.10–12 Replicative senescence is an irreversible growth arrest at the terminal stage of the in vitro life span of normal cell cultures characterized by an enlarged and flattened cellular morphology.13 Replicative senescence also possesses unique molecular features; increase in the number of cytoplasmic micro-filaments, such as F-actin;14,15 increased acidic β-galactosidase activity;16 increased activation of p53 and Rb;17 accumulation of cell cycle inhibitors, p16INK4A and p21WAF1 and a p53 suppressor, p14ARF;10,18,19 and suppression of the transcription of an early response gene, c-fos.20
Heterogeneous nuclear ribonucleoprotein (hnRNP) family members are the most abundant components of messenger ribo-nucleoprotein complexes (mRNPs) and play a variety of regulatory roles in the biogenesis of mRNA.21,22 Over 24 major proteins designated A1-U (34 kDa–120 kDa) have been identified in hnRNP complexes.23 The proteins found in the core hnRNP complex have been identified as hnRNP’s A, B and C and are in the 30–43 kDa range.23 A member of the hnRNP A/B subfamily, hnRNP A1, is highly abundant and has been shown to be involved in pre-mRNA and mRNA processes such as alternative splicing, mRNA export, splice site selection, mRNA turnover and translation.23–31 In addition, hnRNP A1 has nucleocytoplasmic shuttling activity32 and has been shown to be required for cell proliferation, differentiation and the survival of certain normal and transformed cells.33 hnRNP A2, which shares 69% amino acid identity with hnRNP A1, has similar biochemical properties of hnRNP A1 in RNA metabolism.23,34 hnRNP A2 is also been reported to play a critical role in cell proliferation.35
Recent studies have shown that osmotic shock or UVC irradiation induce cytoplasmic accumulation of hnRNP A1. The cytoplasmic accumulation is concomitant with an increase in its phosphorylation and requires p38 MAPK.36 We have previously demonstrated that hnRNP A1 protein levels show diminished expression and altered subcellular distribution in senescent HS74 fibroblasts.37,38 These findings raise the possibilities that there is a relationship between hnRNP A1 and p38 MAPK proteins and suggest that hnRNP A1 may play a significant role in cellular senescence under the control of p38 MAPK pathway. However, the precise molecular mechanisms by which this pathway might regulate hnRNP A1 have yet to be identified.
In this study, we demonstrate that hnRNP A1 and p38 MAPK interact in vivo and that p38 MAPK regulates the the expression level and subcellular distribution of hnRNP A1. We have also found that the phosphorylation level of hnRNP A1 was elevated in senescent cells. Moreover, siRNA inhibition of hnRNP A1 and/or hnRNP A2 expression induced a senescence-like morphology. Our studies suggest that the phosphorylation status of hnRNP A1 may have important consequences for gene expression during senescence.
Results
Age-dependent change in hnRNP A1 expression level in IMR-90 cells
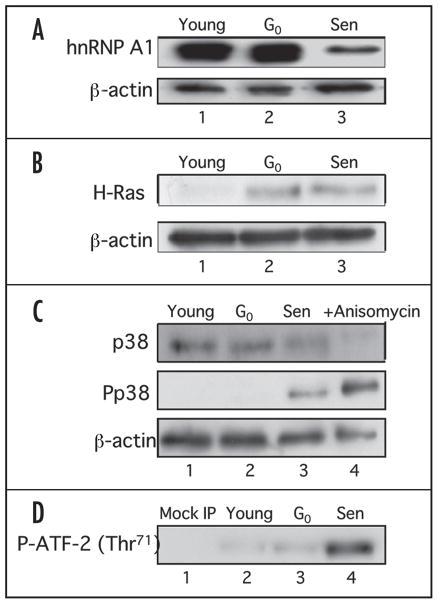
We had previously shown in HS74 human diploid fibroblasts (derived from fetal bone marrow) that the levels of hnRNP A1 protein were lower in senescent cells as compared to young cells.37 However, we had not yet determined if the change in the protein level during senescence was common among fibroblasts from different tissues. Therefore, we performed the same experiment with IMR-90 human fibroblasts (derived from fetal lung) since the similarity of gene expression and biological properties of these two fibroblasts have been well documented. We examined A1 protein levels in IMR-90 cells by western blot analysis and found that the protein levels were highly diminished in senescent cells compared to young and G0-arrested cells, (Fig. 1A, lanes 1–3) as was originally observed in HS74 fibroblasts as well as in WI-38 fibroblasts (data not shown). These results suggest that the change in the level of hnRNP A1 protein during senescence may be a common feature in different fibroblast cell lines.
Figure 1.
Western analysis of hnRNP A1 and the Ras signaling pathway in IMR-90 young, G0-arrested and senescent cells. (A) Western analysis of hnRNP A1. 10 μg of whole cell lysates from young (lane 1), G0-arrested (lane 2) and senescent (lane 3) cells were electrophoresed on a 12% SDS-PAGE and probed with 4B10 monoclonal antibody specific for hnRNP A1. The blot was then stripped and reprobed to detect total β-actin levels. (B) Western analysis of c-H-Ras levels. Fifty micrograms of whole cell lysates from young (lane 1), G0-arrested (lane 2) and senescent (lane 3) cells were subjected to 15% SDS-PAGE and c-H-Ras levels were determined by immunoblot analysis using H-Ras antibody. The membrane was stripped and reprobed with β-actin antibody as a loading control. (C) Western analysis of p38 MAPK and phospho-p38 MAPK levels. 25 μg of whole cell lysates from young (lane 1), G0-arrested (lane 2) and senescent (lane 3) cells were electrophoresed on a 12% SDS-PAGE and probed with antibodies for p38 MAPK, phospho-p38 MAPK (P-p38 MAPK) and β-actin. Anisomycin treated lysates were used as positive controls (lane 4). (D) p38 MAPK activity in IMR-90 cells. 400 μg of whole cell lysates from young (lane 2), G0-arrested (lane 3) and senescent (lane 4) fibroblasts were incubated with immobilized anti-phosphorylated p38 MAPK beads and the kinase activity was measured using a fusion ATF-2 protein with a single phosphorylation site at Thr71. A mock IP reaction containing antibody and beads only was included as a negative control (lane 1).
Upregulation of p38 MAPK in IMR-90 cells
We had previously reported that both hnRNP A1 and A2 had cytoplasmic accumulation in senescent fibroblasts.38 Recent studies have demonstrated that the phosphorylation of hnRNP A1 by the p38 MAPK pathway induced cytoplasmic accumulation.36,41,42 It has also been shown in senescent WI-38 fibroblasts and rabbit chondrocytes, that there is an upregulation of c-H-ras mRNA with a concomitant activation of p38 MAPK.39,40 These reports raised the possibility that an age-dependent change in hnRNP A1 localization and protein levels could be regulated by the Ras-p38 MAPK pathway. To assess this, we first determined the endogenous protein levels of c-H-ras and the phosphorylation level and activity of p38 MAPK in young, G0-arrested and senescent IMP-90 cells by western blot analysis. The protein level of c-H-ras was elevated in G0-arrested cells and senescent cells as compared to young cells (Fig. 1B, lanes 1–3). These results are consistent with a previous study.39 The phosphorylation level of p38 MAPK was highly elevated only in senescent cells, although the total protein levels of p38 MAPK were relatively unchanged (Fig. 1C, lanes 1–3).
We next examined the kinase activity of p38 MAPK by assessing the phosphorylation of the transcription factor, ATF-2. ATF-2 is known to be phosphorylated by activated p38 MAPK and is frequently used as a substrate to measure p38 MAPK activity.43 The kinase activity of p38 towards ATF-2 was markedly elevated in senescent cells compared to young and G0-arrested cells (Fig. 1D, lanes 2–4). The relative kinase activities in G0-arrested cells and senescent cells normalized to that in young cells were 1.8 and 4.5, respectively (data not shown). The elevated phosphorylation and the activity of p38 MAPK observed in senescent cells were equivalent to previous reports with WI-38 cells and rabbit chondrocytes,40,44 suggesting that IMR-90 cells upregulates the Ras-p38 MAPK pathway during senescence as well as other cells. As shown in Figure 1C, lane 2 and D, lane 3, the phosphorylation and activity of p38 MAPK in G0-arrested cells were not elevated, although c-H-ras protein levels was upregulated (Fig. 1B, lane 2). These results indicate that the phosphorylation and activity of p38 MAPK, but not c-H-ras levels might be specifically regulated in senescent cells.
Inhibition of p38 MAPK activity modulates hnRNP A1 expression
We then inhibited p38 MAPK activity to confirm that this kinase was active in our system. We blocked the kinase activity of p38 MAPK using the pyridinyl imidazole, SB203580 (IC50 value = 600 nM for in vivo inhibition), a well characterized inhibitor of p38 MAP kinase in vivo.45 We treated young, G0-arrested and senescent IMR-90 cells with 2.5 mM SB203580 or an equivalent volume of DMSO for 1 h. To determine the effect of SB203580 on the kinase activity, we immunoprecipitated each cell lysate with antibody beads to phospho-p38 MAPK followed by a kinase assay with ATF-2 (Thr71) as the substrate (Fig. 2A). The kinase activity in all cells was partially blocked compared to cells treated with DMSO (Fig. 2A, lanes 2, 4 and 6). As a control, we monitored the expression levels of hnRNP A1 (Fig. 2B). Surprisingly, the endogenous levels of hnRNP A1 were upregulated in SB203580 treated young and G0-arrested cells compared to DMSO treated cells (Fig. 2B, lanes 1–4). Of note, young cells showed a significant increase in the level. When we normalized hnRNP A1 levels to β-actin by densitometric analysis, we found a 1.46-fold (average) increase of A1 in SB203580 treated young and G0-arrested cells (Fig. 2B). The hnRNP A1 levels in senescent cells remain relatively unaffected by SB203580 compared to DMSO treated cells (Fig. 2B, lanes 5 and 6). These results suggest that p38 MAPK activity negatively regulates hnRNP A1 protein expression in young and G0-arrested cells. This is the first observation that hnRNP A1 protein levels can be regulated by the p38 MAPK pathway. This modulation appears to be specific to young cells as we did not observe a change in hnRNP A1 expression in SB203580 treated senescent cells. The downregulation of hnRNP A1 during senescence may be due to other mechanisms.
Figure 2.

Effect of p38 MAPK inhibition on p38 MAPK activity and hnRNP A1 expression. (A) p38 MAPK kinase assays. Two hundred micrograms of total cell lysates prepared from young (lanes 2 and 3), G0-arrested (lanes 4 and 5) and senescent IMR-90 cells (lanes 6 and 7) treated with 2.5 μM SB203580 (SB) or an equivalent volume of DMSO (DM) were immuno-precipitated with P-p38 MAPK antibody and kinase assay was performed using a synthetic phospho-ATF-2 (Thr71) peptide as the substrate. (B) Western blot analysis of total hnRNP A1 protein levels. 20 mg of lysates prepared from young (lanes 1 and 2), G0-arrested (lanes 3 and 4) and senescent cells (lanes 5 and 6) treated with 15 μM SB203580 (lanes 2, 4 and 6) or an equivalent volume of DMSO (lanes 3, 5 and 7) were subjected to 12% SDS-PAGE and probed with the 4B10 hnRNP A1 antibody. Membranes were stripped and reprobed with β-actin as a loading control. hnRNP A1/β actin levels were measured by densitometric analysis.
hnRNP A1 forms a complex with P-p38 MAPK
Because hnRNP A1 was observed to be phosphorylated by the p38 MAPK pathway,41 we determined whether there is a physical interaction between P-p38 MAPK and hnRNP A1 by coimmunoprecipitation assays. Cell lysates from young and G0-arrested IMR-90 cells were immunoprecipitated with a P-p38 MAPK antibody and then the membrane was probed with the 4B10 hnRNP A1 monoclonal antibody. A 34 kDa band was detected in the complex, representing the presence of hnRNP A1 (Fig. 3A, lanes 2 and 3). We also tested the ability of hnRNP A1 to co-precipitate with P-p38 MAPK in senescent IMR-90 lysates, but the 34 kDa band was not detected (data not shown). The lack of detectable signal may be accounted for by the diminished protein levels of hnRNP A1 in senescent cells (Fig. 1A). We next performed the reciprocal analysis to determine whether P-p38 MAPK co-immunoprecipitated with hnRNP A1. Both p38 MAPK and P-p38 MAPK co-immunoprecipitated with hnRNP A1 as indicated by a 38 kDa band (Fig. 3B, parts a and c, lane 2). These results indicate a reciprocal interaction between p38 MAPK and hnRNP A1. The very low levels of co-precipitated P-p38 MAPK may reflect its diminished protein levels observed in young IMR-90 cells (Fig. 1C, lane 1). We also confirmed that P-p38 MAPK co-precipitated with an exogenous GFP-hnRNP A1 fusion protein (data not shown). Immunoprecipitation of hnRNP A1 by 4B10 antibody was confirmed by reprobing these membranes with antibodies specific for hnRNP A1 (Fig. 3B, parts b and d, lane 2).
Figure 3.
hnRNP A1 forms a complex with p38 MAPK in vivo. (A) 300 μg of young and G0-arrested lysates (lanes 2 and 3) were immunoprecipitated with 20 ml Immobilized Phospho-p38 MAPK 180/182 (Thr/Tyr) monoclonal antibody beads, subjected to 12% SDS-PAGE and subsequently probed with the 4B10-hnRNP A1 antibody. Mock IP reactions without lysates were included to establish the molecular weight of the antibody heavy and light chain, 55 and 25 kDa, respectively (lane 1). (B) Co-immunoprecipitation of p38 MAPK and phospho-p38 MAPK with hnRNP A1. 300 μg of total lysates isolated from young IMR-90 cells were immunoprecipitated using the 4B10-hnRNP A1 antibody, subjected to 12% SDS-PAGE and probed with either p38 MAPK (Part a, lane 2) or P-p38 MAPK (Part c, lane 2) antibodies. Both blots were stripped and reprobed with the 4B10 monoclonal antibody to detect total hnRNP A1 (Parts b and d, lane 2). (C) Binding specificity of P-p38 MAPK. 300 μg of total cell lysates from young IMR-90 cells were immunoprecipitated with immobilized phospho-p38 MAPK antibody beads and probed with either GAPDH or Bax (lane 1) antibodies. Positive controls for each protein were included (lane 2). (D) Effect of p38 MAPK inhibition on its interaction with hnRNP A1. Each 200 μg of total cell lysates isolated from 2.5 μM SB203580 (lane 2) or DMSO (lane 1) treated young IMR-90 cells was immunoprecipitated with P-p38 MAPK and probed with the 4B10-hnRNP A1 antibody. 5 μg of collected supernatants from each reaction were subjected to western analysis to determine β-actin levels.
We immunoprecpitated P-p38 MAPK from the lysates of young cells with antibodies for GAPDH and Bax (Fig. 3C) to assess specific binding. Neither GAPDH nor BAX was co-precipitated with P-p38 MAPK (Fig. 3C, lanes 1), indicating that P-p38 MAPK specifically binds to hnRNP A1.
We next determined if the inhibition of p38 MAPK affected its binding to hnRNP A1. We treated young IMR-90 cells with 2.5 μM SB203580 or an equivalent volume of DMSO and immunoprecpitated with P-p38 MAPK antibody. Unexpectedly, the levels of hnRNP A1 co-precipitated with P-p38 MAPK were increased in SB203580-treated cells as compared to DMSO-treated cells (Fig. 3D, lanes 1 and 2). Densitometric analysis indicated more than 4.0-fold increase in the level of co-precipitated hnRNP A1. This increase is due to the increase in the hnRNP A1 expression level by SB203580 treatment (Fig. 2B, lane 2) or a structural change in hnRNP A1 protein when it is hypophosphorylated.
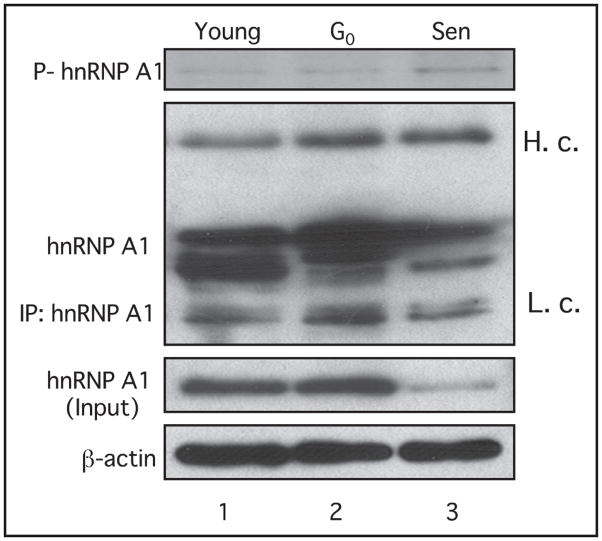
Increased phosphorylation of hnRNP A1 in senescent cells
Recent studies have shown that hnRNP A1 is phosphorylated by kinases downstream of p38 MAPK under various stress conditions.36,41,42,46 Since we have found that there is an elevated activity of p38 MAPK in senescent cells as well as an interaction between hnRNP A1 and p38 MAPK, we assessed whether the phosphorylation level of hnRNP A1 was also altered during senescence. In vivo phosphorylation of hnRNP A1 was determined by metabolic labeling in HS74 cells. HS74 fibroblasts have a similar senescence-associated expression pattern to IMR-90 cells.37 Young, G0-arrested and senescent HS74 fibroblasts were incubated over-night with 32P-ortho-phosphate, lysed, immunoprecipitated with the 4B10 monoclonal antibody and subjected to SDS-PAGE. hnRNP A1 proteins in young and G0-arrested cells were phosphorylated at very low levels (Fig. 4, lanes 1 and 2), while the phosphorylation of hnRNP A1 was increased in senescent fibroblasts (Fig. 4, lane 3). Since the total protein levels of hnRNP A1 were diminished in senescent cells (Fig. 4, lane 3), these results suggest that the majority of hnRNP A1 is in a phosphorylated form in senescent fibroblasts. This increase in phosphorylation might reflect the elevated p38 MAPK activity in senescent cells (Fig. 1D, lane 4). We examined the level of phosphorylated hnRNP A1 in young, G0-arrested and senescent cells following inhibition of p38MAPK by SB203580. However, we did not observe a detectable decrease in signal of phosphorylated hnRNP A1 using an anti-phosphoserine antibody in western analyses (data not shown). These results are consistent with those of van der Houven van Oordt et al. who reported that although hnRNP A1 accumulation was due to activation of the p38MAPK pathway, p38MAPK itself did not directly phosphorylate hnRNP A1.36
Figure 4.
In vivo 32P radiolabeling of young, G0 arrest and senescent HS74 fibroblasts. Cells were incubated overnight in 0.5 mCi/ml 32P-ortho-phosphate. Total protein was isolated and endogenous hnRNP A1 was immunoprecipitated using 4B10 antibody. hnRNP A1 phosphorylation levels were determined by autoradiography following 12% SDS-PAGE. 300 μg of total lysates isolated from IMR-90 cells were immunoprecipitated using the 4B10-hnRNP A1 antibody, subjected to 12% SDS-PAGE and probed with the 4B10-hnRNP A1 antibody. For input, total lysates were subjected directly to western analysis. Membranes were stripped and reprobed with β-actin as a loading control.
p38 MAPK is required for the subcellular distribution of hnRNP A1
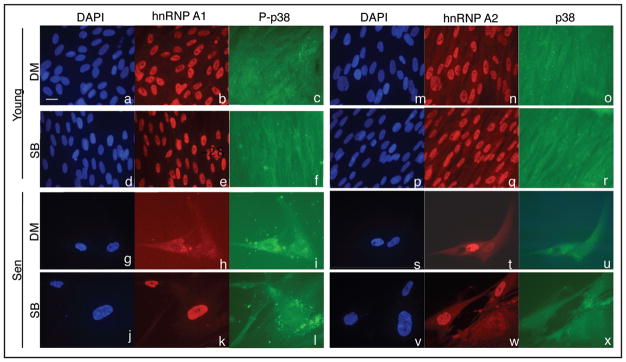
We have previously observed a diffuse distribution of hnRNP A1 protein in the cytoplasm in senescent HS74 cells compared to its predominant nuclear distribution in young cells.38 Van der Houven van Oordt et al. demonstrated that p38 MAPK activation was necessary and sufficient for the induction of hnRNP A1 cytoplasmic accumulation.36 Thus, we assessed whether the activation of p38 MAPK detected in senescent IMR-90 cells elicited a subcellular redistribution of hnRNP A1 protein. We treated young (PD 33.5) and senescent IMR-90 cells with SB203580 for 8 days and determined the subcellular localization pattern of hnRNP A1 by immunocytochemistry. Control young cells showed a predominant nuclear localization of the protein (Fig. 5, part B). Control senescent cells showed cytoplasmic accumulation (Fig. 5, part h). Young cells treated with SB203580 did not show any change in their nucleo-specific localization pattern compared to the control cells (Fig. 5, part e). In contrast, SB203580 treatment inhibited cytoplasmic accumulation of hnRNP A1 as the majority of the protein was localized in the nucleus of treated senescent cells (Fig. 5, part k). These data indicate that the accumulation of hnRNP A1 into the cytoplasm in senescent cells requires the kinase activity of p38 MAPK. We next determined the subcellular distribution pattern of hnRNP A2, which has overlapping biochemical properties of hnRNP A1.21,24 hnRNP A2 also showed similar subcellular distribution to A1 in both young and senescent control cells (Fig. 5, parts n and t). However, the SB203580 treatment did not affect its localization pattern in young and senescent cells (Fig. 5, parts q and w). These results suggest that the subcellular distribution of hnRNP A1, but not A2 is specifically regulated by p38 MAPK.
Figure 5.
Effect of p38 MAPK inhibition on the subcellular distribution of hnRNP A1 and A2. Young and senescent IMR-90 cells were treated with 10 μM SB203580 (Parts d–r and j–x) or an equal volume of DMSO (Parts a–o and g–u) for 8 days and then subjected to in situ immunocytochemistry for hnRNP A1, A2, p38 MAPK and phospho-p38 MAPK (P-p38). All fields are shown at a magnification of 400×. Nuclei were identified by DAPI staining. Identical fields were taken for DAPI, hnRNP A1 and P-p38 MAPK, or DAPI, hnRNP A2 and p38 MAPK signals. The scale bar represents 20 μM.
We then examined the subcellular localization pattern of P-p38 MAPK and p38 MAPK. Both proteins were uniformly present in young control cells (Fig. 5, parts c and o) and mainly in the cytoplasm in the control senescent cells (Fig. 5, parts i and u). The treatment with SB203580 did not induce any change in the localization patterns in either growth state (Figs. 5, parts f, l and 6, part r, 5, part x). These results indicate that the kinase activity of p38 MAPK does not regulate its own subcellular localization. Our results also showed that SB203580 treatment prohibited the cytoplasmic distribution of hnRNP A1 in senescent cells without affecting the localization pattern of P-p38 MAPK.
Inhibition of hnRNP A1/A2 expression induces a senescence-like morphology
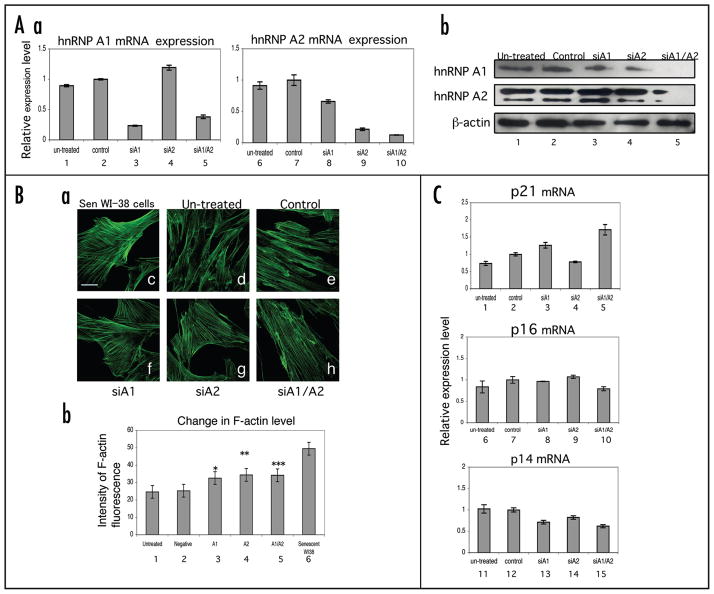
In these studies, we have shown that there were alterations in the expression, phosphorylation and subcellular distribution of hnRNP A1 in senescent cells. This raises the possibility that hnRNP A1 levels might maintain the senescence phenotype. To assess this likelihood, we performed siRNA knockdown analysis of hnRNP A1 and A2 to induce senescence. We transfected hnRNP A1 and A2 siRNA oligonucleotides in young IMR-90 cells followed by incubation for three days. We then determined the mRNAs and protein levels of hnRNP A1 and A2 by real-time PCR and western blot analysis, respectively (Fig. 6A, parts a and b). We observed dramatic reductions in both the mRNA and protein levels, especially when the expression of both A1 and A2 were simultaneously inhibited (Figs. 6A, part a, lanes 5 and 10 and part b, lane 5), indicating that we were able to successfully downregulate the expression of the target genes. Interestingly, the inhibition of hnRNP A2 reduced the protein level of hnRNP A1 (Fig. 6A, part b, lane 4). However, the levels of hnRNP A2 increased when A1 was inhibited (Fig. 6A, part b, lane 3). Therefore, inhibiting hnRNP A2 might reduce A1, but not vice versa.
Figure 6.
siRNA inhibition of hnRNP A1 and A2 expression in IMR-90 fibroblasts. (A) Young IMR-90 cells were transfected with siRNA against hnRNP A1 and/or A2 overnight and incubated in fresh regular media for three days. Control cells were treated with oligofectamine in the absence of siRNA. The cells were harvested followed by isolation of RNA and protein separately for real-time PCR (Part a) and western blot (Part b) analysis, respectively. (Part a) For real-time PCR analysis, RPLPO was used as a control to quantify the amount of hnRNP A1/A2 mRNAs. The relative expression level of these mRNAs (lanes 1, 3–5 or 6, 8–10) was calculated by normalization using the mRNA level in control cells (lanes 2 and 7). Three independent experiments were done, but error bars shown represent the standard errors of triplicate values from one representative experiment. (Part b) For western blot analysis, each 5 μg of each total cell lysate was loaded on 12.5% SDS-PAGE gels and the transferred membranes were probed with 4B10 and 10D1 antibodies for hnRNP A1 and A2, respectively. (B) F-actin fiber formation in siRNA treated IMR-90 fibroblasts. (Part a) F-actin staining was performed 7 days post-transfection of siRNA oligos for hnRNP A1 and A2 in IMR-90 cells (Parts d–h). Senescent WI-38 cells were used as a positive control for F-actin staining (Part c). Images were taken using a Zeiss LSM 510 confocal microscope at a magnification of 600X. The scale bar represents 20 μm. (Part b) The mean F-actin fluorescent intensity in 15 randomly selected fields for each condition were quantified and subjected to statistical analysis (lanes 1–6). The asterisks represent the level of significance using paired t-test values when comparing the mean intensity of each condition to the negative control (mock); *p = 0.000280763, **p = 0.000189 and ***p = 0.001402. (C) Effect of siRNA inhibition of hnRNP A1/A2 on the expression of senescence associated gene expression. Real-time PCR was performed with probes for p21, p16INK4a and p14ARF mRNAs.
We next stained siRNA transfected cells with fluorescein phalloidin in order to measure the level of F-actin, which is a marker that increases in replicative senescence.14,15 All siRNA transfected cells exhibited a senescence-like morphology characterized by an enlarged shape (Fig. 6B, parts a, f, g and h). Measurement of the intensity of F-actin fluorescence showed that the levels of F-actin were significantly increased in all of the cells transfected with A1 and A2 siRNAs (Fig. 6B, part b, lanes 3–5). However, the levels did not attain that of senescent WI38 cells (positive control) (Fig. 6B, part b, lane 6). siRNAs with scrambled sequences for hnRNP A1 and A2 did not produce morphological changes nor reduce the mRNA levels of these proteins (data not shown). We were unable to obtain a signal in any siRNA condition for senescence-associated β-galactosidase activity (data not shown). These results suggest that downregulation of hnRNP A1/A2 expression contributes to a partial senescence-like morphology.
We have previously shown that the overexpression of hnRNP A1 and A2 modulated the splice site usage of the INK4a locus, which contains the genes of cell cycle regulators, p16INK4a and p14ARF, indicating that A1 and A2 have the ability to control the cell cycle.38 We isolated total RNA from siRNA transfected cells and performed real-time PCR with probes for p21, p16INK4a and p14ARF mRNAs (Fig. 6C). The expression level of a cyclin-dependent kinase (cdk) inhibitor, p21 was increased when both A1 and A2 siRNAs were transfected (Fig. 6C, lane 5). On the other hand, the expression level of another cdk-inhibitor, p16INK4a (a senescence marker), was not increased dramatically in any siRNA conditions (Fig. 6C, lanes 8–10). The expression level of the tumor suppressor and senescence-associated gene, p14ARF,19 was decreased in all siRNA conditions (Fig. 6C). These results indicate that downregulation of hnRNP A1/A2 partially contributes to senescence but is not sufficient to account for all of its genetic features.
Effect of p38 MAPK inhibition on cell life span
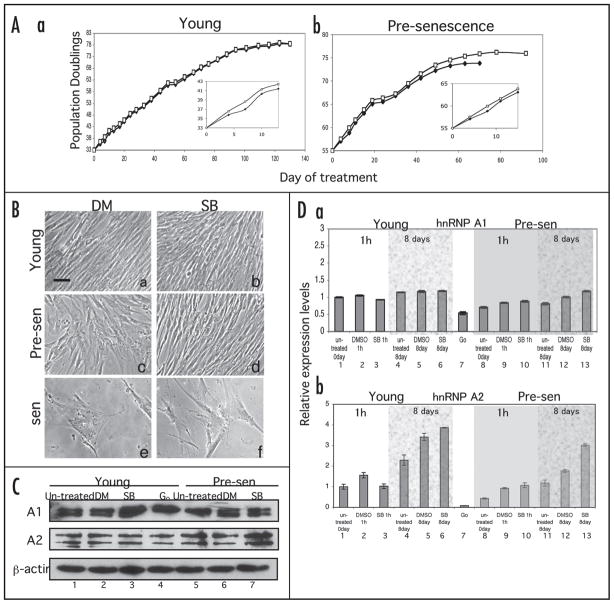
Recently, it has been demonstrated that the inhibition of the p38 MAPK pathway delays the onset of senescence.14,40,44 Our studies showed that downregulation of hnRNP A1/A2 by siRNA transfection induced a senescence-like morphology (Fig. 6B) indicating that hnRNP A1 may play a key role in the regulation of cellular senescence. To determine if hnRNP A1 or A2 could be responsible for the initiation of senescence downstream of the p38 MAPK pathway, we cultured young (PD 33) and pre-senescent (PD55) IMR-90 cells in medium with continual daily supplementation of 10 μM SB203580 or an equal volume of DMSO (control) until the cells reached senescence. PD 55 has mixed populations of young and senescent cells. We measured the in vitro life span of cultures and hnRNP A1 and A2 protein levels. Long-term inhibition of p38 MAPK did not affect the life span of young cells when compared to untreated young cells (Fig. 7A, part a). In contrast, the life span of SB203580 treated pre-senescent cells was increased (by PD 2 or 4 times the cell number) compared to untreated pre-senescent cells (Fig. 7A, part b). Continual SB203580 treatment of senescent cells showed no significant change in the life span (data not shown). These results suggest that pre-senescent cells were the most susceptive to the induction of senescence by the p38 MAPK pathway. We also observed that the proliferation of both the young and pre-senescent cells was promoted during the initial period of the treatment when compared to control cells (Fig. 7A, parts a and b, insets). While treated young and senescent cells did not show any significant change in the morphology (Fig. 7B, parts b and f), the morphology of the pre-senescent cells reverted to that of younger cells characterized by thinner and uniform cell shapes (Fig. 7B, part d). After the initial period, the proliferation of both growth states was no longer affected by the treatment. Thus, inhibition of the p38 MAPK signaling pathway was effective in increasing growth and delaying senescent morphological changes in pre-senescence cells.
Figure 7.
Long-term inhibition of p38 MAPK. (A) Lifespan assays. Young (PD 33) and pre-senescent (PD 55) IMR-90 cells were supplemented daily with complete media and 10 μM SB203580 (open squares) or an equal volume of DMSO (closed diamonds) until cells reached senescence (Parts a and b, respectively). Cell number was determined at each passage using a hemacytometer. Insets show higher-magnifications of the initial period of the treatments. (B) Change in morphology by p38 MAPK inhibition. Young (Parts a and b), pre-senescent (Parts c and d) and senescent (Parts e and f) IMR-90 cells were treated with 10 μM SB203580 (Parts b, d and f) or an equivalent volume of DMSO (Parts a, c and e) for 6–9 days. The scale bar represents 10 μm. (C) Effect of p38 MAPK inhibition on expression of the proteins involved in cellular senescence and cell cycle regulation. After 8 days of SB203580 treatment, 5 μg of total cell lysate was loaded on 12.5% SDS-PAGE gels and western blot analysis was performed with the antibodies specific for hnRNP A1 and A2. Cell lysates of G0-arrested cells were used as a control (lane 4). (D) Effect of p38 MAPK inhibition on mRNA expression. Real-time PCR was performed to determine the transcription level of the proteins. The mRNA levels after one hour of treatment were also determined to confirm the effect of the treatment itself on gene expression. The procedure for real-time PCR is discussed in the legend of Figure 6.
We simultaneously collected cells for each condition one hour and eight days following treatment and performed western blot analysis and real-time PCR to determine the effect of p38 MAPK inhibition on the expression levels of hnRNP A1/A2 (Fig. 7C and D). We observed elevated hnRNP A1 protein expression in SB203580-treated young cells on day eight compared to un-treated and DMSO-treated control cells (Fig. 7C, lane 3). However, no change in hnRNP A1 expression was observed in SB203580-treated pre-senescent cells on day eight (Fig. 7C, lane 7). These observations were similar to that of our short-term treatment of young and senescent cells (Fig. 2B). These results underscore that additional factors, besides p38 MAPK, may be required for the reduction in the expression levels of hnRNP A1 during senescence. In contrast, hnRNP A2 expression was increased only in SB203580-treated pre-senescent cells (Fig. 7C, lane 7). hnRNP A2 has been reported to be required for cell proliferation.35 Our results suggest that the suppression of hnRNP A2 level by p38 MAPK might contribute to the cell growth retardation characteristic of pre-senescent cells. While the protein level of hnRNP A1 expression was increased in young cells on SB203580 treatment, its mRNA level was unchanged (Fig. 7D, part a). These results suggest that hnRNP A1 expression may be subject to post-transcriptional regulation. hnRNP A2 mRNA expression was increased as well as its protein level in SB203580-treated pre-senescent cells (Fig. 7D, part b), suggesting that hnRNP A2 expression may be subject to transcriptional regulation.
Discussion
A member of heterogeneous nuclear ribonucleoprotein (hnRNP) family, hnRNP A1 is known to function in the biogenesis of mRNA. We previously showed age-dependent changes in the expression level and subcellular distribution of hnRNP A1.37,38 However, the molecular mechanisms that regulate these changes have not been elucidated. In this study, we demonstrate that the p38 MAPK pathway regulates hnRNP A1 protein levels. The p38 MAPK pathway has been shown to play an essential role in induction of senescence and our studies suggest that there is an interplay between this pathway and these RNA binding proteins. We have also shown that hnRNP A1 and A2 may have roles in establishing a partial senescence morphology.
Inhibition of p38 MAPK increased the level of hnRNP A1 protein expression in young and G0-arrested IMR-90 cells (Figs. 2B, 7C, lane 3), suggesting a p38 MAPK-dependent regulation. In addition, this regulation of the expression may be subject to either post-transcriptional control or increased protein stability since inhibition of p38 MAPK in young cells even up to eight days did not show any significant change in the level of hnRNP A1 mRNA (Fig. 7D, part a). Our findings of decreased protein expression and increased phosphorylation of hnRNP A1 during senescence (Figs. 1A and 4) suggests that the p38 MAPK pathway might regulate the stability of hnRNP A1 protein via phosphorylation. Previous studies support this idea. It has been shown that osmotic stress induced phosphorylation of cyclin D1 and D2 ubiquitination and degradation of these proteins.47,48 Overexpression of c-Jun mediates the ubiquitin mediated proteolysis of hnRNP A1.49
We did not detect any significant changes in hnRNP A1 protein levels in senescent cells and pre-senescent cells on p38 MAPK inhibition (Figs. 2B and 7C). We proposed two possibilities to explain these results. Firstly, the amount of the inhibitor used may not have been sufficient to suppress the high level of p38 MAPK activity in senescent cells. Secondly, the effect of the treatment may have been counteracted by hnRNP A1’s auto-regulation mechanism, whereby hnRNP A1 modulates its expression level by controlling splicing of its own pre-mRNA.50
We have found elevated phosphorylation of hnRNP A1 in senescent cells (Fig. 4), which correlated with the increased activity of p38 MAPK in senescent cells (Fig. 1D). While p38 MAPK does not appear to regulate hnRNP A1 protein levels during senescence, we observed that the inhibition of p38 MAPK blocked the senescence associated cytoplasmic accumulation of hnRNP A1 (Fig. 5). These results suggest that phosphorylation of hnRNP A1 by p38 MAPK is required for the subcellular distribution of hnRNP A1 during senescence. The recent finding that the phosphorylation of the F-peptide in the C-terminus of hnRNP A1 regulated the cytoplasmic accumulation of hnRNP A1 by reducing its interaction with transportin Trn1 provides support for our findings as well as those by van der Houven van Oordt et al.36,41
The elevated phosphorylation level of hnRNP A1 in senescent cells might contribute to alter its binding activity with target pre-mRNAs. Hamilton et al. showed that modulation of serine-threonine phosphorylation of hnRNP A1 regulated its binding to AU-rich elements (ARE’s), which are cis-acting elements that modulate mRNA stability and turnover.51 In addition, our previous studies had demonstrated that the in vitro binding activities of hnRNP A1 to telomeric DNA increased in senescent fibroblasts as opposed to its RNA binding activity to a model RNA substrate which diminished.37 Reconstituting hnRNP A1 expression has been shown to elicite telomere elongation and telomerase activity.52,53 Taken together, these findings suggest that the alteration of the phosphorylation level of hnRNP A1 may be required for pre-mRNA and telomere regulation in senescent cells.
While our observations show that the p38 MAPK regulates hnRNP A1 protein levels in young and not in senescent cells, other protein kinases may regulate hnRNP A1 protein levels during senescence. The protein kinases, Mnk, Akt, Protein Kinase C (PKC), Protein Kinase A (PKA) and Casein Kinase II, (CKII), have been reported to directly phosphorylate hnRNP A1,42,54,55 whereas, Protein Phosphatase 2A (PP2A) dephosphorylates the serine residues of hnRNP A1.54 Further studies are needed to determine if these kinases directly phosphorylates hnRNP A1 during senescence and regulate its functions. MNK1 may play a significant role as it is a substrate for p38 MAPK and directly phosphorylates hnRNP A1 in vitro and in vivo causing a decrease of hnRNP A1 binding to TNFα mRNA 3′UTR leading to the de-repression of TNFα translation.42 Jo et al. have recently demonstrated that hnRNP A1 was phosphorylated by Akt which induced its binding to the internal ribosome entry sites (IRESs) of both cyclin D1 and c-myc mRNAs, negatively regulating protein translation of these proteins.56 In addition, phosphorylation of hnRNP A1 by PKC and PKA inhibited its strand annealing activity.54,55
Our studies indicate that downregulation of hnRNP A1 and/or A2 by siRNA inhibition induced a senescence-like morphology and caused an increase in the level of actin stress fibers (F-actin), which are known markers for replicative senescence14,15 (Fig. 6B). However, the level of the intensity of F-actin fluorescence did not reach the level observed in control senescent WI-38 cells (Fig. 6B, part b), suggesting that a simple downregulation of hnRNP A1/A2 is not sufficient to induce a complete senescence morphology. This idea is supported by our qRT-PCR gene expression studies that show only an upregulation of p21 mRNA expression (Fig. 6C). For further studies, we will identify target pre-mRNAs that hnRNP A1/A2 bind to in senescent cells by RNA co-immunoprecipitation assays to determine the exact role of hnRNP A1/A2 in cellular senescence.
Our previous observations show that both hnRNP A1 and A2 have diminished expression and increased cytoplasmic accumulation in senescent cells.37,38 In the studies presented, both hnRNP A1 and A2 have similar effects on cellular morphology, F-actin formation, and p21 mRNA level by siRNA transfection analysis, suggesting that hnRNP A1 and A2 may have important roles for gene expression during cellular senescence. On the other hand, the inhibition of p38 MAPK affected the subcellular distribution of hnRNP A1, but not A2 (Fig. 5) as well as modulated both mRNA and protein levels of hnRNP A2 in contrast to hnRNP A1, where only its protein level was changed (Fig. 7C and D, part a). He et al. demonstrated that suppression of hnRNP A2, but not A1 or B1, slows cell growth in cancer cells.35 These observations suggest that hnRNP A1 and A2 possess distinct roles in cellular processes including cellular senescence under the control of different signaling pathways and is substantiated by reports identifying factors that regulate hnRNP A2, but not hnRNP A1.57,58
We have shown that the long-term inhibition of p38 MAPK using SB203580 extended the life span of pre-senescent cells by 2–4 PD (Fig. 7A) and that the cell shape and growth rate were similar to that of young cells (Fig. 7B, part d). Our results are similar to those of previous observations of p38 MAPK inhibition in Werner Syndrome fibroblasts.14 The effects we detected of p38 MAPK inhibition on life span was not dramatic, but are consistent with previous studies that show that individual inhibition of the p53, Rb or ERK pathways had minimal consequences on life span.59
Finally, we observed that inhibiting p38 MAPK had differential effects on the expression level of both hnRNP A1 and A2 in young and pre-senescent cells (Fig. 7C). Other studies have demonstrated links between hnRNP A1/A2 and factors involved in cellular senescence. The p53 target protein, Wig-1, binds to hnRNP A2/B1 protein dependant on the presence of RNA,60 suggesting an involvement of the p53 pathway in the regulation of hnRNP A2. Both p38 MAPK and ERK pathway regulate TNFα expression via Mnk phosphorylation of hnRNP A1 which decreases its binding to TNFα mRNA in T cells,42 suggesting that p38 MAPK might cooperate with ERK in the regulation of hnRNP A1.
Our present studies establish a novel link between hnRNP A1 and p38 MAPK. In addition, we have identified a novel function for hnRNP A1/A2 in promoting the senescence morphology. Our working model is the following: Factors leading to the induction of senescence activate p38 MAPK. This induction alters the subcellular distribution of hnRNP A1 via phosphorylation by the p38 MAPK pathway and in cooperation with other factors. Although p38 MAPK is not involved in the subcellular distribution of hnRNP A2, it may regulate hnRNP A2 expression levels in a signal-dependent manner. These changes in the localization and expression levels of hnRNP A1/A2 proteins could modulate the biogenesis of target pre-mRNAs and have profound effects in maintaining cellular senescence.
Previous studies have demonstrated that hnRNP A1 and A2 proteins are elevated in tumor cells and A2 is especially known as an early marker for lung cancer.61,62 Considering the fact that cellular senescence acts as a tumor suppression mechanism in vivo, these proteins might be ideal targets for cancer therapy. Further study is required to reveal the precise role of hnRNP A1 and A2 in cellular senescence under the control of p38 MAPK.
Materials and Methods
Cell strains and culture conditions
HS74 fetal bone marrow fibroblasts63 and IMR-90 fetal lung fibroblasts64 were cultured as previously described.65 Briefly, cells were cultured at 37°C in DMEM-HAM media (Mediatech, Manassas, VA) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (Mediatech, Gibco-Life technologies). Cultures were serially passaged until terminal passage was reached.65 Life span determinations have been previously described.66 Growth arrested cells were generated by replacing 10% FBS supplemented media with 0 or 0.5% FBS supplemented media for 72 h.
Cell lysis and protein quantitation
Each culture was rinsed with ice cold 1X phosphate buffered saline (PBS), pH 7.4 and 0.5 mL of cold 1% Empigen BB buffer in 1X PBS containing 1 mM EDTA, 0.1 mM dithiothreitol (DTT), 10 mM sodium fluoride and 100 μL of Phosphatase Inhibitor Cocktail I (Sigma, St. Louis, MO) was added to each 100 mm culture dish. Cells were scraped and lysed on ice by sonication three times for five sec each. Lysates were cleared at 14,000 ×g for 10 min. Protein concentration was quantified in triplicate using the Bradford-Lowry protein assay (Bio-Rad™).
Western blot analysis
Western blot analysis was performed using standard western procedures.67 Briefly, all extracts and immunoprecipitates were suspended in protein sample loading buffer,68 separated by 12% SDS-PAGE, and electrophoresed at 120 V 50 mA for 4 h. 0.5 mL of lysates were used for IP reactions. Proteins were electrophoretically transferred to PVDF membranes in 25 mM Tris base, 190 mM glycine and 20% ethanol. Nonspecific binding was blocked by incubation of membranes for one hour in blocking buffer (5% nonfat dry milk in 1X TBS and 0.1% Tween-20). Membranes were incubated in the appropriate dilution of primary antibodies in 3% bovine serum albumin (BSA), 0.1% Tween-20 in 1X PBS overnight at 4°C. Membranes were washed three times for five minutes each in wash buffer (0.1% Tween-20 in 1X PBS) followed by incubation in a 1:1,000 dilution of secondary anti-mouse or rabbit horseradish peroxidase (HRP)-conjugated antibodies (Amersham, Buckinghamshire, UK) in blocking buffer for one hour at room temperature. Membranes were then washed three times for ten min each in 1X PBS containing 0.1% Tween-20, followed by detection of protein bands using enhanced chemiluminescence (ECL-Plus™) from Amersham. For a positive control for p38 MAPK expression, lysates were treated with 5 μM Anisomycin (Sigma), a well-known activator of p38 MAPK69 for 30 min.
The primary antibodies used: 4B10 for hnRNP A1 and 10D4 for hnRNP A2 were generously provided by Dr. Serafin Pinol-Roma; anti-H-ras antibody (Santa Cruz Biotechnology, Santa Cruz, CA); anti-p38 antibody (Cell Signaling™); anti-phospho-p38 antibody (Cell Signaling™); anti-GAPDH antibody (CHEMICON, Temecula, CA); anti-Bax antibody (Cell Signaling™) Densitometric analysis was performed using ImageJ 1.41o software (National Institutes of Health).
Immunoprecipitation
Whole cell lysates were imunoprecipitated for hnRNP A1 proteins using the 4B10 monoclonal antibody. 8.5 μg 4B10 antibody was pre-incubated with 20 μL of Protein A-sepharose beads (Roche, Diagnostics, Mannheim, Germany) for one hour at 4°C. Complexes were then incubated with cell lysates for one hour at 4°C. Immunoprecipitates were centrifuged at 14,000 ×g for five min, washed three times with 1% Empigen BB buffer in 1X PBS containing 1 mM EDTA, 0.1 mM dithiothreitol (DTT), 10 mM sodium fluoride, then resuspended in 1X sample buffer (62.5 mM Tris-HCl, pH 6.8 at 25°C, 2% w/v sodium dodecyl sulfate (SDS), 10% glycerol, 50 μM dithiothreitol (DTT), 0.01% bromophenol blue), and immediately subjected to 12% SDS-PAGE.
For co-immunoprecipitation analysis, cells were lysed in 1X cell lysis buffer from Cell Signaling (20 mM Tris (pH 7.5), 150 mM sodium chloride, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM sodium vanadate, 1 μg/mL leupeptin and 1 mM PMSF). Lysates were incubated overnight at 4°C in 20 μL of resuspended Immobilized Phospho-p38 MAP kinase (Thr180/Tyr182) beads (Cell Signaling™). The immune complex was microcentrifuged at 14,000 ×g for 5 min, washed 4X with 500 mL 1X lysis buffer, resuspended in 1X SDS sample buffer and subjected to 12% SDS-PAGE. Proteins in the immunocomplexes were identified by western analysis.
Metabolic labeling studies
HS74 cells were metabolically labeled overnight with 0.5 mCi/ml 32P-ortho-phosphate and lysed with RIPA buffer (1% NP-40, 1% sodium deoxycholate, 0.1% SDS, 0.15 M NaCl) containing 10 μg/mL of each leupeptin and pepstatin, 1 mM phenylmethylsulfonyl fluoride, 0.25 mM orthovanadate, 20 mM β-glyerophosphate and 10 mM sodium fluoride. Endogenous hnRNP A1 was immunoprecipitated using the 4B10 monoclonal antibody, then subjected to 12% SDS-PAGE. Phosphorylated protein was determined by autoradiography.
p38 MAP kinase assay
Cells were cultured as described above and lysed in 1X cell lysis buffer from Cell Signaling™ (20 mM Tris (pH 7.5), 150 mM sodium chloride, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM sodium vanadate, 1 μg/mL leupeptin and 1 mM PMSF). Two-hundred micrograms of total protein lysate was incubated with 20 μL of resuspended immobilized Phospho-p38 MAP (P-p38) kinase (Thr180/Tyr182) antibody (Cell Signaling™) overnight at 4°C. The immune complex was washed two times with 500 μL 1X lysis buffer, 2 times with 500 μL 1X kinase buffer and resuspended in 50 μL 1X kinase buffer (as provided by Cell Signaling™ Assay Kit) supplemented with 200 μM ATP and 2 μg ATF-2 fusion protein. The reactions were incubated for 30 min at 30°C and terminated with 25 μL 1X SDS sample buffer. Reactions were subjected to 12% SDS-PAGE followed by western blot analysis with primary antibody to Phos-ATF-2 (Thr71) (Cell Signaling™). The protein bands were visualized using ECL-Plus™ (Amersham).
Inhibition of p38 MAP kinase activity
Young, G0-arrested, pre-senescent (PD 55) and senescent IMR-90 cells were treated with 2.5 μM or 10 μM SB203580, a p38 MAPK inhibitor70 or an equivalent volume of dimethyl sulfoxide (DMSO) as a control for one hour or longer at 37°C. For lifespan assays, the media with the inhibitor was changed daily until the cells reached senescence. The cell number was simultaneously counted at each passage using a hemacytometer (Hausser Scientific, Horsham, PA). Following treatment, immunoprecipitation, western blot analysis or immunocytochemistry were performed.
Immunocytochemistry
IMR-90 cells treated with SB203580 were fixed with 4% paraformaldehyde and made permeable with 0.2% triton X-100 for ten min. Cells were washed with 1X PBS for five min twice, blocked with 10% serum in PBS for one hour at room temperature. Then, anti-hnRNP A1 4B10 monoclonal antibodies, anti-phospho-p38 MAPK (Thr180/Tyr182) monoclonal antibody (Cell Signaling™), or anti p38 MAPK antibody (Cell Signaling™) were added at dilutions of 1:1,000, 1:300 and 1:500, respectively and incubated overnight at 4°C. The cells were washed with 1X PBS for five min three times and incubated with the appropriate secondary antibody at a 1:1,000 dilution in blocking solution for one hour at room temperature in the dark. For the secondary antibodies, either Alexa Fluor® 488 anti-rabbit antibody (Invitrogen), or Alexa Fluor® 594 anti-mouse antibody (Invitrogen) were used. Cells were washed with 1× PBS for ten min four times in the dark and mounted in ProLong® Gold antifade reagent with DAPI (Invitrogen). Fluorescence signals were detected using a Zeiss Axioplan fluorescence microscope at a magnification of 400×. Images were then processed using ADOBE Photoshop version 7.0 (Adobe Systems, Mountain View, CA).
siRNA transfection
siRNA inhibition for hnRNP A1 and A2 was done using the following sequences: 5′-AGC AAG AGA UGG CUA GUG CUU-3′ and 5′-UGA AGA GCU UCC UCA GCU GUU-3′ for hnRNP A1; 5′-CGU GCU GUA GCA AGA GAG GUU-3′ and 5′-GAG CUU ACG GAA CUG UUC CUU-3′ for hnRNP A2 (He et al.35 and Kashima et al.71 respectively). siRNA oligonucleotides were purchased from Dharmacom Research, Inc., (Lafayette, CO). Young IMR-90 cells (PD 33) were seeded at a density of 2.5 × 105 cells in a 60 mm plate. After 24 h of incubation when the cells reached approximately 30% confluence, transfection of cells with RNA oligos for hnRNP A1 and A2 was performed using oligofectamine (Invitrogen) according to the manufacturer’s instructions. Cells treated without siRNA oligonucleotides were used as a control.
RNA extraction and reverse transcription
Total RNA was isolated from young and senescent IMR-90 cells and hnRNP A1-overexpressing young IMR-90 cells using TRIZOL (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Total RNA was then treated with DNA-free™ (Ambion, Austin, TX) to remove possible DNA contamination and 1 μg of RNA in a 20 μL reaction mixture was reverse transcribed to cDNA using the Retroscript kit (Ambion) according to the manufacturer’s instructions.
qReal time PCR
For SYBR green assays the primers for hnRNP A1, hnRNP A2, p14ARF, p16INK4a and p21, and the primers for human acidic ribosomal phosphoprotein PO (RPLPO) were previously described by Zhu et al. and Bièche et al. respectively.38,72 The 7500 Real Time PCR System (Applied Biosystems, Foster City, CA) was used for assays. For every assay, cycling conditions were 95°C for ten seconds, followed by 40 cycles at 95°C for 15 sec and 60°C for one min. 22 μL PCR mixture contained 11 μL Power SYBR® green PCR Master Mix (Applied Biosystems), 1 μL of RT reaction mixture and 0.36 μM of each primer. Each sample was prepared in triplicate.
Detection of F-actin stress fibers
Fluorescent Phallotoxins have been used to label F-actin (stress fibers). Stress fibers tend to increase with age and have been used as a marker of replicative senescence.14,15 Early passage siRNA-treated IMR-90 and senescent WI-38 cells were cultured in 4-well chamber slides (Fisher Scientific, Suwanee GA). Slides were washed with 1X PBS and fixed with 2% paraformaldehyde for 15 min at room temperature. Slides were then washed in 1X PBS and permeabilized with 0.1% Triton X-100 (Fisher Scientific) for 5 min at room temperature. Slides were washed in 1× PBS. Slides were blocked in 10% bovine serum albumin (BSA) for 20 min and Alexa Fluor®488 Phalloidin (Molecular Probes, Eugene, OR) was added for 20 min in the dark. Slides were then mounted with SlowFade Antifade solution (Molecular Probes). Images were captured using a Zeiss LSM510 Confocal Microscope (Zeiss, Germany) at a magnification of 600X under an Argon laser. The average F-actin intensity in 15 randomly fields was determined using Adobe photoshop version 7.0 software (Adobe Systems, Mountain View, CA) and paired t-test values were calculated.
Acknowledgments
We thank Dr. Serafin Pinol-Roma for hnRNP A1 and A2 specific antibodies as well as Dr. Bor Jang Hwang for his assistance in the use of a Real Time PCR machine. This research was supported by NIH/NCI CA096299, NIH/NIGMS MBRS GM08168 and NIH/NIGMS RCMI RR03060 to K.H.
Abbreviations
- hnRNP
heterogeneous nuclear ribonucleoproteins
References
- 1.Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–85. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 2.Waskiewicz AJ, Cooper JA. Mitogen and stress response pathways: MAP kinase cascades and phosphatase regulation in mammals and yeast. Curr Opin Cell Biol. 1995;7:798–805. doi: 10.1016/0955-0674(95)80063-8. [DOI] [PubMed] [Google Scholar]
- 3.Su B, Karin M. Mitogen-activated protein kinase cascades and regulation of gene expression. Curr Opin Immunol. 1996;8:402–11. doi: 10.1016/s0952-7915(96)80131-2. [DOI] [PubMed] [Google Scholar]
- 4.Lee JC, Young PR. Role of CSB/p38/RK stress response kinase in LPS and cytokine signaling mechanisms. J Leukoc Biol. 1996;59:152–7. doi: 10.1002/jlb.59.2.152. [DOI] [PubMed] [Google Scholar]
- 5.Freshney NW, Rawlinson L, Guesdon F, Jones E, Cowley S, Hsuan J, et al. Interleukin-1 activates a novel protein kinase cascade that results in the phosphorylation of Hsp27. Cell. 1994;78:1039–49. doi: 10.1016/0092-8674(94)90278-x. [DOI] [PubMed] [Google Scholar]
- 6.Derijard B, Raingeaud J, Barrett T, Wu IH, Han J, Ulevitch RJ, et al. Independent human MAP-kinase signal transduction pathways defined by MEK and MKK isoforms. Science. 1995;267:682–5. doi: 10.1126/science.7839144. [DOI] [PubMed] [Google Scholar]
- 7.Raingeaud J, Gupta S, Rogers JS, Dickens M, Han J, Ulevitch RJ, et al. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J Biol Chem. 1995;270:7420–6. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- 8.Han J, Jiang Y, Li Z, Kravchenko VV, Ulevitch RJ. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature. 1997;386:296–9. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- 9.Tibbles LA, Woodgett JR. The stress-activated protein kinase pathways. Cell Mol Life Sci. 1999;55:1230–54. doi: 10.1007/s000180050369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 11.Wang W, Chen JX, Liao R, Deng Q, Zhou JJ, Huang S, et al. Sequential activation of the MEK-extracellular signal-regulated kinase and MKK3/6-p38 mitogen-activated protein kinase pathways mediates oncogenic ras-induced premature senescence. Mol Cell Biol. 2002;22:3389–403. doi: 10.1128/MCB.22.10.3389-3403.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng Q, Liao R, Wu BL, Sun P. High intensity ras signaling induces premature senescence by activating p38 pathway in primary human fibroblasts. J Biol Chem. 2004;279:1050–9. doi: 10.1074/jbc.M308644200. [DOI] [PubMed] [Google Scholar]
- 13.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 14.Davis T, Baird DM, Haughton MF, Jones CJ, Kipling D. Prevention of accelerated cell aging in Werner syndrome using a p38 mitogen-activated protein kinase inhibitor. J Gerontol A Biol Sci Med Sci. 2005;60:1386–93. doi: 10.1093/gerona/60.11.1386. [DOI] [PubMed] [Google Scholar]
- 15.Cho KA, Ryu SJ, Oh YS, Park JH, Lee JW, Kim HP, et al. Morphological adjustment of senescent cells by modulating caveolin-1 status. J Biol Chem. 2004;279:42270–8. doi: 10.1074/jbc.M402352200. [DOI] [PubMed] [Google Scholar]
- 16.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–7. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ben-Porath I, Weinberg RA. The signals and pathways activating cellular senescence. Int J Biochem Cell Biol. 2005;37:961–76. doi: 10.1016/j.biocel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 18.Hara E, Tsurui H, Shinozaki A, Nakada S, Oda K. Cooperative effect of antisense-Rb and antisense-p53 oligomers on the extension of life span in human diploid fibroblasts, TIG-1. Biochem Biophys Res Commun. 1991;179:528–34. doi: 10.1016/0006-291x(91)91403-y. [DOI] [PubMed] [Google Scholar]
- 19.Dimri GP, Itahana K, Acosta M, Campisi J. Regulation of a senescence checkpoint response by the E2F1 transcription factor and p14(ARF) tumor suppressor. Mol Cell Biol. 2000;20:273–85. doi: 10.1128/mcb.20.1.273-285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seshadri T, Campisi J. Repression of c-fos transcription and an altered genetic program in senescent human fibroblasts. Science. 1990;247:205–9. doi: 10.1126/science.2104680. [DOI] [PubMed] [Google Scholar]
- 21.Krecic AM, Swanson MS. hnRNP complexes: composition, structure and function. Curr Opin Cell Biol. 1999;11:363–71. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- 22.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 23.Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 24.Caceres JF, Stamm S, Helfman DM, Krainer AR. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–9. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 25.Mayeda A, Krainer AR. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–75. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- 26.Yang X, Bani MR, Lu SJ, Rowan S, Ben-David Y, Chabot B. The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc Natl Acad Sci USA. 1994;91:6924–8. doi: 10.1073/pnas.91.15.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izaurralde E, Jarmolowski A, Beisel C, Mattaj IW, Dreyfuss G, Fischer U. A role for the M9 transport signal of hnRNP A1 in mRNA nuclear export. J Cell Biol. 1997;137:27–35. doi: 10.1083/jcb.137.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonnal S, Pileur F, Orsini C, Parker F, Pujol F, Prats AC, et al. Heterogeneous nuclear ribonucleoprotein A1 is a novel internal ribosome entry site trans-acting factor that modulates alternative initiation of translation of the fibroblast growth factor 2 mRNA. J Biol Chem. 2005;280:4144–53. doi: 10.1074/jbc.M411492200. [DOI] [PubMed] [Google Scholar]
- 29.Svitkin YV, Ovchinnikov LP, Dreyfuss G, Sonenberg N. General RNA binding proteins render translation cap dependent. EMBO J. 1996;15:7147–55. [PMC free article] [PubMed] [Google Scholar]
- 30.Hamilton BJ, Nagy E, Malter JS, Arrick BA, Rigby WF. Association of heterogeneous nuclear ribonucleoprotein A1 and C proteins with reiterated AUUUA sequences. J Biol Chem. 1993;268:8881–7. [PubMed] [Google Scholar]
- 31.Henics T, Sanfridson A, Hamilton BJ, Nagy E, Rigby WF. Enhanced stability of interleu-kin-2 mRNA in MLA 144 cells. Possible role of cytoplasmic AU-rich sequence-binding proteins. J Biol Chem. 1994;269:5377–83. [PubMed] [Google Scholar]
- 32.Pinol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–2. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 33.Iervolino A, Santilli G, Trotta R, Guerzoni C, Cesi V, Bergamaschi A, et al. hnRNP A1 nucleocytoplasmic shuttling activity is required for normal myelopoiesis and BCR/ABL leukemogenesis. Mol Cell Biol. 2002;22:2255–66. doi: 10.1128/MCB.22.7.2255-2266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayeda A, Munroe SH, Caceres JF, Krainer AR. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 1994;13:5483–95. doi: 10.1002/j.1460-2075.1994.tb06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He Y, Brown MA, Rothnagel JA, Saunders NA, Smith R. Roles of heterogeneous nuclear ribonucleoproteins A and B in cell proliferation. J Cell Sci. 2005;118:3173–83. doi: 10.1242/jcs.02448. [DOI] [PubMed] [Google Scholar]
- 36.van der Houven van Oordt W, Diaz-Meco MT, Lozano J, Krainer AR, Moscat J, Caceres JF. The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J Cell Biol. 2000;149:307–16. doi: 10.1083/jcb.149.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hubbard K, Dhanaraj SN, Sethi KA, Rhodes J, Wilusz J, Small MB, et al. Alteration of DNA and RNA binding activity of human telomere binding proteins occurs during cellular senescence. Exp Cell Res. 1995;218:241–7. doi: 10.1006/excr.1995.1152. [DOI] [PubMed] [Google Scholar]
- 38.Zhu D, Xu G, Ghandhi S, Hubbard K. Modulation of the expression of p16INK4a and p14ARF by hnRNP A1 and A2 RNA binding proteins: implications for cellular senescence. J Cell Physiol. 2002;193:19–25. doi: 10.1002/jcp.10147. [DOI] [PubMed] [Google Scholar]
- 39.Rittling SR, Brooks KM, Cristofalo VJ, Baserga R. Expression of cell cycle-dependent genes in young and senescent WI-38 fibroblasts. Proc Natl Acad Sci USA. 1986;83:3316–20. doi: 10.1073/pnas.83.10.3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang S, Jung M, Kim CW, Shin DY. Inactivation of p38 kinase delays the onset of senescence in rabbit articular chondrocytes. Mech Ageing Dev. 2005;126:591–7. doi: 10.1016/j.mad.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 41.Allemand E, Guil S, Myers M, Moscat J, Caceres JF, Krainer AR. Regulation of heterogenous nuclear ribonucleoprotein A1 transport by phosphorylation in cells stressed by osmotic shock. Proc Natl Acad Sci USA. 2005;102:3605–10. doi: 10.1073/pnas.0409889102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buxade M, Parra JL, Rousseau S, Shpiro N, Marquez R, Morrice N, et al. The Mnks are novel components in the control of TNFalpha biosynthesis and phosphorylate and regulate hnRNP A1. Immunity. 2005;23:177–89. doi: 10.1016/j.immuni.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 43.Morton S, Davis RJ, Cohen P. Signalling pathways involved in multisite phosphorylation of the transcription factor ATF-2. FEBS Lett. 2004;572:177–83. doi: 10.1016/j.febslet.2004.07.031. [DOI] [PubMed] [Google Scholar]
- 44.Iwasa H, Han J, Ishikawa F. Mitogen-activated protein kinase p38 defines the common senescence-signalling pathway. Genes Cells. 2003;8:131–44. doi: 10.1046/j.1365-2443.2003.00620.x. [DOI] [PubMed] [Google Scholar]
- 45.Davies SP, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guil S, Long JC, Caceres JF. hnRNP A1 relocalization to the stress granules reflects a role in the stress response. Mol Cell Biol. 2006;26:5744–58. doi: 10.1128/MCB.00224-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Casanovas O, Miro F, Estanyol JM, Itarte E, Agell N, Bachs O. Osmotic stress regulates the stability of cyclin D1 in a p38SAPK2-dependent manner. J Biol Chem. 2000;275:35091–7. doi: 10.1074/jbc.M006324200. [DOI] [PubMed] [Google Scholar]
- 48.Kida A, Kakihana K, Kotani S, Kurosu T, Miura O. Glycogen synthase kinase-3beta and p38 phosphorylate cyclin D2 on Thr280 to trigger its ubiquitin/proteasome-dependent degradation in hematopoietic cells. Oncogene. 2007;26:6630–40. doi: 10.1038/sj.onc.1210490. [DOI] [PubMed] [Google Scholar]
- 49.Perrotti D, Iervolino A, Cesi V, Cirinna M, Lombardini S, Grassilli E, et al. BCR-ABL prevents c-jun-mediated and proteasome-dependent FUS (TLS) proteolysis through a protein kinase CbetaII-dependent pathway. Mol Cell Biol. 2000;20:6159–69. doi: 10.1128/mcb.20.16.6159-6169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chabot B, Blanchette M, Lapierre I, La Branche H. An intron element modulating 5′ splice site selection in the hnRNP A1 pre-mRNA interacts with hnRNP A1. Mol Cell Biol. 1997;17:1776–86. doi: 10.1128/mcb.17.4.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hamilton BJ, Burns CM, Nichols RC, Rigby WF. Modulation of AUUUA response element binding by heterogeneous nuclear ribonucleoprotein A1 in human T lymphocytes. The roles of cytoplasmic location, transcription and phosphorylation. J Biol Chem. 1997;272:28732–41. doi: 10.1074/jbc.272.45.28732. [DOI] [PubMed] [Google Scholar]
- 52.LaBranche H, Dupuis S, Ben-David Y, Bani MR, Wellinger RJ, Chabot B. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat Genet. 1998;19:199–202. doi: 10.1038/575. [DOI] [PubMed] [Google Scholar]
- 53.Zhang QS, Manche L, Xu RM, Krainer AR. hnRNP A1 associates with telomere ends and stimulates telomerase activity. Rna. 2006;12:1116–28. doi: 10.1261/rna.58806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cobianchi F, Calvio C, Stoppini M, Buvoli M, Riva S. Phosphorylation of human hnRNP protein A1 abrogates in vitro strand annealing activity. Nucleic Acids Res. 1993;21:949–55. doi: 10.1093/nar/21.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Idriss H, Kumar A, Casas-Finet JR, Guo H, Damuni Z, Wilson SH. Regulation of in vitro nucleic acid strand annealing activity of heterogeneous nuclear ribonucleoprotein protein A1 by reversible phosphorylation. Biochemistry. 1994;33:11382–90. doi: 10.1021/bi00203a037. [DOI] [PubMed] [Google Scholar]
- 56.Jo OD, Martin J, Bernath A, Masri J, Lichtenstein A, Gera J. Heterogeneous nuclear ribonucleoprotein A1 regulates cyclin D1 and c-myc internal ribosome entry site function through Akt signaling. J Biol Chem. 2008;283:23274–87. doi: 10.1074/jbc.M801185200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bosser R, Faura M, Serratosa J, Renau-Piqueras J, Pruschy M, Bachs O. Phosphorylation of rat liver heterogeneous nuclear ribonucleoproteins A2 and C can be modulated by calmodulin. Mol Cell Biol. 1995;15:661–70. doi: 10.1128/mcb.15.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pan H, Luo C, Li R, Qiao A, Zhang L, Mines M, et al. Cyclophilin A is required for CXCR4-mediated nuclear export of heterogeneous nuclear ribonucleoprotein A2, activation and nuclear translocation of ERK1/2, and chemotactic cell migration. J Biol Chem. 2008;283:623–37. doi: 10.1074/jbc.M704934200. [DOI] [PubMed] [Google Scholar]
- 59.Cristofalo VJ, Lorenzini A, Allen RG, Torres C, Tresini M. Replicative senescence: a critical review. Mech Ageing Dev. 2004;125:827–48. doi: 10.1016/j.mad.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 60.Prahl M, Vilborg A, Palmberg C, Jornvall H, Asker C, Wiman KG. The p53 target protein Wig-1 binds hnRNP A2/B1 and RNA Helicase A via RNA. FEBS Lett. 2008;582:2173–7. doi: 10.1016/j.febslet.2008.04.065. [DOI] [PubMed] [Google Scholar]
- 61.Xu X, Joh HD, Pin S, Schiller NI, Prange C, Burger PC, et al. Expression of multiple larger-sized transcripts for several genes in oligodendrogliomas: potential markers for glioma subtype. Cancer Lett. 2001;171:67–77. doi: 10.1016/s0304-3835(01)00573-0. [DOI] [PubMed] [Google Scholar]
- 62.Fielding P, Turnbull L, Prime W, Walshaw M, Field JK. Heterogeneous nuclear ribonu-cleoprotein A2/B1 upregulation in bronchial lavage specimens: a clinical marker of early lung cancer detection. Clin Cancer Res. 1999;5:4048–52. [PubMed] [Google Scholar]
- 63.Smith HS, Owens RB, Hiller AJ, Nelson-Rees WA, Johnston JO. The biology of human cells in tissue culture I. Characterization of cells derived from osteogenic sarcomas. Int J Cancer. 1976;17:219–34. doi: 10.1002/ijc.2910170211. [DOI] [PubMed] [Google Scholar]
- 64.Nichols WW, Murphy DG, Cristofalo VJ, Toji LH, Greene AE, Dwight SA. Characterization of a new human diploid cell strain, IMR-90. Science. 1977;196:60–3. doi: 10.1126/science.841339. [DOI] [PubMed] [Google Scholar]
- 65.Hubbard K, Ozer HL. Senescence & immortalization of human cells. In: Studsinski GP, editor. Cell Growth, Differentiation & Senescence. New York: Oxford University Press; 1999. pp. 281–300. [Google Scholar]
- 66.Neufeld DS, Ripley S, Henderson A, Ozer HL. Immortalization of human fibroblasts transformed by origin-defective simian virus 40. Mol Cell Biol. 1987;7:2794–802. doi: 10.1128/mcb.7.8.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harlow E, Lane DP. In: Antibodies: A Laboratory Manual. Harlow E, Lane DP, editors. New York: Cold Spring Harbor Laboratory Press; 1988. p. 359. [Google Scholar]
- 68.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 69.Cano E, Hazzalin CA, Mahadevan LC. Anisomycin-activated protein kinases p45 and p55 but not mitogen-activated protein kinases ERK-1 and -2 are implicated in the induction of c-fos and c-jun. Mol Cell Biol. 1994;14:7352–62. doi: 10.1128/mcb.14.11.7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cuenda A, Rouse J, Doza YN, Meier R, Cohen P, Gallagher TF, et al. SB203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 1995;364:229–33. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- 71.Kashima T, Manley JL. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet. 2003;34:460–3. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 72.Bieche I, Nogues C, Paradis V, Olivi M, Bedossa P, Lidereau R, et al. Quantitation of hTERT gene expression in sporadic breast tumors with a real-time reverse transcription-polymerase chain reaction assay. Clin Cancer Res. 2000;6:452–9. [PubMed] [Google Scholar]